Abstract
The genome DNA of Escherichia coli is associated with about 10 DNA-binding structural proteins, altogether forming the nucleoid. The nucleoid proteins play some functional roles, besides their structural roles, in the global regulation of such essential DNA functions as replication, recombination, and transcription. Using a quantitative Western blot method, we have performed for the first time a systematic determination of the intracellular concentrations of 12 species of the nucleoid protein in E. coli W3110, including CbpA (curved DNA-binding protein A), CbpB (curved DNA-binding protein B, also known as Rob [right origin binding protein]), DnaA (DNA-binding protein A), Dps (DNA-binding protein from starved cells), Fis (factor for inversion stimulation), Hfq (host factor for phage Qβ), H-NS (histone-like nucleoid structuring protein), HU (heat-unstable nucleoid protein), IciA (inhibitor of chromosome initiation A), IHF (integration host factor), Lrp (leucine-responsive regulatory protein), and StpA (suppressor of td mutant phenotype A). Intracellular protein levels reach a maximum at the growing phase for nine proteins, CbpB (Rob), DnaA, Fis, Hfq, H-NS, HU, IciA, Lrp, and StpA, which may play regulatory roles in DNA replication and/or transcription of the growth-related genes. In descending order, the level of accumulation, calculated in monomers, in growing E. coli cells is Fis, Hfq, HU, StpA, H-NS, IHF*, CbpB (Rob), Dps*, Lrp, DnaA, IciA, and CbpA* (stars represent the stationary-phase proteins). The order of abundance, in descending order, in the early stationary phase is Dps*, IHF*, HU, Hfq, H-NS, StpA, CbpB (Rob), DnaA, Lrp, IciA, CbpA, and Fis, while that in the late stationary phase is Dps*, IHF*, Hfq, HU, CbpA*, StpA, H-NS, CbpB (Rob), DnaA, Lrp, IciA, and Fis. Thus, the major protein components of the nucleoid change from Fis and HU in the growing phase to Dps in the stationary phase. The curved DNA-binding protein, CbpA, appears only in the late stationary phase. These changes in the composition of nucleoid-associated proteins in the stationary phase are accompanied by compaction of the genome DNA and silencing of the genome functions.
The genome DNA of Escherichia coli forms nucleoprotein complexes, often called the nucleoid, together with 5 to 10 major DNA-binding proteins (nucleoid-associated proteins are hereafter referred to nucleoid proteins), among which Fis (factor for inversion stimulation), H-NS (histone-like nucleoid structuring protein), HU (heat-unstable nucleoid protein), and IHF are believed to be the major molecular species (for reviews, see references 8, 42, and 44). In addition to these major nucleoid proteins, the DNA polymerases, the proteins involved in recombination and repair of DNA, RNA polymerase, and about 100 species of the transcription factor (for a review, see reference 19) are associated with the nucleoid at some point during their functions. Several lines of evidence indicate that the intracellular levels of the nucleoid proteins and their localization along the genome DNA influence not only the conformation of nucleoid but also DNA functions such as replication, recombination, repair, and transcription (2, 9, 15, 21, 59). In certain cases, the regulatory roles of nucleoid proteins in DNA functions are attributed to not only modulation of the genome conformation as a whole but also a more direct effect on the local conformation of specific DNA regions or even direct interaction with protein components. For instance, the association of some nucleoid proteins near specific promoters affects the association of RNA polymerase to promoters or the formation of open complexes for transcription initiation (for a review, see reference 19). Since the recent identification of contact surfaces of transcription factors on the RNA polymerase, our understanding of the molecular mechanisms of transcription regulation by gene- or regulon-specific transcription factors has much advanced (19). In contrast, however, the molecular mechanisms underlying the global regulation of transcription of about 4,000 genes on the E. coli genome by the nucleoid proteins have been left mostly unsolved, because of the lack of our knowledge of the intracellular concentrations and localization of these nucleoid proteins in E. coli.
As an initial attempt toward this ultimate goal, we have purified 12 nucleoid proteins from E. coli, including the 4 major species, Fis, H-NS, HU, and IHF (integration host factor protein), and 8 other DNA-binding proteins, CbpA (curved DNA-binding protein A), CbpB (curved DNA-binding protein B or Rob [right origin binding protein]), DnaA (DNA-binding protein A), Dps (DNA-binding protein from starved cells), Hfq (host factor for phage Qβ replication), IciA (inhibitor of chromosome initiation A), Lrp (leucine-responsive regulatory protein), and StpA (suppressor of td mutant phenotype A). Their activity and specificity of DNA binding and possible roles in global regulation of transcription are being analyzed in our laboratory (54).
In this study, we performed for the first time a systematic determination of the intracellular concentrations of these 12 species of the nucleoid protein in E. coli W3110 at different growth phases, by a quantitative Western immunoblot analysis with various antibody probes. Results indicate that the most abundant protein components constituting the nucleoid in growing cells are the three proteins Fis, Hfq, and HU but that only a single protein, Dps, predominates in the stationary-phase nucleoid.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strain used for the analysis of DNA-binding proteins was the A-type lineage of E. coli W3110, which carries the intact forms of both ςS and ςF (24). Cells were grown at 37°C under aeration in Luria-Bertani (LB) broth. Growth was monitored by measuring turbidity with a Klett-Summerson photometer. The culture conditions were fixed as follows. A few colonies from overnight cultures on LB agar plates were inoculated into 3 ml of fresh LB medium. At a cell density of 250 Klett units (the early stationary phase), the culture was diluted 170-fold by adding fresh LB medium, and the culture was continued at 37°C at a constant shaking rate of 160 rpm.
Determination of the total number of cells.
Aliquots of the culture were taken at various time intervals, and cells were fixed with formalin at a final concentration of 1%. The cells were counted with a Coulter Multisizer II (Coulter Electronics Limited, Luton, Bedfordshire, England) equipped with a 30-μm orifice and a 100-μl volume. Gain and current settings of 2.5 and 5.5, respectively, were used for all measurements. Coulter counter readings were maintained in the 3 × 104 to 10 × 104 range by dilution with Coulter Multisizer II specific isotonic buffer.
Microscopic observation of cells and nucleoids.
To observe clearly the shape and size of cells and nucleoids by microscopy, cells were stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) solution, which binds specifically to DNA, by the method developed by Hiraga et al. (16).
Preparation of cell lysates.
Cells were collected by centrifugation and resuspended in 40 mM Tris-HCl (pH 8.1) containing 25% sucrose at 4°C. After treatment with 1 mM EDTA and 500 μg of lysozyme per ml at 0°C for 10 min, cells were lysed by adding 0.1% NP-40. The cell lysate was supplemented with 0.01 M MgCl2 and 0.2 M KCl, digested at 37°C for 10 min with 20 μg of RNase A per ml and 100 μg of DNase I per ml in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF), and sonicated for 1 min with a Cosmo Bio Bioruptor. The cell lysates were used directly for all measurements. The protein concentration of cell lysates was determined with a Bio-Rad protein assay kit.
