Abstract
Sixteen exopolysaccharide (EPS)-producing Lactococcus lactis strains were analyzed for the chemical compositions of their EPSs and the locations, sequences, and organization of the eps genes involved in EPS biosynthesis. This allowed the grouping of these strains into three major groups, representatives of which were studied in detail. Previously, we have characterized the eps gene cluster of strain NIZO B40 (group I) and determined the function of three of its glycosyltransferase (GTF) genes. Fragments of the eps gene clusters of strains NIZO B35 (group II) and NIZO B891 (group III) were cloned, and these encoded the NIZO B35 priming galactosyltransferase, the NIZO B891 priming glucosyltransferase, and the NIZO B891 galactosyltransferase involved in the second step of repeating-unit synthesis. The NIZO B40 priming glucosyltransferase gene epsD was replaced with an erythromycin resistance gene, and this resulted in loss of EPS production. This epsD deletion was complemented with priming GTF genes from gram-positive organisms with known function and substrate specificity. Although no EPS production was found with priming galactosyltransferase genes from L. lactis or Streptococcus thermophilus, complementation with priming glucosyltransferase genes involved in L. lactis EPS and Streptococcus pneumoniae capsule biosynthesis could completely restore or even increase EPS production in L. lactis.
Many gram-positive bacteria produce significant amounts of capsular polysaccharides (CPSs) or exopolysaccharides (EPSs). Most molecular studies have focused on the CPSs from strains of Streptococcus pneumoniae, group B streptococci, and Staphylococcus aureus (23). These CPSs have unique structures that determine the serotype and virulence of these pathogens. Their biosynthesis is encoded by large clusters of genes that often show unidirectional organization, are transcribed into single polycistronic mRNAs, and appear to be coordinately expressed (15, 16, 20, 26). In these clusters, the serotype-specific genes encoding the glycosyltransferases (GTFs) are flanked by genes that are common to all serotypes and are likely to be involved in processes like chain length determination, polymerization, and export (12, 15, 16, 27). Several lactic acid bacteria are known to produce EPSs that are of industrial importance, as they are beneficial for the structure of dairy products (2). Recently, the genes encoding EPS production in the dairy starters Streptococcus thermophilus Sfi6 and Lactococcus lactis NIZO B40 were characterized and their organization was found to be similar to that of the CPS biosynthesis gene clusters of the gram-positive pathogens (29, 34). Functional analysis of the NIZO B40 eps genes demonstrated that the epsDEF genes are functional homologues of the cps14EFG genes from S. pneumoniae serotype 14 and code for GTFs that are involved in identical steps of the polysaccharide biosynthesis route (35). In general, the GTF involved in linking the first sugar of the repeating unit to the lipid carrier, here referred to as the priming GTF, is highly homologous in gram-positive bacteria, while other GTFs are often unique or have very little homology to others (12, 15, 27, 29, 34).
In spite of the increasing sequence information on the CPS or EPS gene clusters in gram-positive cocci, very little is known about the function of the predicted GTF genes and even less is known about their specificities. By investigation of the GTF genes expressed in Escherichia coli, the substrate specificities of GTFs involved in the biosynthesis of S. pneumoniae serotype 14, L. lactis NIZO B40, and S. thermophilus Sfi6 were determined (12, 30, 34). However, it was reported that GTF genes expressed in a heterologous host could result in a different composition of the EPS (30). Therefore, we have used a recently developed homologous expression system to demonstrate the substrate specificity of the epsDEFG genes of L. lactis NIZO B40 (35). Here we describe a screening approach used to identify new GTF genes in L. lactis and show the diversity of GTF genes in L. lactis and their EPSs, resulting in a classification of three major groups. Two new priming GTF genes were selected, and their function and substrate specificity were determined. Finally, a transcomplementation of a knockout of the NIZO B40 epsD gene encoding the priming GTF was realized by controlled expression of several homologous GTF genes derived from different gram-positive cocci.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli was grown in L-broth-based medium at 37°C (24). L. lactis was grown at 30°C in M17 broth (Difco Laboratories) supplemented with 0.5% glucose (GM17) or in a chemically defined medium (22). If appropriate, the media contained chloramphenicol (10 μg/ml), erythromycin (10 μg/ml), or ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | 9 | |
| L. lactis | ||
| NIZO B35 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| NIZO B36 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| NIZO B39 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| NIZO B40 | Lac+ Eps+ multiplasmid strain harboring pNZ4000 | 34 |
