Abstract
The bacterial flagellar motor is a molecular machine that converts ion flux across the membrane into flagellar rotation. The coupling ion is either a proton or a sodium ion. The polar flagellar motor of the marine bacterium Vibrio alginolyticus is driven by sodium ions, and the four protein components, PomA, PomB, MotX, and MotY, are essential for motor function. Among them, PomA and PomB are similar to MotA and MotB of the proton-driven motors, respectively. PomA shows greatest similarity to MotA of the photosynthetic bacterium Rhodobacter sphaeroides. MotA is composed of 253 amino acids, the same length as PomA, and 40% of its residues are identical to those of PomA. R. sphaeroides MotB has high similarity only to the transmembrane region of PomB. To examine whether the R. sphaeroides motor genes can function in place of the pomA and pomB genes of V. alginolyticus, we constructed plasmids including both motA and motB or motA alone and transformed them into missense and null pomA-paralyzed mutants of V. alginolyticus. The transformants from both strains showed restored motility, although the swimming speeds were low. On the other hand, pomB mutants were not restored to motility by any plasmid containing motA and/or motB. Next, we tested which ions (proton or sodium) coupled to the hybrid motor function. The motor did not work in sodium-free buffer and was inhibited by phenamil and amiloride, sodium motor-specific inhibitors, but not by a protonophore. Thus, we conclude that the proton motor component, MotA, of R. sphaeroides can generate torque by coupling with the sodium ion flux in place of PomA of V. alginolyticus.
Bacteria swim by rotating helical flagella toward favorable stimuli in liquid environments. The motor complex generating torque is embedded in the cytoplasmic membrane at the base of the flagella, and it converts a specific ion flux (either a proton or a sodium ion) to motor rotation (7).
Most studies have centered on the proton-driven motors of Escherichia coli and Salmonella typhimurium. The rotor part of the motor is composed of FliG, FliM, and FliN (28, 47), which form the C-ring structure on the cytoplasmic face of the MS ring (13, 50). The C ring is also called the switch complex, and it determines the direction of flagellar rotation, counterclockwise (CCW) or clockwise (CW). On the other hand, the stator complex consists of MotA and MotB, which interact via their transmembrane regions and function as a proton-conducting channel (7, 8, 44). MotA has four transmembrane regions and a large cytoplasmic loop. Recently, it was proposed that some charged residues in this loop interact with oppositely charged residues in FliG when they generate torque (52). MotB has a single transmembrane region including an important Asp, which seems to be involved in proton transfer (43). Moreover, a peptidoglycan binding motif is conserved in the C-terminal region of MotB and plays the role of anchoring the motor to the cell wall (10, 11).
The photosynthetic bacterium Rhodobacter sphaeroides has a single flagellum extending from the center of the cell body. The cell is motile under a wide range of growth conditions, from pH 6 to 9 (37), and at speeds of up to 100 μm/s (38). The rotation of the R. sphaeroides flagellum is unidirectional in the CW direction, and it stops and restarts periodically (1). The motor part is composed of MotA and MotB, which are similar to those of the proton-driven motors (40, 41). The flagellar motor of R. sphaeroides is active in the absence of sodium ions and is inhibited by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). Amiloride, a specific inhibitor of the sodium-driven flagellar motor, inhibits motility; however, the effect is nonspecific. These results indicate that the motor of R. sphaeroides is the proton-driven type (37).
The marine bacterium Vibrio alginolyticus has two types of flagella, lateral (Laf) and polar (Pof). The lateral flagella, which have proton-driven motors, are expressed when cells are transferred to high-viscosity environments. The polar flagella have sodium-driven motors and work better for swimming in low-viscosity environments. The rotation of polar flagella is very fast, about 1,700 rps in 300 mM NaCl at 35°C (29, 30, 35). The sodium-driven motor consists of four components, PomA, PomB, MotX, and MotY, all of which are essential for torque generation (2, 31, 32, 36). Of these, MotX and MotY, which are predicted to be single transmembrane proteins, do not have similarity to proton motor components or any other proteins, except for a C-terminal region of MotY which contains a peptidoglycan-binding motif. MotY and MotX are thought to make a complex in the inner membrane, and MotX is inferred to be a sodium channel component (31, 32). On the other hand, PomA and PomB are similar to MotA and MotB, respectively, of proton-type motor components which are thought to form a proton channel. It is proposed that PomA has four transmembrane regions and a large cytoplasmic loop and that PomB spans the membrane once near the N terminus and has a conserved peptidoglycan-binding motif in the C-terminal region (2). Thus, we thought that PomA and PomB may have similar structure and function to the proton-type MotA and MotB from R. sphaeroides and that it might be possible to compare the coupling mechanisms of the proton and sodium ion flux for force generation. In addition, mutations conferring resistance to phenamil, a specific inhibitor of a sodium-driven motor or sodium channels (5), are found in both pomA and pomB (25). The phenamil-resistant mutants have also been isolated in V. parahaemolyticus, which is closely related to V. alginolyticus, and the mutations have been mapped in the homologous genes of either of pomA and pomB (17). These results strongly support the notion that PomA and PomB form a sodium-conducting channel.
