Abstract
Simple Summary
An analysis of the presence of mutations of 105 genes in appendiceal cancers through the lens of the reviewed literature supports the view that in most of them, the inactivation of tumor suppressor genes, such as TP53 and SMAD4, is required in parallel with the reactivation of genes with oncogenic potentials, such as KRAS, GNAS, and BRAF, which support the main tumor processes, cell proliferation, angiogenesis, and evasion of apoptosis. Of all appendiceal cancers, the most mutated genes are reported in mucinous neoplasms of the appendix, not including those in the RAS–RAF–MEK–ERK signaling pathway, followed by low-grade appendiceal mucinous neoplasms, appendiceal goblet cell adenocarcinomas, and mucinous adenocarcinomas of the appendix, in which this signaling pathway is most frequently affected, showing its importance in their tumorigenesis. Microsatellite instability rarely occurs in appendix cancers, being reported only in adenocarcinomas.
Abstract
In appendiceal cancers, the most frequently mutated genes are (i) KRAS, which, when reactivated, restores signal transduction via the RAS–RAF–MEK–ERK signaling pathway and stimulates cell proliferation in the early stages of tumor transformation, and then angiogenesis; (ii) TP53, whose inactivation leads to the inhibition of programmed cell death; (iii) GNAS, which, when reactivated, links the cAMP pathway to the RAS–RAF–MEK–ERK signaling pathway, stimulating cell proliferation and angiogenesis; (iv) SMAD4, exhibiting typical tumor-suppressive activity, blocking the transmission of oncogenic TGFB signals via the SMAD2/SMAD3 heterodimer; and (v) BRAF, which is part of the RAS–RAF–MEK–ERK signaling pathway. Diverse mutations are reported in other genes, which are part of secondary or less critical signaling pathways for tumor progression, but which amplify the phenotypic diversity of appendiceal cancers. In this review, we will present the main genetic mutations involved in appendix tumors and their roles in cell proliferation and survival, and in tumor invasiveness, angiogenesis, and acquired resistance to anti-growth signals.
Keywords: appendix tumor, KRAS genes, point mutations, signaling pathways, adenocarcinomas
1. Introduction
The vermiform appendix is a cylindrical organ with an average length of 9 cm (5–35 cm) and a transverse diameter of 6 mm, attached to the cecum about 2 cm from its junction with the jejunum. Its origin is located near the iliocecal valve, in the cecal fundus or the posteromedial border of the cecum, and its apex hangs freely in the abdomen, with different orientations [1,2]: retrocecal/retrocolic, pelvic, post-ileal, subcecal, pre-ileal, and paracecal [3], in most cases adopting the retrocecal position [2]. Although considered an atavic organ, the vermiform appendix secretes about 2–3 mL of mucinous fluid each day and appears to have an immunoprotective and lymphatic function, especially in childhood, contributing to the maturation of B lymphocytes and to the production of immunoglobulin A, a function of recolonization of the colon with beneficial bacteria, which it stores, and an endocrine function, through the production and secretion of certain amines and hormones [1,2].
Throughout life, the appendix can be the site of various pathologies, categorized into inflammatory pathologies (acute appendicitis), pathologies related to congenital anomalies of the appendix and other related diseases, and tumors of the appendix. Tumors of the appendix are very rare pathologies and are classified by the World Health Organization [4,5] into Hyperplastic polyps, Sessile serrated lesion without dysplasia, Serrated dysplasia (low or high grade), Appendiceal mucinous neoplasms (low or high grade), Appendiceal adenocarcinomas NOS (mucinous and signet ring cell), Undifferentiated carcinomas NOS; Goblet cell adenocarcinoma, and Appendiceal neuroendocrine neoplasms (well-differentiated neuroendocrine tumors: neuroendocrine tumors NOS, neuroendocrine tumor, grades 1–3, L-cell tumor, glucagon-like peptide-producing tumor, PP/PYY-producing tumor, enterochromaffin-cell carcinoid, and serotonin-producing carcinoid; poorly differentiated neuroendocrine carcinomas or neuroendocrine carcinoma NOS, with large or small cells; and mixed neuroendocrine–non-neuroendocrine neoplasms). In these tumors, the most common mutations occur in the KRAS gene, in addition to which several mutations are common and considered necessary in the epidemiology of appendiceal cancers, including those that occur in TP53, GNAS, SMAD4, and BRAF genes, which occur with a frequency of more than 10% in cases of appendiceal cancer. Other mutations occur less frequently or are mentioned only in single cases. Some genes are predominantly active during the embryonic period, playing critical roles in cell proliferation and evading the apoptotic pathway, thus contributing to the growth and development of the embryo and fetus. However, after birth, they must diminish or cease their function, partially, totally, or conditionally (by various physiological processes) silenced. Their reactivation turns them into proto-oncogenes and is essential in initiating tumor processes. In cancer, genes whose expression products are involved in the uptake of extracellular biological signals and their transmission to the nucleus, activating factors that stimulate cell proliferation while inhibiting apoptotic signals are essential. In the case of tumor growth beyond the size that allows oxygenation and feeding by diffusion, it is necessary to activate signaling pathways that inhibit cell differentiation and stimulate angiogenesis, invasiveness, and tumor metastasis. After the chemotherapeutic treatment has started, cell clones resistant to the chemical agent are selected.
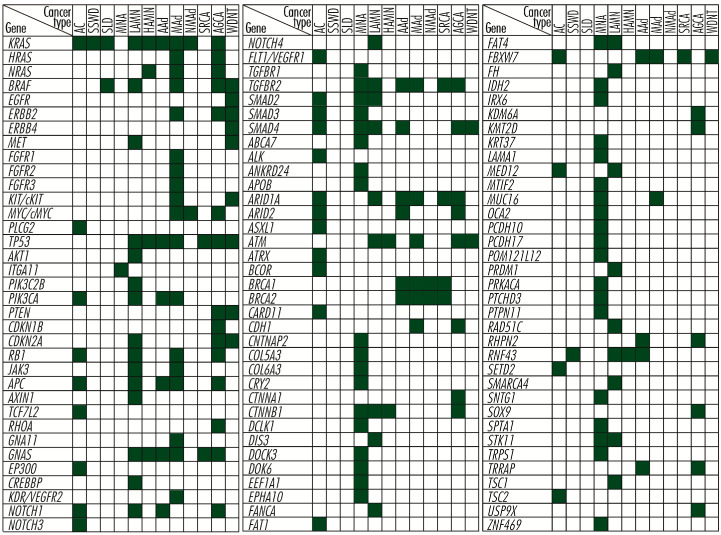
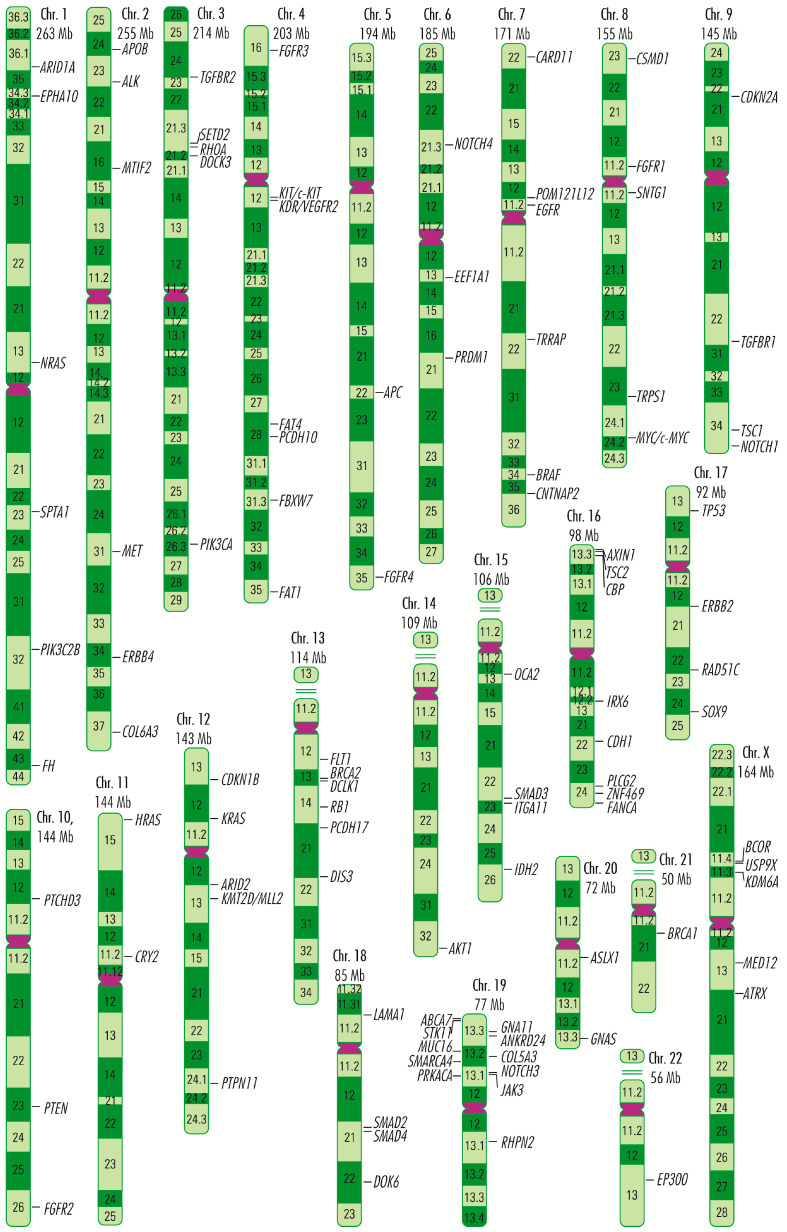
Strictly in appendiceal cancers, most mutations tend to occur in genes whose products are involved in stimulating cell proliferation, such as the activating mutations in RAS and RAF gene families; in evading apoptosis, such as the activating mutations in GNAS gene and RAS and RAF gene families; as well as silencing mutations in TP53 or RB1 genes, or in acquiring resistance to anti-growth signals, such as the mutations in SMAD genes family [6,7,8,9,10,11,12,13,14,15,16]. The description of these genes is made by grouping them, generally, according to the biological processes in which their products are involved, without exhausting the range of functions they perform in the body. As is well known, some proteins perform several functions in a manner dependent on the signaling pathway through which they transmit biological signals, so placing them in one category or another is not likely to include them exclusively in these. Thus, RAS, RAF, EGFR/ERBB, and FGFR gene families, and MET, KIT/cKIT, MYC/c-MYC, PLCG2, TP53, AKT1, ITGA11, PIK3C2B, PIK3CA, PTEN, CDKN1B, CDKN2A, RB1, JAK3, APC, AXIN1, and TCF7L2 genes are involved principally in cell proliferation; the RHOA gene, in cell survival and tumor invasiveness/metastasis; GNA11, GNAS, EP300, CPB/CREBBP, KDR/VEGFR2, NOTCH1, NOTCH3, NOTCH4, and FLT1/VEGFR1 genes, in angiogenesis; and SMAD2, SMAD3, SMAD4, SMADA, TGFBR1, and TGFBR2 genes, in acquiring insensitivity to anti-growth signals. A vast number of genes, ABCA7, ALK, ANKRD24, APOB, ARID1A, ARID2, ASXL1, ATM, ATRX, BCOR, BRCA1, BRCA2, CARD11, CDH1, CNTNAP2, COL5A3, COL6A3, CRY2, CTNNA1, CTNNB1, DCLK1, DIS3, DOCK3, DOK6, EEF1A1, EPHA10, FANCA, FAT1, FAT4, FBXW7, FH, IDH2, IRX6, KDM6A, KMT2D, KRT37, LAMA1, MED12, MLL2, MTIF2, MUC16, OCA2, PCDH10, PCDH17, POM121L12, PRDM1, PRKACA, PTCHD3, PTPN11, RAD51C, RHPN2, RNF43, SETD2, SMARCA4, SNTG1, SOX9, SPTA1, STK11, TRPS1, TRRAP, TSC1, TSC2, USP9X, and ZNF4699 (Figure 1), which in appendiceal cancers occur mutated less frequently or in single cases, are involved in other processes related to tumor growth and development and are grouped separately, but without considering them less important.
Figure 1.
The 105 genes in which mutations are reported in appendiceal cancers. AC, appendiceal carcinoma; SSWD, sigmoid sinus wall dehiscence; SLD, single-lineage dysplasia; MNA, myelocytomatosis amplification; LAMN, low-grade appendiceal mucinous neoplasm; HAMN, high-grade appendiceal mucinous neoplasm; AAd, appendiceal adenocarcinoma; MAd, mucinous adenocarcinoma; SRCA, extragastric signet ring cell adenocarcinoma; AGCA, advanced gastric cancer; WDNT, well-differentiated neuroendocrine tumor. Green, genes reported in appendiceal cancers; white, genes not associated with appendiceal cancers.
In this review, we will present the genetic mutations involved in appendix tumors, including genes whose proteins are involved in cell proliferation and survival; tumor invasiveness, angiogenesis, and acquired insensitivity to anti-growth signals; and different signaling pathways which are modified in appendix cancer.
2. Mutations in Genes Whose Proteins Are Involved in Cell Proliferation
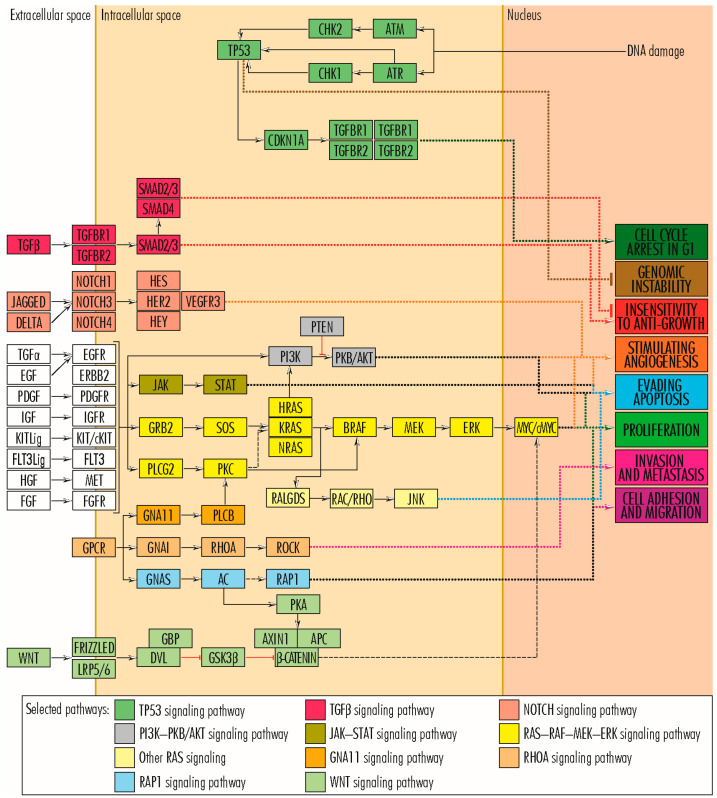
Cancer cell proliferation is stimulated primarily by the RAS–RAF–MEK–ERK signaling pathway and secondarily by other pathways such as PI3K–PKB/AKT and WNT (Figure 2). In the RAS–RAF–MEK–ERK signaling pathway, which comprises the sequence of proteins EGF/TGFA–EGFR(EGF–ERBB2/PDGF–PDGFR/KITLG–KIT/FGF–FGFR/FLT3LG–FLT3/IGF–IGF1R/HGF–MET)–GRB2–SOS–RAS–RAF–MEK–ERK–c-FOS/c-JUN/c-MYC/ETS1–cyclin D1, a central role is played by members of the RAS gene family (KRAS–Kirsten ras oncogene homolog, HRAS–HRAS proto-oncogene and NRAS–Neuroblastoma RAS Viral Oncogene Homolog), which encodes GTPases involved in extracellular signal transduction between the cell membrane and the Golgi apparatus. They link the RAS–RAF–MEK–ERK signaling pathway to other signaling pathways, such as PI3K–PKB/AKT, involved in cell proliferation and evading cell apoptosis. In addition to RAS family proteins, the RAS–RAF–MEK–ERK signaling pathway also includes members of the RAF, EGFR, and FGFR families and the MET, KIT, MYC, and PLCG2 proteins, which can bare mutations in some appendiceal cancers (Figure 2). Physiologically, the WNT signaling pathway is an essential mechanism in regulating tissue morphogenesis and repair during embryogenesis. However, when it functions abnormally in adults, it is associated with several types of cancers, including colorectal, breast, lung, oral, cervical, and hematopoietic neoplasms [17]. Abnormalities of the WNT signaling pathway may include mutations in APC, AXIN1, RHOA, and TCF7L2 genes, reported in some appendiceal cancers. On the other hand, the TP53 protein plays a significant role in ensuring the fate of cells whose genetic material has been altered and which, inactivated by mutations, can leave the cell proliferation pathway open. Other genes, such as RB1, typically have anti-tumor action by arresting the cell in G-phases. However, by acquiring mutations and losing the function of the encoded protein, they leave the way open for tumor proliferation [14].
Figure 2.
The signaling pathways important in cancer (adapted from KEGG Pathways, 2020 [14]).
2.1. RAS–RAF–MEK–ERK Signaling Pathway
2.1.1. RAS Gene Family
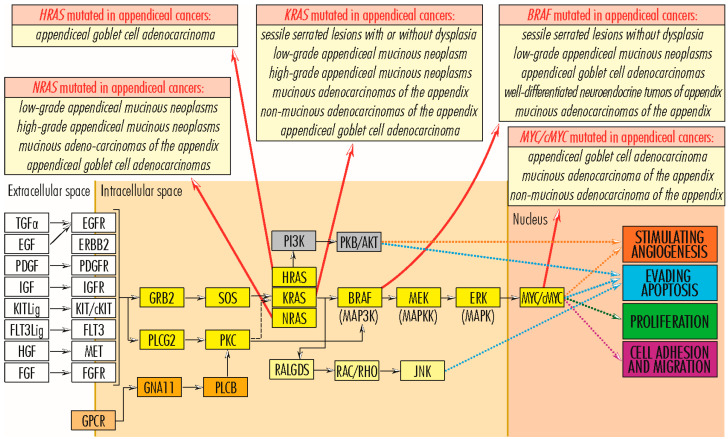
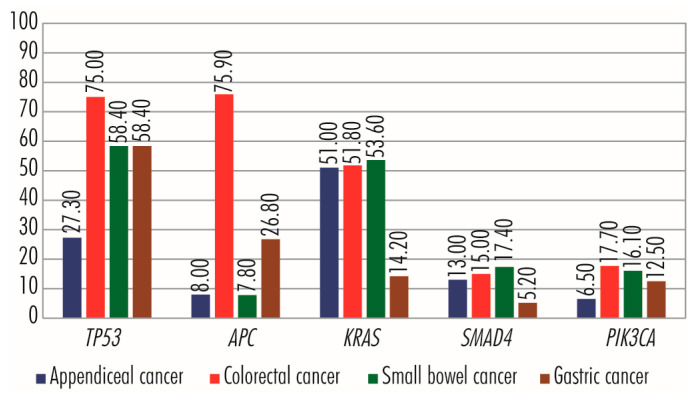
Of the three members of the RAS gene family, in appendiceal cancers, the most common mutations occur in the KRAS gene and have an activating effect. The KRAS gene (12p12.1) encodes the membrane GTP-ase protein KRAS (189 amino acids; 21,656 Da), which is involved in cell division and proliferation [14]. Inactive in adults, the KRAS gene is reactivated by single-amino-acid substitution mutations (missense mutations), and the altered KRAS protein promotes uncontrolled cell proliferation. Regaining KRAS gene function due to mutations is common in many tumor types [18], particularly those of the gastrointestinal tract [19,20]. In appendiceal tumors, KRAS gene mutations are identified in more than 50% of cases [13], being present in some cases of epithelial tumors: sessile serrated lesions with or without dysplasia [15,16,21,22,23], low-grade appendiceal mucinous neoplasm (LAMN) [7,8,9,24,25,26,27,28,29,30,31,32,33,34], high-grade appendiceal mucinous neoplasms (HAMN) [27,29], mucinous adenocarcinomas of the appendix [12,35], non-mucinous adenocarcinomas of the appendix [35], and appendiceal goblet cell adenocarcinoma [36,37], but is completely lacking in neuroendocrine tumors of the appendix (Figure 3). Among the single-amino-acid substitution (missense) mutations of the KRAS protein identified in appendix cancers are Gly12Asp/Val/Ser/Arg/Cys and Gly13Asp/Arg/Cys [28,29]. In appendiceal cancers, mutations of the other two family members, HRAS and NRAS, occur much less frequently. The HRAS gene (11p15.5), which encodes the HRAS protein (189 amino acids; 21,298 Da), with GTP-ase activity, acquires mutations in Costello syndrome and in several tumor types, such as melanomas, follicular thyroid cancers, bladder cancers, oral squamous cell carcinomas [37,38], and sparsely in certain appendiceal cancers: mucinous adenocarcinomas of the appendix and appendiceal adenocarcinomas, especially in appendiceal goblet cell adenocarcinomas [9]. Mutations in the NRAS gene (1p13.2), which encodes the NRAS protein (189 amino acids; 21,229 Da), are associated with Noonan and autoimmune lymphoproliferative syndromes, rectal somatic and follicular thyroid cancers, and juvenile myelomonocytic leukemia [39]. NRAS gene mutations occur in less than 5% of appendiceal cancers, being present in some cases of low- and high-grade appendiceal mucinous neoplasms, in which missense mutation C>A is identified in codon 181, with amino acid replacement Gln61Lys [29], in mucinous adenocarcinomas of the appendix [23] and appendiceal goblet cell adenocarcinomas [26,37]. Raghav and colleagues performed a retrospective review of 607 patients with appendiceal adenocarcinomas. A total of 149 patients underwent molecular testing for activating mutations in KRAS, protein expression of c-KIT or COX-2, or microsatellite instability (MSI) status by immunohistochemistry. COX-2 expression and KRAS mutations were seen in 61% of patients. High MSI was seen in 6% of patients. COX-2 expression and the presence of KRAS mutation did not impact overall survival. Notably, KRAS mutations were seen more commonly in well or moderately differentiated tumors than poorly differentiated histology ones. Additionally, clinical–pathologic variables such as age, histologic grade, stage, signet ring cells, and completeness of cytoreduction score are significant prognostic factors for appendiceal adenocarcinomas [40]. Liu et al. conducted a clinical study including 535 patients diagnosticated with colorectal cancer to compare the expression of immune-related genes and the abundance of tumor-infiltrating immune cells in the tumor microenvironment between KRAS-mutant and KRAS wild-type patients. They observed that NF-κB and T-cell receptor signaling pathways were significantly inhibited in KRAS-mutant CRC patients. They concluded that KRAS mutation in CRC was associated with suppressed immune pathways and immune infiltration [41]. Matas et al. investigated 24 patients without colorectal adenocarcinoma (CRC) undergoing colonoscopic screening or surveillance and 23 patients with a newly diagnosed primary invasive colorectal adenocarcinoma undergoing surgical resection. They revealed that the normal colon of patients with and without CRC carry mutations in common colorectal cancer genes, but these mutations are more abundant in patients with cancer. Oncogenic KRAS mutations were observed in the normal colon of about one-third of patients with CRC but in none of the patients without CRC. Most mutations in the normal colon were different from the driver mutations in tumors, suggesting that independent clones with pathogenic KRAS mutations are a common event in the colon of individuals who develop CRC [42].
Figure 3.
RAS–RAF–MEK–ERK signaling pathway in cancer, pointing the appendiceal cancer types in which members of RAS (KRAS, HRAS, and NRAS) and RAF (BRAF) gene families, and the MYC/cMYC gene are mutated (adapted from KEGG Pathways, 2020 [14]).