| NIZO B891 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| NIZO B1136 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| NIZO B1137 | Lac+ Eps+ multiplasmid strain | NIZO collection |
| SBT 0495 | Lac+ Eps+ multiplasmid strain | 17 |
| H414 | Lac+ Eps+ multiplasmid strain | 8 |
| SD8 | Lac+ Eps+ multiplasmid strain | 19 |
| SD11 | Lac+ Eps+ multiplasmid strain | 19 |
| VI6 | Lac+ Eps+ multiplasmid strain | 19 |
| VI8 | Lac+ Eps+ multiplasmid strain | 19 |
| MLT1 | Lac+ Eps+ multiplasmid strain | Quest collection |
| MLT2 | Lac+ Eps+ multiplasmid strain | Quest collection |
| MLT3 | Lac+ Eps+ multiplasmid strain | Quest collection |
| NZ3900 | pepN::nisRnisK | 4 |
| Plasmids | ||
| pNZ4080 | Apr, 3.8-kb pUC19 derivative carrying NIZO B35 orfU | This study |
| pNZ4081 | Apr, 5.5-kb pUC19 derivative carrying NIZO B35 orfU and epsD | This study |
| pNZ4082 | Apr, 4.0-kb pUC18 derivative carrying NIZO B35 epsD | This study |
| pNZ4083 | Cmr, 4.4-kb pNZ8020 derivative carrying NIZO B35 epsD | This study |
| pNZ4085 | Apr, 6.3-kb p119HE derivative carrying NIZO B891 epsD | This study |
| pNZ4086 | Apr, 7.2-kb p119HE derivative carrying NIZO B891 epsDEF | This study |
| pNZ4087 | Cmr, 4.2-kb pNZ8020 derivative carrying NIZO B891 epsD | This study |
| pNZ4090 | Cmr, 4.5-kb pNZ8020 derivative carrying cps14E | This study |
| pNZ4091 | Cmr, 5.8-kb pNZ8020 derivative carrying cps14EFG | This study |
| pNZ4055 | Eryr Eps−, pNZ4000ΔepsD derivative carrying ery from pIL253 | This study |
| pNZ4030 | Eryr Eps+, 27-kb pNZ4000 derivative carrying ery from pIL253 | 34 |
| pNZ4070 | Cmr, 4.6-kb pNZ8020 fragment carrying NIZO B40 epsD | 35 |
Lac+, lactose-fermenting phenotype; Eps+, EPS-producing phenotype; Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Eryr, erythromycin resistant.
DNA isolation and manipulation.
Isolation of E. coli plasmid DNA and standard recombinant DNA techniques were performed as described by Sambrook et al. (24). Large-scale isolation of E. coli plasmid DNA for nucleotide sequence analysis was performed with JetStar columns by following the instructions of the manufacturer (Genomed). Isolation and transformation of L. lactis plasmid DNA were performed as previously described (6). Southern blots were hybridized with eps gene probes at 45°C and washed with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 45°C before exposure.
Nucleotide sequence analysis.
Automatic double-stranded DNA sequence analysis was performed on both strands with an ALFred DNA sequencer (Pharmacia Biotech). Sequencing reactions were accomplished by using the AutoRead sequencing kit, initiated by using Cy5-labelled universal and reverse primers, and continued with synthetic primers in combination with Cy5-13-dATP by following the instructions of the manufacturer (Pharmacia Biotech). Sequence data were assembled and analyzed by using the PC/GENE program, version 6.70 (IntelliGenetics).
Construction of plasmids.
For expression of the NIZO B35 eps genes in E. coli, a 1.0-kb ScaI-HincII fragment containing orfU, a 2.7-kb ScaI-KpnI fragment containing orfU-epsD, and a 1.3-kb ScaI fragment containing epsD were cloned under control of the lac promoter in pUC18 or pUC19 (41). To express the NIZO B891 eps genes in E. coli, a 1.0-kb ScaI-BalI fragment containing epsD and a 1.9-kb ScaI-EcoRI fragment containing epsDEF were cloned under control of the lac promoter in pJF119HE (7). For expression of the NIZO B35 and NIZO B891 epsD genes in L. lactis, a 1.3-kb ScaI fragment and a 1.0-kb ScaI-BalI fragment, respectively, were cloned under control of the nisA promoter in pNZ8020 (5). To express the streptococcal cps14 GTF genes in L. lactis, a 1.3-kb XbaI-PvuII fragment containing the GTF part of cps14E and a 2.6-kb XbaI fragment containing cps14EFG were cloned from pMK100 (10) under control of the nisA promoter in pNZ8020. To express the streptococcal epsE GTF gene in L. lactis, a 1.8-kb EcoRV-XbaI fragment containing epsE was cloned from pFS30 (29) under control of the nisA promoter in pNZ8020. To construct a NIZO B40 epsD gene disruption, a PCR was used to clone the flanking regions containing epsC (by using the primers 5′-AGCAGCAAGCTTTTCAAGTTATATATTGA-3′ and 5′-TTCAGAGGATCCCTCAAAAACTTCCAT-3′) and epsEF (by using the primers 5′-CTACATGGATCCGATGCTTATTAAAGTAA-3′ and 5′-ATTATTGAATTCATCAGAATAATTCCCCTA-3′) in pUC18, making use of the EcoRI, BamHI, and HindIII sites of the primers (underlined). The ery gene of pIL253 was cloned from pUC18Ery (34) into the BamHI site between the epsC and epsEF fragments in the same orientation as the eps genes. The complete EcoRI-HindIII insert was transferred to pG+host8 (14), resulting in a tetracycline-resistant (Tetr), erythromycin-resistant (Eryr) construct containing a temperature-sensitive replicon which is not functional at 37°C. The resulting plasmid was transformed to strain NZ4010 harboring EPS plasmid pNZ4000 (34), and transformants were subsequently cultured at 37°C. A Tets Eps− Eryr double-crossover mutant of pNZ4000 was obtained in which epsD was exchanged for the ery gene (pNZ4055). The pUC, pJF119HE, and pG+host derivatives were constructed in E. coli DH5α, and the pNZ8020 derivatives were constructed in L. lactis NZ3900.
