Abstract
Expression of the peroxide stress genes alkyl hydroperoxide reductase (ahpC) and catalase (katA) of the microaerophile Campylobacter jejuni is repressed by iron. Whereas iron repression in gram-negative bacteria is usually carried out by the Fur protein, previous work showed that this is not the case in C. jejuni, as these genes are still iron repressed in a C. jejuni fur mutant. An open reading frame encoding a Fur homolog (designated PerR for “peroxide stress regulator”) was identified in the genome sequence of C. jejuni. The perR gene was disrupted by a kanamycin resistance cassette in C. jejuni wild-type and fur mutant strains. Subsequent characterization of the C. jejuni perR mutants showed derepressed expression of both AhpC and KatA at a much higher level than that obtained by iron limitation, suggesting that expression of these genes is controlled by other regulatory factors in addition to the iron level. Other iron-regulated proteins were not affected by the perR mutation. The fur perR double mutant showed derepressed expression of known iron-repressed genes. Further phenotypic analysis of the perR mutant, fur mutant, and the fur perR double mutant showed that the perR mutation made C. jejuni hyperresistant to peroxide stress caused by hydrogen peroxide and cumene hydroperoxide, a finding consistent with the high levels of KatA and AhpC expression, and showed that these enzymes were functional. Quantitative analysis of KatA expression showed that both the perR mutation and the fur mutation had profound effects on catalase activity, suggesting additional non-iron-dependent regulation of KatA and, by inference, AhpC. The PerR protein is a functional but nonhomologous substitution for the OxyR protein, which regulates peroxide stress genes in other gram-negative bacteria. Regulation of peroxide stress genes by a Fur homolog has recently been described for the gram-positive bacterium Bacillus subtilis. C. jejuni is the first gram-negative bacterium where non-OxyR regulation of peroxide stress genes has been described and characterized.
Campylobacter jejuni is a gram-negative, microaerophilic enteric pathogen of humans, causing gastroenteritis. The bacterium is one of the most frequently isolated causes of bacterial diarrhea and can therefore be considered a major public health and economic problem (16). As a microaerophile, C. jejuni needs to protect itself from reactive oxygen species that are the result of aerobic metabolism. A coordinated response of oxidative-stress genes is a necessity of the (micro)aerobic lifestyle.
Two of the most important bacterial oxidative-stress genes in the defense against peroxide stress inducers such as hydrogen peroxide and alkyl hydroperoxides are catalase (KatA) and alkyl hydroperoxide reductase (AhpC). Both of these enzymes have been identified in C. jejuni (3, 15). In most bacteria including Escherichia coli, Salmonella typhimurium (10), Haemophilus influenzae (18), and Mycobacterium leprae (12), KatA and/or AhpC expression is regulated by the OxyR regulator in response to oxidative stress. In C. jejuni, however, AhpC and KatA expression is transcriptionally repressed in response to increasing environmental iron concentration (3, 31). The Fur protein usually mediates bacterial iron-responsive gene regulation. When the intracellular Fe2+ concentration is high, a complex consisting of a Fur dimer and Fe2+ binds to control sequences (Fur boxes) overlapping Fur-regulated promoters (11, 25). The presence of Fur box-like sequences in the promoters of both ahpC and katA indicated that iron regulation of these genes could be carried out by Fur. Unexpectedly, we found that in C. jejuni iron regulation of AhpC and KatA is Fur independent (31), and therefore we hypothesized that iron regulation of AhpC and KatA is mediated by another regulator.
In the gram-positive bacterium Bacillus subtilis, expression of AhpC and KatA is regulated by iron (6), and the presence of a repressor regulating AhpC and KatA has been predicted (9). Analysis of the B. subtilis genome sequence revealed the presence of three open reading frames (ORFs) encoding Fur homologs. One homolog was shown to function as the iron uptake regulator (Fur) (7), one as the zinc uptake regulator (Zur) (14), and the third, designated PerR, was responsible for the regulation of AhpC and KatA expression (7). In B. subtilis, therefore, PerR is a functional analog of the OxyR regulator. It was predicted that in other gram-positive bacteria peroxide stress genes and iron uptake might also be regulated by separate Fur homologs, as multiple Fur homologs have been described for several other gram-positive bacteria (7).
Here we report the identification of a C. jejuni PerR homolog and describe the effects of a perR mutation. This is the first gram-negative organism where PerR-like regulation has been described. A C. jejuni perR fur double mutant is also characterized where all iron regulation was abolished, and we show that both the perR and fur mutations influence expression of KatA.