Purification of DNA-binding proteins and preparation of antibodies.
All 12 DNA-binding proteins, CbpA, CbpB (Rob), DnaA, Dps, Fis, Hfq, H-NS, HU, IciA, IHF, Lrp, and StpA, were overexpressed with pCU60, pMK19, pSY567, pDPS1, pRJ1077, pHFQ607, pHOP11, pLhupAhupB, pICS1, pSA5hiphimA, pMWD1, and pT7stpA, respectively, and purified to apparent homogeneity as described previously (54). Antibodies against each protein were produced in rabbits by injecting the purified proteins as described previously (23).
Quantitative Western blot analysis.
For the measurement of each DNA-binding protein in E. coli W3110 cell lysates, a quantitative Western blot analysis (23, 25) was employed with the polyclonal anti-CbpA, anti-CbpB (or anti-Rob), anti-DnaA, anti-Dps, anti-Fis, anti-Hfq, anti-H-NS, anti-HU, anti-IciA, anti-IHF, anti-Lrp, and anti-StpA antibodies. In brief, cell lysates were treated with a sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 1% 2-mercaptoethanol, 10% glycerol, 0.025% bromophenol blue) and separated by SDS-polyacrylamide gel electrophoresis (PAGE) with 10% (DnaA), 12.5% (CbpA, CbpB, and IciA), or 16.5% (Dps, Fis, Hfq, H-NS, HU, IHF, Lrp, and StpA) polyacrylamide gels. Protein in the gels was directly electroblotted onto polyvinylidene difluoride membranes (Nippon Genetics). Blots were blocked overnight at 4°C in 3% bovine serum albumin in phosphate-buffered saline, probed with the specific antibodies against each protein, washed with 0.5% Tween 20 in phosphate-buffered saline, and incubated with goat anti-rabbit immunoglobulin G conjugated with hydroxyperoxidase (Cappel). The blots were developed with 3,3′-diaminobenzidine tetrahydrochloride (Dojindo). Quantitation of band intensities was performed by scanning the immunostaining band and analyzing the image with NIH Image software (version 1.61). For some standard proteins, the intensity of immunostaining was slightly different in the presence or absence of crude extracts prepared from the respective mutant strains, but the difference was not corrected in this study.
For accurate measurement of the levels of each of the 12 proteins, we took care to do the following. (i) The protein range was determined for each protein, where a linear relationship existed between the protein concentration and the immunostaining intensity. (ii) The determination was carried out with several volumes of cell lysates, which contained the test proteins in the concentration range determined as described above. (iii) Standard samples of known concentrations were always included in the assays. (iv) The determination was repeated for at least three independent cell extracts for each DNA-binding protein. The number of molecules per cell was obtained from the total number of cells used for the preparation of cell lysates (measured as described above), the amount of total proteins in the cell lysates used, and the amount of each DNA-binding protein (measured as described above). The molecular mass of each DNA-binding protein was assumed to be as follows: CbpA, 33.4 kDa; CbpB (Rob), 33.0 kDa; DnaA, 53.0 kDa; Dps, 19.0 kDa; Fis, 11.2 kDa; Hfq, 11.2 kDa; H-NS, 15.4 kDa; HU subunit, 9.2 and 9.5 kDa (average, 9.4 kDa); IciA, 33.5 kDa; IHF subunit, 11.2 and 10.7 kDa (average, 11.0 kDa); Lrp, 19.0 kDa; and StpA, 15.3 kDa.
RESULTS AND DISCUSSION
Growth-dependent changes in the pattern of total proteins.
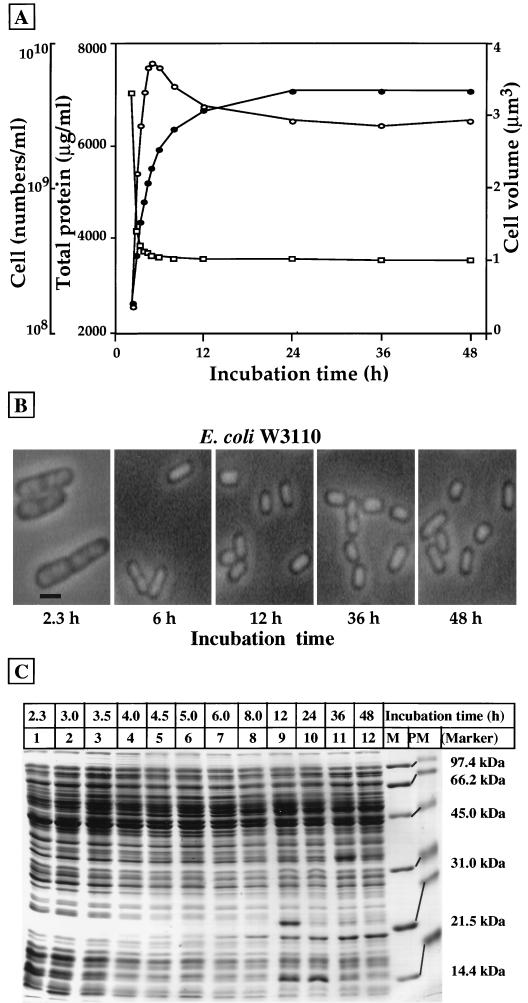
The intracellular levels of the 12 species of E. coli nucleoid protein (CbpA, CbpB, DnaA, Dps, Fis, Hfq, H-NS, HU, IciA, IHF, Lrp, and StpA) were determined for E. coli W3110 type A (24) at various phases of cell growth in LB medium. In the routine assays herewith described, an early-stationary-phase culture was diluted 170-fold with fresh LB medium, and the culture was continued at 37°C with shaking at a rate of 160 rpm. Growth was monitored by measuring turbidity with a Klett-Summerson photometer. At various times, aliquots of the culture were removed and used for determination of the number and size of cells, measured with a Coulter Multisizer II; the concentrations of total proteins and the amounts of each nucleoid protein were measured by Western blot analysis with specific antibodies. The total number of cells, the cell volume, and the concentration of total proteins are summarized in Fig. 1.
FIG. 1.