EPS purification and characterization.
L. lactis was grown in 50 ml of defined medium containing 2% glucose for 48 h at 30°C, and after pelleting of the cells, EPS was purified by dialysis and lyophilization and quantified by gel permeation high-performance liquid chromatography analysis using dextran 500 as a standard as described before (34). Sugar analysis was performed by high-performance liquid chromatography analysis of the monosaccharide units after complete hydrolysis with 4 N HCl (37). To analyze the EPS in overproducing strain NZ3900 harboring pNZ4055 and pNZ8020 derivatives, induction was performed with nisin A (1 ng ml−1) at an optical density at 600 nm of 0.5 (4).
GTF activity assays and TLC analysis.
GTF activity assays and thin-layer chromatography (TLC) analysis were performed with permeabilized E. coli cells as described before (34). Permeabilized L. lactis cells were prepared like those of E. coli after a 30-min incubation with lysozyme (10 mg ml−1) on ice. After incubation with UDP-[14C]glucose and/or UDP-[14C]galactose, the extracted lipid fractions were subjected to complete and mild acid hydrolysis and analyzed by TLC and autoradiography to detect 14C-labelled monosaccharides (complete acid hydrolysis) and oligosaccharides (mild acid hydrolysis), respectively.
Nucleotide sequence accession numbers.
The nucleotide sequences of the NIZO B35 and NIZO B891 eps gene cluster fragments are available under GenBank accession no. AF100297 and AF100298.
RESULTS
Diversity of lactococcal GTF genes and EPSs.
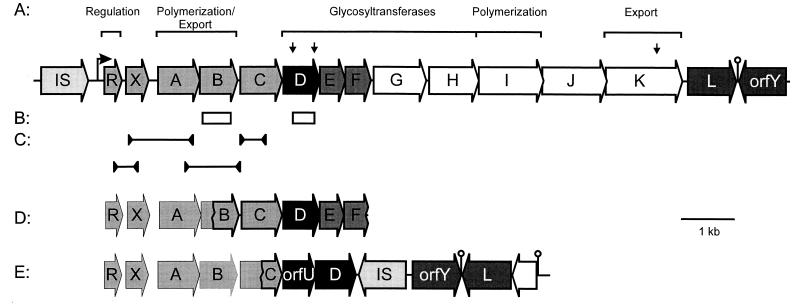
In a search for new GTF genes, we screened a collection of 16 different EPS-producing L. lactis strains at the genetic and biochemical levels. To localize putative eps gene clusters, DNA from the strains was probed with an internal fragment of the L. lactis NIZO B40 epsB gene (Fig. 1B), which is highly conserved and has homologues in all studied EPS or CPS gene clusters of gram-positive cocci (34). All of the L. lactis strains tested contained a single plasmid (>20 kb) that hybridized with the epsB probe (results not shown). This confirms previous suggestions that EPS production in L. lactis is plasmid encoded (19, 38, 39). The diversity of the plasmid-encoded GTF genes was studied by analyzing their hybridization to the NIZO B40 epsB and epsD genes (Fig. 1B). This epsD gene codes for the priming glucosyltransferase and shows homology to other priming GTF genes (34). For this purpose, plasmid DNA of all strains was digested with SstI, which has three sites within the NIZO B40 eps gene cluster, two of which are present in the epsD gene (Fig. 1A). All strains hybridized with both epsB and epsD probes, but the sizes of the hybridizing SstI bands differed considerably, allowing genetic differentiation (Table 2).
FIG. 1.
(A) Genetic map of the eps gene cluster of L. lactis NIZO B40. The SstI recognition sites are indicated by downward-pointing arrows. The predicted functions of the gene products are depicted above the map (34). (B) DNA fragments of the NIZO B40 eps gene cluster used for hybridization. (C) PCR fragments generated by the primers indicated by the arrowheads used to determine the order of the conserved eps genes of various strains (see text). (D and E) Genetic maps of the eps gene cluster of L. lactis NIZO B891 and NIZO B35 based on DNA sequences of cloned fragments and on PCR analysis.
TABLE 2.
Hybridization patterns of SstI-digested lactococcal plasmid DNA and sugar composition of EPS produced
| Strain | Fragment size (kb)a
|
Molar ratiob
|
|||
|---|---|---|---|---|---|
| epsB | epsD | Gal | Glc | Rha | |
| Group I | |||||
| NIZO B40 | 7.5 | 0.4 | 1.00 | 1.82 | 0.88 |
| SBT 0495 | 7.5 | 0.4 | 1.00 | 1.70 | 0.82 |
| NIZO B1136 | 7.5 | 0.4 | 1.00 | 1.70 | 0.82 |
| VI6 | 7.5 | 0.4 | 1.00 | 1.46 | 0.73 |
| VI8 | 7.5 | 0.4 | 1.00 | 1.80 | 0.89 |
| MLT3 | 7.5 | 0.4 | 1.00 | 1.46 | 0.73 |
| Group II | |||||
| NIZO B35 | 7.5 | 15 | 1.00 | ||
| NIZO B36 | 7.5 | 15 | 1.00 | ||
| H414 | 7.5 | 15 | 1.00 | ||
| SD8 | 7.5 | 13 | 1.00 | ||
| SD11 | 7.5 | 13 | 1.00 | ||
| Group III | |||||
| NIZO B891 | 6.5 | 12 | 1.00 | 1.50 | |
| MLT1 | 6.5 | 12 | 1.00 | 1.46 | |
| MLT2 | 6.5 | 12 | 1.00 | 1.44 | |
| Unique | |||||
| NIZO B39 | 7.5 | 12 | 1.00 | 0.60 | 0.55 |
| NIZO B1137 | 15 | 18 | 1.00 | 1.79 | |
Approximate sizes of fragments hybridizing with NIZO B40 probes are listed.