MATERIALS AND METHODS
Media and growth conditions.
C. jejuni strains were maintained on Mueller-Hinton (MH) media (Unipath) under microaerophilic conditions in a Variable Atmosphere Incubator (Don Whitley) containing 85% N2, 10% CO2, and 5% O2. Media were routinely supplemented with 10 μg of vancomycin and 5 μg of trimethoprim per ml. Iron-restricted conditions were achieved by supplementing MH media with the iron chelator deferoxamine mesylate (desferal; Sigma Chemical Co.) to a final concentration of 20 μM. Iron-replete conditions were achieved by adding Fe(III)SO4 to MH media at a final concentration of 40 μM. E. coli was grown aerobically in Luria-Bertani medium (23) at 37°C. When antibiotic selection was necessary, growth media were supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (20 μg/ml). C. jejuni strains were tested for resistance to the peroxide stress inducers cumene hydroperoxide (CHP) and hydrogen peroxide by methods described previously (3).
Bacterial strains and plasmids.
C. jejuni and E. coli strains and plasmids used in this study are listed in Table 1. The region encoding perR and upstream and downstream sequences was amplified with primers 5′-CGC-GGTACC-TAT- TGC-TTT-GCG-TTA-TCC-TAG-A and 5′-CGC-GGATCC-ATT-GGA-ACT-ATC-CAA-AGT-TGG-AA. These primers contain a 5′ KpnI or BamHI restriction enzyme site (underlined) for cloning in pBluescript; thus, the polylinker HindIII and EcoRV sites were removed during cloning, allowing subsequent insertional mutagenesis of perR.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| NCTC 11168 | Parental strain | National Collection of Type Cultures |
| AV63 | 11168 perR::Kanr | This study |
| AV42 | 11168 fur::Cmr | 31 |
| AV67 | 11168 perR::Kanrfur::Cmr | This study |
| E. coli DH5α | F− φ80dlacZΔM15 | Gibco BRL |
| Plasmids | ||
| pBluescript II SK− | Cloning vector, Apr | Stratagene |
| pJMK30 | C. coli kanamycin (Kanr) resistance cassette in pUC19 | 31 |
| pAV205 | 1.4-kb PCR-amplified fragment of C. jejuni NCTC 11168 containing the perR region in pBluescript | This study |
| pAV214 | SmaI-cut Kanr cassette of pJMK30 cloned in the EcoRV site in perR in pAV205; the Kanr cassette is in the reverse orientation with respect to perR | This study |
Recombinant DNA techniques.
Restriction enzymes and T4 DNA ligase were purchased from Gibco BRL. All enzymes were used according to the manufacturer’s instructions. Standard protocols were used for manipulation of DNA and transformation of E. coli (2, 23) and C. jejuni (30). Genomic DNA of C. jejuni was prepared by the method described by Ausubel et al. (2). Plasmid DNA was prepared with affinity columns (Qiagen). PCR was carried out with Expand Polymerase Mix (Boehringer). DNA sequencing was performed with an Applied Biosystems model 377 DNA sequencing system and a Taq Dye Deoxy Terminator Cycle Sequencing Kit (Applied Biosystems).
Protein manipulation and catalase assays.
C. jejuni were fractionated by a technique described previously (31). Briefly, C. jejuni cells were subjected to osmotic shock to release the periplasm. Subsequently, the spheroplasts were disrupted by sonication, and cytoplasm and crude membranes were separated by ultracentrifugation. Inner membranes were solubilized in Sarkosyl, and outer membranes were pelleted by ultracentrifugation. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently stained with Coomassie brilliant blue. N-terminal amino acid sequences were determined from proteins transferred to Fluorotrans membranes (Flowgen Laboratories) by using Edman degradation on an ABI 476 sequencer (Applied Biosystems).
Catalase activity was measured by a technique described by Beers and Sizer (4). Briefly, C. jejuni cells were disrupted by sonication, and insoluble particles were pelleted by ultracentrifugation for 10 min at 100,000 × g. The soluble fraction was subsequently used for quantitative analyses. The protein concentration was measured as described by Bradford (5). Catalase activity was measured by monitoring the enzymatic breakdown of hydrogen peroxide at 240 nm, by using 50 mM phosphate buffer with a hydrogen peroxide concentration of 19.6 mM.