In the case of another rotary machine, FoF1 ATPase (the enzyme that couples ion flux to ATP synthesis), the coupling ions can be proton or sodium, as with the bacterial flagellar motor. The Fo part of the structure is embedded in the membrane and consists of a, b, and c subunits (12). It has been suggested that the c subunits determine the ion specificity (19). The proton-type c subunits of E. coli and the sodium-type subunits of Propionigenium modestum have 25% identity (22). It has been found that a hybrid enzyme composed of the Fo part from the sodium type and the F1 part from the proton type is functional and shows different ion specificities depending on the conditions (18, 20, 27). Moreover, the ion recognition sites have been proposed based on comparison of amino acid sequences between the c subunits (49). Recently, it has been suggested that the coupling ion selectivity of FoF1 ATPase also involves the a subunit of the Fo part (21).
Ultimately, we hope to determine the torque-generating region or ion recognition sites of flagellar motors by comparison between R. sphaeroides MotA and V. alginolyticus PomA. As a precursor to this work, we sought to find the effects of introducing the entire R. sphaeroides motA and motB genes into pomA or pomB mutants of V. alginolyticus, respectively.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and media.
The strains and plasmids used are shown in Table 1. V. alginolyticus cells were cultured at 30°C in VC medium (0.5% Polypepton, 0.5% yeast extract, 0.4% K2HPO4, 3% NaCl, 0.2% glucose) or VPG medium (1% Polypepton, 0.4% K2HPO4, 3% NaCl, 0.5% glycerol). When necessary, chloramphenicol and kanamycin were added to final concentrations of 2.5 and 100 μg/ml, respectively. E. coli cells were cultured at 37°C in LB medium (1% Bacto Tryptone, 0.5% yeast extract, 0.5% NaCl). For E. coli, chloramphenicol and kanamycin were added to final concentrations of 25 and 50 μg/ml, respectively. The swimming speed was measured in Tris motility buffer TMN300 (300 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose), TMK300 (300 mM KCl, 50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose) or mixtures of TMN300 and TMK300 to vary the sodium concentration.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| V. alginolyticus strains | ||
| VIO5 | VIK4 laf (Rifr Pof+ Laf−) | 36 |
| VIO586 | VIO5 pomA (Rifr Laf− Pom−) | 2 |
| NMB190 | VIO5 ΔpomA (211-bp deletion) (Rifr Laf− Pom−) | This work |
| NMB152 | NMB201 pomB (Rifr Laf− Pom− Mpar) | 2 |
| NMB161 | NMB201 pomB (Rifr Laf− Pom− Mpar) | 25 |
| YM19 | 138-2 pof (Pof−) | 23 |
| E. coli strains | ||
| DH5α | F− λ−recA1 hsdR17 endA1 supE44 thi-1 relA1 gyrA96 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | 15 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Km λpirRK6 | 34 |
| Plasmids | ||
| pSU41 | kan (Kmr) Plac lacZα | 6 |
| pKY704 | Suicide vector (Cmr) | 46 |
| pYA301 | pSU41 0.8-kb BamHI-BamHI fragment (pomA+) | 25 |
| pMK201-M2 | pSU21 0.6-kb BamHI-BamHI fragment (pomA); 211-bp deletion in pomA | 26 |
| pYA303 | pSU41 1.9-kb BamHI-SacI fragment (pomAB+) | 25 |
| pRED1 | pUC19 2.5-kb SphI-SalI fragment (motAB+) | 41 |
| pYA603 | pSU41 2.5-kb HindIII-EcoRI fragment from pRED1 (native promoter region and motAB+) | This work |
| pYA6031 | pSU41 1.6-kb HindIII-SmaI fragment (native promoter and motA+) | This work |
| pYA701 | pSU41 0.8-kb BamHI-EcoRI fragment (motA+) | This work |
| pYA303-M2 | pSU41 1.7-kb BamHI-SacI fragment (pomA pomB+); 211-bp deletion in pomA | This work |
| pYA801 | pKY704 1.7-kb XbaI-SacI fragment (pomA pomB+) from pYA303-M2 | This work |
Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Mpar, motility resistant to phenamil; Rifr, rifampin resistant; Plac, lac promoter.
DNA manipulations.
Routine DNA manipulations were carried out by standard procedures (39). Restriction endonucleases and other enzymes for DNA manipulations were purchased from Takara Shuzo (Siga, Japan) and New England Biolabs (Beverly, Mass.).
Plasmid construction and site-directed mutagenesis.
The inserted SphI-SalI fragment of pRED1 (41) contains the whole motAB operon including the putative ς54-dependent promoter region. At first, we inserted the HindIII-EcoRI fragment of pRED1 (including that SphI-SalI fragment and parts of the multiple-cloning site) into vector pSU41, which can be maintained in Vibrio. The resulting plasmid was named pYA603 (see Fig. 1b). Next, we digested pYA603 with SmaI and ligated it to delete the latter half of motB. The multiple-cloning site of pSU41 does not have a SmaI site, but the transferred parts of the multiple-cloning site from pRED1 have such a site. This deletion derivative was named pYA6031. We constructed a plasmid, pYA701, including the PCR-amplified motA fragment from 40 bp upstream of the start codon to 3 bp downstream of the stop codon. The PCR amplification was done as described previously (25). In the reaction, we used the end primers RsmotA.B1 and RsmotA.E2, as well as pYA6031, as templates. RsmotA.B1 has a BamHI site and is homologous to the sense strand of the upstream RsmotA gene, 5′-GCGGGATCCTGCCGCTCCGGACCTGGATGA-3′. RsmotA.E2 including an EcoRI site is complementary to the antisense downstream of the gene, 5′-CTCGAATTCGCCTCACGCCGCCTTGCGCT-3′. This PCR-amplified fragment was inserted into pSU41 between the BamHI site and the EcoRI site. The motA gene of pYA701 is under the lac promoter directly and does not contain the native promoter.