Abbreviations: Rha, rhamnose; Gal, galactose; Glc, glucose. Molar ratios relative to galactose content are listed.
The biochemical diversity of the EPSs isolated from the 16 strains was studied by determining the nature and molar ratio of the sugar monosaccharides (Table 2). No sugars other than glucose, galactose, or rhamnose were present in these polymers. Based on the genetic and biochemical diversity of the putative GTF genes and the EPSs, the L. lactis strains could be classified into three main groups (Table 2). Group I contains six strains that produced EPS containing the monosaccharides galactose, glucose, and rhamnose and includes strains SBT 0495 and NIZO B40, which produce EPSs with repeating units consisting of →4)-[α-l-Rhap-(1→2)][α-d-Galp-1-PO4-3]- β-d-Galp-(1→4)-β-d-Glcp-(1→4)-β-d-Glcp-(1→ (18, 31, 34). Group II comprises five strains that produced EPS with only galactose and includes strain H414, the EPS repeating unit of which is known to be →4)-[β-d-Galp-(1→3)-β-d-Galp-(1→3)]-α-d-Galp-(1→4)-β-d-Galp-(1→3)-β-d-Galp-(1→ (8). This group shows restriction fragment length polymorphism for epsD. Group III contains three strains that produced EPS composed of both galactose and glucose in a molar ratio of approximately 2 to 3. In addition to these three major groups, there are two strains (NIZO B39 and NIZO B1137) that show a unique combination of hybridization pattern and EPS sugar composition.
Genetic variety of eps gene clusters.
From the three major groups of EPS-producing lactococci, strains NIZO B40, NIZO B35, and NIZO B891 were selected as representatives and further characterized together with the unique strains NIZO B39 and NIZO B1137, as the structure of their EPS is known (NIZO B40) or is being analyzed (NIZO B35 and NIZO B891) (32, 33). Plasmid DNA of these strains was analyzed by Southern blot analysis with specific probes for each of the genes of the epsRXABCDEFGHIJKLorfY operon from NIZO B40 plasmid pNZ4000. The genes epsR, epsX, epsA, epsB, epsC, and epsD hybridized with the EPS plasmids of all five strains, and epsL and orfY hybridized with those of NIZO B40, NIZO B35, NIZO B39, and NIZO 1137, indicating their conservation in all gene clusters. The other eps genes of NIZO B40 only hybridized with NIZO B40 plasmid pNZ4000.
To further determine the organization of the different eps gene clusters, specific primers based on the NIZO B40 eps gene cluster were used for PCRs to detect fragments overlapping epsRX, epsXA, epsAB, or epsBC (Fig. 1C). For the epsRX, epsAB, and epsBC fragments, all of the strains yielded PCR products identical in size (results not shown). For the epsXA fragments, NIZO B39, NIZO B891, and NIZO B1137 yielded PCR products that were 165 bp larger than those of NIZO B35 and NIZO B40 (results not shown). These results confirm the homologies found by the Southern blot analysis and indicate that all of the gene clusters contain a conserved region with the same organization i.e., epsRXABC.
NIZO B35 and NIZO B891 eps genes.
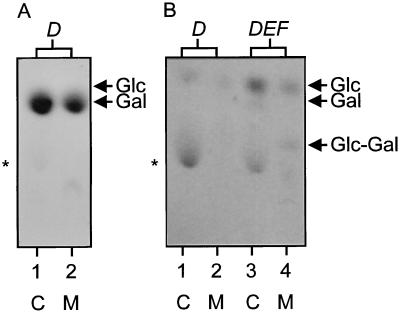
To study the function of the priming GTF genes, strains NIZO B35 and NIZO B891 were selected because they represent the two major groups with an EPS structure that differs markedly from that of strain NIZO B40 (Table 2). Overlapping fragments of the eps gene clusters of NIZO B35 and NIZO B891 that hybridized with the NIZO B40 epsD probe were cloned and sequenced (Fig. 1). The homologies of the deduced gene products are listed in Table 3. Unexpectedly, the NIZO B35 gene cluster contained two different genes that are homologous to NIZO B40 epsD (orfU and epsD, respectively). To test which of these epsD-like genes encodes the priming GTF activity, each of these was cloned under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lac promoter in pUC18 and GTF activities were determined in E. coli. When NIZO B35 epsD was induced in E. coli, galactosyltransferase activity could be detected (Fig. 2A). However, when orfU was induced, no GTF activity could be detected (data not shown). Simultaneous induction of both orfU and epsD from NIZO B35 resulted in the same galactosyltransferase activity as that found with NIZO B35 epsD alone (data not shown). These results indicate that NIZO B35 epsD encodes a priming GTF activity and orfU is either not involved in these synthetic steps, poorly expressed, or unstable.