Expression patterns of E. coli W3110 proteins at various growth phases. (A) An early-stationary-phase culture of E. coli W3110 (A type) in LB medium at 37°C was diluted 170-fold with fresh LB medium, and the shaking culture was continued at 37°C. The total number of cells per milliliter (solid circles) and the average volume of cells (open squares) were measured with a Coulter Multisizer II. The protein concentration of cell lysates (open circles) was determined by staining with a Bio-Rad protein assay kit. (B) The same E. coli W3110 (A type) culture for the indicated times was stained with DAPI and observed for cell morphology. The bar in the 2.3-h sample represents 1 μm. (C) At the indicated time points (above the lane markers), aliquots of the culture were removed for the preparation of cell lysates. Portions of the cell lysates containing 15 μg of total proteins were subjected to SDS–15% PAGE. Gels were stained with Coomassie brilliant blue. The molecular mass markers (M, standard markers; PM, prestained markers) used were as follows: phosphorylase b, 97.4 kDa; bovine serum albumin, 66.2 kDa; ovalbumin, 45.0 kDa; carbonic anhydrase, 31.0 kDa; soybean trypsin inhibitor, 21.5 kDa; and lysozyme, 14.4 kDa.
In the experiment whose results are shown in Fig. 1A, the analysis was continued for 48 h after transfer of the early-stationary-phase culture into a fresh medium. The growth, measured by turbidity, increased for up to 5 to 6 h after the cell inoculation (data not shown). The total number of cells measured with a Coulter counter indeed increased until 5 to 6 h had passed (samples 6 and 7) and thereafter stayed at a constant level. The cell volume reached a maximum (3.5 μm3) during the exponential-growth phase and then decreased to one-third to one-fourth of the volume of growing cells in the stationary phase. The rapidly growing cells contain more than one nucleoid, as observed by DAPI staining (Fig. 1B), but the stationary-phase cells contain a single nucleoid of compact size. The concentration of total proteins reached a maximum at the end of cell growth or at the early stationary phase (4.5 to 5 h; samples 5 and 6) and then decreased, gradually reaching three-fourths of the maximum level at 24 h (Fig. 1A). Thus, a delay exists between the decrease in cell volume and the decrease in total number of proteins, suggesting that the compaction of the nucleoid proceeds earlier than the decrease in cytoplasmic volume.
Phase-contrast microscopic observation indicated significant changes in cell morphology (Fig. 1B). In the early stationary phase, cells became smaller and rounder, but in the late stationary phase, a small population of cells became elongated and rod shaped, suggesting inhibition of the cell division. However, the possibility is not ruled out that a small number of cells regained growth because the level of DnaA protein required for the initiation of chromosome replication also increased in the late stationary phase (see below). The superhelical density of plasmid DNA (and probably the chromosomal DNA) in E. coli decreases in the late stationary phase (12, 47). Under essentially the same culture conditions in LB medium, the superhelical density of plasmid DNA decreases in the stationary phase to about one-half of that in the exponential phase (31). Some promoters associated with the stationary-phase-specific genes are activated concomitantly with the decrease in DNA superhelicity (31).
The composition of total cellular proteins was analyzed by SDS-PAGE. For this purpose, cell lysates were prepared at different growth phases, and aliquots containing 15 μg of total proteins were subjected to SDS-PAGE as shown in Fig. 1C. The gel-staining patterns of cell lysates from the 2.3- to 3.0-h cultures (samples 1 and 2) were essentially identical to the typical pattern of growing E. coli cells, but significant changes were detected at 4.0 to 5.0 h (samples 4 to 6) when cell growth entered the late exponential or early stationary phase. Upon reaching the saturation point of cell growth at 5.0 to 6.0 h (samples 6 and 7), several stationary-phase-specific proteins such as the fast-migrating proteins (including Dps) were already identified, concomitantly with the decrease or disappearance of some growth-related proteins. The relative levels of the stationary-phase-specific proteins changed, depending on the stage of cell growth. Moreover, an order in the appearance of stationary-phase-specific proteins exists, supporting the concept that stationary-phase-specific genes are expressed in a sequential order (for reviews, see references 20, 21 and 58).
Intracellular concentration of each DNA-binding protein.
For measurement of the intracellular concentrations of 12 species of the nucleoid protein in E. coli, all of these proteins were expressed at high levels with the respective cloned genes and purified to apparent homogeneity (54), and polyclonal antibodies were raised in rabbits against the purified proteins (Table 1). For accurate measurement of the DNA-binding proteins by the quantitative Western blot method, we first made a standard curve representing the immunostaining of each of the purified proteins and determined the range where linearity exists between the protein concentration and the intensity of the immunostaining. Linearity was observed between 0.40 and 13.3 ng for all of the 12 purified proteins examined (data not shown). On the basis of these standard curves, we then analyzed several different volumes of each cell lysate and identified the optimum volume that contained this range of proteins. Using the optimum volumes of cell lysates thus estimated, we finally repeated the measurement of individual proteins for at least three independent cell lysates, always in parallel with the determination of six to eight different concentrations of the respective purified protein as the assay standard. The maximum fluctuation among different measurements described in this report was about 20%, if any. The minimum level of detection was 0.10 ng of nucleoid protein per μg of total cell lysate proteins. Typical immunoblot patterns of each of the 12 DNA-binding proteins are shown in Fig. 2. Some antibodies cross-reacted against proteins other than the target proteins used for immunization, but after SDS-PAGE of whole cell lysates, these cross-reactive proteins could be separated, allowing accurate measurement of all 12 nucleoid proteins. The results of calculation for each protein are summarized below.
TABLE 1.
Purification of E. coli DNA-binding proteins and preparation of antibodies
| Protein | Plasmid for expression | Source of plasmid | Source of antibodies |
|---|---|---|---|
| CbpA | pCU60 | T. Mizuno, Nagoya, Japan | T. Mizuno, Nagoya, Japan |
| CbpB | pMK19 | T. Mizuno, Nagoya, Japan | This study |
| DnaA | pSY567 | S. Yasuda, Mishima, Japan | S. Yasuda, Mishima, Japan |
| Dps | pDPS1 | This laboratory | This study |
| Fis | pRJ1077 | R. C. Johnson, Los Angeles, Calif. | This study |
| Hfq | pHFQ607 | This laboratory | This laboratory |
| H-NS | pHOP11 | T. Mizuno, Nagoya, Japan | T. Mizuno, Nagoya, Japan |
| HU | pLhupAhupB | N. Goshima, Hiroshima, Japan | N. Goshima, Hiroshima, Japan |
| IciA | pICS1 | D. S. Hwang, Seoul, Korea | This study |
| IHF | pSA5hiphimA | A. B. Oppenheim, Jerusalem, Israel | This study |
| Lrp | pMWD1 | D. Low, Salt Lake City, Utah | This study |
| StpA | pT7stpA | M. Belfort, Albany, N.Y. | This study |
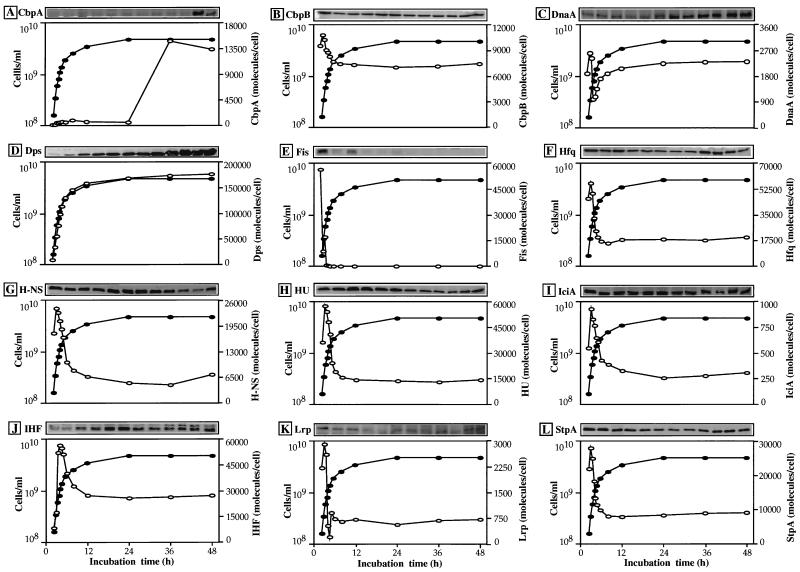
FIG. 2.