FIG. 1.
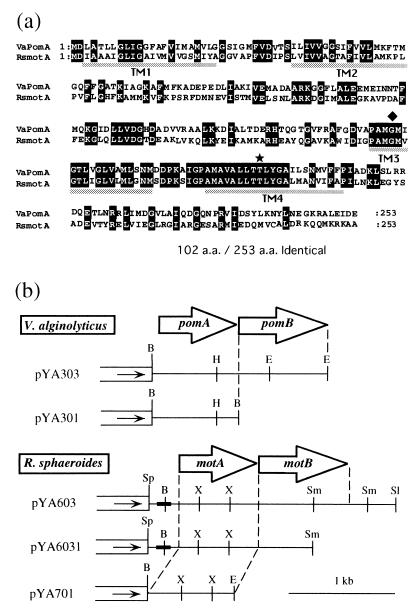
(a) Amino acid alignments of V. alginolyticus PomA and R. sphaeroides MotA. White letters in black boxes, diamond, and star show identical residues, the mutation site of the PomA mutant VIO586 (G154R), and the threonine residue that is highly conserved among all species of MotA, respectively. Dotted bars indicate putative transmembrane regions. (b) Restriction map of plasmids. White boxes and the direction of the arrowheads indicate the vector part of pSU41 and the direction of translation from the lac promoter, respectively. Insertional fragments are indicated by solid lines. The bold lines in the inserted fragments indicate the region of the native promoter. The open arrows show the coding regions of PomA, PomB, MotA, and MotB. Abbreviations: B, BamHI; H, HindIII; E, EcoRI; Sp, SphI; X, XhoI; Sm, SmaI; Sl, SalI.
To introduce mutations into pYA701, we used the two-step PCR method (25). We synthesized pairs of mutant primers, RsmotAT186C.1, designed for the sense strand (5′-GCTGCTCACGTGCCTCTACGGCG-3′), and RsmotAT186C.2, designed for the antisense strand (5′-CGCCGTAGAGGCACGTGAGCAGC-3′). In addition to these primers, we used both of the end primers and amplified the full-length R. sphaeroides motA gene. This mutated motA fragment was cloned into pSU41 (6), and the sequence was checked.
Transformation of the Vibrio cells.
Transformation of Vibrio cells was carried out by electroporation as described previously (24). The cells were subjected to osmotic shock and washed thoroughly with 20 mM MgSO4. Electroporation was carried out with the Gene Pulser electroporation apparatus (Japan Bio-Rad Laboratories, Tokyo, Japan) at an electric field strength between 5.0 and 7.5 kV/cm.
Construction of a pomA null mutant mutated on the chromosome.
The 0.4-kb HindIII fragment of a mutant pomA gene (allele 2 in reference 26) from pMK201-M2, which has a 211-bp deletion in the gene, was replaced with the 0.6-kb HindIII fragment of pomA in pYA303, and the resultant plasmid was named pYA303-M2. The XbaI-SacI fragment in pYA303-M2 was moved to a suicide vector, pKY704 (46). The resultant suicide plasmid (pYA801) was transformed into E. coli SM10λpir (34), and conjugal transfer was done to introduce pYA801 into V. alginolyticus VIO5. The suicide plasmids are expected to integrate into the chromosomal pomA gene. Transconjugants which showed chloramphenicol resistance were inoculated on a semisolid agar VPG plate to select motility mutants. Three Pom− colonies were found in 100 chloramphenicol-resistant transconjugants. Next, these Cmr Pom− cells were cultured in liquid broth without antibiotics and inoculated into new broth every day, and single colonies were isolated on a chloramphenicol-free VC hard agar plate. On day 4, we found one Cms Pom− colony. We confirmed the deletion mutation in the chromosomal pomA gene by the amplified fragment size of PCR and the complementation test with pYA301 and pYA303. This mutant was named NMB190(ΔpomA).
Detection of R. sphaeroides MotA protein by antipeptide antibody.
Preparation of cells and immunoblotting were done as described previously (48). For the first antibody of immunoblotting, we used antipeptide antibody against R. sphaeroides MotA (RsMotA) or V. alginolyticus PomA (VaPomA). The production and affinity purification of anti-RsMotA antibody were carried out by Sawady Technology (Tokyo, Japan). The selected peptide fragment was synthesized artificially, with the sequence NH2-CIEDQMVCALDRKQQMKRKAA-COOH, which is located at the C terminus of MotA. A cysteine was added to the N terminus, and the synthesized fragment was conjugated to keyhole limpet hemocyanin by it. Rabbits were immunized by the conjugated keyhole limpet hemocyanin. PomA91 antibody was used as an anti-VaPomA antibody, generated against three synthetic peptides designed for the different regions of PomA (48). For the second antibody, we used an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody.
Measurement of swimming speed.