TABLE 3.
Homologies of the L. lactis NIZO B35 and NIZO B891 eps gene products to those from L. lactis NIZO B40
| Strain and gene | No. of amino acids of protein | Proposed function of gene producta | Homology (% identity) to:
|
||
|---|---|---|---|---|---|
| NIZO B40 | S. thermophilus Sfi6 | S. pneumoniae serotype 14 | |||
| NIZO B35 | |||||
| epsCb | 125 | Unknown | EpsC (93.6) | EpsB (33.9) | Cps14B (33.0) |
| orfU | 199 | Unknown | EpsD (85.1) | EpsE (42.1) | Cps14E (39.1) |
| epsD | 251 | Gal-P-TF | EpsD (39.2) | EpsE (41.9) | Cps14E (36.3) |
| orf982 | 296 | Transposase | orf982 (98.0) | ||
| orfY | 300 | Unknown | OrfY (95.7) | ||
| epsL | 300 | Unknown | EpsL (88.6) | ||
| NIZO B891 | |||||
| epsBb | 155 | Chain length determination | EpsB (93.5) | EpsD (31.8) | Cps14D (33.1) |
| epsC | 230 | Unknown | EpsC (96.9) | EpsB (26.7) | Cps14B (29.2) |
| epsD | 228 | Glc-P-TF | EpsD (88.1) | EpsE (40.0) | Cps14E (39.1) |
| epsE | 149 | Gal-TFc | EpsE (40.4) | Cps14F (83.8) | |
| epsFd | 150 | Gal-TF | EpsF (36.5) | Cps14G (53.0) | |
Gal-P-TF, priming galactosyltransferase; Glc-P-TF, priming glucosyltransferase; Gal-TF, galactosyltransferase.
Incomplete at 5′ end.
Accessory function to EpsF (see text).
Incomplete at 3′ end.
FIG. 2.
TLC of 14C-labelled intermediates isolated from the lipid fraction of permeabilized E. coli cells. (A) E. coli expressing NIZO B35 epsD incubated with UDP-[14C]galactose. (B) E. coli expressing NIZO B891 epsD (1, 2) or NIZO B891 epsDEF incubated with a combination of UDP-[14C]glucose and UDP-[14C]galactose (3, 4). The positions of the standard sugars glucose (Glc), galactose (Gal), and lactose (Glc-Gal) are indicated on the right. The products which are nonspecific for lactococcal GTF activity are indicated by the asterisk on the left. C, complete acid hydrolysis; M, mild acid hydrolysis.
The products of the NIZO B891 epsD, epsE, and epsF genes are expected to be the GTFs involved in the first two steps of EPS biosynthesis in this strain, as they are homologous to NIZO B40 epsD, epsE, and epsF. Fragments containing NIZO B891 epsD and epsDEF were cloned under control of the lac promoter in medium-copy-number expression vector pJF119HE, since attempts to clone them in pUC18 were unsuccessful. When NIZO B891 epsD was expressed in E. coli, only glucosyltransferase activity could be detected (Fig. 2B, lanes 1 and 2). When epsDEF was expressed, both glucosyltransferase and galactosyltransferase activities could be detected (Fig. 2B, lane 3) and the lipid-linked oligosaccharide had the same mobility on TLC as lactose (Fig. 2B, lane 4). The incorporation of 14C-labelled sugars was approximately fivefold lower than that of cells expressing NIZO B40 or NIZO B35 eps genes (data not shown), and this lower GTF activity resulted in an increase in the appearance of a product in the complete acid hydrolysates which is nonspecific for lactococcal GTF activity (Fig. 2B, lanes 1 and 3). These results demonstrate that NIZO B891 epsD encodes a glucosyltransferase linking glucose to the lipid carrier and epsE and/or epsF encode a galactosyltransferase linking galactose via a β-1,4 linkage to lipid-linked glucose, resulting in lipid-linked lactose. Methylation analysis of NIZO B891 EPS has confirmed the presence of 1,4-linked glucose and galactose residues (32). In analogy to the homologous pneumococcal proteins Cps14F and Cps14G (11), EpsF is expected to contain GTF activity while EpsE is expected to have an accessory function.
Homologous and heterologous complementation of a NIZO B40 epsD mutant.