Growth phase-dependent variation in the intracellular levels of 12 DNA-binding proteins in E. coli W3110. For measurement of the concentrations of each DNA-binding protein, aliquots of cell lysates containing 15 μg of total proteins were subjected to SDS-PAGE (with a 10% gel for DnaA; a 12.5% gel for CbpA, CbpB, and IciA; and a 16.5% gel for Dps, Fis, Hfq, H-NS, HU, IFH, Lrp, and StpA) and proteins were electroblotted onto polyvinylidene difluoride membranes. The blots were probed with anti-CbpA (A), anti-CbpB or anti-Rob (B), anti-DnaA (C), anti-Dps (D), anti-Fis (E), anti-Hfq (F), anti-H-NS (G), anti-HU (H), anti-IciA (I), anti-IHF (J), anti-Lrp (K), or anti-StpA (L) antibodies. After immunostaining, the intensity of stained bands was measured by scanning the gels with an NIH Image analyzer system (version 1.61). To increase the accuracy of the calculation of protein molecules, the Western blot analysis was repeated at least three times for all samples. Solid circles represent the total number of cells measured with a Coulter Multisizer II, while open circles represent the total number of protein molecules per cell, calculated as averages of three independent measurements.
CbpA.
CbpA, a curved DNA-binding protein which displays a high level of amino acid sequence homology with DnaJ, was first isolated as a DNA-binding protein that preferentially recognizes a curved DNA sequence (57). Using the purified CbpA protein, we indeed demonstrated that CbpA is a non-sequence-specific curved DNA-binding protein (54). An aliquot of E. coli W3110 cell lysate containing 15 μg of total proteins was separated by SDS–12.5% PAGE, and the gel was subjected to Western blot analysis with polyclonal anti-CbpA antibodies. The immunostained band was not detected until the late-stationary-phase lysate, i.e., 36 h after the start of cell culture (Fig. 2A, sample 11), indicating that CbpA is a unique protein that is synthesized only in the late stationary phase. From the staining intensity, the level of CbpA protein at 36 h was estimated to be about 3% of the total proteins or 15,000 molecules per cell. Previously, Yamashino et al. (61) suggested that the level of CbpA protein increased at the stationary phase or after phosphate starvation and that the expression of cbpA is dependent on the function of stationary-phase-specific ςS of RNA polymerase.
CbpB (Rob).
CbpB was identified after random screening of curved DNA-binding proteins from E. coli (29), but after sequencing, this protein was found to be identical with the previously identified Rob (51). Rob is also involved in transcription regulation of a group of genes by interaction with the RNA polymerase (22). The amount of CbpB in W3110 cells was measured by quantitative Western immunoblotting with highly specific anti-CbpB antibodies. The antisera used recognized only the CbpB protein in whole-cell lysates, showing no cross-reaction against other E. coli proteins (data not shown). The maximum level of CbpB in the growing cells was found to be about 10,000 molecules per cell (Fig. 2B). This value is about twice the value of 5,000 molecules per cell estimated by Skarstad et al. (51). The difference might be due to differences in the cell growth phase or the strains used. The relative amount of CbpB stays constant from the exponential to the stationary phase, but after correction for the content of total proteins per cell, the total number of CbpB molecules in a stationary-phase cell was found to decrease to about 60% of the maximum level in the exponential-phase cells.
DnaA.
DnaA is the sequence-specific DNA-binding protein and plays a key role in the initiation of chromosomal DNA replication in vivo and in vitro (11). DnaA also functions as an activator or a repressor of transcription of many genes, including the genes in the ori region and the dnaA gene itself (34). As shown in Fig. 2C, the intracellular level of DnaA protein showed a unique pattern of variation, reaching a maximum first in the mid-exponential-growth phase, in agreement with the high rate of DNA replication in rapidly growing cells and a second peak at the late stationary phase.
Previously, Sakakibara and Yuasa (45) reported that by the analysis of 14C-amino-acid-labeled DnaA protein on two-dimensional gel electrophoresis, the cellular abundance of DnaA protein remained constant during the transition from the growth phase to stationary phase. Sekimizu et al. (49) reported that the amount of DnaA protein in E. coli W3100 cells was relatively constant, at a range of about 800 to 2,100 molecules per cell from the exponential growth to the stationary phase. Our results indicate a growth-dependent fluctuation in the level of DnaA protein within the range of 900 to 2,700 molecules per cell. The second rise in DnaA level in the stationary phase may also represent the resumption of cell growth in a population of stationary-phase cells.
Dps.
Dps was identified as a starvation-inducible DNA-binding protein in E. coli (1). High levels of Dps are also produced under conditions of nutritional or oxidative stress. The purified Dps binds to DNA with no apparent sequence specificity (54), and thus Dps is classified as a member of the bacterial nucleoid-associated or histone-like protein family which includes HU, H-NS, IHF, and Fis (14, 46). Upon binding Dps, the nucleoid DNA is transformed into a compact configuration (14). A systematic Western immunoblot analysis of W3110 cell lysates indicated that about 6,000 Dps molecules per cell exist in the exponential growth phase and thereafter begin to increase gradually, reaching a peak of about 180,000 molecules per cell at the late stationary phase (Fig. 2D), the most abundant DNA-binding protein so far identified in E. coli (Fig. 3). The level measured by quantitative Western blot analysis is in good agreement with the value, 150,000 to 200,000 molecules per cell, estimated indirectly from the DNA-binding activity assay of cell lysates from an overnight E. coli culture (33). Since the level of Dps continued to increase for at least 48 h (Fig. 2D, sample 12), it may further increase after prolonged culture.