2.1.2. RAF Gene Family
The RAF (rapidly accelerated fibrosarcoma) gene family comprises three independent mammalian genes, Raf-1/c-Raf, B-Raf, and A-Raf, which use MEK1/2 kinases as substrates [43]. In intrauterine life, RAF family members are very active, supporting the growth and development of the embryo and fetus. However, as intense growth processes slow down in extrauterine life, their activity is significantly reduced, activated only by mutations, as in various tumorigenic processes. Of the RAF gene family, in appendiceal cancers, the most mutated gene is the BRAF (rapidly accelerated fibrosarcoma B) gene (7q34). It encodes the serine/threonine kinase BRAF (766 amino acids; 84,437 Da) and frequently undergoes the missense mutation Val600Glu, which contributes to its activation and being associated with cardiofaciocutaneous, Noonan, and Costello syndromes, as well as with several cancers, including non-Hodgkin’s lymphoma, colorectal cancer, thyroid carcinoma, non-small-cell lung carcinoma, hairy cell leukemia, and adenocarcinoma of the lung [44]. In tumors of the appendix, the most common BRAF gene mutation is V600E and it is found in less than 5% of cases of sessile serrated lesions without dysplasia [22], low-grade appendiceal mucinous neoplasms [23], appendiceal goblet cell adenocarcinomas [37], well-differentiated neuroendocrine tumors of appendix [37], and mucinous adenocarcinomas of the appendix. In the X chromosome exists a pseudogene of BRAF. Ueda et al. conducted a prospective observational study including 215 patients with right-side colon cancer. BRAFV600E mutations of cancer tissue and plasma were detected using droplet digital PCR. They identified BRAFV600E mutations in right-side colon cancer at high frequency [45]. Asako et al. performed a retrospective cohort study of the medical records and frozen tissue samples of 268 consecutive patients with stage I-III CRC. This study aimed to investigate the relationship of RAS mutations on an exon basis with clinicopathological features and prognosis in CRC. The RAS mutation rate was significantly associated with age and histology. Patients with KRAS exon 2 mutations exhibited shorter recurrence-free survival than those with KRAS wild-type, KRAS exon 3 mutations, KRAS exon 4 mutations, and NRAS mutation [46].
2.1.3. EGFR/ERBB Gene Family
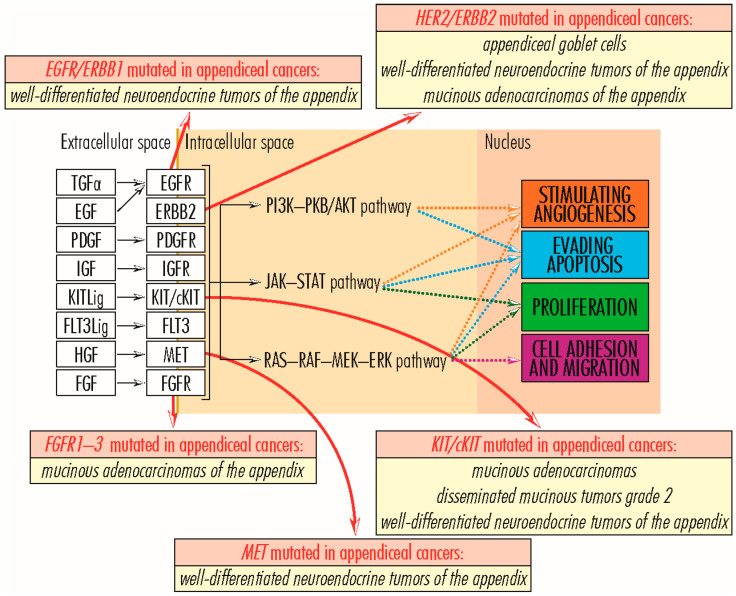
The EGFR/ERBB (Receptor tyrosine-protein kinase erbB) gene family comprises four members (EGFR/ERBB1—Receptor tyrosine-protein kinase erbB-1, HER2/ERBB2—Receptor tyrosine-protein kinase erbB-2, HER3/ERBB3—Receptor tyrosine-protein kinase erbB-3 and HER4/ERBB4—Receptor tyrosine-protein kinase erbB-4), with tyrosine kinase function and structurally related to epidermal growth factor receptor (EGFR). Anchored in the plasma membrane, EGFR/ERBB family members transmit signals via the RAS–RAF–MEK–ERK, PI3K–PKB/AKT, JAK–STAT, and PLC signaling pathways, sustaining cell proliferation, angiogenesis, and evading apoptosis [14]. Mutated or overexpressed proteins are associated with several types of malignancies of the lung, breast, stomach, head, neck, colorectal cancer, pancreatic carcinomas, glioblastoma, and some appendiceal cancers, with the latter being associated with mutations in EGFR/ERBB1 (7p11.2), HER2/ERBB2 (17q12), and HER4/ERBB4 (2q34) genes, unlike other family members. The EGFR/ERBB1 protein (1210 amino acids; 134,277 Da) is a component of the cytokine storm associated with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [47]. The HER2/ERBB2 protein (1255 amino acids; 137,910 Da) has a not-yet-known ligand, but forms a heterodimer with the kinase-deficient protein HER3/ERBB3, which constitutes the most potent signaling complex in the EGFR/ERBB family [48,49], while the HER4/ERBB4 protein (1308 amino acids; 146,808 Da) causes the formation of kinase-active hetero-oligomers [50,51]. In appendiceal cancers, mutations in the EGFR/ERBB1 gene are found in well-differentiated neuroendocrine tumors of the appendix [37]; mutations in HER2/ERBB2 gene are associated with the adenocarcinomas of the appendiceal goblet cells, well-differentiated neuroendocrine tumors of the appendix, and mucinous adenocarcinomas of the appendix; and those in the HER4/ERBB4 gene in well-differentiated neuroendocrine tumors of the appendix [37].
2.1.4. MET Gene
MET gene—MNNG HOS transforming gene (7q31.2) encodes the MET protein (1390 amino acids; 155,541 Da), a class IV receptor tyrosine kinase family member, expressed on the surface of epithelial cells. Through interaction with hepatocyte growth factor (HGF), the MET protein dimerizes, becoming active and supporting cell proliferation, cell motility, and invasiveness via the RAS–RAF–MEK–ERK signaling pathway; angiogenesis and invasion via the JAK–STAT signaling pathway; apoptosis evasion and cell survival via the PI3K–PKB/AKT signaling pathway; and embryogenesis. During the embryo–fetal period, the HGF–MET pair is involved in trophoblast formation, liver formation, myoblast migration, kidney development, nerve wiring, small airways segmentation, melanocyte migration, differentiation of erythroid progenitors, and mammary gland formation; after birth, it participates in tissue repair and wound healing, as well as a protective agent against pulmonary fibrosis, cirrhosis, myocardial ischemia, and acute kidney injury. Overexpression of the two proteins is common in solid tumors; MET mutations are associated with papillary renal cell carcinoma, hepatocellular carcinoma, and several types of tumors formed in the head and neck region [14,52,53]. In low-grade appendiceal mucinous neoplasms, mutations in the MET gene are insignificant and occur rarely [26,30], acquiring greater importance and occurring more frequently in well-differentiated neuroendocrine tumors of the appendix [37] (Figure 4). Li et al. conducted a study aiming to assess the clinical utility of TP53 mutation and MET amplification in ctDNA as biomarkers for monitoring advanced gastric cancer disease progression. They performed mutation detection on circulating-tumor DNA in 23 patients with advanced gastric cancer and identified the top 20 mutant genes. The five most frequently mutated genes were TP53 (55%), EGFR (20%), ERBB2 (20%), MET (15%), and APC (10%). TP53 was the most common mutated gene (55%), and MET had a higher frequency of mutations (15%). They concluded that TP53 mutation and MET amplification in circulating-tumor DNA could predict the disease progression of advanced gastric cancer patients [54].
Figure 4.
Receptors of PI3K–PKB/AKT, JAK–STAT, and RAS–RAF–MEK–ERK signaling pathways in cancer, pointing out the appendiceal cancer types in which the most important members are mutated (adapted from KEGG Pathways, 2020 [14]).
2.1.5. FGFR Gene Family
The FGFR gene family consists of four transmembrane tyrosine kinase receptors: FGFR1—fibroblast growth factor receptor 1 (8p11.23), which encodes FGFR1 protein (822 amino acids; 91,868 Da); FGFR2—fibroblast growth factor receptor 2 (10q26.13), which encodes FGFR2 protein (821 amino acids; 92,025 Da); FGFR3—fibroblast growth factor receptor 3 (4p16.3), which encodes FGFR3 protein (806 amino acids; 87,710 Da); and FGFR4—fibroblast growth factor receptor 4 (5q35.2), which encodes FGFR3 protein (802 amino acids; 87,954 Da), with highly conserved structure between members during evolution, but with differences in ligand affinity and tissue distribution. FGFRs have 22 ligands; activate the RAS–RAF–MEK–ERK, JAK–STAT, PI3K–PKB/AKT, and PLC signaling pathways; and are involved in cell proliferation and survival by evading apoptosis and in stimulating angiogenesis [14,55]. Genomic alterations of FGFRs (single-nucleotide variants, gene fusions, and copy number amplifications) are identified, on average, in 5–10% of cancers, with higher frequencies of 10–30% in urothelial cancer and intrahepatic cholangiocarcinoma. Single-nucleotide variations occur less frequently in the FGFR1 gene than other family members and have missense and activating effects when they occur in the kinase domains (Met546Lys and Lys656Glu) or unknown when they affect the Ser125Leu locus. In the FGFR2 gene, single-nucleotide variants express mutations with an activating effect more frequently in the transmembrane (Tyr375Cys, Cys382Tyr/Arg) and extracellular domains (Ser252Trp, Trp290Cys, Pro253Arg) and more rarely in the kinase domain (Asn549His/Lys, Lys659Glu). In the FGRF3 gene, activating mutations are present in 10 to 60% of urothelial cancers and about 5% of cervical cancers, more frequently in low-grade tumors and often affecting the extracellular domain (Arg248Cys and Ser249Cys) and the transmembrane domain (Gly370Cys and Tyr373Cys). In contrast, in the FGFR4 gene, missense mutations Val550Glu and Asn535Lys occur in approximately 7–8% of rhabdomyosarcoma cases, causing autophosphorylation and constitutive activation of the kinase domain, and Tyr367Cys in the human breast cancer cell line, MDA-MB453, in which constitutive activation of the kinase domain causes receptor activation in a ligand-independent manner. Gene fusions of FGFR family members (FGFR1–3) with various other partner genes are identified in several cancers, including breast, carcinomas, and adenocarcinomas. Most copy number amplification events occur in the FGFR1 gene [49], being associated with several cancers, particularly breast cancers, to which they indicate poor prognosis and recurrence, and FGFR4 genes [56], but they are also present in other family members, with those of FGFR2 gene being found in gastric and breast tumors [57,58]. In appendiceal cancers, mutations in FGFR1–3 genes are present in mucinous adenocarcinomas of the appendix [59]. Jogo et al. examined whether ctDNA helps detect FGFR2 amplification and co-occurring resistance mechanisms in advanced gastric cancer in a nationwide ctDNA screening study. In addition, they examined patients with FGFR2-amplified advanced gastric cancer identified by ctDNA sequencing who received FGFR inhibitors. FGFR2 amplification was more frequently detected by ctDNA sequencing in 28 (7.7%) of 365 patients with advanced gastric cancer than by tissue analysis alone (2.6–4.4%). FGFR2 amplification profiling of paired tissue and plasma revealed that FGFR2 amplification was detectable only by ctDNA sequencing in 6 of 44 patients, which was associated with a worse prognosis. The researchers observed that ctDNA sequencing identifies FGFR2 amplification missed by tissue testing in patients with advanced gastric cancer [60].
2.1.6. KIT/cKIT Gene
KIT/c-KIT gene—V-Kit Hardy-Zuckerman 4 Feline Sarcoma Viral Oncogene-Like Protein (4q12) encodes KIT/cKIT protein (976 amino acids; 109,865 Da), with function as a membrane tyrosine kinase receptor and involved in the transduction of the activating signal for cell proliferation, via the RAS–RAF–MEK–ERK signaling pathway, for cell survival by evading apoptosis, via the PI3K–PKB/AKT and PLC signaling pathways, and for angiogenesis, via the JAK–STAT and RAS–RAF–MEK–ERK signaling pathways [14]. KIT/c-KIT gene activation is oncogenic and associated with several tumor types, such as melanoma, lung cancer, and gastrointestinal stromal cancers [37]. In cancers of the appendix, KIT/c-KIT gene activating mutations occur rarely and are identified in a small number of cases of mucinous adenocarcinomas [12] and in disseminated mucinous tumors grade 2, whereas in well-differentiated neuroendocrine tumors of the appendix, mutations in the KIT/c-KIT gene appear to have a higher frequency [37].
2.1.7. MYC/c-MYC Gene
The MYC/c-MYC gene (8q24.21) encodes the MYC/c-MYC protein (439 amino acids; 48,804 Da), with essential functions in cell growth and cell metabolism. Through the RAS–RAF–MEK–ERK signaling pathway, the MYC/c-MYC protein is involved in cell proliferation and angiogenesis; through the WNT, ESR/JUP–FOS/JUN/SP1/NCOA/ESR, and MYC/c-MYC–CCND–RB1–E2F signaling pathways, it is involved in cell proliferation; and through MYC/c-MYC–E2F signaling pathway, it plays a role in blocking cell differentiation [14,61], which are very important in the progression of tumor processes. On the other hand, the MYC/c-MYC protein appears to induce apoptosis, which counteracts its pro-tumor effects. In non-transformed cells, MYC/c-MYC gene expression is regulated by developmental factors or mitogenic signals and the short lifespan of mRNA and MYC/c-MYC protein. However, in several cancers, MYC/c-MYC genes and those downstream of signaling pathways that stimulate cell proliferation almost always become overexpressed through mutations in the MYC/c-MYC gene or, more commonly, by induction of its expression due to activation of an upstream gene (e.g., ERK), or by its life extension due to mutations, such as that in the Thr58 phosphorylation site, which reduce the efficiency of its phosphorylation-dependent ubiquitination. This process is commonly seen in human lymphomas. Mutations in the MYC/c-MYC gene, which presumably have a stimulatory effect on its function, are also reported in less than 10% of cases of appendiceal goblet cell adenocarcinoma and mucinous/non-mucinous adenocarcinoma of the appendix [62,63], being a secondary mutation in appendiceal cancers. Lin et al. performed a retrospective analysis including 494 patients with breast cancer. Genomic alterations were determined using next-generation sequencing. Survival analysis was applied to assess the effects of genetic alterations on relapse-free survival. Additionally, they used logistic regression to identify the factors associated with pathological complete response after neoadjuvant chemotherapy. Patients with TP53/MYC co-alteration exhibited higher grade and stage, more positive HER2 status, and higher Ki67 levels but fewer luminal A subtypes. They also had more mutations in genes involved in ERBB and TGF-β signaling pathways, as well as exclusive FANCG/CDKN2B/QKI copy number amplifications and SUFU/HIST3H3/ERCC4/JUN/BCR mutations. Concurrent TP53 and MYC alterations independently increased relapse hazards, conferring unfavorable prognoses [64].
2.1.8. PLCG2 Gene
PLCG2 gene (16q24.1) encodes PLCγ2–Phospholipase C Gamma 2 protein (1265 amino acids; 147,870 Da), a transmembrane signaling enzyme, which, in the presence of calcium ions, catalyzes the transition of 1-phosphatidyl-1D-myo-inositol 4, 5-bisphosphate to 1D-myo-inositol 1, 4, 5-trisphosphate (IP3) and diacylglycerol (DAG), which function as secondary messengers for growth factors (TGFα/EGF to EGFR/ERBB1 and PDGF to PDGFR) and transmembrane immune receptors for various cellular functions, including proliferation and angiogenesis; through the RAS/RAF–MEK–ERK signaling pathway, proliferation and evading apoptosis; and through RAS–PI3K pathway, endocytosis and calcium efflux [14]. PLCG2 gene dysfunctions are associated with several pathologies, including neurodegenerative diseases, immune disorders (familial cold autoinflammatory syndrome three and antibody deficiencies), and cancer [65,66]. Peng and colleagues investigated a 7-year-old patient with APLAID, a rare primary immunodeficiency caused by gain-of-function mutations in the PLCG2 gene. The patient carried a novel de novo missense mutation c.2534T>C in exon 24 of the PLCG2 gene that causes a leucine-to-serine amino acid substitution (p.Leu845Ser). Bioinformatics analysis revealed that this mutation harmed the structure of the PLCγ2 protein, which is highly conserved in many other species. Also, immunophenotyping by flow cytometry revealed that in addition to the typical decrease in circulating memory B cells, the levels of myeloid dendritic cells (mDCs) in the children’s peripheral blood were significantly lower, as were the CD4+ effector T cells induced by their activation [67]. Thus, in solid colon tumors and soft tissue sarcoma, PLCG2 gene expression positively correlates with immune cell infiltration into the tumor microenvironment and favorable prognosis [68], while PLCG2 gene mutations produce resistance of some tumors to chemotherapy. In appendiceal tumors, a small number of cases carry mutations in the PLCG2 gene with no reference to their effect [37].
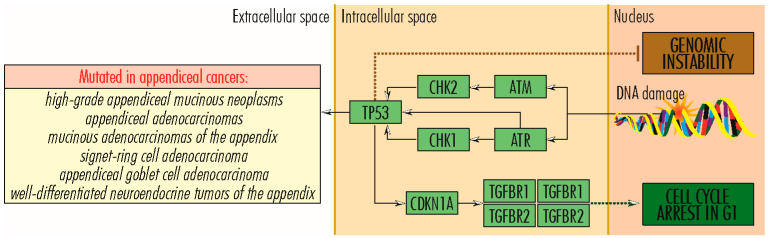
2.2. TP53 Signaling Pathway
The TP53 gene (17p13.1) encodes P53–protein 53 or TP53–tumor protein 53 (393 amino acids; 43,653 Da), which, because of the essential functions it performs in the proper functioning of cells, is nicknamed the guardian of the genome. Protein 53 occupies a central place in the TP53 signaling pathway (Figure 5). It plays a vital role in protecting DNA integrity and regulating DNA repair in cell division, differentiation, and senescence; regulating metabolism; and suppressing tumor development. Protein 53 is a transcription factor located in the nucleus of all cells in the body, attaches directly to genetic material, is activated by cellular stress signals (oncogene activation, DNA damage, DNA replication stress), controls stress-specific gene transcription, and is involved in determining cell fate [69,70] and in signaling pathways in cancer, in which the relationship with HIF1A is essential. HIF1A gene expression is induced by reduced intracellular oxygen pressure, and the HIF1A protein causes TP53 activation, which plays a role in stopping the tumor process. TP53 is degraded in proteasomes by a mechanism ubiquitin-dependent/independent mediated by MDM2–Human Homolog of Mouse Double Minute 2. Mutations in the TP53 gene, which lead to abnormal protein synthesis, are found in various cancers, including Li Fraumeni syndrome, which predisposes to tumors occurring at a young age [70]. In appendiceal tumors, TP53 mutations rarely occur in low-grade appendiceal mucinous neoplasms [23], in which they are a marker of progression to high-grade tumors, and high-grade appendiceal mucinous neoplasms [23,33], and with higher frequency in appendiceal adenocarcinomas, mucinous adenocarcinomas of the appendix [16,23,33], signet ring cell adenocarcinoma [33], and appendiceal goblet cell adenocarcinoma [37], whose aggressive properties are due, to a reasonable extent, to TP53 inactivation by mutations [33], as well as in well-differentiated neuroendocrine tumors of the appendix [33]. Oka and colleagues investigated a case of early appendiceal adenocarcinoma in a female who presented at the hospital because of an enlarged appendix noted by contrast-enhanced CT performed for hematuria. They observed that the atypical epithelium in a small area at the tip was particularly strong in nuclear atypia and showed a strong positive diffusely in p53, which was an image of well-differentiated tubular adenocarcinoma [71]. Yanai et al. investigated 51 appendiceal mucinous tumors using a next-generation sequencing (NGS) cancer hotspot panel. They detected p53 mutation in 31 cases, mainly in low-grade mucinous appendiceal neoplasms [72]. Yan and colleagues determined the p53 levels in pseudomyxoma peritonei of appendiceal origin and correlated the levels with clinicopathological characteristics and overall survival in 141 patients. They concluded that the analysis of aberrant p53 might provide the basis for evaluating the biological behavior of pseudomyxoma peritonei and predicting clinical outcomes [73].
Figure 5.
TP53 signaling pathway in cancer, pointing out the appendiceal cancer types in which TP53 gene is mutated (adapted from KEGG Pathways, 2020 [14]).
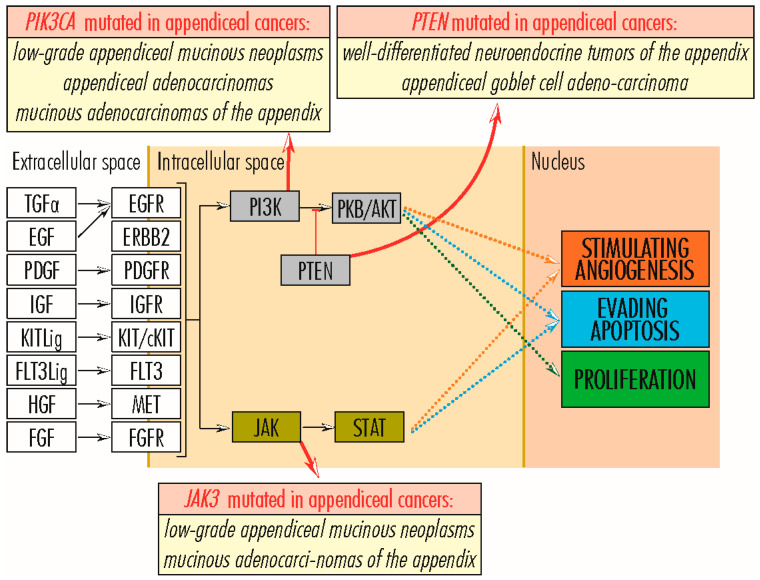
2.3. PI3K–PKB/AKT Signaling Pathway
2.3.1. AKT1 Gene
The AKT1 gene (14q32.33) encodes the AKT1–AKT Serine/Threonine Kinase 1 protein (480 amino acids; 55,686 Da), one of the core pillars of the PI3K–PKB/AKT signaling pathway and its most active member. Unlike other signaling pathways, PI3K–PKB/AKT is often activated by gene mutations encoding its proteins, triggering cascade signaling for cell proliferation and survival by preventing apoptosis and angiogenesis. The AKT1 protein receives signals from the PI3K, which in turn transmits signals from numerous proteins, including RAS, which links to the RAS/RAF–MEK–ERK signaling pathway, and further transduces them to numerous targets [14,74,75]. AKT1 gene mutations occur with some frequency in cancer, particularly the one in the pleckstrin homology domain, Glu17Lys, which enhances its binding to the PI3K ligand and relocalization to the plasma membrane, intensifying cell migration and resistance to chemotherapeutic treatment in breast cancers, as well as selective destruction of chemotherapy-resistant tumor cells and those with low levels of AKT gene expression [75,76,77]. Activating mutations of the AKT1 gene (Glu17Lys and Glu49Lys) are associated in a small number of cases with breast cancers, head and neck squamous cell carcinomas, endometrial cancers, non-small-cell lung cancers, renal cancers, and appendix cancers, the latter of which are occasionally reported in low-grade appendiceal mucinous neoplasms [76].
2.3.2. ITGA11 Gene
The ITGA11 gene (15q23) encodes the ITGA11–Integrin Subunit Alpha 11 protein (1188 amino acids; 133,470 Da) forms a heterodimer with the beta subunit, binding collagen, and is involved in muscle cells attaching to the extracellular matrix, as well as transducing signals for cell proliferation, evasion of apoptosis, and enhancement of angiogenesis, mainly through the PI3K–PKB/AKT signaling pathway [14]. Also, overexpression of the ITGA11 gene in fibroblasts associated with breast cancer and non-small-cell lung cancers indicates high grades and poor prognosis [78,79,80]. On the other hand, mutations in the ITGA11 gene have been reported only occasionally in appendiceal mucinous neoplasms, with no correlation with the degree of differentiation or tumor progression [7].