To analyze the function of the GTFs in a gram-positive host, we constructed pNZ4055, a pNZ4000 derivative in which the epsD gene was replaced with an erythromycin resistance (ery) gene. This was achieved through a double crossover with a pGhost8 derivative containing the ery gene from pIL252 flanked by NIZO B40 epsC and epsEF. The ery gene has no terminator, ensuring expression of the downstream genes (34). L. lactis harboring pNZ4055 was erythromycin resistant and produced no EPS. To test whether the epsD knockout could be complemented, the pNZ8020 derivative pNZ4070 carrying the NIZO B40 epsD gene under control of the lactococcal nisA promoter was cotransformed with pNZ4055 into L. lactis NZ3900, which allows the use of the NICE (nisin-controlled expression) system (5, 13). Upon induction with nisin A, the EPS production of the resulting heteroplasmid strain was even higher than that of the wild-type strain, demonstrating that controlled overexpression of the epsD gene was achieved (Table 4). To test their heterologous complementation ability, various priming GTF genes from L. lactis, S. thermophilus, and S. pneumoniae were cloned in pNZ8020. The EPS produced by cultures of L. lactis harboring pNZ4055 and pNZ8020 derivatives was quantified, and the monosaccharide composition was determined (Table 4). The NIZO B40 and NIZO B891 genes encoding glucosyltransferases were able to complement the EPS-deficient phenotype. While expression of the NIZO B40 epsD gene restored EPS production completely, the amount of EPS produced by expression of NIZO B891 epsD was dramatically lower. A low GTF activity of the NIZO B891 EpsD compared to that of NIZO B40 EpsD was also found in E. coli (see above). In contrast, complete restoration of wild-type EPS production by heterologous complementation was achieved by using the cps14E gene of S. pneumoniae type 14 (Table 4). This gene is involved in pneumococcal capsule synthesis, encoding the priming glucosyltransferase (11), and is homologous to NIZO B40 epsD (34). Expression of the NIZO B35 epsD or the S. thermophilus Sfi6 epsE gene (30), both encoding a galactosyltransferase, did not complement the EPS-deficient phenotype (Table 4), indicating that a matching sugar specificity is required for transcomplementation. Although expression of cps14E restored EPS production completely, complementation with the pneumococcal cps14EFG genes resulted in reduced production of wild-type EPS compared to complementation with cps14E alone. The products of the cps14F and cps14G genes are involved in the second step of serotype 14 CPS biosynthesis linking galactose to lipid-linked glucose (11). Therefore, it is likely that they will compete for the lipid-linked glucose as the acceptor molecule with the products of the NIZO B40 epsE and epsF genes that link glucose to it, resulting in lipid-linked cellobiose (35). If so, it may be assumed that the lipid-linked lactose resulting from Cps14F and Cps14G activity cannot be used for NIZO B40 EPS biosynthesis, hence lowering NIZO B40 EPS production. These results demonstrate that functional expression of gram-positive GTFs in L. lactis is possible and may result in heterologous complementation when the enzymes are alike in sugar specificity.
TABLE 4.
Functional expression of streptococcal GTF genes to complement an epsD knockout in L. lactis NZ3900
| Original host | Gene(s) | Specificitya | EPS production (mg liter−1)b |
|---|---|---|---|
| L. lactis NIZO B40 | epsD | Glc | 133 |
| L. lactis NIZO B891 | epsD | Glc | 7.5 |
| L. lactis NIZO B35 | epsD | Gal | <0.5 |
| S. pneumoniae type 14 | cps14E | Glc | 102 |
| S. pneumoniae type 14 | cps14EFG | Glc + Gal | 3.9 |
| S. thermophilus Sfi6 | epsE | Gal | <0.5 |
Glc, glucosyltransferase; Gal, galactosyltransferase.
Amounts of EPS are the mean values of data from two independent cultures. The EPS production of L. lactis NZ3900 harboring pNZ4030 (wild type) is 113 mg liter−1, and that of NZ3900 harboring pNZ4055 (ΔepsD) is <0.5 mg liter−1. All EPSs had a monosaccharide composition identical to that produced by the wild type.
DISCUSSION
We have analyzed the diversity of GTF genes of 16 different ropy L. lactis strains and the EPSs they produced, allowing division into three major groups and two individual strains. The grouping observed is in agreement with the known structural EPS information, as the EPSs produced by group I strains NIZO B40 and SBT 0495 are identical and differ from those of strains H414 (group II) and NIZO B891 (group III) (8, 18, 32, 34). Furthermore, methylation analysis of the EPS produced by strain NIZO B35 (group II) demonstrated that it contains the same galactose linkages as the H414 EPS and it is expected to have an identical EPS repeating unit (33). The sugar specificity of the GTFs needed for EPS biosynthesis in the different groups can be predicted according to the sugars present in the EPSs. The results suggest that EPS biosynthesis in all groups requires active galactosyltransferases, while groups I and III also need glucosyltransferases and only group I needs rhamnosyltransferases.
The genetic organization of the lactococcal eps gene clusters is conserved with respect to the first genes epsRXABC, which seem to be highly homologous for all strains. Furthermore, these genes share the most homology with those of other gram-positive polysaccharide biosynthesis gene clusters, including those of S. aureus, S. pneumoniae, S. agalactiae, and S. thermophilus (34). These homologies are confirmed for the NIZO B891 epsB and epsC and NIZO B35 epsC gene products by analysis of their nucleotide sequences, demonstrating that these genes are common to gene clusters involved in the biosynthesis of many gram-positive polysaccharide types (Table 3). It is likely that they will be involved in general functions and not directly related to the composition of the polymer produced (16, 25, 29).
The epsL and orfY genes have homologues in all of the lactococcal gene clusters tested. The function of these genes is unknown. OrfY is homologous to the regulator protein LytR from Bacillus subtilis (34). NIZO B40 epsL can be disrupted by single crossover using an internal gene fragment or overproduced without any effect on EPS production (36). Nonetheless, epsL- and orfY-like genes are also found at the end of the eps gene cluster from S. thermophilus CNRZ368 adjacent to an IS element (1).