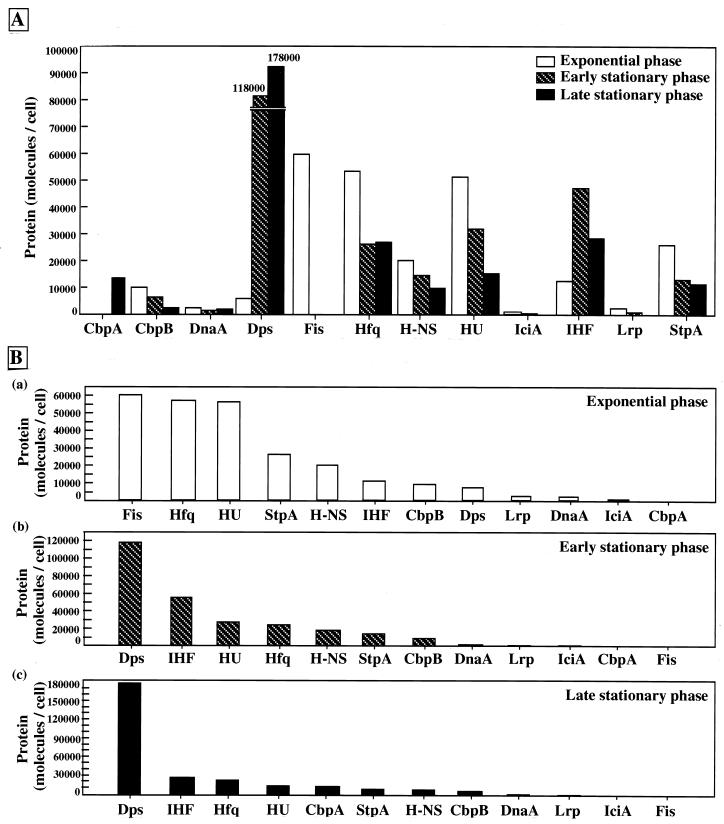
FIG. 3.
Growth phase-dependent variation in the intracellular levels of 12 DNA-binding proteins in E. coli W3110. The measurement of each DNA-binding protein was carried out as described in Materials and Methods. (A) Total numbers of each protein molecule at three distinct growth phases are shown. Exponential phase, 2.3 to 3.0 h; early stationary phase, 5.0 h; late stationary phase, 48 h (see Fig. 1 for the growth curve). (B) The order of the total number of molecules among the 12 DNA-binding proteins.
Fis.
Fis is a small basic DNA-binding protein that was identified in E. coli as a factor involved in site-specific DNA recombination (26, 30, 56). Several lines of evidence indicate that Fis also participates in other processes of DNA functions such as transcription of the growth-related genes and DNA replication (for a review, see reference 9). The intracellular level of Fis protein in exponential growth phase cells of E. coli W3110 was found to be about 60,000 molecules per cell at maximum, but thereafter it decreased to undetectable levels in the stationary phase (Fig. 2E). Fis is the most abundant nucleoid-associated protein in growing E. coli cells (Fig. 3). The growth-dependent change in the Fis level is in good agreement with the observations that Fis is needed for transcription of growth-related genes, such as those for rRNA and tRNA, and for DNA replication.
Our measurement of the intracellular Fis level agrees well with the observations that the Fis level in E. coli MC1000 growing under various conditions fluctuates within a range of less than 100 in the stationary phase to over 50,000 molecules per cell in the early-exponential growth phase (3). The regulation of fis is essentially the same in Salmonella typhimurium, even though the autoregulation is less efficient (40). The timing of the Fis peak occurs prior to the first cell division of cell growth after recovery from the stationary phase. If the DNA-bound Fis remains a homodimer, Fis may bind every 200 to 300 bp of DNA, on average, along the genome DNA in growing cells of E. coli (but the Fis sites are not regularly distributed along the genome DNA). Highly expressed Fis represses its own synthesis by binding to the fis promoter region. Upon entry into the stationary phase, Fis synthesis is switched off, resulting in a decrease in intracellular level by 500- to 1,000 fold (reference 3 and this report).
Hfq.
Hfq was identified as a host factor, designated HF-I, which is required for the replication of phage Qβ RNA (27) with binding activity to both DNA and RNA (28). Hfq binds preferentially to curved DNA in a non-sequence-specific manner (54). Previously, it was reported that the rate of synthesis of Hfq in E. coli W3350 cells at the exponential growth phase accounts for its intracellular concentration of about 30,000 to 60,000 molecules per cell (28).
Here, we measured the intracellular concentration of Hfq by Western immunoblot analysis with specific antibodies. In good agreement with a previous estimation (28), the Hfq level at the exponential growth phase was found to be about 55,000 molecules per cell (Fig. 2F). If all the Hfq molecules are associated with the nucleoid, the Hfq level is close to that of the two major nucleoid components, Fis and HU, in exponential growth phase cells (Fig. 3B), but Hfq is also associated with ribosomes (28). In fact, the Hfq protein controls the translation of some mRNA, including those of the rpoS ς factor and the DNA repair gene mutS (35, 48). During the transition from the growth phase to the stationary phase, the level of Hfq decreased gradually, reaching a plateau of about one-third of the maximum level. Results indicate that Hfq is maintained at a level characteristic of the rate of cell growth, supposedly playing a role in control of the expression of growth-related genes.
H-NS.
H-NS is a well-characterized nucleoid-associated protein that functions as a global repressor of transcription, affecting more than 35 genes or operons in E. coli (for a review, see reference 2). The quantitative Western blot analysis was carried out with anti-H-NS antibodies. Although H-NS and StpA, the recently identified H-NS homolog (63), migrated to the same position on SDS-PAGE gels, the anti-H-NS antibodies did not cross-react with StpA (however, the anti-StpA antibodies cross-reacted with H-NS, albeit at about one-tenth of the level of StpA) and thus the comigrating StpA did not interfere with the measurement of H-NS (data not shown). Results indicated that the number of H-NS molecules reached a maximum of about 20,000 molecules per cell in the exponential phase of cell growth, but thereafter decreased to 40% of the maximum at the late stationary phase (Fig. 2G). It is noteworthy that the level of H-NS and the pattern of its growth-dependent variation are similar to those of StpA (see below).
The pattern of growth-dependent change in the H-NS level is, however, different from that reported by Spassky et al. (52). By two-dimensional gel analysis of total proteins, they estimated that the total number of H-NS protein molecules in the exponential-phase cells was about 4,000, which increased about fivefold at the stationary phase. The disagreement in growth phase-dependent change of H-NS may be due to comigration of an unrelated protein with H-NS on a two-dimensional gel, because the immunological detection of H-NS always indicates a decrease in H-NS levels in stationary-phase cells (references 10 and 62 and this study).
HU.