2.3.3. PIK3C2B and PIK3CA Genes
PIK3C2B (1q32.1) and PIK3CA (3q26.32) genes encode two phosphoinositide 3-kinases (PI3Ks), PIK3C2B–Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Beta (1634 amino acids; 184,768 Da) and PIK3CA–Phosphatidylinositol-4, 5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (1068 amino acids; 124,284 Da), with catalytic activity and phosphorylating inositol lipids. They are important nodes in the PI3K–PKB/AKT signaling pathway, integrating biological signals from a wide variety of sources, including the entire range of membrane receptors of the RAS/RAF–MEK–ERK signaling pathway, directly or via the RAS protein, or FAK/PTK2–protein tyrosine kinase 2, a ligand of membrane integrins, and further transducing them to the PKB/AKT protein, to stimulate cell proliferation and angiogenesis, and being involved in cell survival by evading apoptosis [14,81]. Mutations in the PIK3C2B gene, including Arg564Cys, occur in a wide variety of cancers [82], such as endometrial, breast, ovarian, colorectal, bladder, lung, cervical, head and neck, prostate, esophageal, liver, and renal cancers [83]; in a case of low-grade appendiceal mucinous neoplasms; and some cases of glioblastoma, melanoma, sarcoma, and megalencephaly. In leukemias, increased expression of the PIK3C2B gene can lead to resistance to chemotherapeutic agents [84]. On the other hand, activating mutations of the PIK3CA gene shift tumor behavior, enhancing cell proliferation and migration and tumor invasiveness and metastasis [84], being reported in breast cancers [83], colorectal cancers, head and neck squamous cell carcinoma, and appendiceal cancer, including low-grade appendiceal mucinous neoplasms [12,30], appendiceal adenocarcinomas [85], and mucinous adenocarcinomas of the appendix [33]. Activating mutations of PIK3CA are correlated with poor prognosis, even in patients whose tumor is wholly excised [86]. Also, in a wide variety of neoplasms, PIK3C genes appear amplified, whereas in prostate cancer, the PIK3CA gene is overexpressed [85,87] (Figure 6).
Figure 6.
PI3K–PKB/AKT and JAK–STAT signaling pathways in cancer, pointing out the appendiceal cancer types in which PIK3CA, PTEN, and JAK3 genes are mutated (adapted from KEGG Pathways, 2020 [14]).
2.3.4. PTEN Gene
PTEN gene (10q23.31) encodes PTEN–phosphatase and tensin homolog protein (403 amino acids; 47,166 Da), with antitumor function by preferentially dephosphorylating 3-position on inositol head groups and reversing the reaction catalyzed by PI3Ks, inhibiting cell proliferation and migration and tumor invasiveness and metastasis, by inactivating the transmission of biological signals through the PI3K–PKB/AKT pathway [14,86,88]. Mutations and inactivation of the PTEN gene frequently occur in numerous tumors, including well-differentiated neuroendocrine tumors of the appendix [37] and appendiceal goblet cell adenocarcinoma, sometimes in association with activating mutations in the PIK3CA gene [86].
2.4. AKT-mTOR Signaling Pathway
The CDKN1B (12p13.1) and CDKN2A (9p21.3) genes encode the CDKN1B–Cyclin-Dependent Kinase Inhibitor 1B (198 amino acids; 22,073 Da) and CDKN2A–Cyclin-Dependent Kinase Inhibitor 2A (132 amino acids; 13,903 Da) proteins. CDKN1B protein inhibits the activity of cyclin CDK2 bound to cyclin A or E; inhibits or activates cyclin D–CDK4 complexes concerning microenvironment variables; contributes to the realization, stabilization, modulation, and activation of the CCND1–CDK4 complex; and inhibits cell proliferation, by blocking the cell cycle at G1 stage. Mutations in the CDKN1B gene occur only in some types of cancers, including breast, prostate, and small intestine neuroendocrine tumors [14,89,90] and appendiceal goblet cell adenocarcinoma. On the other hand, the CDKN2A gene encodes two transcripts, p16INK4A and p14ARF, with differences in the first exons, inhibiting CDK4 activity, being involved in stopping the degradation of TP53 protein by the E3 ubiquitin-protein ligase MDM2, to which it binds and sequesters. Blocking the cell cycle at the G1 stage, the CDKN2A protein inhibits cell proliferation [14,91,92]. In many types of cancers, such as adenocarcinomas of the uterus, bladder, stomach, colorectal, cervical, brain and papillary thyroid cancers, head and neck squamous cell carcinoma, malignant peripheral nerve sheath tumor, neurilemmoma, pituitary macroadenoma, multiple myeloma, neuroblastoma, osteochondroma, low-grade neuroepithelial tumor, non-Hodgkin’s lymphoma, glioblastoma multiforme, astrocytoma, meningioma [91], low-grade appendiceal mucinous neoplasms, appendiceal goblet cell adenocarcinoma, and well-differentiated neuroendocrine tumors of the appendix [37], a wide variety of inactivating mutations are reported.
2.5. RB1 Signaling Pathway
The RB1 gene (13q14.2) encodes the RB1–Retinoblastoma-Associated Protein or RB Transcriptional Corepressor 1 protein (928 amino acids; 106,159 Da), which, by methylating histones and stabilizing heterochromatin, maintains the overall chromatin structure and negatively regulates the cell cycle, preventing cell proliferation. In the dephosphorylated or hypo-phosphorylated state, RB1 protein interacts with members of the E2F–Retinoblastoma-Associated Protein 1 family and blocks transcription of many of E2F-responsive genes required for progression through S-phase, mitosis, and cytokinesis, thus preventing G1/S transition of the cell cycle. Phosphorylation mediated by cyclin-dependent kinases CDK4/6 and dissociation of the RB1–E2F heterodimer by their and cyclins activity leads to inactivation of RB1 and expression of downstream genes that determine cell progression through the S phase. Through phosphorylation and CDK3/cyclin C-induced activation, RB1 initiates the G0-G1 transition. The degradation of RB1 in proteasomes in a ubiquitin-dependent and -independent manner is promoted by MDM2–Human Homolog of Mouse Double Minute 2. The loss of RB1 gene function through mutation or other mechanisms leads to retinoblastoma, rare cancer that is the primary type of ocular neoplasia in children and is also found in small cell cancer of the lung, endometrial cancer, bladder cancer, and osteogenic sarcoma [14,93,94,95,96]. In appendiceal cancers, RB1 gene mutations are reported in some isolated cases of appendiceal cancers without further specification, low-grade appendiceal mucinous neoplasms [15,30], mucinous adenocarcinomas of the appendix, and appendiceal goblet cell adenocarcinoma, without being defined for them.
2.6. JAK3 Signaling Pathway
The JAK3 gene (19p13.11) encodes the JAK3–Janus Kinase 3 protein (1124 amino acids; 125,099 Da), a critical JAK–STAT signaling pathway member. The JAK3 protein collects signals from many cytokine receptors, including activating RAS/RAF–MEK–ERK signaling pathway receptors. It transmits them to VEGFs (Vascular Endothelial Growth Factors) via STAT (Signal Transducer and Activator of Transcription) proteins to amplify angiogenic processes [14]. The JAK3 gene is predominantly expressed in immune system cells. Its underexpression is associated with autosomal SCID (Severe Combined Immunodeficiency Disease) [97] and oncogenic activation with natural killer/T-cell lymphoma, a rare and aggressive non-Hodgkin’s lymphoma [98], and in malignant T cells in cutaneous T cell lymphoma [99]. In appendiceal cancers, JAK3 gene mutations occur occasionally and are reported in low-grade appendiceal mucinous neoplasms [15,30] and mucinous adenocarcinomas of the appendix.
2.7. Wnt/β-Catenin Signaling Pathway
2.7.1. APC Gene
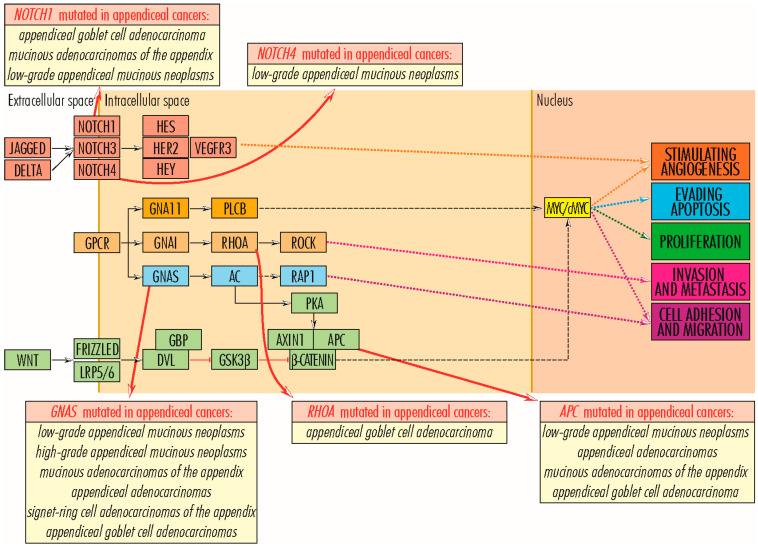
The APC gene (5q22.2) encodes the APC–Adenomatosis Polyposis Coli Tumor Suppressor protein (2843 amino acids; 311,646 Da), with antagonistic regulatory function of the WNT signaling pathway [14,100]. The WNT signaling pathway is actively involved in tissue morphogenesis and repair and is physiologically functional in the embryo–fetal stage. In the adult, it is blocked through β-catenin inactivation due to the formation of a complex between GSK3–Glycogen Synthase Kinase-3, APC–Adenomatosis Polyposis Coli, AXIN–Axis Inhibitor, and CKIa–Casein Kinase Ia [17], with mutations that inactivate genes encoding proteins of the complex being likely to inhibit its formation and allow the canonical (β-catenin-dependent) WNT signaling pathway to function. In blocking the WNT signaling pathway, APC protein plays a vital role in inhibiting cell proliferation and survival (by evading apoptosis); tumor processes stimulated by it; and cell migration, adhesion, differentiation, and chromosome segregation [82]. Mutations in the APC gene inhibit its function and are generally associated with cancers of the digestive tract, such as stomach, colon/colorectal [101,102], and some appendiceal cancers, such as low-grade appendiceal mucinous neoplasms, appendiceal adenocarcinomas [16,33,85], mucinous adenocarcinomas of the appendix, and appendiceal goblet cell adenocarcinoma [30].
2.7.2. AXIN1 Gene
The AXIN1 gene (16p13.3) encodes the AXIN1–Axis Inhibitor 1 or Axis Inhibition Protein 1 protein (862 amino acids; 95,635 Da), which, together with APC, GSK3, and CKIa, enters the structure of the complex to inactivate β-catenin and thereby block the WNT signaling pathway, inhibiting cell proliferation and survival, by evading apoptosis [14,103]. Inactivating mutations in the AXIN1 gene are reported in hepatocellular carcinoma, hepatoblastomas, ovarian endometrioid adenocarcinomas, medulloblastomas [103], prostate cancers [104], and colorectal cancers [105]. In appendiceal cancers, the missense mutation (Arg484Cys) is reported in a case of low-grade appendiceal mucinous neoplasm with no known significance.
2.7.3. TCF7L2 Gene
The TCF7L2 gene (10q25.2-q25.3) encodes the transcription factor TCF7L2–Transcription Factor 7-Like 2 (T-Cell Specific, HMG-Box) (619 amino acids; 67,919 Da) in intestinal cells. It comprises a high-mobility group (HMG). It plays an essential role in transmitting mitogenic signals required for cell proliferation and those required for cell survival by inhibiting apoptosis. TCF7L2 is an essential node in the WNT signaling pathway, receiving signals from β-catenin and positively regulating the MYC/c-MYC proto-oncogene and the BIRC5–Baculoviral IAP Repeat Containing five proteins. TCF7L2 protein also blocks the cell cycle inhibitors CDKN2C/CDKN2D. On the other hand, TCF7L2 manifests an anti-tumor function by suppressing cell motility and invasiveness and by directly suppressing the activity of the pro-metastatic RUNX2–Runt-related transcription factor 2, its loss of function stimulating tumor malignancy [14,106,107]. TCF7L2 protein is also involved in blood glucose homeostasis, with some defects in it associated with an increased risk of developing type 2 diabetes or nonspecific syndromic intellectual disability [106]. Polymorphisms of this gene are reported in colorectal cancer [107,108] and a small fraction of appendiceal cancers without specifying their type.
3. Mutations in Genes Whose Proteins Are Involved in Cell Survival and Tumor Invasiveness/Metastasis
The survival of transformed cells, through insensitivity to apoptotic signals and escape from programmed cell death, is essential for tumor progression and is ensured through several signaling pathways, including RAS–RALGDS–RHO–JNK, in which RHO family genes and PI3K–PKB/AKT play an important role. On the other hand, RHO family members are involved in tumor invasiveness and metastasis in angiotensinogen-initiated signaling pathways [14].
RHOA Gene
The RHOA gene (3p21.31) encodes the RHOA–Ras Homolog Gene Family protein, Member A (193 amino acids; 21,768 Da), a member of the RHO family of small GTPases that function as molecular switches in various signaling pathways, including MEMO1–RHOA–DIAPH1, WNT11, and RAS–RAF–MEK–ERK. RHOA protein stimulates actin filament reorganization in the cytoskeleton and regulates cell adhesion and motility. Via the JNK protein, it promotes cell survival by evading apoptosis, and via ROCK–Rho protein associated coiled-coil containing protein kinases, by evading tumor invasiveness and metastasis [109]. Overexpression and mutations of this gene are associated with tumor cell proliferation and metastasis in germinal center-derived B cell lymphomas, angioimmunoblastic T cell lymphoma, and gastric adenocarcinomas [110]; invasiveness and poor prognosis in colorectal cancers [111]; and reduced RHOA gene expression, in parallel with amplified signaling by the cytokines CCR5 and CXCR4, is associated with breast cancer metastasis [112]. It is unclear whether the RHOA protein functions as a promoter or suppressor of tumorigenesis. In appendiceal cancers, two possible pathogenic mutations (Tyr42Cys and Asp49His) are identified in a few cases of appendiceal goblet cell adenocarcinoma.
4. Mutations in Genes Regulating the Angiogenesis
Angiogenesis is the main mechanism by which tumors larger than 2–4 mm in diameter are supplied with oxygen and nutrients. The main signaling pathways by which new blood vessel formation is stimulated from pre-existing ones are GNA11–PLCβ–RAS–RAF–MEK–ERK, BCR/ABL/JAK–STAT–VEGF, PI3K–PKB/AKT, β-catenin–TCF/LEF, NOTCH, and HIF–VEGF–TGF [14].
4.1. GNA11 and GNAS Genes
The GNA11 and GNAS genes encode guanine nucleotide-binding proteins (G proteins), which modulate or transmit many biological signals and consist of three units: alpha, beta, and gamma. The GNA11–Alpha subunit 11 (359 amino acids; 42,123 Da), encoded by the GNA11 gene (19p13.3), activates the beta subunit of phospholipase C, further transmitting signals to RAS and RAF proteins in the RAS/RAF–MEK–ERK signaling pathway, whereby it stimulates cell proliferation and survival by evading apoptosis, and angiogenesis via the RAS–RAF–MEK–ERK pathway [14,113]. The 626A>C (Gln209Pro) and 627G>C (Gln209His) mutations are reported in some benign hemangiomas [114] and a case of mucinous adenocarcinoma of the appendix. On the other hand, the GNAS gene (20q13.32) shows a complex expression pattern with several alternatively spliced variants, one of which forms the ubiquitously expressed GNAS–G Protein Subunit Alpha S or Secretogranin VI (626 amino acids; 67,948 Da), with an essential function in the action of numerous hormones and other molecules and involved in cAMP activation, which in turn links to several signaling pathways. Thus, the GNAS gene product indirectly stimulates, via PKA–protein kinase A, the formation of the AXIN1–APC complex in the WNT signaling pathway, with an inhibitory function on β-catenin and cell proliferation and promoting cell death through apoptosis. Also, through RAPGEF2–Rap guanine nucleotide exchange factor 2, GNAS links the cAMP pathway to the RAS/RAF–MEK–ERK signaling pathway, promoting cell proliferation and angiogenesis, and through phospholipase C, GNAS participates in the activation of RAP1–Ras-related protein Rap-1A, involved in gene activation and cell proliferation, survival, adhesion, migration, and polarity. The GNAS gene is biallelically expressed in most tissues and is specific-paternally silenced in a small number of tissues, leading to differential transmission of parental phenotypes in GNAS mutations [115,116]. Mutations at this locus are associated with pseudohypoparathyroidism (PHP) type I, progressive osseous heteroplasia, McCune–Albright Syndrome, and breast cancer, with no known role in their progression [116,117]. In appendiceal cancers, GNAS mutations are reported with high frequency in low- and high-grade appendiceal mucinous neoplasms [15,16,30,33], in codons 601 (c.601 C>T, p. Arg201His) and 602 (c.602 G>A, p. Arg201Cys), in which they often occur together with mutations in the KRAS gene as early as G1 and G2 [15] stages and are associated with abundant mucin production, without affecting patient survival [27], and in mucinous adenocarcinomas of the appendix. With reduced frequency, mutations in the GNAS gene also occur in appendiceal adenocarcinomas, signet ring cell adenocarcinomas of the appendix, and appendiceal goblet cell adenocarcinomas (Figure 7).
Figure 7.
WNT, angiogenesis, and NOTCH signaling pathways in cancer, pointing out the appendiceal cancer types in which GNAS, RHOA, NOTCH1, NOTCH4, and APC genes are mutated (adapted from KEGG Pathways, 2020 [14]).
4.2. EP300 and CPB/CREBBP Genes
The EP300 (22q13.2) and CBP/CREBBP (16p13.3) encode two histone acetyltransferase (HAT) paralogs, EP300–E1A Binding Protein P300 (2414 amino acids; 264,161 Da) and CBP/CREBBP–Cyclic adenosine monophosphate Response Element Binding Protein or CREB-binding protein (2442 amino acids; 265,351 Da), which act as cofactors in stimulating the transcription of numerous genes, including the MYB, JUN, FOS, E1A, and E6 oncogenes, and the tumor-suppressor proteins TP53, E2F, RB, SMADs, RUNX, and BRCA1 [118]. EP300 and CBP/CREBBP act mainly on histone H3, which acetylates the lysine residue at position 27 (H3Lys27ac), and, in vitro, on the other three histone types, neutralizing their charge and weakening their DNA binding [118,119], or on non-histone targets. Both genes have high sequence similarity, the proteins exhibiting 63% amino acid sequence homology [118,120] and have partially overlapping regulatory functions, functioning as co-activators of HIF1A–Hypoxia-inducible factor 1 alpha, which stimulates angiogenesis and tumor growth via the HIF-1–VEGF–TGFB signaling pathway. Inhibition of CBP/CREBBP and EP300 activity in vitro and in vivo blocks tumor growth in neuroblastoma, pancreatic cancer, and acute myeloid leukemia [121]. Through chromatin remodeling, the EP300 gene product plays a vital role in cell proliferation and differentiation, including hematopoietic stem cells, cell cycle regulation, apoptosis, and DNA damage response. EP300 gene abnormalities are associated with Rubinstein–Taybi syndrome, characterized by a predisposition to malignancies from childhood; epithelial cancers [118,120], such as high-risk pediatric neuroblastomas, in which the EP300 protein is currently required for H3Lys27ac acetylation [122]; esophageal squamous carcinoma, which, in more than 10% of cases, the EP300 gene is mutated in numerous ways (37 mutations, of which almost 50% are missense), a situation correlated with poor prognosis [123]; or in a very small number of appendiceal cancers, without specifying their type or without details of their prognosis. The CBP/CREBBP protein is critically involved in embryonic development, growth control, and homeostasis [121], with CBP/CREBBP gene mutations causing Rubinstein–Taybi syndrome, while chromosomal translocations affecting it are associated with acute myeloid leukemia [119]. Unlike EP300, in pediatric neuroblastomas, CBP/CREBBP plays a limited role [122] but stimulates lung tumorigenesis via MAPK and CPSF4 signaling pathways, reducing survival and disease-free survival. In appendiceal cancers, mutations in the CBP/CREBBP gene are rarely identified in low-grade appendiceal mucinous neoplasms, with no known biological effect [121].
4.3. KDR/VEGFR2 Gene
KDR/VEGFR2 gene (4q12) encodes KDR/VEGFR2–Kinase insert domain receptor/Vascular endothelial growth factor receptor 2 protein (1356 amino acids; 151,527 Da), which functions as a surface receptor for VEGFC, VEGFD, and especially VEGFA proteins, sustaining angiogenic (endothelial proliferation, survival, migration, tubular morphogenesis, and blood vessel sprouting) and tumor (tumor growth, invasion, and therapeutic resistance) processes via HIF-1–VEGF–TGFB, PLCγ–PKC–MEK–MAPK, and PI3K/AKT signaling pathways [124,125]. Overexpression of KDR/VEGFR2 is reported in breast/chest wall angiosarcomas through activating mutations affecting amino acids 717 (Asp717Val) and 1065 (Ala1065Thr) [126] in colorectal cancer, correlating with vascularization and increased metastatic potential of tumors [124,127], with the 889C>T mutation affecting the efficacy of the chemotherapeutic agent bevacizumab [127], in non-small-cell lung cancer [128,129], being correlated with poor clinical outcomes, and, in the case of activation due to copy number gains, with in vitro resistance to platinum-compounds used in chemotherapy, with aggressive phenotypes and with activation of signaling pathways promoting tumor invasiveness [128], and in oropharyngeal neoplasms (own data, unpublished). In appendiceal cancers, a missense mutation p. Gln472His in the KDR gene is reported in mucinous adenocarcinomas of the appendix without knowing its nature. On the other hand, some mutations in the KDR gene seem to be correlated with better responses in non-small-cell lung cancer patients undergoing immunotherapy with pan-cancer for immune checkpoint inhibitors [130].
4.4. NOTCH Gene Family
The NOTCH gene family comprises four members, encoding the type I transmembrane receptor NOTCH1–4 (Translocation-Associated Notch Protein TAN-1–4), which initiates the NOTCH pro-angiogenic signaling pathway (NOTCH–HER/ERBB2–HES/HEY–VEGFR3). Although NOTCH genes are located on different chromosomes (NOTCH1 on chromosome 1, NOTCH2 on chromosome 9, NOTCH3 on chromosome 19, and NOTCH4 on chromosome 6), the proteins encoded by them share common structural features, including the extracellular sequence with several epidermal growth factor (EGF)-like repeats, to which three Delta-like proteins (DLL1, DLL2, and DLL4) and two Jagged proteins (JAG1 and JAG2) bind, and the intracellular sequence with several different domains, allowing the binding of different targets. The NOTCH signaling pathway is highly conserved throughout evolution, regulates intercellular interactions, and is involved in processes related to cell fate specification, differentiation, proliferation, angiogenesis, and survival [131]. Depending on the context, NOTCH proteins can stimulate or inhibit tumor development [132]. In appendiceal cancers, mutations in NOTCH1, NOTCH3, and NOTCH4 genes are reported [133]. NOTCH1 gene (9q34.3) encodes the NOTCH1 protein (2555 amino acids; 272,505 Da), which, through the attachment of the JAG1 ligand, participates in upper airway organogenesis, nephron development, stimulation of cell proliferation in the intestinal crypts, and proliferation of glandular epithelium in the endometrium. In an additive manner with NOTCH3, NOTCH1 is involved in the regulation of endocrine cell size and number. Mutations in the NOTCH1 gene are associated with aortic valve disease, Adams–Oliver syndrome, T-cell acute lymphoblastic leukemia, chronic lymphocytic leukemia, and head and neck squamous cell carcinoma [132]. In lung cancers, the NOTCH1 protein plays a dual role as a suppressor of tumorigenesis in adenocarcinomas, where its overexpression is correlated with favorable progression, and as a stimulator of tumorigenesis in lung squamous cell carcinoma [132]. In appendiceal cancers, NOTCH1 gene mutations are identified in some cases of appendiceal goblet cell adenocarcinoma [10], mucinous adenocarcinomas of the appendix, and low-grade appendiceal mucinous neoplasms, and in some cases of appendiceal cancers, without further specification [15]. The NOTCH3 gene (19p13.12) encodes the NOTCH3 protein (2321 amino acids; 243,631 Da), which is expressed in fetal lung mesenchyme, endothelial, and other cell types, stimulating upper airway organogenesis, endometrial luminal epithelial proliferation during the proliferative phase, and mammary gland development. NOTCH3 is a stimulator of tumorigenesis in lung adenocarcinomas and lung squamous cell carcinoma; its overexpression is associated with resistance to chemotherapeutic treatment and reduced survival. On the other hand, regarding cell adhesion, epithelial–mesenchymal transition, and motility, NOTCH3 is a tumor suppressor in small-cell lung cancer and a tumor promoter in non-small-cell lung cancer [132,133]. Diseases associated with abnormalities in the NOTCH3 gene include cerebral arteriopathy, autosomal dominant, subcortical infarcts and leukoencephalopathy type 1, and lateral meningocele syndrome. Overexpression of the NOTCH3 gene, mainly due to mutations, is associated with activation of tumor stem cells, leading to tumor growth, increased resistance to chemotherapy, lymph node metastasis, poor prognosis, reduced relapse-free survival and poor overall survival in bladder, colorectal, gastric, head and neck, kidney, liver, lung, ovarian, appendix, and pancreatic cancers, and in leukemia, whereas decreased/blocked expression correlates with reduced proliferation, inhibition of tumor growth, and better disease-free survival in colorectal cancer and reduced proliferation and tumor growth in bladder cancer [134]. The NOTCH4 gene (6p21.32) encodes the NOTCH4 protein (2003 amino acids; 209,622 Da), expressed predominantly in endothelial cells and stimulating their proliferation; vascular, renal, and hepatic development; and angiogenesis. In the embryonic period, abnormalities in NOTCH4 gene function due to mutations are associated with vascular defects and abnormal development of several organs, whereas in adulthood, they are identified in schizophrenia, hemangiomas, and breast cancers [135,136], and repeated allergen exposure leads to overexpression of NOTCH4, which contributes to asthma triggering. In cancer, NOTCH4 activity is ambiguous, being a favorable marker in some cancers and associated with poor prognosis in others [136]. Thus, NOTCH4 underexpression increases disease-free survival in long squamous cell carcinoma, and mutations in the NOTCH4 gene appear to lead to improved survival rates in non-small-cell lung cancer [137]. In appendix cancers, mutations of the NOTCH4 gene are identified in some low-grade appendiceal mucinous neoplasms.