The genetic organization of the NIZO B35 eps gene cluster differs from that of NIZO B40 and NIZO B891 by an interruption of the gene cluster by an IS982 element after the first GTF gene. An almost identical IS element is located upstream of the NIZO B40 eps gene cluster (Fig. 1A). Furthermore, the NIZO B35 gene cluster differs by containing two epsD-like genes, of which only one is actively involved in the first step of EPS biosynthesis, as was shown by the analysis of the products formed in the GTF activity assays of E. coli cells expressing NIZO B35 epsD, orfU, or epsD and orfU. A possible explanation for the differences in organization of the NIZO B35 eps gene cluster is that it has undergone rearrangement mediated by the IS element and received an additional epsD gene from another eps gene cluster. Horizontal gene transfer of parts of polysaccharide gene clusters has been observed in various bacteria, including S. pneumoniae (3).
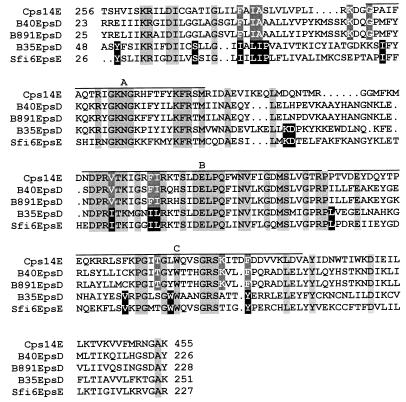
All 16 of the L. lactis strains studied carry an epsD homologue which was cloned and subjected to functional analysis for strains NIZO B35 and NIZO B891. The product of the NIZO B891 epsD gene is a glucosyltransferase that is more homologous to NIZO B40 EpsD than to the product of the NIZO B35 epsD gene, which is a galactosyltransferase (Table 3). Sequence alignment of several EpsD-like proteins from different polysaccharide biosynthesis systems with known glucosyl- or galactosyltransferase activity showed three blocks that are conserved in all of the proteins (40). An alignment of the EpsD-like gram-positive GTFs with known sugar specificity shows that the three blocks are also conserved in these proteins (Fig. 3). Blocks A and B are predicted to interact with the lipid carrier, and block C is supposed to contain specific conserved residues for each type of transferase (40). From these, only a galactosyltransferase-specific tyrosine was observed (Fig. 3) and different residues appeared to be conserved for the gram-positive GTFs, demonstrating that the previously reported residues are not critical in determining sugar specificity. GTF activity involves amino acids that can catalyze an acid-base reaction. Hydrophobic cluster analysis of various β-GTFs has shown two aspartic acid residues with a spacing of approximately 50 amino acids to be conserved, and these are predicted to be the catalytic residues (28). Four conserved aspartate residues (D) and two conserved glutamate residues (E) were found for the gram-positive GTFs (Fig. 3), two of which are likely to be the catalytic residues. Two possible candidates are the conserved E residue in block C in combination with the conserved D residue in the C terminus just outside block C, which are separated by 50 amino acids (51 in Cps14E). The amino acid sequence of NIZO B35 OrfU lacks 30 amino acids at its C terminus compared to the other priming GTFs, including this conserved aspartate.
FIG. 3.
Multiple-sequence alignment of priming GTFs with known sugar specificity from gram-positive bacteria. Cps14E, B40EpsD, and B891EpsD are glucosyltransferases from S. pneumoniae serotype 14 (11) and L. lactis NIZO B40 and NIZO B891, respectively. B35EpsD and Sfi6EpsE are galactosyltransferases from L. lactis NIZO B35 and S. thermophilus Sfi6 (30), respectively. Residues conserved in all five sequences, residues conserved only in glucosyltransferases, and residues conserved only in galactosyltransferases are shaded light grey, dark grey, and black, respectively. The three conserved blocks (A, B, and C) described by Wang et al. (40) are indicated.
Disruption of the NIZO B40 epsD gene could be complemented by homologous expression of NIZO B40 epsD and heterologous expression of NIZO B891 epsD or the streptococcal capsule biosynthesis gene cps14E, which is known to be involved in a similar reaction (10). The use of a controlled expression system enabled the expression of GTFs that did not complement the mutation and could be toxic to the cell as a result of the accumulation of lipid-linked intermediates (NIZO B35 epsD, S. thermophilus epsE, and S. pneumoniae cps14EFG), as has been reported for the heterologous expression of several gram-negative GTFs (21). Moreover, to the best of our knowledge, this is the first demonstration of functional heterologous expression of a GTF gene in a gram-positive host allowing the expression of GTF genes from different origins by the shotgun or directed-cloning approach in L. lactis. Furthermore, these results demonstrate that the enzymes involved in the biosynthesis of different polysaccharides can be functionally coupled, although the eps genes are located on different transcriptional units. The possibility of constructing clean deletion mutations in the lactococcal eps gene cluster combined with the use of the NICE expression system, enabling induced expression of GTF genes, opens the way to polysaccharide engineering in L. lactis and provides a new approach to the study of polysaccharide biosynthesis genes of gram-positive cocci.
ACKNOWLEDGMENTS
We thank Ingeborg Boels for assistance with the cloning of Sfi6 epsE and EPS quantifications. We are grateful to Peter Vandenbergh for providing us with strains MLT1, MLT2, and MLT3; Marc Kolkman for providing us with plasmid pMK100; Francesca Stingele for providing us with plasmid pFS30; and Willemiek van Casteren for sharing her results prior to publication. We thank Dick van den Berg and Roland Siezen for critically reading the manuscript.