A DNA-binding protein, HU, has long been considered a prokaryotic homolog of eukaryotic histones (43, 55) that restrains DNA supercoils in the nucleoid (for a review, see reference 8), but it is more analogous in function to the eukaryotic HMG proteins (39, 41). HU exists in solution as a heterodimer consisting of two similar subunits. Like Hfq (27), a certain fraction of HU is also associated with ribosomes (53). Results of the quantitative Western blot analysis with antibodies against a mixture of two HU subunits indicated that about 30,000 to 55,000 HU molecules per cell exist in the exponentially growing cells of E. coli W3110 (Fig. 2H), indicating that HU, together with Fis, forms a major nucleoid component in the growing E. coli cell (see Fig. 3).
Our measurement of HU abundance agrees well with the two independent estimations of about 30,000 HU dimers (or 60,000 monomers) in E. coli WM433(dnaA204) (7, 13). At saturation, HU dimers may be associated, on average, every 300 to 400 bp of the E. coli genome. Upon entry into the stationary phase, the HU level started to decrease gradually, decreasing to less than one-third of the maximum level in the late stationary phase (Fig. 2H). The overall pattern of the growth-dependent change in HU levels is similar to that of Hfq, H-NS, and StpA (see Fig. 2F, 2G, and 2L).
IciA.
IciA is known to bind to three repeat sequences 13 nucleotides in length located near the replication origin (oriC) and to inhibit the initiation of DNA replication in vitro by blocking the opening of the oriC region driven by the initiator DnaA protein (12, 18). The quantitative Western immunoassay, whose results are shown in Fig. 2I, indicates that about 800 molecules (or 400 dimers) of IciA exist in exponentially growing cells of E. coli W3110, and then the level decreases to about 500 molecules (or 250 dimers) per cell in the early stationary phase. After prolonged culture, IciA decreases to less than one-third of the maximum level in the late stationary phase. Among the 12 DNA-binding proteins examined in this study, IciA was the second-least-abundant protein, next to CbpA, in growing cells. The IciA level in growing cells is close to the reported value of about 200 molecules per cell (17). However, it was also reported that the cellular abundance of IciA increased fourfold (800 molecules per cell) at the stationary phase, although DnaA decreases (17). The disagreement between these two measurements remains unresolved.
IHF.
IHF is one of the most abundant sequence-specific DNA-binding protein in E. coli, which was first identified and isolated as a host factor for integrative recombination of phage λ (5, 36). IHF is now recognized as a factor of global regulation in the transcription of many genes (for a review, see reference 19). The native form of IHF is a heterodimer of two different subunits with similar amino acid sequences. The quantitative Western immunoblot analysis of E. coli W3110 cell lysates was carried out with antibodies against both IHF subunits. Results indicate that in exponential growth phase cells the sum of IHF monomers is about 12,000 molecules per cell (Fig. 2J).
The level of IHF increased when the cell growth started to decrease and finally reached a maximum of about 55,000 monomers per cell in the early stationary phase (Fig. 2J), generally in agreement with published observations (6). In the transition from the growth phase to the stationary phase, IHF becomes the second-most-abundant protein (see Fig. 3), suggesting that IHF plays a key role in the structural and functional conversion of nucleoid during the phase transition of cell growth. After prolonged culture, however, the IHF level decreased again to less than one-half of the maximum level.
Lrp.
Lrp is a global transcription factor which regulates, positively or negatively, more than 75 genes in E. coli (for reviews, see references 4, 19, 37, and 38). The cellular abundance of Lrp in crude extracts of E. coli W3110 was estimated by quantitative Western immunoblot analysis with newly prepared anti-Lrp antibodies. Results indicated that the intracellular concentration of Lrp was 2,500 molecules per cell in the exponential phase (Fig. 2K). If all the Lrp molecules in E. coli exist as homodimers, the intracellular concentration of Lrp should be about 1,200 to 1,300 dimers per cell at the growing phase. Upon entry into the stationary phase, the Lrp level decreased rapidly to less than one-tenth of the maximum level. A similar pattern of Lrp accumulation in different growth phases has been reported (32, 60).
The Lrp level is influenced by the composition of culture medium. The expression level in minimal medium is three- to fourfold higher (approximately 3,200 dimers per cell) than in rich medium. Since the Lrp level decreases in relA and spoT mutants defective in ppGpp accumulation, the expression of lrp seems to be under the positive control of ppGpp (32). However, this does not mean that the synthesis of Lrp is under the direct control of ppGpp. Taken together, it appears that Lrp functions as a positive factor for transcription enhancement of the genes which are induced under starved conditions.
StpA.
StpA, a putative homolog of H-NS protein in E. coli, was first identified as a multicopy suppressor of a td mutant phenotype of phage T4 (63). More recently, Shi and Bennett (50) identified the stpA gene as a multicopy suppressor that could transcomplement hns mutants with respect to the expression of the arginine decarboxylase gene. The high sequence identity (58%) between StpA and H-NS suggested that the two proteins could have similar functions. In a previous study, the sequence recognition specificity and DNA-binding affinity of both proteins under the same experimental conditions were reported for the first time (54). Here, we determined the intracellular concentration of StpA by Western blot analysis with newly prepared anti-StpA antibodies. Since the anti-StpA antibodies cross-reacted with H-NS at a rate of about one-tenth of the reactivity with StpA, the Western blot intensity was corrected for the background level of H-NS, which was determined with the anti-H-NS antibodies (see above). The growing cells of E. coli W3110 contain about 25,000 molecules per cell at the exponential phase (Fig. 2L). Upon entry into the stationary phase, the level began to decrease, finally reaching about 8,000 to 10,000 molecules per cell. Both the intracellular level and the pattern of its growth phase-coupled variation are similar to those of H-NS, supporting the assumption that these two proteins have similar functions.
Relative levels of 12 species of DNA-binding proteins.
The abundant nucleoid proteins in E. coli have been recognized as the structural components for compaction of the genome DNA. Several lines of recent study, however, indicate that these nucleoid-associated proteins also play certain roles in controlling DNA functions such as replication, recombination, transposition, and transcription. At the present time, our knowledge of the regulation of synthesis and intracellular localization of these nucleoid proteins is limited. Here, we determined for the first time the intracellular concentrations of 12 molecular species of the nucleoid protein for the same cultures of E. coli W3110 by the same experimental system. As summarized in Fig. 3, these 12 nucleoid proteins showed different patterns of growth phase-dependent accumulation. The level reaches a maximum during the growing phase for nine proteins, CbpB, DnaA, Fis, Hfq, H-NS, HU, IciA, Lrp, and StpA, while the level of three other proteins, CbpA, Dps, and IHF, increases in the stationary phase.