RESULTS
C. jejuni contains a second Fur homolog.
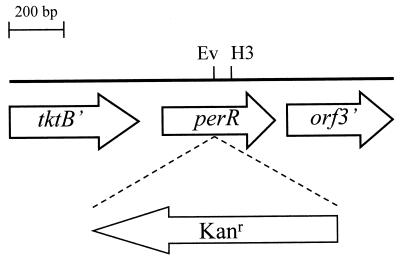
To identify putative regulators of oxidative-stress genes, we screened the C. jejuni genome sequence (24) for OxyR and Fur homologs. This analysis did not reveal the presence of an OxyR homolog but did reveal the presence of an ORF encoding a second Fur homolog approximately 70 kb from fur. The region containing this second Fur homolog was amplified from NCTC 11168 by PCR, and the nucleotide sequence was confirmed to be identical to the sequence determined by the C. jejuni genome project. The 1.4-kb region amplified contained three ORFs (Fig. 1). The product of the one complete ORF, designated PerR, showed significant homology with bacterial Fur homologs. An alignment of the C. jejuni PerR protein with the B. subtilis PerR and the C. jejuni and B. subtilis Fur proteins is shown in Fig. 2. The identity between all Fur and PerR sequences was 17%. However, the identity between the two Fur proteins was 34%, and that between the two PerR proteins was 32%. The ORF upstream of perR encodes a putative transketolase B (tktB) homolog, and the downstream ORF (orf3) did not have any significant homology with sequences deposited in the GenBank and EMBL databases (Fig. 1). The predicted perR gene is 408 bp long, encoding a protein of 15,926 Da with seven cysteine residues and eight histidine residues (Fig. 2). A putative ribosome binding site (AAGGA) was identified 10 bp upstream of the perR gene. There is 86 bp between tktB and perR, and this intergenic region has a very high A+T content of 90.6% and includes a putative stem-loop structure overlapping with the stop codon of tktB. This putative stem-loop structure has a stem of 11 bp and a loop of 5 bp and has a free energy of −8.6 kcal. Downstream of perR a small putative stem-loop structure is found with a stem of 5 bp, a loop of 3 bp, and a free energy of −2.2 kcal. The predicted transcriptional organization of the perR genomic region and the putative stem-loop structures present suggest that perR is transcribed as a monocistronic messenger, unlike C. jejuni fur (8, 29) and thus probably has its own promoter. The PCR primers used to amplify the perR region from C. jejuni NCTC 11168 gave similar-sized products with four other C. jejuni strains, indicating that the organization of the perR region is conserved in other C. jejuni strains (data not shown).
FIG. 1.
Schematic diagram representing the genomic region containing the perR gene. The position and orientation of the inserted antibiotic resistance cassette in perR are shown. Only partial ORFs of tktB and orf3 are indicated. Ev, EcoRV; H3, HindIII.
FIG. 2.
Alignment of C. jejuni (Cj) and B. subtilis (Bs) PerR and Fur proteins. Boxed residues are identical in the PerR and Fur proteins. Asterisks and dots indicate identical residues and conservative substitutions, respectively, in all four proteins.
PerR is the repressor of C. jejuni AhpC and KatA peroxide stress genes.
The perR gene was mutated to determine whether it was the repressor of the ahpC and katA genes in C. jejuni. By using the unique internal EcoRV site (Fig. 1), a kanamycin resistance cassette was inserted into the perR gene, and the disrupted perR gene was subsequently introduced into the C. jejuni NCTC 11168 genome by allelic exchange (17). Mutants were obtained with the antibiotic resistance cassette inserted in either orientation at the same frequency. This is in contrast with the C. jejuni fur gene (31). Correct allelic exchange of the wild-type perR allele with the mutated copy was confirmed by PCR (data not shown). The C. jejuni perR mutant was designated AV63. In addition to the perR mutant, we also constructed a perR fur double mutant by mutating perR in C. jejuni AV42, which is a fur mutant with a chloramphenicol resistance cassette inserted in the same orientation as fur (31). The presence of mutated fur and perR alleles was confirmed by PCR (data not shown). The perR fur double mutant, named AV67, was viable and showed the same growth characteristics as a fur mutant (31). The perR mutant showed the same growth characteristics as the wild-type strain (data not shown).