An overnight culture in VC medium was inoculated into fresh VPG medium at a 100-fold dilution and grown at 30°C to exponential phase. Cells were harvested by centrifugation and suspended in TMN50 (50 mM NaCl, 250 mM KCl, 50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 5 mM glucose). The cell suspension was diluted about 100-fold into Tris motility buffers containing various concentrations of NaCl and 20 mM serine (to suppress the directional change of swimming). Motility of the cells was observed under a dark-field microscope and recorded on videotape. The swimming speed was determined as described previously (3).
Behavioral assay.
Tumbling frequency was determined as described previously (16). Cell suspension as prepared for measurement of swimming speed was diluted about 100-fold into TMN50, and individual chemoeffectors were added. Within 1 min of addition, cell motility was observed under a dark-field microscope and recorded on videotape. Each frame (30 frames in 1 s) on the videotapes was played back on a monitor and the number of times the cell changed direction in 1 s was noted. At least 20 cells were measured for any given condition.
RESULTS
Introduction of R. sphaeroides motA and motB into the pom mutants of V. alginolyticus.
The proton-type MotA of R. sphaeroides is very similar to the sodium-type PomA of V. alginolyticus (2). Both are 253-amino-acid proteins, and they have ca. 40% identity over the entire region (Fig. 1a). Moreover, the transmembrane regions of MotB and PomB have also high similarity (2, 40).
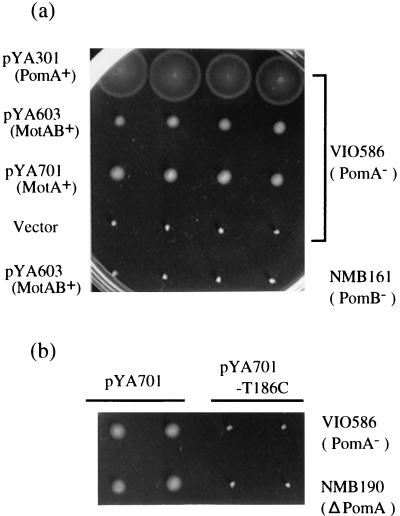
As shown in Fig. 1b, in pYA301, pYA303, and pYA701, the pomA, pomAB, and Rhodobacter motA genes were controlled under the lac promoter directly. pYA603 and pYA6031 include Rhodobacter motAB and motA genes under the native promoter. These plasmids were introduced into the pomA mutant, VIO586, and the pomB mutant, NMB161. Figure 2a shows the motility of the transformants. The transformed VIO586 was restored to motility by pYA603, pYA6031 (data not shown), and pYA701, which contain the motA gene of R. sphaeroides, although the swarm size was smaller than that of cells transformed by pYA301, which contain the pomA gene. On the other hand, the transformed pomB mutants, NMB161 and NMB152, could not have motility restored.
FIG. 2.
Swarming abilities of transformants. Fresh colonies were inoculated in 0.3% agar VPG plates containing 100 μg of kanamycin per ml and incubated at 30°C for 5 h. (a) Motility of the pomA mutant (VIO586) and the pomB mutant (NMB161) as the host strains were transformed with plasmids pYA301, pYA603, pYA701, pSU41, and pYA603. The introduced plasmids and the host strains are noted on the left and right, respectively. (b) Effect of the T186C mutation of R. sphaeroides MotA. Two pomA mutants, VIO586 and NMB190, were transformed with plasmids pYA701 and pYA701-T186C.
We thought that the defective PomA of VIO586 might be suppressed by MotA, and we identified the pomA mutation site of VIO586 by amplifying and sequencing the PCR fragment of the whole pomAB operon. Only the G154R mutation was found (Fig. 1a). If nonfunctional PomA proteins were expressed in VIO586, it was also possible that R. sphaeroides MotA could “suppress,” not complement, this PomA mutation. Thus, we constructed a pomA null mutant to examine the possibility.
The pomA deletion allele has been isolated by Kojima et al. (26). Using this mutant pomA and suicide vector pKY704, we constructed a strain (NMB190) carrying the chromosomal deletion mutation of pomA. Even in this strain, motA of pYA701 could restore motility to the same extent as in VIO586 (Fig. 2b).
The threonine residue in the fourth transmembrane region (Fig. 1a) is conserved among sodium-type PomA and all of the reported proton-type MotAs. This residue corresponds to position 202 in E. coli, and 186 in V. alginolyticus and R. sphaeroides. It was proposed that a mutation such as T202W in E. coli and T186C or I in V. alginolyticus would be nonfunctional (26, 42). We introduced the T186C mutation into pYA701 and examined if the mutated R. sphaeroides MotA could restore motility (Fig. 2b). Wild-type MotA (pYA701) restored motility to both pomA mutants. In contrast, mutant MotA (pYA701-T186C) did not. Other nonfunctional amino acid substitutions at highly conserved residues, T186I and P199L (P199 corresponds to P222 in E. coli) (9, 26, 51), also did not restore motility (data not shown). These results suggested that R. sphaeroides MotA, which is a proton component, could work as a replacement for PomA in V. alginolyticus.
Detection of R. sphaeroides MotA.