4.5. FLT1/VEGFR1 Gene
The FLT1/VEGFR1 gene (13q12.3) encodes the FLT1/VEGFR1–Fms Related Receptor Tyrosine Kinase 1 protein (1338 amino acids; 150,769 Da), which functions as a receptor for VEGF1/VEGFA and VEGFB, some of the most potential factors favorably involved in angiogenesis, and PGF–placental growth factor. FLT1/VEGFR1 is part of the HIF1–VEGF signaling pathway, playing an essential role in the development of embryonic vasculature in limiting the excessive proliferation of embryonic endothelial cells, and in adult PGF-mediated endothelial proliferation, the regulation of angiogenesis, cell survival, cell migration, macrophage function, chemotaxis, and cancer cell invasion. By forming heterodimers with KDR, FLT1/VEGFR1 modulates its function. By excessively binding VEGF1/VEGFA, FLT1/VEGFR1 reduces its availability to the KDR receptor, limiting signal transmission through the VEGF pathway since FLT1/VEGFR1 can activate a large number of signaling pathways. Thus, by activating PLGC, FLT1/VEGFR1 induces activation of protein kinase C and signal transmission through the RAF–MEK–ERK pathway. On the other hand, FLT1/VEGFR1 mediates PIK3R1 phosphorylation and MAPK1/ERK2 and MAPK3/ERK1 activation, contributing to the stimulation of PIK3/AKT and MAPK signaling pathways. FLT1/VEGFR1-associated diseases are pre-eclampsia and eclampsia [138]. The FLT1/VEGFR1 gene is overexpressed in colorectal cancers and is associated with poor prognosis [124,139], whereas in a minimal number of appendiceal cancers, it carries mutations, the nature of which is not specified.
5. Mutations in Genes Whose Proteins Are Involved in Acquiring Insensitivity to Anti-Growth Signals
Acquiring resistance to anti-growth signals through the TGFB–TGFBR–SMAD signaling pathway is a critical step in tumor growth, allowing tumor cells to override the proliferation-inhibitory signals that ensure normal homeostasis. The TGFB–TGFBR–SMAD signaling pathway initiators, TGFB1–Transforming Growth Factor β1, TGFB2–Transforming Growth Factor β2, and TGFB3–Transforming Growth Factor β3, bind to the TGFBR2 receptor, which then recruits and phosphorylates TGFBR1. Activated by phosphorylation, TGFBR1 phosphorylates SMAD2–Mothers Against Decapentaplegic Homolog 2 and SMAD3–Mothers Against Decapentaplegic Homolog 3, which further recruits SMAD4–Mothers Against Decapentaplegic Homolog 4, and, after translocation to the nucleus, regulate transcription of TGFB target genes involved in numerous biological processes, including the regulation of proliferation, migration, differentiation, immune response, apoptosis, and G1-phase cell arrest. In non-transformed cells, TGFB–TGFBR–SMAD signaling inhibits tumor growth and proliferation, but in malignancies, its role becomes the opposite, stimulating neoplastic processes [140]. In appendiceal cancers, mutations in TGFBR and SMAD genes are reported.
5.1. TGFBR Gene Family
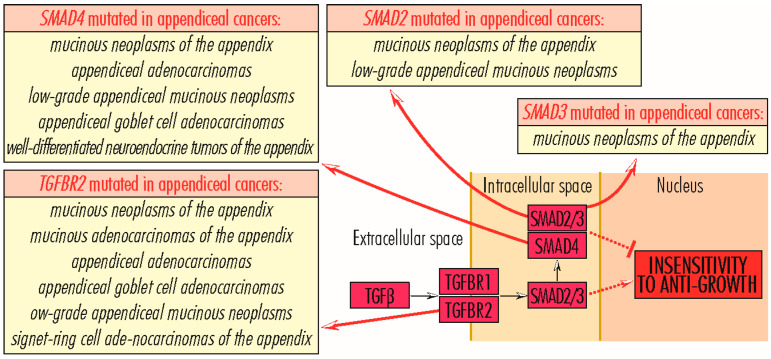
The TGFBR gene family encodes three proteins, TGFBR1, TGFBR2, and TGFBR3, which are receptors for signals transmitted by TGFB proteins. In the TGFB–TGFBR–SMAD signaling pathway, TGFB proteins bind to TGFBR2–Transforming Growth Factor Beta Receptor 2, which, when activated, binds TGFBR1–Transforming Growth Factor Beta Receptor 1, but the reverse process is not possible. The TGFBR2 gene (3p24.1) encodes the TGFBR2 protein (567 amino acids; 64,568 Da), which auto-phosphorylates or recruits and phosphorylates other downstream proteins, including TGFBR1, involved in the regulation of cell proliferation, G1-phase cell cycle arrest, wound healing, immunosuppression, and tumorigenesis. Deficiencies due to mutations in TGFBR2 gene function are associated with Marfan syndrome, Loeys–Deitz aortic aneurysm syndrome, and tumor progression. In cancers, mutations in the TGFBR2 gene are identified in a small number of cases (less than 5%) in lung adenocarcinoma, lung squamous cell carcinoma, esophageal cancer, and head and neck squamous cancer [7]. Some mutations, such as the one leading to the Glu526Gln alteration, lead to loss of kinase activity and signaling functions, since others, including Arg537Pro, amplify kinase activity, with loss of SMAD2/SMAD3 activation and constitutive SMAD1/SMAD5 activation. These mutations are common in carcinoma cells, conferring high mobility and invasiveness [14,141]. Some mutations identified in non-small-cell lung carcinoma confer resistance to treatment with immune checkpoint inhibitors [7,14]. Mutations in the TGFBR2 gene are reported in a small number of cases of mucinous neoplasms of the appendix, with possible loss of function [7], and in less than 10% of cases of mucinous adenocarcinomas of the appendix, appendiceal adenocarcinomas, appendiceal goblet cell adenocarcinomas, low-grade appendiceal mucinous neoplasms, and signet ring cell adenocarcinomas of the appendix. TGFBR1 gene (9q22.33) encodes the TGFBR1 protein (503 amino acids; 55,960 Da). It is phosphorylated by TGFB-activated TGFBR2 and downstream phosphorylates SMAD2 and SMAD3 proteins from the TGFB–TGFBR–SMAD signaling pathway, or TRAF6, involved in activation of the PI3K/AKT and JNK signaling pathways. In these pathways, TGFBR1 is involved in G-phase cell cycle arrest in epithelial and hematopoietic cells, the regulation of cellular apoptosis, epithelial–mesenchymal transition, mesenchymal cell proliferation and differentiation, extracellular matrix synthesis, wound healing, immunosuppression, cancer cell invasion, and tumorigenesis. Defects in TGFBR1 gene function are associated with Loeys–Dietz aortic aneurysm syndrome, multiple self-healing squamous epitheliomas [140,141,142], and the development of some cancers. Thus, TGFBR1*6A polymorphism, consisting of nine-base pair in-frame deletion within exon 1, and IVS7+24G>A polymorphism and TGFBR1 haploinsufficiency reduce TGFBR1 protein functionality, are correlated with tumor cell proliferation and early onset adenocarcinoma, and increase the risk of colon, colorectal, breast, and osteosarcoma cancers [143,144,145,146]. In appendiceal tumors, mutations in the TGFBR1 gene are identified in a few cases of mucinous neoplasms with possible loss of function [147] (Figure 8).
Figure 8.
TGFB signaling pathway in cancer, pointing out the appendiceal cancer types in which TGFBR2, SMAD2, SMAD3, and SMAD4 genes are mutated (adapted from KEGG Pathways, 2020 [14]).
5.2. SMAD Gene Family
The SMAD gene family comprises eight related genes, SMAD1–SMAD8, encoding SMAD-Mothers Against Decapentaplegic Homologues, which transmit signals from TGFBR1 to the nucleus. SMAD genes are located in four chromosomes (4, 13, 15, and 18), and three of them, SMAD2, SMAD4, and SMAD7, are clustered in the 18q21.1 region, frequently deleted in cancers; three of them, SMAD3, SMAD6, and SMAD5, are clustered in chromosome 15 (SMAD3 and SMAD6 in region 15q21–22, and SMAD5 in region 15q31); and SMAD1 and SMAD8 are located in chromosomes 4 and 13. SMAD2 and SMAD3 are regulated by TGFβ/activin; SMAD1, SMAD5, and SMAD8 are activated by BMP receptors; SMAD4 is commonly recruited by SMAD2/SMAD3; and SMAD6 and SMAD7 have inhibitory activity on the TGFB–TGFBR–SMAD signaling pathway. In the absence of SMAD4, SMAD2 and SMAD3 may regulate specific transcriptional pathways of oncogenic signals, probably through interaction with IKK–Inhibitor of Nuclear Factor Kappa B Kinase or TIF1G–Transcriptional Intermediary Factor 1γ. SMAD genes are essential for embryogenesis, expressed throughout embryonic development, with differences in their action, but to a somewhat lesser extent also in adult tissues. Thus, in the embryo, SMAD6 and SMAD7 are highly expressed in the developing cardiovascular system and sporadically expressed in other tissues, such as bone and testis. The SMAD4 gene is also highly expressed during embryonic development, especially in the epithelial crypts of the small intestine. The other SMADs (SMAD1, SMAD5, SMAD8, and SMAD2/SMAD3) are expressed in all tissue types. Their absence during embryonic development is lethal (especially SMAD2, SMAD4, or SMAD5) or leads to defects in gastrulation, mesoderm induction, and endoderm formation; numerous cardiovascular, immune, digestive, and joint abnormalities; as well as metastatic colon cancer or other intestinal tumors [14,148,149]. Mutations in the SMAD2, SMAD3, and SMAD4 genes are associated with cancers of the intestine, pancreas, or appendix. The SMAD2 gene (18q21.1) encodes the SMAD2–SMAD Mothers Against Decapentaplegic Homolog 2 protein (467 amino acids; 52,306 Da). Activin type 1 or TGFBR1 phosphorylates it, subsequently recruits SMAD4 and transmits signals to the nucleus, and is thought to suppress tumor development. Mutations of the SMAD2 gene are rare, leading to congenital heart defects with or without heterotaxy and Loeys–Dietz syndrome 6. In cancers, the frequency of mutations is also low and found in cervical, colorectal, hepatocellular carcinoma, breast, non-small-cell lung, and appendiceal cancers [150,151], including a small number of cases of mucinous neoplasms of the appendix, with possible loss of function, and low-grade appendiceal mucinous neoplasms [148]. The SMAD3 gene (15q22.33) encodes the SMAD3–SMAD Mothers Against Decapentaplegic Homolog 3 protein (425 amino acids; 48, 081 Da), which functions analogously to SMAD2, as a transcription factor and tumor suppressor, through several mechanisms, including apoptosis [152]. Mutations in the SMAD3 gene can disrupt the TGFB–TGFBR–SMAD signaling pathway and are associated with aneurysms–osteoarthritis syndrome and Loeys–Dietz syndrome 3. In cancers, SMAD3 gene mutations are rare and are identified in colorectal adenocarcinomas, choriocarcinomas, and rarely in pancreatic ductal adenocarcinomas and appendiceal cancers [150,153], including mucinous neoplasms of the appendix, with possible loss of function [7]. The SMAD4 gene (18q21.2) encodes the common SMAD4–SMAD Mothers Against Decapentaplegic Homolog 2 protein (552 amino acids; 60,439 Da), recruited by SMAD2/SMAD3, with which it forms a heterotrimeric complex in the nucleus, through which biological signals from the TGFB–TGFBR–SMAD signaling pathway are transmitted. SMAD4 has tumor suppressor activity, and, in its absence, the SMAD2/SMAD3 heterodimer mediates the oncogenic effects of TGFB by activating a SMAD4-independent signaling pathway. SMAD4-associated diseases include Myhre syndrome, juvenile polyposis syndrome, and hereditary hemorrhagic telangiectasia syndrome. Mutations in the SMAD4 gene are present in T lymphocytes and frequently in pancreatic ductal adenocarcinomas (in 50% of cases), in which they are associated with metastasis and poor overall survival [148,154]. Rarely, mutations or deletions of the SMAD4 gene occur in other cancers, such as colorectal cancer [150]; head and neck cancer, in which it is associated with metastasis and in cancers of the appendix, including mucinous neoplasms of the appendix, with possible loss of function [7]; appendiceal adenocarcinomas [15,16,30,33], in which their frequency exceeds 10%; and low-grade appendiceal mucinous neoplasms, appendiceal goblet cell adenocarcinomas, and well-differentiated neuroendocrine tumors of the appendix [37].
6. Mutations in Genes Whose Proteins Are Involved in the Organization of the Extracellular Matrix
Extracellular matrices (ECMs) are multiplicate well-organized three-dimensional architectural networks with critical structural and functional roles in tissue organization and remodeling and the regulation of cellular processes [155,156]. The building blocks of these ultrastructures are collagens, proteoglycans and glycosaminoglycans, elastin and elastic fibers, laminins, fibronectin, and other proteins/glycoproteins such as matricellular proteins [157]. Cells sense the biochemical and mechanical properties of the ECM through specialized transmembrane receptors that include integrins, discoidin domain receptors (DDRs), and syndecans. Once activated, these ECM receptors recognize specific motifs within the ECM molecule that induce conformational changes within the receptor that promote molecular associations between the receptor and intracellular adhesion plaque proteins that activate signaling to influence cell behavior. Diseases such as cancer, defined by disrupted tissue homeostasis and loss of a differentiated phenotype, are accompanied by alterations in the composition, organization, and mechanical properties of the ECM that contribute not only to malignant transformation but also to tumor progression and metastasis [158].
The development of invasive tumors and their subsequent spread to other body parts involve distinct biological changes in the extracellular matrix (ECM). Various steps can be observed during this process. To become invasive, cancer cells must modify the basement membrane by thinning it and reducing the levels of an important protein called laminin-111 [158]. Additionally, around precancerous lesions, there is an increase in the amount and thickness of interstitial collagen [159]. Once tumor cells have invaded the surrounding tissue, their ability to spread is facilitated by a remodeled interstitial ECM. These ECM changes, such as increased stiffness and reorganization into linear bundles of type I collagen, are often accompanied by fibronectin [160]. These altered ECM characteristics create pathways that promote the directed migration of tumor cells toward blood vessels, allowing them to enter the bloodstream efficiently [161]. Importantly, the presence of perpendicular, linearized, and stiffened collagen bundles has been linked to poorer prognosis in breast cancer patients [162]. Moreover, aggressive types of breast cancer, such as HER2+ and triple-negative, exhibit a stiffer invasive stroma with higher levels of linearized collagens [159].
The associations between the tumor ECM and the metastatic cascade not only provide evidence of a clinical connection between ECM remodeling and the development of malignancy and metastasis but also suggest that there is likely a cause-and-effect relationship. This knowledge could be utilized to develop predictive biomarkers or targeted therapeutics to reduce patient mortality. Among the genes involved in extracellular matrix organization, in appendiceal tumors, only COL5A3, COL6A3, and ZNF469 seem to be mutated to a small extent.
The gene products of COL5A3 (19p13.2) and COL6A3 (2q37.3) genes, COL5A3–Collagen Type V Alpha 3 Chain (1745 amino acids; 172,121 Da) and COL6A3–Collagen Type VI Alpha 3 Chain (3177 amino acids; 343,669 Da) proteins, are involved in the organization of the extracellular matrix and thus in cell adhesion. Diseases associated with defects in the COL5A3 gene are Ehlers–Danlos syndrome and colon small cell carcinoma and those associated with abnormalities in the COL6A3 gene, Dystonia 27 and Ullrich congenital muscular dystrophy 1, whereas mutations in these genes are present in a minimal extent in some mucinous neoplasms of the appendix [7,163,164].
The ZNF469 gene (16q24.2) encodes the ZNF469–Zinc Finger Protein 469 (3925 amino acids; 410,202 Da), whose low-percent homology with some collagens suggests its involvement in transcription or regulation of synthesis and organization of collagen fibers. Among the diseases associated with ZNF469 are brittle cornea syndrome 1 and keratoconus 1 [165]. In addition, mutations in this gene are reported in some mucinous neoplasms of the appendix cases [7].
7. Other Genes Showing Mutations in Appendix Cancer
Included in this category are genes whose products are not involved, according to KEGG Pathways, 2020, in critical signaling pathways in cancer, i.e., RAS–RAF–MEK–ERK, PI3K–PKB/AKT, WNT, JAK–STAT, PLCɣ–PKC–MEK–MAPK, ESR/JUP–FOS/JUN/SP1/NCOA/ESR, MYC/c-MYC–CCND–RB1–E2F, RAS–RALGDS–RHO–JNK, NOTCH–HER/ERBB2–HES/HEY–VEGFR3, HIF1–VEGF, and TGFB–TGFBR–SMAD, but this does not reduce their importance in the tumor process. A brief review of these genes will be given below, some being included in certain functional categories (DNA metabolism/expression, cell junctions/adhesion, enzymes) and others having various functions and being included in a separate category.
7.1. Genes Involved in DNA Metabolism/Expression
The ATRX gene (Xq21.1) encodes the ATRX Chromatin Remodeler (2492 amino acids; 282,587 Da), which contains an ATPase/helicase domain, is a member of the SWI/SNF family and is involved in embryonic patterning, cell lineage gene regulation, cell cycle control, and the transcription of specific genes by altering chromatin structure. Among the diseases produced by alterations in the typical structure of the ATRX gene are alpha-thalassemia myelodysplasia syndrome and intellectual disability–hypotonic facies syndrome, X-linked, 1 [166], and its mutations are also present in some appendiceal cancers [16].
The BRCA1 gene (17q21.31), which encodes the BRCA1 DNA Repair-Associated protein (1863 amino acids; 207,721 Da), and the BRCA2 gene (13q13.1), which encodes the BRCA2 DNA Repair-Associated protein (3418 amino acids; 384,230 Da), contribute to maintaining genome stability [167,168]. Defects in these genes lead to Fanconi anemia and complementation group D1, and mutations are present in breast cancer, appendiceal adenocarcinomas, mucinous adenocarcinomas of the appendix, and signet ring cell adenocarcinoma [8].
The FANCA gene (16q24.3) encodes the FANCA–Fanconi Anemia Complementation Group A protein (1455 amino acids; 162,775 Da), a member of the FANC group, which may be involved in DNA repairing processes, interstrand DNA cross-link repair, and maintenance of chromosome stability. Defects in the FANCA gene are identified in Fanconi anemia, complementation group a, and pituitary stalk interruption syndrome [169]. However, this gene is rarely mutated in appendix cancers, as in a single case of low-grade appendiceal mucinous neoplasm [32].
The KDM6A gene (Xp11.3) encodes the KDM6A–Lysine Demethylase 6A protein (1401 amino acids; 154,177 Da), which catalyzes the demethylation of tri/di-methylated histone H3. Defects in the functioning of the KDM6A gene are present in Kabuki syndromes 1 and 2 [170], and mutations in this gene are reported in a few cases of appendiceal goblet cell adenocarcinomas [32].
The KMT2D/MLL2 gene (12q13.12) encodes the KMT2D/MLL2–Lysine Methyltransferase 2D protein (5537 amino acids; 593,389 Da), which methylates the Lys-4 position of histone H3 and coactivates the estrogen receptor. When mutated, it is associated with Kabuki syndrome 1 and choanal atresia–athelia–hypothyroidism–delayed puberty–short stature syndrome [171]. However, in appendiceal cancers, mutations in the KTM2D gene are rarely mutated, with two cases of appendiceal goblet cell adenocarcinomas reported so far [32].
The RAD51C gene (17q22) encodes the RAD51C–RAD51 Paralog C protein (376 amino acids; 42,190 Da), a member of the RAD51 family, involved in the homologous recombination and repair of DNA. The 17q22 region is frequently amplified in breast cancers, suggesting that amplifying the RAD51C gene may affect tumor progression. Defects in this gene are present in familial breast–ovarian cancer and Fanconi anemia, complementation group O [172]. In appendiceal cancers, the RAD51C gene is reported with the pathogenic mutation c.955C>T (Arg319*) in a single case of low-grade appendiceal mucinous neoplasm [32].
The SETD2 gene (3p21.31) encodes the SETD2–SET Domain Containing 2, Histone Lysine Methyltransferase (2564 amino acids; 287,597 Da), involved in chromatin structure modulation during elongation, regulation of DNA mismatch repair in G1 and early S phase, and DNA double-strand break repair in response to DNA damage, through the formation of H3K36me3 and endoderm development. It also mediates protein methylation and is a tumor suppressor [173]. However, its defects are found in some diseases, including Luscan–Lumish and Sotos Syndromes and appendiceal cancers [32].
The TRPS1 gene (8q23.3) encodes the TRPS1–Transcriptional Repressor GATA Binding 1 protein (1281 amino acids; 141,521 Da), which represses GATA-regulated genes and binds to a dynein light chain protein, regulates chondrocyte proliferation and differentiation, and is involved in vertebrate development. Defects in the TRPS1 gene are identified in tricho-rhino-phalangeal syndrome types I–III [174]. In addition, in appendiceal cancers, the TRPS1 gene is mutated in mucinous neoplasms of the appendix [7].
7.2. Genes Involved in Cell Junctions/Adhesion
The CDH1 gene (16q22.1) encodes the CDH1–Cadherin 1 (882 amino acids; 97,456 Da), which acts as a calcium-dependent adhesion protein. Deficiencies in CDH1 gene function lead to blepharocheilodontic syndrome 1 and diffuse gastric and lobular breast cancer syndrome. Loss of CDH1 gene function leads to gastric, breast, colorectal, thyroid, and ovarian cancer [175], and its mutations have been reported in appendiceal goblet cell adenocarcinomas [27] and, very rarely, in mucinous adenocarcinomas of the appendix.