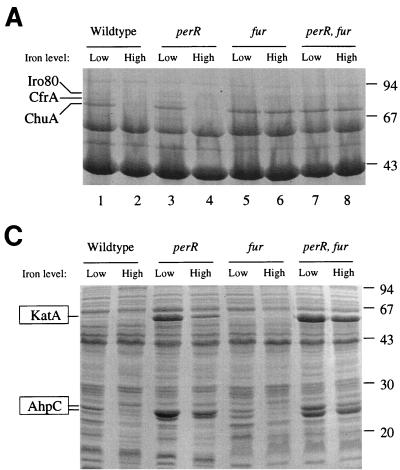
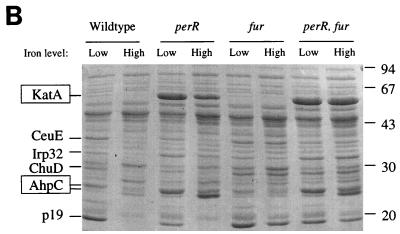
Protein profiles and subsequent protein sequencing were used to determine whether PerR was indeed the iron-responsive repressor of ahpC and katA expression. Bacteria were grown under iron-restricted and iron-replete conditions and fractionated into periplasm, cytoplasm, and outer membranes. This approach has been used previously to identify members of the fur regulon of C. jejuni (31). These protein profiles following separation by SDS-PAGE are shown in Fig. 3; proteins previously identified as Fur repressed are indicated, and these are all still iron repressed in the perR mutant but not in the fur mutant or the double mutant. This shows that the perR mutation does not notably affect Fur-regulated protein expression. Three highly expressed proteins with molecular sizes of approximately 25, 26, and 55 kDa can be seen in the periplasmic and cytoplasmic fractions of the perR mutant and perR fur mutant. Expression of these three proteins is iron repressed in the wild type and fur mutant (Fig. 3). All three proteins were identified by N-terminal amino acid sequencing. The N-terminal amino acid sequence of the 55-kDa protein (MKKLTNDFG) was identical to that of C. jejuni KatA (15). Surprisingly, the N-terminal amino acid sequence of both the 25- and the 26-kDa proteins (MIVTKKALDF) was identical to that of AhpC (3).
FIG. 3.
Protein profiles of fractionated C. jejuni wild type and mutants grown in low-iron and high-iron media, following separation by SDS-PAGE. Iron-repressed proteins are indicated on the left, with PerR-regulated proteins boxed. (A) Cytoplasmic fraction; (B) periplasmic fraction; (C) outer membrane fraction.
The perR mutation makes C. jejuni hyperresistant to peroxide stress.
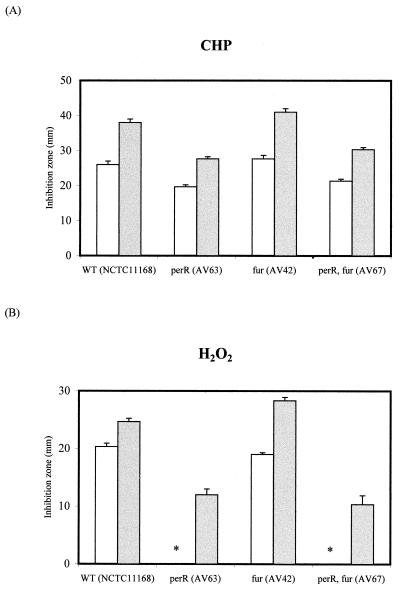
The perR mutation derepressed the katA and ahpC promoters, leading to a very high level of expression of KatA and AhpC. In order to test whether this led to expression of functional enzymes, the level of resistance of wild-type C. jejuni to peroxide stress was compared to those of the perR, fur, and perR fur mutants. The results obtained with two levels of the peroxide stress inducers hydrogen peroxide (cleared by KatA) and CHP (cleared by AhpC) are shown in Fig. 4. The perR mutation induces hyperresistance to both of these peroxide stress inducers, showing that in a perR background both overexpressed enzymes are functional. The fur mutation alone did not have a major effect on resistance of C. jejuni to the peroxide stress inducers.
FIG. 4.
Effects of perR and fur mutations on oxidative-stress resistance of C. jejuni wild type and mutants to CHP (A) and hydrogen peroxide (B). Resistance is expressed as the zone of growth inhibition after overnight growth on agar plates. Resistance was measured against 3% (open bars) and 10% (closed bars) CHP and H2O2. The error bars represent data from three separate plates. The perR mutation derepresses expression of AhpC and KatA and results in the perR mutants showing hyperresistance to CHP and H2O2. The fur mutation on its own slightly decreases resistance to both CHP and H2O2, whereas the fur perR double mutant is more resistant to CHP and H2O2. The perR mutants did not show an inhibition zone with 3% H2O2 (asterisk).