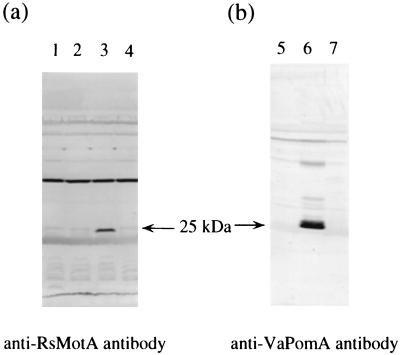
We tried to detect the R. sphaeroides proteins by immunoblotting with anti-RsMotA synthetic peptide antibody (Fig. 3a). A band with a molecular mass of 25 kDa was detected specifically from NMB190 harboring pYA701 (lane 3). The molecular mass (25 kDa) is almost equal to the predicted size of R. sphaeroides MotA (27 kDa). On the other hand, in the case of anti-VaPomA antibody (Fig. 3b), no band appeared in NMB190 harboring pYA701 (lane 7); however, the V. alginolyticus PomA band could be detected in NMB190 harboring pYA301 (lane 6). From these results, we assessed that this anti-RsMotA antibody could detect specifically R. sphaeroides MotA. None of the nonfunctional mutant R. sphaeroides MotA proteins could be detected by our system (for example, T186I in lane 4). The mutant proteins might be degraded more rapidly than the wild type or might be changed in this antigenicity.
FIG. 3.
Detection of the R. sphaeroides MotA protein by the antipeptide antibody. NMB190 cells harboring each plasmid (lanes 1 and 5, pSU41; lanes 2 and 6, pYA301; lanes 3 and 7, pYA701; lane 4, pYA701-T186I) were cultured until the mid-log phase, harvested, and suspended in distilled-deionized water. Immunoblottings were done with anti-RsMotA antibody (a) and anti-PomA antibody (b).
Characterization of the hybrid motor between the H+ and Na+ types.
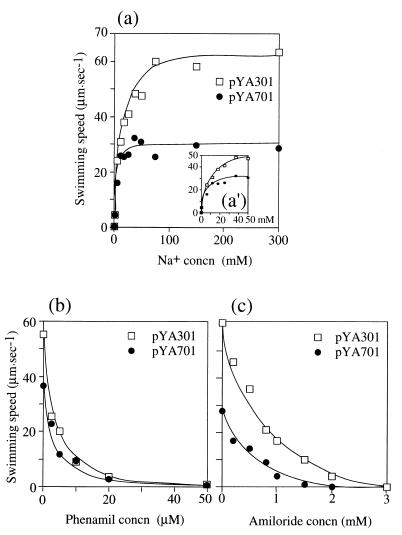
NMB190 with pYA701 should have hybrid motors composed of H+-type MotA and Na+-type PomB, MotX, and MotY. Which ions couple to the flagellar rotation in the motors? We examined this question by performing several experiments. In the first, we asked whether cells can swim in the Na+-free buffer TMK300 and whether the swimming speed of the cells is dependent on the Na+ concentration. As the results (Fig. 4a) show, the hybrid motor (pYA701/NMB190) could not rotate in Na+-free buffer as the Na+-type motor (pYA301/NMB190). Moreover, the speed of swimming generated by the hybrid motor increased with the Na+ concentration, although the maximum speed was low and saturated at lower Na+ concentration than that by the Na+-type motor of pYA301/NMB190. Second, we asked whether the free swimming is inhibited by phenamil, an Na+-driven motor-specific inhibitor. The swimming speeds were measured in various concentrations of phenamil (Fig. 4b). We found that pYA701/NMB190 cells were inhibited in motility strongly with the increase of the phenamil concentration, and stopped almost completely with 50 μM phenamil. pYA301/NMB190 cells showed similar results. They were also inhibited in motility by amiloride (Fig. 4c). The motility inhibition of pYA701/NMB190 cells by amiloride was competitive with sodium ions such as that of pYA301/NMB190 cells, although their initial swimming speeds were different (Table 2). Finally, we tested the effects of a protonophore, CCCP (Table 3). In alkaline Tris motility buffer, TMN300 (pH 8.5), YM19 (Pof− Laf+) cells could swim in the absence of 20 μM CCCP and could stop completely in the presence of CCCP, because lateral flagella have proton-driven motors. On the other hand, pYA301/NMB190 and pYA701/NMB190 cells did not stop in the presence of 20 μM CCCP, but they both stopped completely after the further addition of 20 μM 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO), a specific inhibitor of the Na+ pump (45). We concluded from these results that the hybrid motor of pYA701/NMB190 rotates the flagella by using Na+-motive force.
FIG. 4.
Swimming speed of NMB190 cells harboring pYA301 or pYA701. (a) The cells suspended in Tris motility buffer were diluted 100-fold into buffer containing various concentration of sodium ions, and the swimming speed was measured. (a′) Enlargement of the low-Na+-concentration part of panel a. (b and c) The swimming speed was measured at various concentrations of the specific inhibitor of the sodium motor, phenamil (b) or amiloride (c). The sodium concentration was 50 mM in all measurements.
TABLE 2.
Effect of Na+ concentration on the inhibition of motility by amiloride
| Na+ concn (mM) | Amiloride concn (mM) | Swimming speed (μm · s−1) for:
|
|
|---|---|---|---|
| pYA301/NMB190 | pYA701/NMB190 | ||
| 10 | 0 | 33 | 23 |
| 0.8 | 6 | 2 | |
| 50 | 0 | 54 | 28 |
| 0.8 | 18 | 6 | |
| 300 | 0 | 56 | 28 |
| 0.8 | 27 | 12 | |
TABLE 3.