The CTNNA1 gene (5q31.2), which encodes CTNNA1–Catenin Alpha 1 (906 amino acids; 100,071 Da), and CTNNB1 gene (3p22.1), which encodes CTNNB1–Catenin Beta 1 (781 amino acids; 85,497 Da), are part of the catenin gene family, whose products are involved in the formation of adherens junctions. Defects in the CTNNA1 gene are identified in macular dystrophy, patterned, 2, and butterfly-shaped pigment dystrophy, since an abnormal CTNNB1 gene is detected in pilomatrixoma and colorectal cancer [176,177]. Furthermore, in appendiceal cancers, mutations in the CTNNA1 gene are occasionally found in appendiceal goblet cell adenocarcinomas, and mutations in CTNNB1 are found in mucinous neoplasms of the appendix [7], low and high-grade appendiceal mucinous neoplasms, and appendiceal goblet cell adenocarcinomas [10,178].
The CNTNAP2 gene (7q35-q36.1) encodes the CNTNAP2–Contactin-Associated Protein 2 (1331 amino acids; 148,167 Da), a member of the neurexin family, involved in nervous cell adhesion. CNTNAP2 defects are present in Pitt–Hopkins-like syndrome 1 and autism 15 [179] since its mutations are detected in some mucinous neoplasms of the appendix [7].
The LAMA1 gene (18p11.31) encodes the LAMA1–Laminin Subunit Alpha 1 protein (3075 amino acids; 337,084 Da), part of the laminin gene family. Laminins are heterotrimeric proteins consisting of alpha, beta, and gamma subunits, making the significant component of basement membranes and being involved in cell adhesion, differentiation, migration, signaling, neurite outgrowth, and metastasis. Diseases associated with LAMA1 defects are Poretti–Boltshauser syndrome and myopia [180]. In addition, in appendix cancers, mutations in its structure are identified in a few cases of mucinous neoplasms of the appendix [7].
The SPTA1 gene (1q23.1) encodes the SPTA1–Spectrin Alpha, Erythrocytic 1 protein (2419 amino acids; 280,014 Da), which binds the plasma membrane to the cytoskeleton, in cell shaping, arrangement of transmembrane proteins, and organization of organelles. Disruptions in SPTA1 gene function due to mutations lead to red blood cell disorders, such as elliptocytosis-2, pyropoikilocytosis, and spherocytosis type 3 [181]. In addition, in appendiceal cancers, mutations in this gene are reported in mucinous neoplasms of the appendix [30].
7.3. Genes Encoding for Enzymes
The ALK gene (2p23.2-p23.1) encodes the ALK Receptor Tyrosine Kinase (1620 amino acids; 176,442 Da), which is part of the insulin receptor superfamily and receptor tyrosine kinases (RTKs), with an essential role in the development of the brain and specific neurons in the nervous system and whose abnormalities are present in neuroblastoma 3 and neuroblastoma. Mutations of the ALK gene are also reported in appendiceal cancers without their nature being specified [182].
The ARID1A gene (1p36.11), which encodes the ARID1A–AT-Rich Interaction Domain 1A (2285 amino acids; 242,045 Da), and the ARID2 gene (12q12), which encodes the ARID2–AT-Rich Interaction Domain 2 (1835 amino acids; 197,391 Da), are members of the SWI/SNF family of helicases/ATPases, which act as regulators of embryonic patterning, cell lineage gene regulation, cell cycle control, and transcription of specific genes by altering chromatin structure. Among the diseases associated with defects in this gene are Coffin–Siris syndrome 1 and Coffin–Siris syndrome 2 [183]. Mutations in ARID1A and ARID2 genes are present in appendiceal cancers in general [7], with ARID1A gene mutations reported in appendiceal goblet cell adenocarcinomas [11], appendiceal adenocarcinomas, low-grade appendiceal mucinous neoplasms, signet ring cell adenocarcinoma, and mucinous adenocarcinomas of the appendix, and mutations in the ARID2 gene in appendiceal goblet cell adenocarcinomas and appendiceal adenocarcinomas [11,184].
The ATM gene (11q22.3) encodes the ATM Serine/Threonine Kinase (3056 amino acids; 350,687 Da), which belongs to PI3/PI4-kinase family and is involved in regulating the functions of a large number of proteins, including p53, BRCA1, CHK2, RAD17, RAD9, and NBS1. Among the diseases produced by defects in this gene are ataxia-telangiectasia and mantle cell lymphoma [185]. ATM gene mutations are present in appendiceal goblet cell adenocarcinomas [27], mucinous adenocarcinomas of the appendix, low- and high-grade appendiceal mucinous neoplasms [15,16], and well-differentiated neuroendocrine tumors of the appendix [37].
The CARD11 gene (7p22.2) encodes CARD11–Caspase Recruitment Domain Family Member 11 (1154 amino acids; 133,284 Da), which is part of the membrane-associated guanylate kinase (MAGUK) family, with roles in the assembly of protein complexes in specialized regions of the plasma membrane, and the CARD family of proteins, which carry characteristic CARD caspase-associated recruitment domains. These domains interact with the BCL10 Immune Signaling Adaptor, which positively regulates apoptosis and NF-kappaB activation. Abnormalities in CARD11 gene function are found in B-cell expansion with NfkB and T-cell anergy and immunodeficiency 11 and in appendiceal cancers [8,186].
The DCLK1 gene (13q13.3) encodes the DCLK1–Doublecortin Like Kinase 1 (740 amino acids; 82,224 Da), a member of both the protein kinase superfamily and the doublecortin family, being involved in neurogenesis, neuronal apoptosis, microtubule polymerization, multiple protein–protein interactions, neuronal migration, and retrograde transport. Its defects are present in band heterotopia and Zellweger syndrome [187]. In addition, in some mucinous neoplasms of the appendix cases, there are reported mutations in the DCLK1 gene [7].
The DIS3 gene (13q21.33) encodes the DIS3 Homolog, Exosome Endoribonuclease, and 3′-5′ Exoribonuclease (958 amino acids; 109,003 Da), which is part of the nuclear exosome RNAase complex in the nuclear membrane and cytosol and, by regulating the activity of 3′-5′-exoribonuclease and guanyl-nucleotide exchange factor, is involved in CUT and rRNA catabolism. Therefore, its structure and function defects lead to plasma cell neoplasm and myeloma. At the same time, in appendiceal cancers, mutations in the DIS3 gene have been reported in a single case of low-grade appendiceal mucinous neoplasm [10,188].
The FH gene (1q43) encodes the FH–Fumarate Hydratase protein (510 amino acids; 54,637 Da), which catalyzes the conversion of fumarate to L-malate and promotes energy production in the form of NADH. Deficiencies in the functioning of this gene are present in fumarase deficiency and hereditary leiomyomatosis and renal cell cancer [189]. In appendiceal cancers, the FH gene appears mutated in a single case of low-grade appendiceal mucinous neoplasm [32].
The IDH2 gene (15q26.1) encodes the IDH2–Isocitrate Dehydrogenase (NADP(+)) 2 protein (452 amino acids; 50,909 Da), a member of the IDH family, whose members catalyze the oxidative decarboxylation of isocitrate to 2-oxoglutarate. Defects in the IDH2 gene are associated with d-2-hydroxyglutaric aciduria 2 and multiple enchondromatosis, Maffucci type. At the same time, mutations in it are reported in some cancers, such as sarcomas, hematologic malignancies, colon cancer, and brain cancer [190], also reported in a single case of mucinous neoplasm of the appendix.
The PRKACA gene (19p13.12) encodes the PRKACA–Protein Kinase CAMP-Activated Catalytic Subunit Alpha (351 amino acids; 40,590 Da), which is part of the PKA-protein kinase A structure, the latter phosphorylating, in a cAMP-dependent manner, several proteins involved in differentiation, proliferation, and apoptosis. Defects in the PRKACA gene are present in several hyperplasias and adenomas of the adrenal cortex and in ACTH-independent Cushing’s syndrome, cardioacrofacial dysplasia 1, and primary pigmented nodular adrenocortical disease 4 [191]. In addition, mutations in the PRKACA gene are found in a few cases of mucinous neoplasms of the appendix [7].
The PTPN11 gene (12q24.13) encodes the PTPN11–Protein Tyrosine Phosphatase Non-Receptor Type 11 protein (593 amino acids; 68,011 Da), a member of the protein tyrosine phosphatase (PTP) family, which are signaling molecules in pathways including cell growth, differentiation, mitotic cycle, and oncogenic transformation. The PTPN11 gene is expressed in most tissues and regulates mitogenic activation, metabolic control, transcriptional regulation, and cell migration, and mutations in this gene are found in Noonan syndrome 1, juvenile myelomonocytic leukemia [192], and, less commonly, in some appendicular cancers, including mucinous neoplasms of the appendix [15,15,32].
The RHPN2 gene (19q13.11) encodes the RHPN2–Rhophilin Rho GTPase Binding Protein 2 (686 amino acids; 76,993 Da), a member of the rhophilin family of Ras-homologous (Rho)-GTPase binding proteins, and probably involved in the organization of the actin cytoskeleton. Defects in the RHPN2 gene are identified in diseases including hereditary mixed polyposis syndrome and isolated cleft palate [193]. However, in appendiceal cancers, mutations in the RHPN2 gene are rarely found in appendiceal goblet cell adenocarcinomas and appendiceal adenocarcinomas [8,10].
The SMARCA4 gene (19p13.2) encodes SMARCA4–SWI/SNF-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily A, Member 4 (1647 amino acids; 184,646 Da), a member of the SWI/SNF family of proteins with helicase and ATPase activities, which likely regulates transcription of some genes and is thus involved in some biological processes. Defects in protein function are identified in some diseases, including Coffin–Siris syndrome 4 and predisposition to rhabdoid tumors syndrome 2 [194]. In appendiceal cancers, the Ala1536Ser variant with uncertain significance is reported in a case of low-grade appendiceal mucinous neoplasm [32].
The TRRAP gene (7q22.1) encodes the TRRAP–Transformation/Transcription Domain-Associated Protein (3859 amino acids; 437,600 Da), a member of the phosphoinositide 3-kinase-related kinases (PIKK) family and a standard component of many histone acetyltransferases (HAT) complexes, being involved in transcription and DNA repair. Defects in the TRRAP gene are found in some diseases, including developmental delay with/without dysmorphic facies and autism and autosomal dominant deafness 75, and dysregulations are reported in certain types of cancer, including glioblastoma multiforme [195], appendiceal adenocarcinomas, and appendiceal goblet cell adenocarcinomas [10].
The USP9X gene (Xp11.4) encodes the USP9X–Ubiquitin Specific Peptidase 9 X-Linked protein (2554 amino acids; 290,463 Da), a member of the C19 family of peptidases and similar to ubiquitin-specific proteases. It may play an essential role in regulating protein turnover by preventing protein degradation. Mutations in the USP9X gene are associated with Turner syndrome, syndromic and X-linked female-restricted intellectual developmental disorder, and X-linked intellectual developmental disorder [196], being also reported in a single case of appendiceal goblet cell adenocarcinoma [10].
7.4. Genes with Various Cellular Activities
The ABCA7 gene (19p13.3) encodes the ABCA7–ATP Binding Cassette Subfamily A Member 7 protein (2146 amino acids; 234,350 Da), which plays a role in the transport of molecules across extra- and intra-cellular membranes, is associated with Alzheimer disease 9 and early-onset, autosomal dominant Alzheimer disease, and is mutated in a minimal number of cases of mucinous neoplasms of the appendix [7,197].
The ANKRD24 gene (19p13.3) encodes the ANKRD24–Ankyrin Repeat Domain 24 protein (1146 amino acids; 124,187 Da), whose abnormalities are identified in Spinocerebellar Ataxia 17. In addition, mutations of this gene are reported in a small number of mucinous neoplasms of the appendix [7,198].
The APOB gene (2p24.1) encodes the APOB–Apolipoprotein B (4563 amino acids; 515,605 Da), a ligand for LDL (low-density lipoproteins), and is the primary apolipoprotein of chylomicrons and low-density lipoproteins (LDL). Its abnormalities are present in hypobetalipoproteinemia, familial, 1, and hypercholesterolemia, familial, 2. In addition, mutations in this gene are detected in a few cases of mucinous neoplasms of the appendix [7,199].
The ASXL1 gene (20q11.21) encodes the ASXL1–ASXL Transcriptional Regulator 1 (1541 amino acids; 165,432 Da), involved in embryonic development. Defects of this gene are present in Bohring–Opitz and myelodysplastic syndromes, and some mutations are also identified in appendiceal cancers [7,200].
The BCOR gene (Xp11.4) encodes the BCOR–BCL6 Corepressor (1755 amino acids; 192,189 Da), involved in the germinal center formation and, probably, apoptosis. Defects in the BCOR gene are present in syndromic microphthalmia 1 and 2, and mutations are detected in some appendicular cancers [16,201].
The CRY2 gene (11p11.2) encodes the CRY2–Cryptochrome Circadian Regulator 2 flavin adenine dinucleotide-binding protein (593 amino acids; 66,947 Da), an essential component of the circadian core oscillator complex, involved in the regulation of the circadian clock. Defects in the CRY2 gene lead to advanced sleep phase syndrome and severe congenital neutropenia 2, since mutations are found in some mucinous neoplasms of the appendix [7,202].
The DOCK3 gene (3p21.2) encodes the DOCK3–Dedicator of Cytokinesis 3 guanine nucleotide exchange factor (2030 amino acids; 233,103 Da). It is a member of the DOCK (dedicator of cytokinesis) family. It is expressed predominantly in the central nervous system, stimulating axon growth by stimulating membrane recruitment of the WAVE complex and activating the RAC1 protein. Defects in this gene are associated with neurodevelopmental disorders with impaired intellectual development, hypotonia, and ataxia and hypotonia [203]. In appendiceal cancers, mutations in the DOCK3 gene occur sparsely in some cases of mucinous neoplasms of the appendix [7].
The DOK6 gene (18q22.2) encodes DOK6–Docking Protein 6 (331 amino acids; 38,318 Da), a member of the DOK family of intracellular adaptors that activate retinoate. Among the diseases associated with defects in DOK6 gene function are malignant iris melanoma and Hirschsprung disease 1 [204]. In appendiceal cancers, some mutations occur in a few mucinous neoplasms [7].
The EEF1A1 gene (6q13) encodes the EEF1A1–Eukaryotic Translation Elongation Factor 1 Alpha 1 (462 amino acids; 50,141 Da) isoform of the alpha subunit of the elongation factor-1 complex. Isoform A1 is expressed in the brain, placenta, lung, liver, kidney, and pancreas, where it is involved in the enzymatic delivery of aminoacyl tRNA to the ribosome. Defects in the EEF1A1 gene are identified in Felty syndrome and eukaryotic translation elongation factor 1 alpha-1-like 14 [205]. In addition, in appendiceal cancers, the EEF1A1 gene is sparsely mutated in mucinous neoplasms of the appendix [7].
The EPHA10 gene (1p34.3) encodes the EPHA10–Ephrin Receptor A10 (1008 amino acids; 109,716 Da), a member of the most prominent family of tyrosine kinase receptors, which regulate cell attachment, shape, and mobility of neuronal and epithelial cells. Its defects are associated with hypogonadotropic hypogonadism 6 with or without anosmia [206]. In addition, mutations in the EPHA10 gene are reported in a few cases of mucinous neoplasms of the appendix [7].
The FAT1 gene (4q35.2), which encodes FAT1–FAT Atypical Cadherin 1 or Protocadherin Fat 1 (4588 amino acids; 506,273 Da), and the FAT4 gene (4q28.1), which encodes FAT Atypical Cadherin 4 or Protocadherin Fat 4 (4981 amino acids; 542,687 Da), are members of the protocadherin family. The FAT1 gene is expressed in various fetal epithelia, modulating cellular polarization, cell migration, and cell–cell contact. Defects in the FAT1 gene are present in Myasthenic Syndrome, Congenital, 12, Nephrotic Syndrome, Type 1, and appendiceal cancers [207,208]. The FAT4 gene is involved in the regulation of planar cell polarity. Among the diseases associated with defects in the FAT4 gene are van Maldergem syndrome 2 and Hennekam lymphangiectasia–lymphedema syndrome 2. In appendiceal cancers, mutations in the FAT4 gene are reported in a small number of cases of mucinous neoplasms of the appendix [7] and in a case of low-grade appendiceal mucinous neoplasms.
The FBXW7 gene (4q31.3) encodes the FBXW7–F-Box and WD Repeat Domain Containing 7 (707 amino acids; 79,663 Da), a member of the F-box protein family, which binds and can target cyclin E for its ubiquitin-mediated degradation. Mutations of the FBXW7 gene are reported in ovarian and breast cancer cell lines, as well as in appendiceal cancers [37,209], including appendiceal adenocarcinomas, mucinous adenocarcinomas, and signet ring cell adenocarcinomas of the appendix, where they occur in 5–10% of cases, and in well-differentiated neuroendocrine tumors of the appendix [37].
The IRX6 gene (16q12.2) encodes the IRX6–Iroquois Homeobox Protein 6 (446 amino acids; 48,240 Da), whose predicted functions include cell development, neuron differentiation, and regulation of transcription, and retina morphogenesis in the camera-type eye. Its anomalies lead to transient refractive change and aniseikonia [210] since mutations in the IRX6 gene are found in some cases of mucinous neoplasms of the appendix [7].
The KRT37 gene (17q21.2) encodes KRT37–Keratin 37 (449 amino acids; 49,747 Da), which is found in hair and nails, and whose defects are identified in multiple types of cataract 1 and mucinous neoplasms of the appendix [7,211].
The MED12 gene (Xq13.1) encodes the MED12–Mediator Complex Subunit 12 (2177 amino acids; 243,081 Da), part of the Mediator complex, with a mass of 1.2 MDa and which, after binding to the CDK8 subcomplex, pre-initiates transcription. The MED12 protein is essential for CDK8 activation. Defects of the MED12 gene are present in the X-linked Opitz–Kaveggia and Lujan–Fryns syndromes, and mutations of this gene are reported in appendiceal cancers in general [212] and in a single case of low-grade appendiceal mucinous neoplasm [32].
The MTIF2 gene (2p16.1) encodes the MTIF2–Mitochondrial Translational Initiation Factor 2 protein (727 amino acids; 81,317 Da), essentially initiating protein synthesis and in the hydrolysis of GTP during the formation of the 70S ribosomal complex. Diseases associated with defects in the MTIF2 gene are combined oxidative phosphorylation deficiency 39 and combined oxidative phosphorylation deficiency 3 [213], while mutations in its sequence are reported in some cases of mucinous neoplasms of the appendix [7].
The MUC16 gene (19p13.2) encodes the high-molecular-weight, O-glycosylated MUC16–Mucin 16, Cell Surface-Associated protein (14,507 amino acids; 1,519,175 Da). As a member of the mucin family, MUC16 forms a mucous barrier over the apical surfaces of the epithelia, protecting them against particles and infectious agents. Defects of the MUC16 gene are found in some diseases, including clear-cell adenocarcinoma and cystadenocarcinoma [214]. Together with MUC17, MUC16 is expressed in pancreato-biliary and small intestinal cancers. In appendiceal cancers, mutations in the MUC16 sequence are identified in some cases of mucinous neoplasms of the appendix [7,32]. Furthermore, MUC16 is expressed in mucinous adenocarcinomas of the appendix, showing cytosolic expression and being related to lymph node metastasis since, in appendiceal carcinomas, its expression seems to be lowered [215].
The OCA2 gene (15q12-q13.1) encodes the OCA2 Melanosomal Transmembrane Protein (838 amino acids; 92,850 Da), probably involved, within melanocytes, in the transport of tyrosine, from which melanin, the pigment in the integument of mammals, is derived. The OCA2 gene may be involved in determining eye color in humans. Defects in this gene lead to diseases such as oculo-cutaneous albinism type II and variations in skin/hair/eye pigmentation [216]. In appendiceal cancers, some mutations in the OCA2 gene are reported in mucinous neoplasms of the appendix [7], with no known significance in disease progression.
Protocadherin genes PCDH10 (4q28.3) and PCDH17 (13q21.1) belong to the protocadherin gene family, included in the cadherin superfamily. Protocadherin 10 (1040 amino acids; 112,936 Da) and Protocadherin 17 (1159 amino acids; 126,229 Da) may play roles in specific cell–cell connections in the brain; protocadherin 10 seems to be involved in inhibiting cancer cell motility and cell migration. Defects on these genes are found in some diseases, including developmental and epileptic encephalopathy 9 and Dravet syndrome [217,218], and mutations in both genes are reported in a small number of mucinous neoplasms of the appendix cases [7].
The POM121L12 gene (7p12.1) encodes the POM121L12–POM121 Transmembrane Nucleoporin Like 12 protein (296 amino acids; 31,848 Da), whose predicted activity is to enable nuclear localization sequence binding activity, and, as a nuclear pore structural constituent, it may aid in RNA export from nucleus and protein import inside it. Among diseases related to POM121L12, defects are phosphoserine phosphatase and serine deficiencies [219]. In addition, the POM121L12 gene is mutated in a few cases of mucinous neoplasms of the appendix [7].
The PRDM1 gene (6q21) encodes the PRDM1-PR/SET Domain 1 protein (825 amino acids; 91,771 Da), repressing beta-interferon gene expression and stimulating B cell maturation. However, its defects are present in some diseases, including plasmablastic and B-cell lymphomas [220], and the c.711A>T point mutation in the PRDM1 gene is reported in a single case of low-grade appendicular mucinous neoplasm [32].
The PTCHD3 gene (10p12.1) encodes the PTCHD3–Patched Domain Containing 3 protein (954 amino acids; 107,689 Da). Probably expressed in the sperm midpiece and active in the membrane, the PTCHD3 protein seems to participate in sperm development or function, with no critical involvement in spermatogenesis and fertility. Defects in the PTCHD3 gene are present in some diseases, including kidney leiomyosarcoma and multiple cataracts [221]. In appendiceal cancers, mutations in the gene PTCHD3 are present in mucinous neoplasms of the appendix [7] with no known significance.
The RNF43 gene (17q22) encodes the RNF43–Ring Finger Protein 43 (783 amino acids; 85,722 Da), which probably negatively regulates the WNT signaling pathway, its defects being identified in sessile serrated polyposis cancer syndrome and colorectal cancer, as well as in multiple tumor types, including colorectal and endometrial cancers [222]. For example, in appendiceal cancers, mutations in the RNF43 gene are reported in low-grade appendiceal mucinous neoplasms [23,30], high-grade appendiceal mucinous neoplasms, appendiceal adenocarcinomas, and sessile serrated lesions without dysplasia [23].
The SNTG1 gene (8q11.21) encodes the SNTG1–Syntrophin Gamma 1 protein (517 amino acids; 57,969 Da). As a member of the syntrophin family, the SNTG1 protein mediates dystrophin binding, being involved in gamma-enolase trafficking to the plasma membrane, enhancing its neurotrophic activity and probably organizing the subcellular localization of certain proteins. Some mutations in the SNTG1 gene are identified in various scoliosis types [223] and, in some cases, in mucinous neoplasms of the appendix [7].
The SOX9 gene (17q24.3) encodes the SOX9–SRY-Box Transcription Factor 9 (509 amino acids; 56,137 Da), involved in chondrocyte differentiation. Along with steroidogenic factor 1, SOX9 regulates the AMH-anti-Muellerian hormone gene transcription. Deficiencies in SOX9 gene function are found in some diseases, including campomelic dysplasia and 46, Xy sex reversal 10 [224]. In addition, frameshift mutations of SOX9 are reported in a few appendiceal goblet cell adenocarcinomas [7].
The STK11 gene (19p13.3) encodes the STK11–Serine/Threonine Kinase 11 (433 amino acids; 48,636 Da), a tumor suppressor which regulates cell polarity and energy metabolism. Among the disorders associated with STK11 malfunctions are Peutz–Jeghers syndrome and skin, pancreatic, and testicular cancers [225]. In addition, in appendiceal cancers, mutations in the STK11 gene are observed in low-grade appendiceal mucinous neoplasms [30] and mucinous neoplasms of the appendix [16,32].
The TSC1 (9q34) and TSC2 (16p13.3) genes encode two tumor suppressor subunits, TSC1–Subunit 1 or Hamartin (1164 amino acids; 129,767 Da) and TSC2–Subunit 2 or Tuberin (1807 amino acids; 200,608 Da) of the TSC complex, which negatively regulates anabolic cell growth. Disorders associated with TSC genes include tuberous sclerosis and lymphangioleiomyomatosis [226,227]. In appendiceal cancers, the TSC1 protein bears a His732Tyr modification in a case of low-grade appendiceal mucinous neoplasms [32], and the TSC2 gene is mutated in appendiceal cancers, with no specification about their type.