The nine species of nucleoid protein showing the highest expression in growing cells may be involved in growth-related functions. For instance, CbpB (Rob), DnaA, and IciA are involved in the initiation and regulation of chromosome replication. These proteins are also involved in the transcription regulation of the genes for some components of the DNA replication apparatus. Fis is the only protein that is present in growing cells, but it disappears in the stationary phase, in good agreement with its key role in transcription enhancement of the growth-related genes such as those coding for rRNA, tRNA, and ribosomal proteins (for a review, see reference 9). Lrp is also involved in the regulation of transcription of some growth-related genes (for reviews, see references 4, 37, and 38). The levels of both Fis and Lrp increase in parallel with the increase in amino acids or leucine, respectively, in culture medium. Taking the data together, we propose that these nucleoid-associated transcription factors play key roles in the global control of transcription of the essential genes for cell growth (19–21).
Up to the present time, four proteins, Fis, H-NS, HU, and IHF, were believed to be the major components of the E. coli nucleoid (for reviews, see references 42 and 44). However, we demonstrated in this study that two proteins, Fis and HU, constitute the major components of the E. coli nucleoid in growing cells, each ranging from 55,000 to 60,000 monomer molecules per cell (Fig. 3B). The level of Hfq is as high as those of Fis and HU, but at least some of Hfq may be associated with ribosomes (27). H-NS is a less abundant protein, and the levels of H-NS and its homolog StpA stay within the range of 20,000 to 25,000 molecules per cell. Likewise, IHF is not the major nucleoid component in growing cells but becomes the second-most-abundant nucleoid component (next to Dps) in stationary-phase cells (Fig. 3B). The descending order of abundance in exponentially growing E. coli W3110 cells in LB medium, calculated as monomer proteins, was thus found to be Fis, Hfq, HU, StpA, H-NS, IHF*, CbpB (Rob), Dps*, Lrp, DnaA, IciA, and CbpA* (stars indicate the nucleoid proteins which increase in the stationary phase) (Fig. 3). This order, however, changes when cells are grown in a different medium. Moreover, the number of molecules present in a cell does not represent the number of molecules bound with the nucleoid because different proteins bind DNA with different affinities (54).
Most of these nucleoid proteins (except for two monomeric proteins, CbpB [Rob] and DnaA) form dimers or oligomers under isolated states. CbpA, Fis, H-NS, IciA, Lrp, and StpA form homodimers; HU and IHF form heterodimers; Hfq forms a hexamer; and Dps forms a dodecamer. If all these nucleoid proteins self-assemble in vivo and these nucleoid proteins remain assembled even after binding to DNA, the major nucleoid components should be two proteins, Fis and HU, but the concentration of Hfq hexamer is as low as those of StpA, H-NS, and IHF.
Three nucleoid proteins, CbpA, Dps, and IHF, increase in the stationary phase (Fig. 3). The major structural components of the nucleoid change from Fis and HU (and Hfq) in the exponential phase to Dps and IHF in the early stationary phase (Fig. 3B). In descending order, the level of accumulation in the early stationary phase is Dps*, IHF*, HU, Hfq, H-NS, StpA, CbpB, DnaA, Lrp, IciA, CbpA, and Fis (Fig. 3B). Among the three stationary-phase nucleoid proteins, however, the maximum level was observed at different time periods of the stationary phase, the order of maximum level appearance being IHF, Dps, and CbpA (Fig. 2 and 3). The IHF level starts to increase in the early stationary phase, concomitantly with a decrease in the levels of the major proteins Fis and HU (and Hfq) in growing cells of the nucleoid. Upon entry into the stationary phase, the E. coli nucleoid becomes more compact and DNA superhelicity decreases. Either the association of the two stationary-phase proteins, Dps and IHF, or the decrease in Fis, Hfq, HU, StpA, and H-NS levels must be involved in this process of DNA compaction. In the late stationary phase, the relative level of Dps approaches almost one-half of the level of total nucleoid-bound proteins. The third stationary-phase-specific protein, CbpA, appears only in the late stationary phase (Fig. 2 and 3). Although the function of CbpA is totally unknown, it may be involved in the final stage of DNA compaction, ultimately leading to the formation of a more compact stored form of the genome DNA in the dormant phase. Abundance, in descending order, changes in the late stationary phase to Dps*, IHF*, Hfq, HU, CbpA*, StpA, H-NS, CbpB, DnaA, Lrp, and IciA. However, Fis is virtually undetectable in the late stationary phase. The stage-dependent change in the appearance of stationary-phase nucleoid proteins supports our hypothesis that a hierarchy exists in the order of expression of stationary-phase genes (for reviews, see references 21 and 58).
The level of three stationary-specific DNA-binding proteins, CbpA, Dps, and IHF, in the growing phase varies depending on the culture conditions; the level of all three increases in slowly growing culture in poor medium.
ACKNOWLEDGMENTS
We thank T. Mizuno, S. Yasuda, and N. Goshima for gifts of the antibodies against CbpA and H-NS, DnaA, and HU, respectively. This work was supported by Grants-in-Aid from the Ministry of Education, Science, and Culture of Japan and CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Corporation (JST).