Effect of pH and CCCP on swimming speed
| Na+ concn (mM) | pH | CCCP concn (μM) | Swimming speeda (μm · s−1) for:
|
||
|---|---|---|---|---|---|
| pYA301/NMB190 | pYA701/NMB190 | YM19 | |||
| 300 | 7.5 | 0 | 63 | 28 | 27 |
| 20 | 10 | 8 | 0 | ||
| 8.5 | 0 | 64 | 26 | 20 | |
| 20 | 64 (0) | 23 (0) | 0 | ||
| 0 | 7.5 | 0 | 0 | 0 | 22 |
The swimming speed after HQNO (20 μM final) addition is shown in parentheses.
We tested the chemotaxis of pYA701/NMB190, because pYA701/NMB190 formed a very small swarm on a semisolid agar plate even though its swimming speed in liquid medium was not so low. The tumble frequency of pYA301/NMB190 was increased from the basal level after 2 mM phenol was added as a repellent (Table 4). After the further addition of 20 mM serine as an attractant, the tumble frequency decreased to 0.40 s−1 from 3.10 s−1. In contrast, pYA701/NMB190 showed smooth-biased swimming as a basal condition and little response to either repellent or attractant.
TABLE 4.
Chemotaxis of NMB190 cells having pYA301 or pYA701
| Phenol concn (mM) | Serine concn (mM) | Tumble frequency (s−1) for:
|
|
|---|---|---|---|
| pYA301/NMB190 | pYA701/NMB190 | ||
| 0 | 0 | 1.81 | 0.20 |
| 2 | 0 | 3.10 | 0.25 |
| 2 | 20 | 0.40 | 0.05 |
DISCUSSION
Recently, two sodium-driven motor components, PomA and PomB, were identified in addition to MotX and MotY (2). They are similar to the proton motor components MotA and MotB and different from MotX and MotY. Thus, we thought that PomA and PomB might function as a sodium channel and hoped that it might be possible to study the ion specificity by various chimeric proteins between PomA and MotA. However, surprisingly, we found that the whole R. sphaeroides motA gene could partially complement a pomA mutant of V. alginolyticus. This worked despite the different codon usages of Rhodobacter and Vibrio genes (which may account for the less than 100% complementation that was achieved). We had previously tested for motility restoration of pomA mutants by E. coli MotA, but it did not work (data not shown). This is probably because E. coli MotA has lower similarity to Vibrio PomA than does R. sphaeroides and could not precisely interact with PomB. Thus, the lateral motor components LafT and LafU of Vibrio seem not to be interchangeable with PomA and PomB, because they are highly similar to E. coli MotA and MotB but not PomA and PomB (32). As shown in Fig. 1a, the similarity between the PomA and R. sphaeroides MotA transmembrane regions is very high. The large cytoplasmic loop regions have poor similarity, although some important residues for torque generation were conserved, for example, two proline, arginine, and aspartate residues proposed by Zhou et al. in E. coli MotA (51). It has been also proposed that the cytoplasmic loop of E. coli MotA interacts with the rotor part directly (52). Therefore, we speculate that if the transmembrane regions could form a functional channel, the conformational change coupling with the ion flux could be transmitted to the rotor since some important residues were at least conserved. If this is true, a chimeric protein, which consists of the transmembrane regions of R. sphaeroides MotA and the cytoplasmic loop of V. alginolyticus PomA, might propel the cell at the high speed generated by the native motor. This is currently being studied.
Which ion flux, proton or sodium, was coupled to the force generation in this hybrid motor? We conclude that sodium ions were involved in the torque generation of the hybrid motor for the following reasons. V. alginolyticus has two types of flagella, polar and lateral; the latter is a proton type, and it is known that the mutants defective in polar flagella can swim in sodium-free buffer by using the proton-driven Laf (4, 23) (Table 3). However, the hybrid motor could not rotate in sodium-free motility buffer at pH 7.5, when V. alginolyticus can make a sufficient proton gradient. Phenamil and its analogs, which are potent inhibitors of the sodium channels of various organisms (5), inhibit polar flagellar motors but not lateral ones. In particular, phenamil is a more specific inhibitor for the motor function because target sites of phenamil are found in the sodium motor proteins PomA and PomB (17, 25). Both the hybrid and native motors were inhibited, being severely dependent on the concentration of phenamil (Fig. 4b) and amiloride (Fig. 4c). The motility inhibition of the hybrid motors by amiloride was also competitive with sodium ions (Table 2). This shows that the effect of amiloride is specific in the hybrid motor. Moreover, it has been shown that the sodium-driven polar flagella can rotate even though the proton motive force is abolished by CCCP, because Vibrio has respiration-coupled Na+ pumps that are active only in an alkaline pH range (4, 23). The hybrid motor in pYA701/NMB190 and the sodium motor in pYA301/NMB190 did not stop in the presence of 20 μM CCCP under the alkaline condition. These results suggest that the hybrid motor is not dependent on the proton motive force.