The derepression of KatA expression by the perR mutation was determined by direct enzyme assay. KatA synthesis was measured directly by spectophotometric detection of the rate at which hydrogen peroxide was utilized. The results of this assay are shown in Table 2. In the wild-type strain, KatA activity was almost completely absent under iron-replete conditions. The perR mutation substantially increased KatA activity under iron-replete conditions but particularly under low-iron conditions, where KatA activity was 10-fold higher than that in the wild type. Interestingly, KatA activity was still 2-fold iron repressed in the perR mutant, and in the fur mutant KatA activity was 4-fold lower than that in the wild type under low-iron conditions but higher than that in the wild type under high-iron conditions. Such an effect of the fur mutation on iron-repressed expression of katA suggests that Fur coregulates katA expression with PerR. This hypothesis is supported by the observation that in the perR fur double mutant KatA levels were almost as high as those in the perR mutant, but regulation in response to iron levels is no longer evident (Table 2).
TABLE 2.
Effects of C. jejuni perR and fur mutations on iron-regulated catalase activity
| C. jejuni strain | Catalase activity (U ± SD)
|
|
|---|---|---|
| Low iron | High iron | |
| NCTC 11168 (wild type) | 665.0 ± 94.0 | 0.1 ± 0.0 |
| AV63 (perR) | 7,101.4 ± 1,038.9 | 3,205.8 ± 134.6 |
| AV42 (fur) | 166.7 ± 13.4 | 20.1 ± 5.8 |
| AV67 (perR fur) | 6,435.6 ± 285.6 | 6,902.7 ± 1,215.1 |
DISCUSSION
Reactive oxygen species cause damage to DNA, proteins, and membranes. Bacteria have developed systems which include catalase, alkyl hydroperoxide reductase, and superoxide dismutase to clear these reactive oxygen species. In the Enterobacteriaceae, most oxidative-stress proteins have been identified as being induced by different kinds of oxidative stress and can be subdivided in two separately regulated classes; (i) the O2− (superoxide) stress proteins and (ii) the peroxide stress proteins. The O2− stress proteins include manganese-containing superoxide dismutase (SodA) and endonuclease IV (13). Catalase and alkyl hydroperoxide reductase are members of the peroxide regulon (13).
Oxidative-stress defense proteins are expressed at a basal level under normal nonstressed conditions, but all these systems need to be able to be upregulated in conditions of increased oxidative stress. The regulation of these systems has been extensively investigated in E. coli, where there are two regulatory systems for oxidative-stress genes. The SoxR-SoxS system regulates the superoxide regulon (1), and mutations in soxR or soxS fail to induce the members of their regulon (13). OxyR regulates the OxyR regulon, which is part of the peroxide stress regulon. The OxyR protein is a transcription factor that senses oxidative stress through disulfide bond formation, and, under conditions of oxidative stress, transcription of its regulon is induced following a change in OxyR conformation due to this disulfide bond formation (26, 27, 33). OxyR is considered a global regulator, and H. influenzae oxyR mutants are unable to respond to oxidative stress (18).
Homologs of the oxyR gene have been identified in many gram-negative bacteria, but no oxyR homologs have been identified in the genome sequence of Helicobacter pylori (28), and we did not find a homolog in the C. jejuni genome (24). This indicates that in C. jejuni as well as H. pylori regulation of oxidative-stress defense is organized differently. Three oxidative-stress defense genes—superoxide dismutase (sodB) (21, 22), catalase (katA) (15), and alkyl hydroperoxide reductase (ahpC) (3)—have been identified in C. jejuni and the closely related species C. coli. The transcription of the katA and ahpC genes in C. jejuni is responsive to the iron concentration in the growth medium (3). Iron-responsive regulation is usually mediated by the Fur protein, and putative Fur binding sequences were identified upstream of sodB, katA, and ahpC. However, it was demonstrated that in a C. jejuni fur mutant expression of katA and ahpC was still iron responsive, unlike the expression of iron-uptake systems (31). These findings led us to search the C. jejuni genome sequence (24) for the presence of an ORF encoding a second Fur homolog. We found this second fur homolog (named perR) and have now demonstrated using insertional mutagenesis that the product of perR is responsible for regulating expression of the katA and ahpC genes. Expression of both KatA and AhpC was very high in the perR mutants, such that these proteins were now also detected in the periplasmic fraction. The increase in KatA and AhpC expression is higher than that observed under conditions of iron limitation, indicating that even under iron limitation transcription from the promoters of katA and ahpC is still not maximal. Thus, there are likely to be other environmental stimuli regulating expression of these oxidative-stress defense genes. This agrees with the observation that in C. coli KatA expression is induced under conditions of oxidative stress (19). As yet it is unknown whether other stimuli are important in oxidative-stress responses in Campylobacter.