REFERENCES
- 1.Almirón M, Link A J, Furlong D, Kolter R. A novel DNA binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 2.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 3.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo J M, Matthews R G. Leucine-responsive regulatory protein: a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–498. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig N L, Nash S E. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39:707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 6.Ditto M D, Roberts D, Weisberg R A. Growth phase variation of integration host factor level in Escherichia coli. J Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon N E, Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc Natl Acad Sci USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel S E, Johnson R C. The Fis protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 10.Free A, Dorman C J. Coupling of Escherichia coli hns mRNA levels to DNA synthesis by autoregulation: implications for growth phase control. Mol Microbiol. 1995;18:101–113. doi: 10.1111/j.1365-2958.1995.mmi_18010101.x. [DOI] [PubMed] [Google Scholar]
- 11.Fuller R S, Funnell B E, Kornberg A. The dnaA protein complex with the E. coli chromosome replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 12.Galke V A, Balke V L, Gralla J D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987;169:4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geider K, Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- 14.Grant R A, Filman D J, Finkel S E, Kolter R, Hogle J M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 15.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr Opin Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 16.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. Chromosomal partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang D S, Thony B, Kornberg A. IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J Biol Chem. 1992;267:2209–2213. [PubMed] [Google Scholar]
- 18.Hwang D S, Kornberg A. A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell. 1990;63:325–331. doi: 10.1016/0092-8674(90)90165-b. [DOI] [PubMed] [Google Scholar]
- 19.Ishihama A. Promoter selectivity control of Escherichia coli RNA polymerase. In: Eckstein F, Lilley D, editors. Nucleic acids and molecular biology. 11. Mechanism of transcription. Heidelberg, Germany: Springer-Verlag; 1997. pp. 53–70. [Google Scholar]
- 20.Ishihama A. Adaptation of gene expression in stationary phase bacteria. Curr Opin Genet Dev. 1997;7:582–588. doi: 10.1016/s0959-437x(97)80003-2. [DOI] [PubMed] [Google Scholar]
- 21.Ishihama A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells. 1999;3:135–143. doi: 10.1046/j.1365-2443.1999.00247.x. [DOI] [PubMed] [Google Scholar]
- 22.Jair K-W, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf R E., Jr Transcription activation of promoters of the superoxide and multiple antibiotic resistance regulation by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jishage M, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of ς70 and ς38. J Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jishage M, Ishihama A. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J Bacteriol. 1997;179:959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson R C, Bruist M F, Simon M I. Host protein requirement for in vitro site-specific DNA inversion. Cell. 1986;46:531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- 27.Kajitani M, Ishihama A. Identification and sequence determination of the host factor gene for bacteriophage Qβ. Nucleic Acids Res. 1991;19:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kajitani M, Kato A, Wada A, Inokuchi Y, Ishihama A. Regulation of the Escherichia coli hfq gene encoding the host factor for phage Qβ. J Bacteriol. 1994;176:531–534. doi: 10.1128/jb.176.2.531-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakeda M, Ueguchi C, Yamada H, Mizuno T. An Escherichia coli curved DNA-binding protein whose expression is affected by the stationary phase-specific sigma factor ςS. Mol Gen Genet. 1995;248:629–634. doi: 10.1007/BF02423459. [DOI] [PubMed] [Google Scholar]
- 30.Koch C, Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 31.Kusano S, Ding Q, Fujita N, Ishihama A. Promoter selectivity of Escherichia coli RNA polymerase Eς70 and Eς38 holoenzymes: effect of DNA supercoiling. J Biol Chem. 1996;271:1998–2004. doi: 10.1074/jbc.271.4.1998. [DOI] [PubMed] [Google Scholar]
- 32.Landgraf J R, Wu J, Calvo J M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messer W, Weigel C. DnaA initiator—also a transcription factor. Mol Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- 35.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 36.Nash H A, Robertson C A. Purification and properties of the Escherichia coli protein factor required for lambda integration recombination. J Biol Chem. 1981;256:9246–9253. [PubMed] [Google Scholar]
- 37.Newman E B, D’Ari R, Lin R T. The leucine-Lrp regulon in E. coli: a global response in search of a raison d’etre. Cell. 1992;68:617–619. doi: 10.1016/0092-8674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- 38.Newman E B, Lin R T. Leucine-responsive regulatory protein, a global regulator of gene expression in E. coli. Annu Rev Microbiol. 1995;49:747–775. doi: 10.1146/annurev.mi.49.100195.003531. [DOI] [PubMed] [Google Scholar]
- 39.Oberto J, Drlica K, Rouviere-Yaniv J. Histones, HMG, HU, IHF: meme combat. Biochimie. 1994;76:901–908. doi: 10.1016/0300-9084(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 40.Osuna R, Lienau D, Hughes K T, Johnson R C. Sequence, regulation, and functions of fis in Salmonella typhimurium. J Bacteriol. 1995;177:2021–2032. doi: 10.1128/jb.177.8.2021-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul T T, Haykinson M J, Johnson R C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 42.Pettijohn D E. The nucleoid. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 158–166. [Google Scholar]
- 43.Pontiggia A, Negri A, Beltrame M, Bianchi M E. Protein HU binds specifically to kinked DNA. Mol Microbiol. 1993;7:343–350. doi: 10.1111/j.1365-2958.1993.tb01126.x. [DOI] [PubMed] [Google Scholar]
- 44.Rouviere-Yaniv J, Yaniv M, Germond J E. Escherichia coli DNA-binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 45.Sakakibara Y, Yuasa S. Continuous synthesis of the dnaA gene product of Escherichia coli in the cell cycle. Mol Gen Genet. 1982;186:87–94. doi: 10.1007/BF00422917. [DOI] [PubMed] [Google Scholar]
- 46.Schmid M B. More than just ‘histone-like’ proteins. Cell. 1990;63:451–453. doi: 10.1016/0092-8674(90)90438-k. [DOI] [PubMed] [Google Scholar]
- 47.Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 48.Schuppli D, Miranda G, Tsui H C, Winkler M E, Sogo J M, Weber H. Altered 3′-terminal RNA structure in phage Qβ adapted to host factor-less Escherichia coli. Proc Natl Acad Sci USA. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekimizu K, Yat-ming Yung B, Kornberg A. The DnaA protein of Escherichia coli: abundance, improved purification, and membrane binding. J Biol Chem. 1988;263:7136–7140. [PubMed] [Google Scholar]
- 50.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skarstad K, Thony B, Hwang D S, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 52.Spassky A, Rimsky S, Garreau H, Buc H. H1a, an E. coli DNA-binding protein which accumulates in stationary phase, strongly compacts DNA in vitro. Nucleic Acids Res. 1984;12:5321–5340. doi: 10.1093/nar/12.13.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suryanarayana T, Subramanian A-R. Specific association of two homologous DNA-binding proteins to the native 30S ribosomal subunits of Escherichia coli. Biochim Biophys Acta. 1978;520:342–357. doi: 10.1016/0005-2787(78)90232-0. [DOI] [PubMed] [Google Scholar]
- 54.Talukder, A. A., and A. Ishihama. Twelve species of nucleoid-associated protein from Escherichia coli: sequence recognition specificity and DNA binding affinity. J. Biol. Chem., in press. [DOI] [PubMed]
- 55.Tanaka H, Goshima N, Khono K, Kano Y, Imamoto F. Properties of DNA-binding of HU heterotypic and homotypic dimers from Escherichia coli. J Biochem. 1993;113:568–572. doi: 10.1093/oxfordjournals.jbchem.a124084. [DOI] [PubMed] [Google Scholar]
- 56.Thompson J F, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathways. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 57.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperon in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wada A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells. 1998;3:203–208. doi: 10.1046/j.1365-2443.1998.00187.x. [DOI] [PubMed] [Google Scholar]
- 59.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1987;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 60.Willins D A, Ryan C W, Platko J V, Calvo J M. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J Biol Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 61.Yamashino T, Kakeda M, Ueguchi C, Mizuno T. An analogue of the DnaJ molecular chaperone whose expression is controlled by sigma S during the stationary phase and phosphate starvation in Escherichia coli. Mol Microbiol. 1994;13:475–483. doi: 10.1111/j.1365-2958.1994.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 62.Yasuzawa K, Hayashi N, Goshima N, Kohno K, Imamoto F, Kano Y. Histone-like proteins are required for cell growth and restraint of supercoils in DNA. Gene. 1992;122:9–14. doi: 10.1016/0378-1119(92)90026-l. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Z, Belfort M. Nucleotide sequence of a newly identified Escherichia coli gene, stpA, encoding an H-NS-like protein. Nucleic Acids Res. 1992;20:6734. doi: 10.1093/nar/20.24.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]