This raises the question of which components decide ion selectivity. The most potent candidate seems to be PomB. Its N-terminal transmembrane region is thought to form a sodium channel with PomA, and it includes the important Asp that seems to be involved in proton transfer. The similarity between MotB and PomB in the N-terminal transmembrane region is very high. In this study, we showed that R. sphaeroides MotB could not suppress the pomB mutants (Fig. 2a). Thus, it is not clear whether PomB decides ion selectivity. The other candidates are MotX and MotY, sodium-type specific components (31, 32). Actually, it was suggested that MotX was a channel component of a sodium motor from the evidence that overproduction of MotX was lethal to E. coli at some sodium concentrations and that this effect was suppressed by the addition of amiloride (31). From the present information, we speculate that PomA, PomB, MotX, and MotY act together to form a sodium channel complex in Vibrio motor; moreover, multiple ion binding sites are located in these motor complexes and may recognize sodium ions.
It has been shown that chemotactic signals are transduced to the C ring of the flagellar rotor by phospho-CheY and that the binding of phospho-CheY changes the rotor conformation, although little is known about how the direction is determined. As shown in Table 4, pYA701/NMB190 cells rarely change direction, compared with pYA301/NMB190 cells. To explain this phenotype, we suggest that the MotA cytoplasmic loop interacts poorly with the C ring in hybrid motors because it has poor similarity to the PomA cytoplasmic loop and that this imperfect rotor-stator interaction is enough to generate torque but not to change the direction of flagellar rotation properly. It was reported that a single mutation in the E. coli MotA periplasmic loop resulted in a CW-biased phenotype (14), and the mutation in periplasmic space is supposed to affect the structure of the cytoplasmic loop and change the interaction with the switch complex of the rotor.
This work has shown that a component of a proton-driven flagellar motor can substitute for a component of a sodium-driven motor. It has opened the way for further studies on chimeric motor proteins, which should determine how different ions can be used to power flagellar rotation and should lead to a greater understanding of the whole flagellar rotation and switching processes themselves.
ACKNOWLEDGMENTS
We thank K. Yamamoto for plasmids and R. Macnab for critically reading the manuscript.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to I.K. and M.H.) and the Japan Society for the Promotion of Science (to Y.A.) and by grants from the Royal Society (to R.E.S.).
REFERENCES
- 1.Armitage J P, Macnab R M. Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol. 1987;169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M. Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolitycus. J Bacteriol. 1996;178:5024–5026. doi: 10.1128/jb.178.16.5024-5026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 5.Atsumi T, Sugiyama S, Cragoe E J, Jr, Imae Y. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J Bacteriol. 1990;172:1634–1639. doi: 10.1128/jb.172.3.1634-1639.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartolomé B, Jubete Y, Martínez E, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 7.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 8.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 9.Blair D F, Berg H C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 10.Chun S Y, Parkinson J S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 11.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 12.Fillingame R H. Molecular mechanics of ATP synthesis by F1Fo-type H+-translocating ATP synthases. In: Krulwich T A, editor. The bacteria. Vol. 12. New York, N.Y: Academic Press, Inc.; 1990. pp. 345–391. [Google Scholar]
- 13.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 14.Garza A G, Biran R, Wohlschlegel J A, Manson M D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 15.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homma M, Oota H, Kojima S, Kawagishi I, Imae Y. Chemotactic responses to an attractant and a repellent by the polar and lateral flagellar systems of Vibrio alginolyticus. Microbiology. 1996;142:2777–2783. doi: 10.1099/13500872-142-10-2777. [DOI] [PubMed] [Google Scholar]
- 17.Jaques S, Kim Y K, McCarter L L. Mutations conferring resistance to phenamil and amiloride, inhibitors of sodium-driven motility of Vibrio parahaemolyticus. Proc Natl Acad Sci USA. 1999;96:5740–5745. doi: 10.1073/pnas.96.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaim G, Dimroth P. Construction, expression and characterization of a plasmid-encoded Na+-specific ATPase hybrid consisting of Propionigenium modestum Fo-ATPase and Escherichia coli F1-ATPase. Eur J Biochem. 1994;222:615–623. doi: 10.1111/j.1432-1033.1994.tb18904.x. [DOI] [PubMed] [Google Scholar]
- 19.Kaim G, Dimroth P. A double mutation in subunit c of the Na+-specific F1Fo-ATPase of Propionigenium modestum results in a switch from Na+ to H+-coupled ATP synthesis in the Escherichia coli host cells. Signal transduction schemes of bacteria. J Mol Biol. 1995;253:726–738. doi: 10.1006/jmbi.1995.0586. [DOI] [PubMed] [Google Scholar]
- 20.Kaim G, Dimroth P. Formation of a functionally active sodium-translocating hybrid F1Fo ATPase in Escherichia coli by homologous recombination. Eur J Biochem. 1993;218:937–944. doi: 10.1111/j.1432-1033.1993.tb18450.x. [DOI] [PubMed] [Google Scholar]