8. Microsatellite Instability in Appendix Tumors
In addition to genetic mutations identified in all types of appendiceal cancers, there are reports of microsatellite instability (MSI) and small repetitive segments of one, two, or three nucleotides spread in non-coding regions between or within genes. Microsatellite instability (MSI) is a condition caused by the alteration of DNA mismatch repair (MMR) mechanisms, resulting in the accumulation of mutations in nucleotide repeats. These mutations primarily occur in the coding regions of cancer-related genes like TGFβRII, PTEN, and BAX [228].
The DNA mismatch repair (MMR) system plays a crucial role in maintaining the integrity of the genome. It involves several proteins, such as MLH1, MLH3, MSH2, MSH6, MSH3, PMS2, PMS1, and Exo1. MMR proteins work together to identify errors in DNA, which can occur through various mechanisms. The detection of DNA errors is carried out by MSH2/MSH6 and MSH2/MSH3 heterodimers. These heterodimers recognize and bind to mismatches in the DNA sequence. Subsequently, the MLH1/PMS2 complex is recruited to the error site and introduces nicks in the DNA strand, initiating the repair process [229].
Loss of the primary partners (MLH1 and MSH2) results in the loss of the entire heterodimer, while loss of the secondary partners (PMS2 and MSH6) does not lead to the loss of the heterodimer. Mutations or alterations in the MMR proteins can lead to microsatellite instability (MSI), characterized by the accumulation of errors in microsatellite regions of the DNA [230].
Microsatellite instability can arise from different causes. It can be caused by point mutations in MMR genes, such as MLH1 and MSH2, which disrupt their normal function. Errors during DNA replication, where the DNA polymerase slips and introduces errors in repetitive DNA sequences, can also contribute to MSI. Additionally, insertions or deletions of bases in microsatellite regions can lead to MSI [231].
Aside from genetic factors, other aberrations can contribute to the MSI phenotype. For example, hypermethylation of the MLH1 promoter can silence MLH1 gene expression and result in MSI. Epigenetic inactivation of MSH2 or MLH1, downregulation of MMR genes by microRNAs, and slipped strand mating errors (SSMs) are other factors associated with MSI [229].
Microsatellite instability is particularly associated with Lynch syndrome (LS), an inherited genetic disorder caused by germline mutations in MMR genes. LS is commonly observed in colorectal and endometrial cancer. However, it can also affect other types of cancer, including ovarian, gastric, hepatobiliary tract, upper urinary tract, pancreatic, brain, and skin cancer [232,233].
Microsatellite instability can manifest in both familial and sporadic colorectal cancer (CRC). Familial cases, comprising approximately 2–3% of all CRC cases [234], are associated with inherited mutations in DNA mismatch repair genes, namely, MLH1, MSH2, MSH6, or PMS2. In rare instances, a deletion in the last exon of the EPCAM gene can cause MSH2 methylation, resulting in heritable MSI.
In contrast, sporadic cases of MSI-related CRC are more common and primarily arise from epigenetic silencing of the MLH1 gene through promoter hypermethylation. This hypermethylation often occurs alongside more generalized methylation of CpG islands, a phenomenon known as the CpG island methylator phenotype (CIMP) [234,235]. Detected in several types of human cancers, most prominently gastrointestinal and endometrial cancers [234,236], MSI is also reported in some appendiceal cancers, such as appendiceal adenocarcinomas [237] and non-mucinous [234] and mucinous adenocarcinomas of the appendix [238]. In cancers, MSI appears to result in a more favorable disease course and prognosis than microsatellite stability. However, the response to 5 fluorouracil (5- FU)-based chemotherapy is poorer [234]. Emile and colleagues performed a retrospective cohort analysis including 1681 patients, 211 of whom were positive for MSI. This study aimed to assess the impact of MSI on the overall survival of patients with appendiceal adenocarcinoma, stratified by disease stage, tumor histology, and patient demographics. They concluded that the MSI status of appendiceal adenocarcinomas did not significantly impact survival, except for stage IV disease, in which a survival benefit of MSI was noted [239].
9. Comparison of Genetic Aberrations in Appendiceal, Colorectal, Small Bowel, and Gastric Cancers
Frequently, appendiceal cancers are treated in a similar manner to colorectal cancers, although the former category represents a distinct and unique pathogenetic entity. Thus, mutations in the TP53 and APC genes are present in 27.3% and ~8% of appendiceal cancers, 75% and 75.9% of colorectal cancers, 58.4% and 7.8% of small bowel cancers, and 58.4% and 26.8% of gastric cancers. On the other hand, the frequency of KRAS mutations in appendiceal cancers (51%; with a maximum of 56% in adenocarcinomas) is close to that in colorectal cancers (51.8%) and small bowel cancers (53.6%), but much higher compared to gastric cancers (14.2%). The SMAD4 gene is mutated in about 13% of appendiceal cancers, which is close to that found in colorectal cancers (15%) and small bowel tumors (17.4%), but higher than in gastric cancers (5.2%). PIK3CA gene mutations occur with higher frequency in colorectal (17.7%), small bowel (16.1%), and gastric (12.5%) cancers and with lower frequency in appendiceal cancers (~6.5%) [9,13,240] (Figure 9). An interesting feature is the co-occurrence or mutual exclusion of mutations in significant pairs of genes in appendiceal cancers, depending on the time of disease onset. Thus, in cancers that occur early in individuals’ lives (age < 50 years), mutations in SMAD4 and PIK3CA tend to co-occur, whereas mutations in TP53 and GNAS and mutations in SOX9 and KRAS or TP53 tend to be mutually exclusive. In late-onset cancers (age ≥ 50 years), only mutations in the GNAS and TP53 gene pair are mutually exclusive, while mutations in GNAS and KRAS or TP53 genes, in PIK3CA and APC or KRAS genes, in TP53 and KRAS or SMAD4 genes, and in ATM and SMAD4 genes tend to occur together [13] (Table 1).
Figure 9.
Comparative analysis of gene mutation frequency in appendiceal, colorectal, small bowel, and gastric cancers. According to it, the frequency of mutations in TP53 and PIK3CA genes is lower in appendiceal tumors compared to the other tumor types, and the frequency of mutations in APC gene in appendiceal cancers is comparable to that in small bowel tumors, differentiating appendiceal tumors from tumors of the middle and lower digestive tract. However, the frequency of mutations in the KRAS and SMAD4 genes is comparable to the frequency of mutations in these genes in colorectal and small bowel cancers, indicating the possibility of common genetic causes for all three tumors.
Table 1.
Significant pairs of genes whose mutations co-occur or are mutually exclusive in early and late-onset appendiceal cancers.
| First Mutated Gene | Second Mutated Gene | Relation |
|---|---|---|
| Early-onset appendiceal cancers | ||
| P53 | GNAS | Mutual exclusivity |
| SOX9 | KRAS/TP53 | Mutual exclusivity |
| SMAD4 | PIK3CA | Co-occurrence |
| Late-onset appendiceal cancers | ||
| GNAS | KRAS | Co-occurrence |
| PI3KCA | APC/KRAS | Co-occurrence |
| TP53 | KRAS | Co-occurrence |
| GNAS | TP53 | Mutual exclusivity |
| SMAD4 | TP53/ATM | Co-occurrence |
10. Discussion
Through a review of the literature published up to October 2022, 105 genes have been identified to have mutations in appendiceal cancers (Figure 1).
Of these, 42 genes are included in nine essential signaling pathways in cancer (Figure 2), the rest being more or less involved in tumor progression. Thus, 13 genes, KRAS, HRAS, NRAS, BRAF, EGFR, ERBB2, ERBB4, MET, FGFR13, KIT/cKIT, MYC/cMYC, and PLCG2, are part of the RAS–RAF–MEK–ERK signaling pathway, which in cancer stimulates proliferation and angiogenesis. Physiologically, genes in this signaling pathway are only/predominantly active during the embryo–fetal period, when they drive tissue growth and differentiation. However, once these processes become much slower in adulthood, these genes are more or less silenced. Due to mutations or epigenetic events, their activation transforms them into oncogenes and contributes to tumor development. This occurs both at transmembrane receptors such as EGFR, ERBB2, ERBB4, MET, FGFR1–3, and KIT/cKIT, which take up extracellular signals, and at proteins in the cytosol such as KRAS, HRAS, NRAS, BRAF, MYC/cMYC, and PLCG2, responsible for their transduction to the nucleus, with KRAS and BRAF genes undergoing activating mutations in a huge number of appendiceal cancers. Most mutations in genes constituting the RAS–RAF–MEK–ERK signaling pathway are reported in mucinous adenocarcinomas of the appendix (nine genes), followed by appendiceal goblet cell adenocarcinomas and well-differentiated neuroendocrine tumors of the appendix, with six genes each. None of the genes in this pathway appear mutated in mucinous neoplasms of the appendix, appendiceal adenocarcinomas, and signet ring cell adenocarcinoma.
Frequently, mutations in the KRAS gene occur concomitantly with mutations in the GNAS gene in the RAP1 signaling pathway, which involves numerous biological processes, including invasiveness, adhesion, cell migration and polarization, and tumor metastasis. Frequent mutations in appendiceal cancers are also reported in the TP53 gene, this time being inactivating. The TP53 gene helps determine the fate of cells whose genetic material is damaged, and its inactivation leads to cells remaining on the path of tumor transformation.
Another pathway mutated in appendiceal cancers is PI3K–PKB/AKT, in which eight genes, AKT1, ITGA11, PIK3C2B, PIK3CA, PTEN, CDKN1B, CDKN2A, and RB1, in addition to those affecting receptors typical to the RAS–RAF–MEK–ERK signaling pathway, are mutated. The eight genes encode cytosolic proteins, which transduce signals from transmembrane receptors to the nucleus through the range of molecules they activate. They are involved in the most critical tumor processes, proliferation, evading apoptosis, and angiogenesis. Among appendiceal cancers, some low-grade appendiceal mucinous neoplasms carry mutations in five genes, and appendiceal goblet cell adenocarcinomas carry mutations in four genes.
In the JAK–STAT signaling pathway, in addition to molecules that function as transmembrane receptors, including the RAS–RAF–MEK–ERK and PI3K–PKB/AKT signaling pathways, the JAK3 gene, its central node, is also mutated in appendiceal cancers, and through which signals necessary for both evasions of apoptosis and support of angiogenesis are transmitted.
Resistant tumors are known to develop alternative pathways for survival so that if one of these is inactivated, the others will take its place. In some appendiceal cancers, the alternative to the PI3K–PKB/AKT signaling pathway is the activation of the WNT pathway, which also signals cell proliferation and evasion of apoptosis. Of the members of this signaling pathway, the APC, AXIN1, TCF7L2, and RHOA genes are most frequently mutated, the latter encoding a low-molecular-mass protein that contributes to cell invasiveness, adhesion, migration, and polarization, and tumor metastasis. This function overlaps with the more frequently mutated GNA11 and GNAS genes of the RAP1 signaling pathway. Members of the WNT signaling pathway are more frequently mutated in low-grade appendiceal mucinous neoplasms and appendiceal goblet cell adenocarcinomas.
Tumor growth requires an increased supply of oxygen and nutrients, the lack of which activates the HIF1–VEGF signaling pathway to neo-form blood vessels. In tumors, these have an altered structure, with other types of cells, including tumor cells, coexisting in their walls alongside normal endothelial cells, or lacunae, which allow the transformed cells to enter and metastasize elsewhere in the body. In appendiceal cancers activating mutations in the HIF1–VEGF signaling pathway are reported in three genes, EP300, CREBBP, and VEGFR2. In addition to the classical HIF1–VEGF signaling pathway, tumor angiogenesis is stimulated by two alternative mechanisms. One is through the NOTCH pathway. The NOTCH1, NOTCH3, NOTCH4, and FLT1/VEGFR1 genes are mutated/reactivated in appendiceal cancers, and the other is through inactivation of the tumor suppressor genes SMAD2–4 and TGFBR1–2 in the TGFβ signaling pathway. While in the case of members of the NOTCH signaling pathway, mutations are rare and not prevalent in specific tumor types, in the case of genes in the TGFβ signaling pathway, they are mutated in some cases of mucinous neoplasms of the appendix and low-grade appendiceal mucinous neoplasms.
The other genes, which are not included in the nine major cancer signaling pathways, are sporadically mutated, although those in the RNF43 sequence occur more frequently. Most mutations in these genes are present in mucinous neoplasms of the appendix, the generic name for mucinous tumors, and of these, in low-grade appendiceal mucinous neoplasms.
In the analysis of the total number of mutated genes in various types of appendiceal cancers, mucinous neoplasms of the appendix are ranked first (Figure 10), with mutations reported in 38 genes out of 105, i.e., ITGA11, TGFBR1, TGFBR2, SMAD2, SMAD3, SMAD4, ABCA7, ANKRD24, APOB, CNTNAP2, COL5A3, COL6A3, CRY2, CTNNB1, DCLK1, DOCK3, DOK6, EEF1A1, EPHA10, FAT4, IDH2, IRX6, KRT37, LAMA1, MTIF2, MUC16, OCA2, PCDH10, PCDH17, POM121L12, PRKACA, PTCHD3, PTPN11, SNTG1, SPTA1, STK11, TRPS1, and ZNF469, of which low-grade appendiceal mucinous neoplasms carry mutations in 33 genes: KRAS, BRAF, MET, TP53, AKT1, PIK3C2B, PIK3CA, CDKN2A, RB1, JAK3, APC, AXIN1, GNAS, CREBBP, NOTCH1, NOTCH4, TGFBR2, SMAD2, SMAD4, ARID1A, ATM, CTNNB1, DIS3, FANCA, FAT4, FH, MED12, PRDM1, RAD51C, RNF43, SMARCA4, STK11, and TSC1.
Figure 10.
Number of genes in which mutations occur in appendiceal cancers.
Next are appendiceal goblet cell adenocarcinomas, in which 29 mutated genes, KRAS, HRAS, NRAS, BRAF, ERBB2, MYC/cMYC, TP53, PTEN, CDKN1B, CDKN2A, RB1, APC, RHOA, GNAS, NOTCH1, TGFBR2, SMAD4, ARID1A, ARID2, ATM, CDH1, CTNNA1, CTNNB1, KDM6A, KMT2D, RHPN2, SOX9, TRRAP, and USP9X, and mucinous adenocarcinomas of the appendix, with 25 genes, KRAS, HRAS, NRAS, BRAF, ERBB2, FGFR1, FGFR2, FGFR3, KIT/cKIT, MYC/cMYC, TP53, PIK3CA, RB1, JAK3, APC, GNA11, GNAS, KDR/VEGFR2, TGFBR2, ARID1A, ATM, BRCA1, BRCA2, CDH1, FBXW7, and MUC16, in which mutations are identified. In the other types of appendiceal tumors, the number of genes in which mutations are present is lower. Among the loci that concentrate the most genes in which mutations occur in appendiceal cancers, 19p13 ranks first, with ten genes; other loci such as 3p21.2-3p21.3, 16q24, 16p13.3, and Xp11.3-11.4 each concentrate three genes (Figure 11). These loci could be used for searching possible genetic markers for appendiceal cancers.
Figure 11.
Chromosome distribution of genes that are mutated in appendiceal cancers.
Chromosome 18q is the location of the DCC, DPC4 (SMAD4; MADH4), and JV-18 (SMAD2; MADH2) genes, and loss is frequent in colorectal adenocarcinoma [241]. The status of chromosome 18q has also been shown in previous studies to be an independent prognostic factor in patients with stage II and III colon carcinomas, and tumors without loss are associated with better clinical outcomes. DPC4 is a tumor suppressor gene with frequent alterations in pancreatic adenocarcinomas [242]. Maru et al. studied the genetic alterations, including loss of chromosome 18q (location of DCC, DPC4, and JV-18 genes) and mutations of the DPC4 (SMAD4) and beta-catenin genes in 28 appendiceal adenocarcinomas. Chromosome 18q loss was present in 57% of appendiceal carcinomas. Mutation of the DPC4 gene was present in 14% (three of 22) of the carcinomas occurring in one tumor with chromosome 18q loss and in two with unassessed chromosome 18q status. The authors concluded that the presence of chromosome 18q loss and DPC4 mutations in appendiceal adenocarcinomas suggests the involvement of DPC4 and nearby genes on chromosome 18q (DCC and/or JV-18) in the pathogenesis of appendiceal adenocarcinomas [243]. Davison et al. performed a study including 109 disseminated appendiceal mucinous neoplasms aiming to correlate SMAD4 immunohistochemical expression with tumor grade and assess the prognostic significance of SMAD4 expression in predicting overall survival. Their results indicate that loss of SMAD4 immunohistochemical expression is associated with loss of heterozygosity at chromosome 18q and is always associated with aggressive histologic features in disseminated appendiceal mucinous neoplasms. SMAD4 immunohistochemistry may be a practical instrument in select cases of disseminated appendiceal neoplasia, in which the distinction between low-grade and high-grade tumors is difficult [241].
11. Future Perspectives
The appendix tumor microenvironment consists of cellular and noncellular components: the former includes the immunocompetent cells, while the latter represents the supporting stroma. Particularly in carcinoids, the immune cell reaction can be explicated by tumor-infiltrating lymphocytes, which, in some circumstances, may arrange around and inside the tumor briskly, influencing the prognosis favorably. This active reaction has to be distinguished from any preexisting inflammatory condition of the appendix and superimposed tumor complications, such as infection or ischemia. In practice, the appendix TME is a complex framework with immunological, mechanic, and metabolic functions, all supported by marked neo-lymphoangiogenesis [244].
Current surgical strategies for localized well-differentiated appendiceal cancers may vary from simple appendicectomy to right hemicolectomy, depending on tumor size and invasion patterns. Because the diagnosis is usually established during the histological examination of an appendectomy specimen, it is essential to identify the subgroup of patients who will require further therapy. Recently, significant advances have been made in the management of appendix cancers, and novel therapeutic options are available for patients with metastatic disease; cytotoxic chemotherapy should be used only for patients with high-grade tumor burden who have no other therapeutic options. Although poorly differentiated appendiceal cancers are rare, their management involves platinum etoposide chemotherapy. Very few trials have included patients with these tumors; the data are scant. Therefore, it is crucial to evaluate many factors, including surgical respectability even in the metastatic setting, the status of somatostatin avidity, histological grade, and tumor burden, to select the appropriate personalized treatment for every patient through multidisciplinary discussion by experts [245]. Surgical intervention is determined in mucinous neoplasm according to the mesoappendix’s pathological subtype and involvement. Regarding the need for additional surgical intervention or medical treatment for patients with tumors, histopathological results must be followed carefully after appendectomy.
The review of these scattered cases raises a question regarding the true incidence of appendiceal involvement in gastric tumors, particularly the asymptomatic metastases, the answer to which leads to re-questioning the benefit–risk ratio of performing appendectomy for every gastric oncology surgery. Given reported patients’ prognoses, guidelines on managing such cases are also needed. Clinical and morphological analyses of cases from the practice of malignant tumors of the appendix (neuroendocrine tumor and adenocarcinoma) will be valuable and exciting for the medical community and should stimulate cancer vigilance in physicians [246].
Traditional cancer models should be generated for rare cancers, even using conventional methods, because the urgent need for rare cancer models has long been neglected. We need targeted and conscious efforts to generate “orphan models” to promote rare cancer research, as we currently have for orphan drugs [247]. Such model creation should be conducted in a non-competitive way. In one example, patient-derived rare cancer models are being generated at the National Cancer Center Research Institute, Tokyo, Japan, in global collaboration with multiple institutions. These models are then characterized and applied to translational research by our group and, in parallel, are delivered to multiple research groups.
Understanding that the paucity of cancer models is the fundamental problem in rare cancer research is essential. It is necessary to intensively promote the development of rare cancer models and to share them freely within the research community. This requires multi-institutional studies, as clinical samples are rare, and the research that single research groups can perform is limited. Such efforts and the models they produce will enable future generations to better understand cancer biology and develop novel biotechnologies and treatments, dramatically improving rare cancer outcomes. By creating novel patient-derived cancer models, we can change the future of rare cancer research.
12. Conclusions
An analysis of the presence of mutations of 105 genes in appendiceal cancers through the lens of the literature reviewed supports the view that in most of them, the inactivation of tumor suppressor genes, such as TP53 and SMAD4, is required in parallel with the reactivation of genes with oncogenic potentials, such as KRAS, GNAS, and BRAF, which support the main tumor processes, cell proliferation, angiogenesis, and evasion of apoptosis. Frequently, genes belonging to the RAS–RAF–MEK–ERK signaling pathway are mutated as the primary mechanism of appendix tumorigenesis, activating alternative pathways such as PI3K–PKB/AKT, NOTCH, RAP1, TGFB, and WNT.
Of all appendiceal cancers, the most mutated genes are reported in mucinous neoplasms of the appendix, not including those in the RAS–RAF–MEK–ERK signaling pathway, followed by low-grade appendiceal mucinous neoplasms, appendiceal goblet cell adenocarcinomas, and mucinous adenocarcinomas of the appendix, in which this signaling pathway is most frequently affected, showing its importance in their tumorigenesis. Microsatellite instability rarely occurs in appendix cancers, reported only in adenocarcinomas.
Author Contributions
M.C. and C.B. conceived, revised, and corrected the manuscript; M.C., C.B., C.M. and L.P. drafted Section 1, Section 2, Section 3, Section 4 and Section 5; A.B., O.A., M.M.M., S.T. and C.O.V. drafted Section 6, Section 7, Section 8 and Section 9; M.C. designed the figures. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data can be found in the text.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by the Romanian Executive Agency for Higher Education, Research, Development, and Innovation (https://uefiscdi.gov.ro/, accessed on 24 February 2023), research project FDI 0690/2023, and the “Analysis of the potential for sustainable use of vegetation specific to the Danube-Danube Delta-Black Sea system” project, awarded by the European Regional Development Fund through the Competitiveness Operational Program 2014–2020, contract no. 108630.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Deshmukh S., Verde F., Johnson P.T., Fishman E.K., Macura K.J. Anatomical variants and pathologies of the vermix. Emerg. Radiol. 2014;21:543–552. doi: 10.1007/s10140-014-1206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumpelick V., Dreuw B., Ophoff K., Prescher A. Appendix and cecum. Embryology, anatomy, and surgical applications. Surg. Clin. N. Am. 2000;80:295–318. doi: 10.1016/S0039-6109(05)70407-2. [DOI] [PubMed] [Google Scholar]
- 3.De Souza S.C., da Costa S.R.M.R., de Souza I.G.S. Vermiform appendix: Positions and length—A study of 377 cases and literature review. J. Coloproctol. 2015;35:212–216. doi: 10.1016/j.jcol.2015.08.003. [DOI] [Google Scholar]
- 4.Amin M.B., Greene F.L., Edge S.B., Compton C.C., Gershenwald J.E., Brookland R.K., Meyer L., Gress D.M., Byrd D.R., Winchester D.P. Appendix: Carcinoma. In: Amin M.B., editor. AJCC Cancer Staging Manual. 8th ed. Springer; New York, NY, USA: 2017. [DOI] [PubMed] [Google Scholar]
- 5.Nagtegaal I.D., Klimstra D.S., Washington M.K. WHO Classification of Tumors: Digestive System Tumors. 5th ed. International Agency for Research on Cancer; Lyon, France: 2019. Tumors of the appendix. [Google Scholar]
- 6.Ramnani D.M., Wistuba I.I., Behrens C., Gazdar A.F., Sobin L.H., Albores-Saavedra J. K-ras and p53 mutations in the pathogenesis of classical and goblet cell carcinoids of the appendix. Cancer. 1999;86:14–21. doi: 10.1002/(SICI)1097-0142(19990701)86:1<14::AID-CNCR4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 7.Alakus H., Babicky M.L., Ghosh P., Yost S., Jepsen K., Dai Y., Arias A., Samuels M.L., Mose E.S., Schwab R.B., et al. Genome-wide mutational landscape of mucinous carcinomatosis peritonei of appendiceal origin. Genome Med. 2014;6:43. doi: 10.1186/gm559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borazanci E., Millis S., Kimbrough J., Doll N., Hoff D., Ramanathan R. Potential actionable targets in appendiceal cancer detected by immunohistochemistry, fluorescent in situ hybridization, and mutational analysis. J. Gastrointest. Oncol. 2017;8:164–172. doi: 10.21037/jgo.2017.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ang C.S., Shen J.P., Hardy-Abeloos C.J., Huang J.K., Ross J.S., Miller V.A., Jacobs M.T., Chen I.L., Xu D., Ali S.M., et al. Genomic landscape of appendiceal neoplasms. JCO Precis. Oncol. 2018;2:PO.17.00302. doi: 10.1200/PO.17.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jesinghaus M., Konukiewitz B., Foersch S., Stenzinger A., Steiger K., Muckenhuber A., Groß C., Mollenhauer M., Roth W., Detlefsen S., et al. Appendiceal goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid are genetically distinct from primary colorectal-type adenocarcinoma of the appendix. Mod. Pathol. 2018;31:829–839. doi: 10.1038/modpathol.2017.184. [DOI] [PubMed] [Google Scholar]
- 11.Johncilla M., Stachler M., Misdraji J., Lisovsky M., Yozu M., Lindeman N., Lauwers G.Y., Odze R.D., Srivastava A. Mutational landscape of goblet cell carcinoids and adenocarcinoma ex goblet cell carcinoids of the appendix is distinct from typical carcinoids and colorectal adenocarcinomas. Mod. Pathol. 2018;31:989–996. doi: 10.1038/s41379-018-0003-0. [DOI] [PubMed] [Google Scholar]
- 12.Liu W., Liu L., Wang R., Gong G., Ding X., Yang B., Bao Y., Wang Z., Zhang B., Zhao D., et al. Bevacizumab combined with oxaliplatin/capecitabine in patient with refractory and recurrent mucinous adenocarcinoma of the appendix: A case report. Front. Oncol. 2019;9:55. doi: 10.3389/fonc.2019.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holowatyj A.N., Eng C., Wen W., Idrees K., Guo X. Spectrum of somatic cancer gene variations among adults with appendiceal cancer by age at disease onset. JAMA Netw. Open. 2021;4:e216703. doi: 10.1001/jamanetworkopen.2020.28644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.KEGG Pathways in Cancer-Reference Pathway. [(accessed on 3 October 2022)]. Available online: https://www.genome.jp/kegg-bin/show_pathway?map05200.