Part of this work was supported by EC research grants 1116/92 1.6 and BIOT-CT96-0498.
REFERENCES
- 1.Bourgoin F. Ph.D. thesis. France: Université Henri Poincaré Nancy I, Vandœvre-lès-Nancy; 1997. [Google Scholar]
- 2.Cerning J. Exocellular polysaccharides produced by lactic acid bacteria. FEMS Microbiol Rev. 1990;87:113–130. doi: 10.1111/j.1574-6968.1990.tb04883.x. [DOI] [PubMed] [Google Scholar]
- 3.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 4.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I J, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruyter P G G A, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine protease. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 7.Fürste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 8.Gruter M, Leeflang B R, Kuiper J, Kamerling J P, Vliegenthart J F G. Structure of the exopolysaccharide produced by Lactococcus lactis subspecies cremoris H414 grown in a defined medium or skimmed milk. Carbohydr Res. 1992;231:273–291. doi: 10.1016/0008-6215(92)84025-n. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Kolkman M A B, Morrison D A, van der Zeijst B A M, Nuijten P J M. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyltransferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 12.Kolkman M A B, Wakarchuk W, Nuijten P J M, van der Zeijst B A M. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol Microbiol. 1997;26:197–208. doi: 10.1046/j.1365-2958.1997.5791940.x. [DOI] [PubMed] [Google Scholar]
- 13.Kuipers O P, de Ruyter P G G A, Kleerebezem M, de Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 14.Maguin E, Prévost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 16.Muñoz R, Mollerach M, López R, García E. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol. 1997;25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 17.Nakajima H, Toyoda S, Toba T, Itoh T, Mukai T, Kitazawa H, Adachi S. A novel phosphopolysaccharide from slime-forming Lactococcus lactis subspecies cremoris SBT 0495. J Dairy Sci. 1990;73:1472–1477. [Google Scholar]
- 18.Nakajima H, Hirota T, Toba T, Itoh T, Adachi S. Structure of the extracellular polysaccharide from the slime-forming Lactococcus lactis subsp. cremoris SBT0495. Carbohydr Res. 1992;224:245–253. doi: 10.1016/0008-6215(92)84110-e. [DOI] [PubMed] [Google Scholar]
- 19.Neve H, Geis A, Teuber M. Plasmid-encoded functions of ropy lactic acid streptococcal strains from Scandinavian fermented milk. Biochimie. 1988;70:437–442. doi: 10.1016/0300-9084(88)90218-0. [DOI] [PubMed] [Google Scholar]
- 20.Ouyang S, Lee C Y. Transcriptional analysis of type 1 capsule genes in Staphylococcus aureus. Mol Microbiol. 1997;23:473–482. doi: 10.1046/j.1365-2958.1997.d01-1865.x. [DOI] [PubMed] [Google Scholar]
- 21.Pollock T J, van Workum W A T, Thorne L, Mikolajczak M J, Yamazaki M, Kijne J W, Armentrout R W. Assignment of biochemical functions to glycosyltransferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol. 1998;180:586–593. doi: 10.1128/jb.180.3.586-593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 28.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stingele F, Neeser J-R, Mollet B. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stingele F, Newell J W, Neeser J-R. Unraveling the function of glycosyltransferases in Streptococcus thermophilus Sfi6. J Bacteriol. 1999;181:6354–6360. doi: 10.1128/jb.181.20.6354-6360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Casteren W H M, Kabel M A, Dijkema C, Schols H A, Beldman G, Voragen A G J. Endoglucanase V and a phosphatase from Trichoderma viride are able to act on modified exopolysaccharide from Lactococcus lactis subsp. cremoris B40. Carbohydr Res. 1999;137:131–144. doi: 10.1016/s0008-6215(99)00072-5. [DOI] [PubMed] [Google Scholar]
- 32.van Casteren, W. H. M., C. Dijkema, H. A. Schols, G. Beldman, and A. G. J. Voragen. Unpublished results.
- 33.van Casteren, W. H. M. Personal communication.
- 34.van Kranenburg R, Marugg J D, Willem N J, van Swam I I, de Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide production in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 35.van Kranenburg R, van Swam I I, Marugg J D, Kleerebezem M, de Vos W M. Exopolysaccharide biosynthesis in Lactococcus lactis NIZO B40: functional analysis of the glycosyltransferase genes involved in the synthesis of the polysaccharide backbone. J Bacteriol. 1999;181:338–340. doi: 10.1128/jb.181.1.338-340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Kranenburg, R. Unpublished results.
- 37.van Riel J, Olieman C. Selectivity control in the anion-exchange chromatographic determination of saccharides in dairy products using pulsed amperometric detection. Carbohydr Res. 1991;215:39–46. [Google Scholar]
- 38.Vedamuthu E R, Neville J M. Involvement of a plasmid in production of ropiness (mucoidness) in milk cultures by Streptococcus cremoris MS. Appl Environ Microbiol. 1986;51:677–682. doi: 10.1128/aem.51.4.677-682.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Von Wright A, Tynkkynen S. Construction of Streptococcus lactis subsp. lactis strains with a single plasmid associated with mucoid phenotype. Appl Environ Microbiol. 1987;53:1385–1386. doi: 10.1128/aem.53.6.1385-1386.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]