- 21.Kaim G, Dimroth P. A triple mutation in the a subunit of the Escherichia coli/Propionigenium modestum F1Fo ATPase hybrid causes a switch from Na+ stimulation to Na+ inhibition. Biochemistry. 1998;37:4626–4634. doi: 10.1021/bi973022f. [DOI] [PubMed] [Google Scholar]
- 22.Kaim G, Ludwig W, Dimroth P, Schleifer K H. Cloning, sequencing and in vivo expression of genes encoding the Fo part of the sodium-ion-dependent ATP synthase of Propionigenium modestum in Escherichia coli. Eur J Biochem. 1992;207:463–470. doi: 10.1111/j.1432-1033.1992.tb17072.x. [DOI] [PubMed] [Google Scholar]
- 23.Kawagishi I, Maekawa Y, Atsumi T, Homma M, Imae Y. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol. 1995;177:5158–5160. doi: 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawagishi I, Okunishi I, Homma M, Imae Y. Removal of the periplasmic DNase before electroporation enhances efficiency of transformation in a marine bacterium Vibrio alginolyticus. Microbiology. 1994;140:2355–2361. [Google Scholar]
- 25.Kojima S, Asai Y, Atsumi T, Kawagishi I, Homma M. Na+-driven flagellar motor resistant to phenamil, an amiloride analog, caused by mutations of putative channel component. J Mol Biol. 1999;285:1537–1547. doi: 10.1006/jmbi.1998.2377. [DOI] [PubMed] [Google Scholar]
- 26.Kojima S, Kuroda M, Kawagishi I, Homma M. Random mutagenesis of the pomA gene encoding a putative channel component of the Na+-driven polar flagellar motor of Vibrio alginolyticus. Microbiology. 1999;145:1759–1767. doi: 10.1099/13500872-145-7-1759. [DOI] [PubMed] [Google Scholar]
- 27.Laubinger W, Deckers H G, Altendorf K, Dimroth P. A hybrid adenosinetriphosphatase composed of F1 of Escherichia coli and Fo of Propionigenium modestum is a functional sodium ion pump. Biochemistry. 1990;29:5458–5463. doi: 10.1021/bi00475a008. [DOI] [PubMed] [Google Scholar]
- 28.Macnab R. Flagellar switch. In: Hoch J A, Silhavey T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 181–199. [Google Scholar]
- 29.Magariyama Y, Sugiyama S, Muramoto K, Kawagishi I, Imae Y, Kudo S. Simultaneous measurement of bacterial flagellar rotation rate and swimming speed. Biophys J. 1995;69:2154–2162. doi: 10.1016/S0006-3495(95)80089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magariyama Y, Sugiyama S, Muramoto K, Maekawa Y, Kawagishi I, Imae Y, Kudo S. Very fast flagellar rotation. Nature. 1994;371:752. doi: 10.1038/371752b0. [DOI] [PubMed] [Google Scholar]
- 31.McCarter L L. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarter L L. MotY, a component of the sodium-type flagellar motor. J Bacteriol. 1994;176:4219–4225. doi: 10.1128/jb.176.14.4219-4225.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarter L L, Wright M E. Identification of genes encoding components of the swarmer cell flagellar motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J Bacteriol. 1993;175:3361–3371. doi: 10.1128/jb.175.11.3361-3371.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muramoto K, Kawagishi I, Kudo S, Magariyama Y, Imae Y, Homma M. High-speed rotation and speed stability of the sodium-driven flagellar motor in Vibrio alginolyticus. J Mol Biol. 1995;251:50–58. doi: 10.1006/jmbi.1995.0415. [DOI] [PubMed] [Google Scholar]
- 36.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Packer H L, Harrison D M, Dixon R M, Armitage J P. The effect of pH on the growth and motility of Rhodobacter sphaeroides WS8 and the nature of the driving force of the flagellar motor. Biochim Biophys Acta. 1994;1188:101–107. doi: 10.1016/0005-2728(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 38.Poole P S, Sinclair D R, Armitage J P. Real time computer tracking of free-swimming and tethered rotating cells. Anal Biochem. 1988;175:52–58. doi: 10.1016/0003-2697(88)90359-4. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Habor Laboratory Press; 1989. [Google Scholar]
- 40.Shah D S, Armitage J P, Sockett R E. Rhodobacter sphaeroides WS8 expresses a polypeptide that is similar to MotB of Escherichia coli. J Bacteriol. 1995;177:2929–2932. doi: 10.1128/jb.177.10.2929-2932.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah D S, Sockett R E. Analysis of the motA flagellar motor gene from Rhodobacter sphaeroides, a bacterium with a unidirectional, stop-start flagellum. Mol Microbiol. 1995;17:961–969. doi: 10.1111/j.1365-2958.1995.mmi_17050961.x. [DOI] [PubMed] [Google Scholar]
- 42.Sharp L L, Zhou J, Blair D F. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp L L, Zhou J, Blair D F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 44.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokuda H, Unemoto T. Characterization of the respiration-dependent Na+ pump in the marine bacterium Vibrio alginolyticus. J Biol Chem. 1982;257:10007–10014. [PubMed] [Google Scholar]
- 46.Xu M, Yamamoto K, Honda T. Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh) J Bacteriol. 1994;176:4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yorimitsu T, Sato K, Asai Y, Kawagishi I, Homma M. Functional interaction between PomA and PomB, the Na+-driven flagellar motor components of Vibrio alginolyticus. J Bacteriol. 1999;181:5103–5106. doi: 10.1128/jb.181.16.5103-5106.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Fillingame R H. Changing the ion binding specificity of the Escherichia coli H+-transporting ATP synthase by directed mutagenesis of subunit c. J Biol Chem. 1995;270:87–93. doi: 10.1074/jbc.270.1.87. [DOI] [PubMed] [Google Scholar]
- 50.Zhao R H, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 51.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 52.Zhou J, Lloyd S A, Blair D F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]