- 15.Kang D.W., Kim B.H., Kim J.M., Kim J., Chang H.J., Chang M.S., Sohn J.H., Cho M.Y., Jin S.Y., Chang H.K., et al. Gastrointestinal pathology study group of the Korean society of pathologists. Standardization of the pathologic diagnosis of appendiceal mucinous neoplasms. J. Pathol. Transl. Med. 2021;55:247–264. doi: 10.4132/jptm.2021.05.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Copur M., Cushman-Vokoun A., Padussis J., Whitney W., Schroeder C., Herold D., Lintel N., Horn A. Mucinous adenocarcinoma of the appendix with histologic response to neoadjuvant chemotherapy: Review of histologic and clinical spectrum of epithelial neoplastic mucinous lesions of the appendix. Oncology. 2021;35:335–340. doi: 10.46883/ONC.2021.3506.0335. [DOI] [PubMed] [Google Scholar]
- 17.Patel S., Alam A., Pant R., Chattopadhyay S. Wnt signaling and Its significance within the tumor microenvironment: Novel therapeutic insights. Front. Immunol. 2019;10:2872. doi: 10.3389/fimmu.2019.02872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I.C., Kratz C.P., Zenker M., Wittinghofer A., Ahmadian M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobre M., Dinu D.E., Panaitescu E., Bîrlă R.D., Iosif C.I., Boeriu M., Constantinoiu S., Ivan R.N., Ardeleanu C.M., Costache M. KRAS gene mutations—Prognostic factor in colorectal cancer? Rom. J. Morphol. Embryol. 2015;56:671–678. [PubMed] [Google Scholar]
- 20.KRAS Gene—KRAS Proto-Oncogene, GTPase. [(accessed on 14 September 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KRAS&keywords=KRAS.
- 21.Yantiss R.K., Panczykowski A., Misdraji J., Hahn H.P., Odze R.D., Rennert H., Chen Y.T. A comprehensive study of nondysplastic and dysplastic serrated polyps of the vermiform appendix. Am. J. Surg. Pathol. 2008;32:175. doi: 10.1097/PAS.0b013e31806bee6d. [DOI] [PubMed] [Google Scholar]
- 22.Pai R.K., Hartman D.J., Gonzalo D.H., Lai K.K., Downs-Kelly E., Goldblum J.R., Liu X., Patil D.T., Bennett A.E., Plesec T.P., et al. Serrated lesions of the appendix frequently harbor KRAS mutations and not BRAF mutations indicating a distinctly different serrated neoplastic pathway in the appendix. Hum. Pathol. 2014;45:227–235. doi: 10.1016/j.humpath.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Munari G., Businello G., Mattiolo P., Pennelli G., Sbaraglia M., Borga C., Pucciarelli S., Spolverato G., Mescoli C., Galuppini F., et al. Molecular profiling of appendiceal serrated lesions, polyps and mucinous neoplasms: A single-centre experience. J. Cancer Res. Clin. Oncol. 2021;147:1897–1904. doi: 10.1007/s00432-021-03589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zauber P., Berman E., Marotta S., Sabbath-Solitare M., Bishop T. Ki-ras gene mutations are invariably present in low-grade mucinous tumors of the vermiform appendix. Scand. J. Gastroenterol. 2011;46:869–874. doi: 10.3109/00365521.2011.565070. [DOI] [PubMed] [Google Scholar]
- 25.Shetty S., Thomas P., Ramanan B., Sharma P., Govindarajan V., Loggie B. Kras mutations and p53 overexpression in pseudomyxoma peritonei: Association with phenotype and prognosis. J. Surg. Res. 2013;180:97–103. doi: 10.1016/j.jss.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 26.Liu X., Mody K., de Abreu F.B., Pipas J.M., Peterson J.D., Gallagher T.L., Suriawinata A.A., Ripple G.H., Hourdequin K.C., Smith K.D., et al. Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin. Chem. 2014;60:1004–1011. doi: 10.1373/clinchem.2014.225565. [DOI] [PubMed] [Google Scholar]
- 27.Singhi A.D., Davison J.M., Choudry H.A., Pingpank J.F., Ahrendt S.A., Holtzman M.P., Zureikat A.H., Zeh H.J., Ramalingam L., Mantha G., et al. GNAS is frequently mutated in both low-grade and high-grade disseminated appendiceal mucinous neoplasms but does not affect survival. Hum. Pathol. 2014;45:1737–1743. doi: 10.1016/j.humpath.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 28.Davison J.M., Choudry H.A., Pingpank J.F., Ahrendt S.A., Holtzman M.P., Zureikat A.H., Zeh H.J., Ramalingam L., Zhu B., Nikiforova M., et al. Clinicopathologic and molecular analysis of disseminated appendiceal mucinous neoplasms: Identification of factors predicting survival and proposed criteria for a three-tiered assessment of tumor grade. Mod. Pathol. 2014;27:1521–1539. doi: 10.1038/modpathol.2014.37. [DOI] [PubMed] [Google Scholar]
- 29.Noguchi R., Yano H., Gohda Y., Suda R., Igari T., Ohta Y., Yamashita N., Yamaguchi K., Terakado Y., Ikenoue T., et al. Molecular profiles of high-grade and low-grade pseudomyxoma peritonei. Cancer Med. 2015;4:1809–1816. doi: 10.1002/cam4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pengelly R.J., Rowaiye B., Pickard K., Moran B., Dayal S., Tapper W., Mirnezami A., Cecil T., Mohamed F., Carr N., et al. Analysis of mutation and loss of heterozygosity by whole-exome sequencing yields insights into Pseudomyxoma peritonei. J. Mol. Diagn. 2018;20:635–642. doi: 10.1016/j.jmoldx.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Schrader L., Choo E. 330 low-grade mucinous neoplasm of the appendix in the setting of a CDKN2A gene mutation: A potential association risk factor. Am. J. Clin. Pathol. 2018;149:142. doi: 10.1093/ajcp/aqx127.329. [DOI] [Google Scholar]
- 32.King M.C., Munoz-Zuluaga C., Ledakis P., Studeman K., Sittig M., Gushchin V., Sardi A. Germline and somatic genetic alterations in two first-degree relatives with appendiceal low-grade mucinous carcinoma peritonei. Clin. Case. Rep. 2020;8:3168–3177. doi: 10.1002/ccr3.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikaeel R.R., Young J.P., Tapia Rico G., Hewett P.J., Hardingham J.E., Uylaki W., Horsnell M., Price T.J. Immunohistochemistry features and molecular pathology of appendiceal neoplasms. Crit. Rev. Clin. Lab. Sci. 2021;58:369–384. doi: 10.1080/10408363.2021.1881756. [DOI] [PubMed] [Google Scholar]
- 34.Flatmark K., Torgunrud A., Fleten K.G., Davidson B., Juul H.V., Mensali N., Lund-Andersen C., Inderberg E.M. Peptide vaccine targeting mutated GNAS: A potential novel treatment for pseudomyxoma peritonei. J. Immunother. Cancer. 2021;9:e003109. doi: 10.1136/jitc-2021-003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabbani W., Houlihan P.S., Luthra R., Hamilton S.R., Rashid A. Mucinous and nonmucinous appendiceal adenocarcinomas: Different clinicopathological features but similar genetic alterations. Mod. Pathol. 2002;15:599–605. doi: 10.1038/modpathol.3880572. [DOI] [PubMed] [Google Scholar]
- 36.Arai H., Baca Y., Battaglin F., Kawanishi N., Wang J., Soni S., Zhang W., Millstein J., Johnston C., Goldberg R.M., et al. Molecular characterization of appendiceal goblet cell carcinoid. Mol. Cancer Ther. 2020;19:2634–2640. doi: 10.1158/1535-7163.MCT-20-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ottaiano A., Santorsola M., Perri F., Pace U., Marra B., Correra M., Sabbatino F., Cascella M., Petrillo N., Ianniello M., et al. Clinical and molecular characteristics of rare malignant tumors of colon and rectum. Biology. 2022;11:267. doi: 10.3390/biology11020267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.HRAS Gene—HRas Proto-Oncogene, GTPase. [(accessed on 3 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=HRAS.
- 39.NRAS Gene—NRAS Proto-Oncogene, GTPase. [(accessed on 3 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NRAS&keywords=NRAS.
- 40.Raghav K.P., Shetty A.V., Kazmi S.M., Zhang N., Morris J., Taggart M., Fournier K., Royal R., Mansfield P., Eng C., et al. Impact of molecular alterations and targeted therapy in appendiceal adenocarcinomas. Oncologist. 2013;18:1270–1277. doi: 10.1634/theoncologist.2013-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Huang X., Liu H., Wei C., Ru H., Qin H., Lai H., Meng Y., Wu G., Xie W., et al. Immune landscape and prognostic immune-related genes in KRAS-mutant colorectal cancer patients. J. Transl. Med. 2021;19:27. doi: 10.1186/s12967-020-02638-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matas J., Kohrn B., Fredrickson J., Carter K., Yu M., Wang T., Gui X., Soussi T., Moreno V., Grady W.M., et al. Colorectal cancer is associated with the presence of cancer driver mutations in normal colon. Cancer Res. 2022;82:1492–1502. doi: 10.1158/0008-5472.CAN-21-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matallanas D., Birtwistle M., Romano D., Zebisch A., Rauch J., von Kriegsheim A., Kolch W. Raf family kinases: Old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.BRAF Gene—B-Raf Proto-Oncogene, Serine/Threonine Kinase. [(accessed on 3 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRAF&keywords=BRAF.
- 45.Ueda K., Yamada T., Ohta R., Matsuda A., Sonoda H., Kuriyama S., Takahashi G., Iwai T., Takeda K., Miyasaka T., et al. BRAF V600E mutations in right-side colon cancer: Heterogeneity detected by liquid biopsy. Eur. J. Surg. Oncol. 2022;48:1375–1383. doi: 10.1016/j.ejso.2022.01.016. [DOI] [PubMed] [Google Scholar]
- 46.Asako K., Hayama T., Hashiguchi Y., Miyata T., Fukushima Y., Shimada R., Kaneko K., Nozawa K., Matsuda K., Fukagawa T. Prognostic value of KRAS exon-specific mutations in patients with colorectal cancer. Anticancer Res. 2023;43:1563–1568. doi: 10.21873/anticanres.16306. [DOI] [PubMed] [Google Scholar]
- 47.EGFR Gene—Epidermal Growth Factor Receptor. [(accessed on 4 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EGFR.
- 48.Roskoski R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 49.ERBB2 Gene—Erb-B2 Receptor Tyrosine Kinase 2. [(accessed on 4 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERBB2.
- 50.Arteaga C.L., Engelman J.A. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.ERBB4 Gene—Erb-B2 Receptor Tyrosine Kinase. [(accessed on 4 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERBB4.
- 52.Petrini I. Biology of MET: A double life between normal tissue repair and tumor progression. Ann. Transl. Med. 2015;3:82. doi: 10.3978/j.issn.2305-5839.2015.03.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MET Gene—MET Proto-Oncogene, Receptor Tyrosine Kinase. [(accessed on 5 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MET.
- 54.Li J., Li Z., Ding Y., Xu Y., Zhu X., Cao N., Huang C., Qin M., Liu F., Zhao A. TP53 mutation and MET amplification in circulating tumor DNA analysis predict disease progression in patients with advanced gastric cancer. PeerJ. 2021;9:e11146. doi: 10.7717/peerj.11146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.FGFR1 Gene—Fibroblast Growth Factor Receptor 1. [(accessed on 5 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR1.
- 56.FGFR4 Gene—Fibroblast Growth Factor Receptor 2. [(accessed on 5 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR4.
- 57.Krook M.A., Reeser J.W., Ernst G., Barker H., Wilberding M., Li G., Chen H.Z., Roychowdhury S. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer. 2021;124:880–892. doi: 10.1038/s41416-020-01157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.FGFR2 Gene—Fibroblast Growth Factor Receptor 2. [(accessed on 5 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR2.
- 59.FGFR3 Gene—Fibroblast Growth Factor Receptor 2. [(accessed on 5 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR3.
- 60.Jogo T., Nakamura Y., Shitara K., Bando H., Yasui H., Esaki T., Terazawa T., Satoh T., Shinozaki E., Nishina T., et al. Circulating tumor DNA analysis detects FGFR2 amplification and concurrent genomic alterations associated with FGFR inhibitor efficacy in advanced gastric cancer. Clin. Cancer Res. 2021;27:5619–5627. doi: 10.1158/1078-0432.CCR-21-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller D.M., Thomas S.D., Islam A., Muench D., Sedoris K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MYC Gene—MYC Proto-Oncogene, BHLH Transcription Factor. [(accessed on 6 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MYC.
- 63.Riviere P., Fanta P.T., Ikeda S., Baumgartner J., Heestand G.M., Kurzrock R. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol. Cancer Ther. 2018;17:297–305. doi: 10.1158/1535-7163.MCT-17-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin X., Lin X., Guo L., Wang Y., Zhang G. Distinct clinicopathological characteristics, genomic alteration and prognosis in breast cancer with concurrent TP53 mutation and MYC amplification. Thorac. Cancer. 2022;13:3441–3450. doi: 10.1111/1759-7714.14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jackson J.T., Mulazzani E., Nutt S.L., Masters S.L. The role of PLCγ2 in immunological disorders, cancer, and neurodegeneration. J. Biol. Chem. 2021;297:100905. doi: 10.1016/j.jbc.2021.100905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.PLCG2 Gene—Phospholipase C Gamma 2. [(accessed on 6 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PLCG2.
- 67.Peng Q., Luo D., Yang Y., Zhu Y., Luo Q., Chen H., Chen D., Zhou Z., Lu X. Clinical and immunological features of an APLAID patient caused by a novel mutation in PLCG2. Front. Immunol. 2023;14:1014150. doi: 10.3389/fimmu.2023.1014150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Z., Zhao R., Yang W., Li C., Huang J., Wen Z., Du G., Jiang L. PLCG2 as a potential indicator of tumor microenvironment remodeling in soft tissue sarcoma. Medicine. 2021;100:e25008. doi: 10.1097/MD.0000000000025008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernandez Borrero L.J., El-Deiry W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188556. doi: 10.1016/j.bbcan.2021.188556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.TP53 Gene—Tumor Protein P53. [(accessed on 30 September 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TP53&keywords=TP.
- 71.Oka K., Ueda M., Ikenaga M., Tanida T., Taguchi D., Fukushima S., Yoshida S., Yukimoto R., Iede K., Tsuda Y., et al. A Case of Early Appendiceal Adenocarcinoma Coexisting with High-Grade Appendiceal Mucinous Neoplasm. Cancer Chemother. 2022;49:1714–1716. [PubMed] [Google Scholar]
- 72.Yanai Y., Saito T., Hayashi T., Akazawa Y., Yatagai N., Tsuyama S., Tomita S., Hirai S., Ogura K., Matsumoto T., et al. Molecular and clinicopathological features of appendiceal mucinous neoplasms. Virchows Arch. 2021;478:413–426. doi: 10.1007/s00428-020-02906-5. [DOI] [PubMed] [Google Scholar]
- 73.Yan F., Shi F., Li X., Chang H., Jin M., Li Y. Prognostic significance of CEA, Ki67 and p53 in pseudomyxoma peritonei of appendiceal origin. J. Int. Med. Res. 2021;49:3000605211022297. doi: 10.1177/03000605211022297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.AKT1 Gene—AKT Serine/Threonine Kinase. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=AKT1.
- 75.Chen Y., Huang L., Dong Y., Tao C., Zhang R., Shao H., Shen H. Effect of AKT1 (p. E17K) hotspot mutation on malignant tumorigenesis and prognosis. Front. Cell Dev. Biol. 2020;8:573599. doi: 10.3389/fcell.2020.573599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mundi P.S., Sachdev J., McCourt C., Kalinsky K. AKT in cancer: New molecular insights and advances in drug development. Br. J. Clin. Pharmacol. 2016;82:943–956. doi: 10.1111/bcp.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalinsky K., Hong F., McCourt C.K., Sachdev J.C., Mitchell E.P., Zwiebel J.A., Doyle L.A., McShane L.M., Li S., Gray R.J., et al. Effect of capivasertib in patients with an AKT1 E17K-mutated tumor: NCI-MATCH subprotocol EAY131-Y nonrandomized trial. JAMA Oncol. 2021;7:271–278. doi: 10.1001/jamaoncol.2020.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.ITGA11 Gene—Integrin Subunit Alpha 11. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ITGA11.
- 79.Primac I., Maquoi E., Blacher S., Heljasvaara R., Van Deun J., Smeland H.Y., Canale A., Louis T., Stuhr L., Sounni N.E., et al. Stromal integrin α11 regulates PDGFR-β signaling and promotes breast cancer progression. J. Clin. Investig. 2019;129:4609–4628. doi: 10.1172/JCI125890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iwai M., Tulafu M., Togo S., Kawaji H., Kadoya K., Namba Y., Jin J., Watanabe J., Okabe T., Hidayat M., et al. Cancer-associated fibroblast migration in non-small cell lung cancers is modulated by increased integrin α11 expression. Mol. Oncol. 2021;15:1507–1527. doi: 10.1002/1878-0261.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.PIK3CA Gene—Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PIK3CA.
- 82.Zhang Y., Ng P.K.-S., Kucherlapati M., Chen F., Liu Y., Tsang Y.H., De Velasco G., Jeong K.J., Akbani R., Hadjipanayis A., et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017;31:820–832. doi: 10.1016/j.ccell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thorpe L.M., Yuzugullu H., Zhao J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 2015;15:7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giudice F.S., Squarize C.H. The determinants of head and neck cancer: Unmasking the PI3K pathway mutations. J. Carcinog. Mutagen. 2013:3. doi: 10.4172/2157-2518.S5-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tokunaga R., Xiu J., Johnston C., Goldberg R.M., Philip P.A., Seeber A., Naseem M., Lo J.H., Arai H., Battaglin F., et al. Molecular profiling of appendiceal adenocarcinoma and comparison with right-sided and left-sided colorectal cancer. Clin. Cancer Res. 2019;25:3096–3103. doi: 10.1158/1078-0432.CCR-18-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samuels Y., Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 2010;347:21–41. doi: 10.1007/82_2010_68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.PIK3C2B Gene—Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Beta. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PIK3C2B.
- 88.PTEN Gene—Phosphatase and Tensin Homolog. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTEN.
- 89.Belletti B., Baldassarre G. Roles of CDKN1B in cancer? Aging. 2015;7:529–530. doi: 10.18632/aging.100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.CDKN1B Gene—Cyclin Dependent Kinase Inhibitor 1B. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDKN1B.
- 91.Chan S.H., Chiang J., Ngeow J. CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered. Cancer Clin. Pract. 2021;19:21. doi: 10.1186/s13053-021-00178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.CDKN2A Gene—Cyclin Dependent Kinase Inhibitor 2A. [(accessed on 7 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDKN2A.
- 93.Yun J., Li Y., Xu C.T., Pan B.R. Epidemiology and Rb1 gene of retinoblastoma. Int. J. Ophthalmol. 2011;4:103–109. doi: 10.3980/j.issn.2222-3959.2011.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lan X., Xu W., Tang X., Ye H., Song X., Lin L., Ren X., Yu G., Zhang H., Wu S. Spectrum of RB1 germline mutations and clinical features in unrelated chinese patients with retinoblastoma. Front. Genet. 2020;11:142. doi: 10.3389/fgene.2020.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knudsen E.S., Nambiar R., Rosario S.R., Smiraglia D.J., Goodrich D.W., Witkiewicz A.K. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun. Biol. 2020;3:158. doi: 10.1038/s42003-020-0873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.RB1 Gene—RB Transcriptional Corepressor 1. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RB1.
- 97.JAK3 Gene—Janus Kinase 3. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=JAK3.
- 98.Nairismägi M., Gerritsen M.E., Li Z.M., Wijaya G.C., Chia B.K.H., Laurensia Y., Lim J.Q., Yeoh K.W., Yao X.S., Pang W.L., et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia. 2018;32:1147–1156. doi: 10.1038/s41375-017-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vadivel C.K., Gluud M., Torres-Rusillo S., Boding L., Willerslev-Olsen A., Buus T.B., Nielsen T.K., Persson J.L., Bonefeld C.M., Geisler C., et al. JAK3 is expressed in the nucleus of malignant T cells in cutaneous T cell lymphoma (CTCL) Cancers. 2021;13:280. doi: 10.3390/cancers13020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.APC Gene—APC Regulator of WNT Signaling Pathway. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=APC.
- 101.Palacio-Rúa K.A., Isaza-Jiménez L.F., Ahumada-Rodríguez E., Muñetón-Peña C.M. Genetic analysis in APC, KRAS, and TP53 in patients with stomach and colon cancer. Rev. Gastroenterol. Mex. 2014;79:79–89. doi: 10.1016/j.rgmxen.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 102.Zhang L., Shay J.W. Multiple roles of APC and its therapeutic implications in colorectal cancer. J. Natl. Cancer Inst. 2017;109:djw332. doi: 10.1093/jnci/djw332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.AXIN1 Gene—Axin 1. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=AXIN1.
- 104.Schalken J.A. Editorial comment on: Mutations in the AXIN1 gene in advanced prostate cancer. Eur. Urol. 2009;56:494. doi: 10.1016/j.eururo.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 105.Picco G., Petti C., Centonze A., Torchiaro E., Crisafulli G., Novara L., Acquaviva A., Bardelli A., Medico E. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Mol. Med. 2017;9:293–303. doi: 10.15252/emmm.201606773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.TCF7L2 Gene—Transcription Factor 7 like 2. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TCF7L2.
- 107.Wenzel J., Rose K., Haghighi E.B., Lamprecht C., Rauen G., Freihen V., Kesselring R., Boerries M., Hecht A. Loss of the nuclear Wnt pathway effector TCF7L2 promotes migration and invasion of human colorectal cancer cells. Oncogene. 2020;39:3893–3909. doi: 10.1038/s41388-020-1259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitroi A.F., Leopa N., Dumitru E., Brînzan C., Tocia C., Dumitru A., Popescu R.C. Association of TCF7L2, CASC8 and GREM1 polymorphisms in patients with colorectal cancer and type II diabetes mellitus. Genes. 2022;13:1297. doi: 10.3390/genes13081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.RHOA Gene—Ras Homolog Family Member A. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RHOA.
- 110.Schaefer A., Der C.J. RHOA takes the RHOad less traveled to cancer. Trends Cancer. 2022;8:655–669. doi: 10.1016/j.trecan.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 111.Jeong D., Park S., Kim H., Kim C.J., Ahn T.S., Bae S.B., Kim H.J., Kim T.H., Im J., Lee M.S., et al. RhoA is associated with invasion and poor prognosis in colorectal cancer. Int. J. Oncol. 2016;48:714–722. doi: 10.3892/ijo.2015.3281. [DOI] [PubMed] [Google Scholar]
- 112.Kalpana G., Figy C., Yeung M., Yeung K.C. Reduced RhoA expression enhances breast cancer metastasis with a concomitant increase in CCR5 and CXCR4 chemokines signaling. Sci. Rep. 2019;9:16351. doi: 10.1038/s41598-019-52746-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.GNA11 Gene—G Protein Subunit Alpha 11. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GNA11.
- 114.Jansen P., Müller H., Lodde G.C., Zaremba A., Möller I., Sucker A., Paschen A., Esser S., Schaller J., Gunzer M., et al. GNA14, GNA11, and GNAQ mutations are frequent in benign but not malignant cutaneous vascular tumors. Front. Genet. 2021;12:663272. doi: 10.3389/fgene.2021.663272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.GNAS Gene—GNAS Complex Locus. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GNAS.
- 116.Turan S., Bastepe M. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm. Res. Paediatr. 2013;80:229–241. doi: 10.1159/000355384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jin X., Zhu L., Cui Z., Tang J., Xie M., Ren G. Elevated expression of GNAS promotes breast cancer cell proliferation and migration via the PI3K/AKT/Snail1/E-cadherin axis. Clin. Transl. Oncol. 2019;21:1207–1219. doi: 10.1007/s12094-019-02042-w. [DOI] [PubMed] [Google Scholar]
- 118.Iyer N.G., Ozdag H., Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 119.EP300 Gene—E1A Binding Protein P300. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EP300.
- 120.CREBBP Gene—CREB Binding Protein. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CREBBP.
- 121.Tang Z., Yu W., Zhang C., Zhao S., Yu Z., Xiao X., Tang R., Xuan Y., Yang W., Hao J., et al. CREB-binding protein regulates lung cancer growth by targeting MAPK and CPSF4 signaling pathway. Mol. Oncol. 2016;10:317–329. doi: 10.1016/j.molonc.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Durbin A.D., Wang T., Wimalasena V.K., Zimmerman M.W., Li D., Dharia N.V., Mariani L., Shendy N.A.M., Nance S., Patel A.G., et al. EP300 selectively controls the enhancer landscape of MYCN-amplified Nneuroblastoma. Cancer Discov. 2022;12:730–751. doi: 10.1158/2159-8290.CD-21-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bi Y., Kong P., Zhang L., Cui H., Xu X., Chang F., Yan T., Li J., Cheng C., Song B., et al. EP300 as an oncogene correlates with poor prognosis in esophageal squamous carcinoma. J. Cancer. 2019;10:5413–5426. doi: 10.7150/jca.34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rezaei F.M., Hashemzadeh S., Gavgani R.R., Feizi M.H., Pouladi N., Kafil H.S., Rostamizadeh L., Oskooei V.K., Taheri M., Sakhinia E. Dysregulated KDR and FLT1 gene expression in colorectal cancer patients. Rep. Biochem. Mol. Biol. 2019;8:244–252. [PMC free article] [PubMed] [Google Scholar]
- 125.KDR Gene—Kinase Insert Domain Receptor. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KDR.
- 126.Antonescu C.R., Yoshida A., Guo T., Chang N.E., Zhang L., Agaram N.P., Qin L.X., Brennan M.F., Singer S., Maki R.G. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69:7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang F., Liu G. Influence of KDR genetic variation on the effectiveness and safety of bevacizumab in the first-line treatment for patients with advanced colorectal cancer. Int. J. Gen. Med. 2022;15:5651–5659. doi: 10.2147/IJGM.S362366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nilsson M.B., Giri U., Gudikote J., Tang X., Lu W., Tran H., Fan Y., Koo A., Diao L., Tong P., et al. KDR amplification is associated with VEGF-induced activation of the mTOR and invasion pathways but does not predict clinical benefit to the VEGFR TKI vandetanib. Clin. Cancer Res. 2016;22:1940–1950. doi: 10.1158/1078-0432.CCR-15-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Geng N., Su J., Liu Z., Ding C., Xie S., Hu W. The influence of KDR genetic variation on the efficacy and safety of patients with advanced NSCLC receiving first-line bevacizumab plus chemotherapy regimen. Technol. Cancer Res. Treat. 2021;20:15330338211019433. doi: 10.1177/15330338211019433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cui Y., Zhang P., Liang X., Xu J., Liu X., Wu Y., Zhang J., Wang W., Zhang F., Guo R. Association of KDR mutation with better clinical outcomes in pan-cancer for immune checkpoint inhibitors. Am. J. Cancer Res. 2022;12:1766–1783. doi: 10.2139/ssrn.3991639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.NOTCH1 Gene—Notch Receptor 1. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH1.
- 132.Anusewicz D., Orzechowska M., Bednarek A.K. Notch signaling pathway in cancer-review with bioinformatic analysis. Cancers. 2021;13:768. doi: 10.3390/cancers13040768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.NOTCH3 Gene—Notch Receptor 3. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH3.
- 134.Xiu M., Wang Y., Li B., Wang X., Xiao F., Chen S., Zhang L., Zhou B., Hua F. The role of Notch3 signaling in cancer stemness and chemoresistance: Molecular mechanisms and targeting strategies. Front. Mol. Biosci. 2021;8:694141. doi: 10.3389/fmolb.2021.694141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.NOTCH4 Gene—Notch Receptor 4. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH4.
- 136.Zhou B., Lin W., Long Y., Yang Y., Zhang H., Wu K., Chu Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022;7:95. doi: 10.1038/s41392-022-00934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.D’Assoro A.B., Leon-Ferre R., Braune E.B., Lendahl U. Roles of Notch signaling in the tumor microenvironment. Int. J. Mol. Sci. 2022;23:6241. doi: 10.3390/ijms23116241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.FLT1 Gene—Fms Related Receptor Tyrosine Kinase 1. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FLT1.
- 139.Zhang S.D., McCrudden C.M., Meng C., Lin Y., Kwok H.F. The significance of combining VEGFA, FLT1, and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab. OncoTargets Ther. 2015;8:835–843. doi: 10.2147/OTT.S80518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vander Ark A., Cao J., Li X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018;52:112–120. doi: 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 141.Bharathy S., Xie W., Yingling J.M., Reiss M. Cancer-associated transforming growth factor beta type II receptor gene mutant causes activation of bone morphogenic protein-Smads and invasive phenotype. Cancer Res. 2008;68:1656–1666. doi: 10.1158/0008-5472.CAN-07-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.TGFBR2 Gene—Transforming Growth Factor Beta Receptor 2. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TGFBR2.
- 143.Li T., Wang H., Xu J., Li C., Zhang Y., Wang G., Liu Y., Cai S., Fang W., Li J., et al. TGFBR2 mutation predicts resistance to immune checkpoint inhibitors in patients with non-small cell lung cancer. Ther. Adv. Med. Oncol. 2021;13:17588359211038477. doi: 10.1177/17588359211038477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.TGFBR1 Gene—Transforming Growth Factor Beta Receptor 1. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TGFBR1.
- 145.Wang Y.Q., Qi X.W., Wang F., Jiang J., Guo Q.N. Association between TGFBR1 polymorphisms and cancer risk: A meta-analysis of 35 case-control studies. PLoS ONE. 2012;7:e42899. doi: 10.1371/journal.pone.0042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pasche B., Pennison M.J., Jimenez H., Wang M. TGFBR1 and cancer susceptibility. Trans. Am. Clin. Climatol. Assoc. 2014;125:300–312. [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou R., Huang Y., Cheng B., Wang Y., Xiong B. TGFBR1*6A is a potential modifier of migration and invasion in colorectal cancer cells. Oncol. Lett. 2018;15:3971–3976. doi: 10.3892/ol.2018.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Attisano L., Lee-Hoeflich S.T. The Smads. Genome Biol. 2001;2:REVIEWS3010. doi: 10.1186/gb-2001-2-8-reviews3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.SMAD2 Gene—SMAD Family Member 2. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD2.
- 150.Samanta D., Datta P.K. Alterations in the Smad pathway in human cancers. Front. Biosci. 2012;17:1281–1293. doi: 10.2741/3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fleming N.I., Jorissen R.N., Mouradov D., Christie M., Sakthianandeswaren A., Palmieri M., Day F., Li S., Tsui C., Lipton L., et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013;73:725–735. doi: 10.1158/0008-5472.CAN-12-2706. [DOI] [PubMed] [Google Scholar]
- 152.SMAD3 Gene—SMAD Family Member 3. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD3.
- 153.SMAD4 Gene—SMAD Family Member 4. [(accessed on 14 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD4.
- 154.Bertrand-Chapel A., Caligaris C., Fenouil T., Savary C., Aires S., Martel S., Huchedé P., Chassot C., Chauvet V., Cardot-Ruffino V., et al. SMAD2/3 mediate oncogenic effects of TGF-β in the absence of SMAD4. Commun. Biol. 2022;5:1068. doi: 10.1038/s42003-022-03994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Manou D., Caon I., Bouris P., Triantaphyllidou I.E., Giaroni C., Passi A., Karamanos N.K., Vigetti D., Theocharis A.D. The complex interplay between extracellular matrix and cells in tissues. Methods Mol Biol. 2019;1952:1–20. doi: 10.1007/978-1-4939-9133-4_1. [DOI] [PubMed] [Google Scholar]
- 156.Karamanos N.K., Theocharis A.D., Piperigkou Z., Manou D., Passi A., Skandalis S.S., Vynios D.H., Orian-Rousseau V., Ricard-Blum S., Schmelzer C.E.H., et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021;288:6850–6912. doi: 10.1111/febs.15776. [DOI] [PubMed] [Google Scholar]
- 157.Theocharis A.D., Gialeli C., Bouris P., Giannopoulou E., Skandalis S.S., Aletras A.J., Iozzo R.V., Karamanos N.K. Cell-matrix interactions: Focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J. 2014;281:5023–5042. doi: 10.1111/febs.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kai F., Drain A.P., Weaver V.M. The extracellular matrix modulates the metastatic journey. Dev. Cell. 2019;49:332–346. doi: 10.1016/j.devcel.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., Weaver V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Miroshnikova Y.A., Rozenberg G.I., Cassereau L., Pickup M., Mouw J.K., Ou G., Templeman K.L., Hannachi E.I., Gooch K.J., Sarang-Sieminski A.L., et al. α5β1-Integrin promotes tension-dependent mammary epithelial cell invasion by engaging the fibronectin synergy site. Mol. Biol. Cell. 2017;28:2958–2977. doi: 10.1091/mbc.e17-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Han W., Chen S., Yuan W., Fan Q., Tian J., Wang X., Chen L., Zhang X., Wei W., Liu R., et al. Oriented collagen fibers direct tumor cell intravasation. Proc. Natl. Acad. Sci. USA. 2016;113:11208–11213. doi: 10.1073/pnas.1610347113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Goddard E.T., Bozic I., Riddell S.R., Ghajar C.M. Dormant tumour cells, their niches and the influence of immunity. Nat. Cell Biol. 2018;20:1240–1249. doi: 10.1038/s41556-018-0214-0. [DOI] [PubMed] [Google Scholar]
- 163.COL5A3 Gene—Collagen Type V Alpha 3 Chain. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL5A3.
- 164.COL6A3 Gene—Collagen Type VI Alpha 3 Chain. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL6A3.
- 165.ZNF469 Gene—Zinc Finger Protein 469. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF4692022.
- 166.ATRX Gene—ATRX Chromatin Remodeler. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ATRX.
- 167.BRCA1 Gene—BRCA1 DNA Repair Associated. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRCA1.
- 168.BRCA2 Gene—BRCA2 DNA Repair Associated. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRCA2.
- 169.FANCA Gene—FA Complementation Group A. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FANCA.
- 170.KDM6A Gene—Lysine Demethylase 6A. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KDM6A.
- 171.KMT2D Gene—Lysine Methyltransferase 2D. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KMT2D.
- 172.RAD51C Gene—RAD51 Paralog C. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RAD51C.
- 173.SETD2 Gene—SET Domain Containing 2, Histone Lysine Methyltransferase. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SETD2.
- 174.TRPS1 Gene—Transcriptional Repressor GATA Binding 1. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRPS1.
- 175.CDH1 Gene—Cadherin 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDH1.
- 176.CSMD1 Gene—CUB and Sushi Multiple Domains 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CSMD1.
- 177.CTNNA1 Gene—Catenin Alpha 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CTNNA1.
- 178.CTNNB1 Gene—Catenin Beta 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CTNNB1.
- 179.CNTNAP2 Gene—Contactin Associated Protein 2. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CNTNAP2.
- 180.LAMA1 Gene—Laminin Subunit Alpha 1. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=LAMA1.
- 181.SPTA1 Gene—Spectrin Alpha, Erythrocytic 1. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SPTA1.
- 182.ALK Gene—ALK Receptor Tyrosine Kinase. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ALK.
- 183.ARID1A Gene—AT-Rich Interaction Domain 1A. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ARID1A.
- 184.ARID2 Gene—AT-Rich Interaction Domain 2. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ARID2.
- 185.ATM Gene—ATM Serine/Threonine Kinase. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ATM.
- 186.CARD11 Gene—Caspase Recruitment Domain Family Member 11. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CARD11.
- 187.DCLK1 Gene—Doublecortin Like Kinase 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DCLK1.
- 188.DIS3 Gene—DIS3 Homolog, Exosome Endoribonuclease and 3′-5′ Exoribonuclease. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DIS3.
- 189.FH Gene—Fumarate Hydratase. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FH.
- 190.IDH2 Gene—Isocitrate Dehydrogenase (NADP+) 2. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=IDH2.
- 191.PRKACA Gene—Protein Kinase CAMP-Activated Catalytic Subunit Alpha. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRKACA.
- 192.PTPN11 Gene—Protein Tyrosine Phosphatase Non-Receptor Type 11. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTPN11.
- 193.RHPN2 Gene—Rhophilin Rho GTPase Binding Protein 2. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RHPN2.
- 194.SMARCA4 Gene—SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily A, Member 4. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMARCA4.
- 195.TRRAP Gene—Transformation/Transcription Domain Associated Protein. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRRAP.
- 196.USP9X Gene—Ubiquitin Specific Peptidase 9 X-Linked. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=USP9X.
- 197.ABCA7 Gene—ATP Binding Cassette Subfamily A Member 7. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ABCA7.
- 198.ANKRD24 Gene—Ankyrin Repeat Domain 24. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ANKRD24.
- 199.APOB Gene—Apolipoprotein B. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=APOB.
- 200.ASXL1 Gene—ASXL Transcriptional Regulator 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ASXL1.
- 201.BCOR Gene—BCL6 Corepressor. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BCOR.
- 202.CRY2 Gene—Cryptochrome Circadian Regulator 2. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CRY2.
- 203.DOCK3—Dedicator of Cytokinesis 3. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DOCK3.
- 204.DOK6 Gene—Docking Protein 6. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DOK6.
- 205.EEF1A1 Gene—Eukaryotic Translation Elongation Factor 1 Alpha 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EEF1A1.
- 206.EPHA10 Gene—EPH Receptor A10. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EPHA10.
- 207.FAT1 Gene—FAT Atypical Cadherin 1. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAT1.
- 208.FAT4 Gene—FAT Atypical Cadherin 4. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAT4.
- 209.FBXW7 Gene—F-Box and WD Repeat Domain Containing 7. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FBXW7.
- 210.IRX6 Gene—Iroquois Homeobox 6. [(accessed on 17 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=IRX6.
- 211.KRT37 Gene—Keratin 37. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KRT37.
- 212.MED12 Gene—Mediator Complex Subunit 12. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MED12.
- 213.MTIF2 Gene—Mitochondrial Translational Initiation Factor 2. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MTIF2.
- 214.MUC16 Gene—Mucin 16, Cell Surface Associated. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MUC16.
- 215.Shibahara H., Higashi M., Yokoyama S., Rousseau K., Kitazono I., Osako M., Shirahama H., Tashiro Y., Kurumiya Y., Narita M., et al. A comprehensive expression analysis of mucins in appendiceal carcinoma in a multicenter study: MUC3 is a novel prognostic factor. PLoS ONE. 2014;9:e115613. doi: 10.1371/journal.pone.0115613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.OCA2 Gene—OCA2 Melanosomal Transmembrane Protein. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=OCA2.
- 217.PCDH10 Gene—Protocadherin 10. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PCDH10.
- 218.PCDH17 Gene—Protocadherin 17. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PCDH17.
- 219.POM121L12 Gene—POM121 Transmembrane Nucleoporin Like 12. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=POM121L12.
- 220.PRDM1 Gene—PR/SET Domain 1. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRDM1.
- 221.PTCHD3 Gene—Patched Domain Containing 3 (Gene/Pseudogene) [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTCHD3.
- 222.RNF43 Gene—Ring Finger Protein 43. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RNF43.
- 223.SNTG1 Gene—Syntrophin Gamma 1. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SNTG1.
- 224.SOX9 Gene—SRY-Box Transcription Factor 9. [(accessed on 18 October 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=2022.
- 225.STK11 Gene—Serine/Threonine Kinase 11. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=STK11.
- 226.TSC1 Gene—TSC Complex Subunit 1. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TSC12022.
- 227.TSC2 Gene—TSC Complex Subunit 2. [(accessed on 30 August 2022)]. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TSC2.
- 228.Iacopetta B., Grieu F., Amanuel B. Microsatellite instability in colorectal cancer. Asia Pac. J. Clin. Oncol. 2010;6:260–269. doi: 10.1111/j.1743-7563.2010.01335.x. [DOI] [PubMed] [Google Scholar]
- 229.Amato M., Franco R., Facchini G., Addeo R., Ciardiello F., Berretta M., Vita G., Sgambato A., Pignata S., Caraglia M., et al. Microsatellite Instability: From the Implementation of the Detection to a Prognostic and Predictive Role in Cancers. Int. J. Mol. Sci. 2022;23:8726. doi: 10.3390/ijms23158726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bonneville R., Krook M.A., Chen H.Z., Smith A., Samorodnitsky E., Wing M.R., Reeser J.W., Roychowdhury S. Detection of Microsatellite Instability Biomarkers via Next-Generation Sequencing. Methods Mol. Biol. 2020;2055:119–132. doi: 10.1007/978-1-4939-9773-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li K., Luo H., Huang L., Luo H., Zhu X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020;20:16. doi: 10.1186/s12935-019-1091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dominguez-Valentin M., Sampson J.R., Seppälä T.T., Ten Broeke S.W., Plazzer J.P., Nakken S., Engel C., Aretz S., Jenkins M.A., Sunde L., et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the Prospective Lynch Syndrome Database. Genet. Med. 2020;22:1569. doi: 10.1038/s41436-020-0892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bucksch K., Zachariae S., Aretz S., Büttner R., Holinski-Feder E., Holzapfel S., Hüneburg R., Kloor M., von Knebel Doeberitz M., Morak M., et al. Cancer risks in Lynch syndrome, Lynch-like syndrome, and familial colorectal cancer type X: A prospective cohort study. BMC Cancer. 2020;20:460. doi: 10.1186/s12885-020-06926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Morales-Miranda A., Rosado I.D., Núñez C.C., Montero F.C. Appendiceal carcinoma associated with microsatellite instability. Mol. Clin. Oncol. 2018;8:694–698. doi: 10.3892/mco.2018.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Taggart M.W., Galbincea J., Mansfield P.F., Fournier K.F., Royal R.E., Overman M.J., Rashid A., Abraham S.C. High-level microsatellite instability in appendiceal carcinomas. Am. J. Surg. Pathol. 2013;37:1192–1200. doi: 10.1097/PAS.0b013e318282649b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bonneville R., Krook M.A., Kautto E.A., Miya J., Wing M.R., Chen H.Z., Reeser J.W., Yu L., Roychowdhury S. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017;2017 doi: 10.1200/PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Misdraji J., Burgart L.J., Lauwers G.Y. Defective mismatch repair in the pathogenesis of low-grade appendiceal mucinous neoplasms and adenocarcinomas. Mod. Pathol. 2004;17:1447–1454. doi: 10.1038/modpathol.3800212. [DOI] [PubMed] [Google Scholar]
- 238.Komm M., Kronawitter-Fesl M., Kremer M., Lutz L., Holinski-Feder E., Kopp R. Primary mucinous adenocarcinoma of the vermiform appendix with high grade microsatellite instability. J. Cancer. 2011;2:302–306. doi: 10.7150/jca.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Emile S.H., Horesh N., Garoufalia Z., Gefen R., Zhou P., Wexner S.D. The Prognostic Impact of Microsatellite Instability on the Outcome of Appendiceal Adenocarcinoma: A National Cancer Database Analysis. J. Gastrointest. Surg. 2023;27:354–362. doi: 10.1007/s11605-023-05586-z. [DOI] [PubMed] [Google Scholar]
- 240.Pandya K., Overman M.J., Gulhati P. Molecular Landscape of Small Bowel Adenocarcinoma. Cancers. 2022;14:1287. doi: 10.3390/cancers14051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Davison J.M., Hartman D.A., Singhi A.D., Choudry H.A., Ahrendt S.A., Zureikat A.H., Ramalingam L., Nikiforova M., Pai R.K. Loss of SMAD4 protein expression is associated with high tumor grade and poor prognosis in disseminated appendiceal mucinous neoplasms. Am. J. Surg. Pathol. 2014;38:583–592. doi: 10.1097/PAS.0000000000000194. [DOI] [PubMed] [Google Scholar]
- 242.Singhi A.D., Foxwell T.J., Nason K., Cressman K.L., McGrath K.M., Sun W., Bahary N., Zeh H.J., Levy R.M., Luketich J.D., et al. Smad4 loss in esophageal adenocarcinoma is associated with an increased propensity for disease recurrence and poor survival. Am. J. Surg. Pathol. 2015;39:487–495. doi: 10.1097/PAS.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Maru D., Wu T.T., Canada A., Houlihan P.S., Hamilton S.R., Rashid A. Loss of chromosome 18q and DPC4 (Smad4) mutations in appendiceal adenocarcinomas. Oncogene. 2004;23:859–864. doi: 10.1038/sj.onc.1207194. [DOI] [PubMed] [Google Scholar]
- 244.Roncati L., Gasparri P., Gallo G., Bernardelli G., Zanelli G., Manenti A. Appendix Tumor Microenvironment. Adv. Exp. Med. Biol. 2020;1226:87–95. doi: 10.1007/978-3-030-36214-0_7. [DOI] [PubMed] [Google Scholar]
- 245.Mohamed A., Wu S., Hamid M., Mahipal A., Cjakrabarti S., Bajor D., Selfridge J.E., Asa S.L. Management of Appendix Neuroendocrine Neoplasms: Insights on the Current Guidelines. Cancers. 2022;15:295. doi: 10.3390/cancers15010295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Alhadid D., AlShammari A., Almana H., Aburahmah M. Missed Gastric Cancer Metastasis to the Appendix: Case Report and Literature Review. Am. J. Case. Rep. 2020;21:e920010. doi: 10.12659/AJCR.920010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kondo T. Current status and future outlook for patient-derived cancer models from a rare cancer research perspective. Cancer Sci. 2021;112:953–961. doi: 10.1111/cas.14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data can be found in the text.