The protein profiles showed only two proteins whose expression was affected by the perR mutation, and these proteins were identified as KatA and AhpC. In addition, unlike the fur mutant (31), the C. jejuni perR mutant showed growth characteristics similar to those of its parental strain, indicating that in C. jejuni PerR may not be a global regulator. In contrast, Fur regulates at least 13 proteins in C. jejuni (Fig. 3) (31) and also affects the expression of KatA and possibly AhpC (Fig. 3 and Table 2). The apparently small PerR regulon might explain why the perR fur double mutant was still viable. We also tested the influence of the perR and fur mutations on SodB activity. Although SOD activity was strongly induced under high-iron conditions, neither the perR or fur mutation influenced SOD enzyme activity (data not shown). However, given that SodB is an iron-containing enzyme, under low-iron conditions sodB may be expressed but is not functional due to the iron limitation.
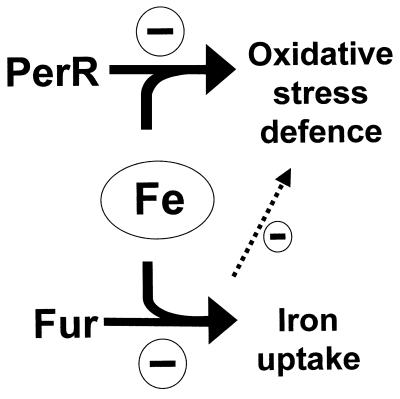
The perR system was first described for B. subtilis, where expression of KatA and AhpC is regulated by metal ions and oxidative stress (9). The PerR regulator was recently shown to be one of three Fur homologs (7), one other being the iron uptake regulator (designated Fur), and the third being the zinc uptake regulator (Zur) (14). Multiple Fur homologs have been described for several other gram-positive organisms and also for P. aeruginosa (32) and E. coli (20). The second E. coli Fur homolog regulates zinc uptake and has been designated Zur (20). The function of the second Fur homolog of P. aeruginosa is still unknown (32). We have now demonstrated the function of a second Fur homolog in C. jejuni, where it can be regarded as a functional analog for OxyR (Fig. 5). It is noteworthy that there is no second Fur or OxyR homolog in H. pylori, and thus, it remains unclear whether and how H. pylori regulates katA and ahpC expression.
FIG. 5.
Model illustrating Fur homolog-mediated negative regulation of oxidative-stress genes and iron transport systems in Campylobacter. The dotted arrow indicates a possible influence of Fur on oxidative-stress gene regulation involving either a direct interaction with oxidative-stress gene promoters or an indirect effect via transport systems on intracellular iron concentrations.
Regulation of gene expression in C. jejuni is relatively poorly understood. This report is the first description of peroxide stress regulation by a second Fur homolog in a gram-negative bacterium. We also describe the first C. jejuni double regulatory mutant and show that both PerR and Fur have an effect on catalase expression, demonstrating regulatory cross-talk in C. jejuni. Future work will focus on the mechanisms of PerR regulation of peroxide stress resistance and further characterization of the perR and perR fur mutants using two-dimensional gel electrophoresis.
ACKNOWLEDGMENTS
This study was supported by the Wellcome Trust and a Royal Society University Research Fellowship to J. M. Ketley. M.-L.A. Baillon was supported by a studentship from the Ministry of Agriculture, Fisheries and Food.
We thank Kathryn Lilley for N-terminal amino acid sequencing and Gina Manning, Andrey Karlyshev, and Diane Newell for helpful discussions.
REFERENCES
- 1.Amabile-Cuevas C F, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 3.Baillon M L A, van Vliet A H M, Ketley J M, Constantinidou C, Penn C W. An iron-regulated alkyl hydroperoxide reductase confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J Bacteriol. 1999;181:4798–4804. doi: 10.1128/jb.181.16.4798-4804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beers R F, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bsat N, Chen L, Helmann J D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen-peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron-uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 8.Chan V L, Louie H, Bingham H L. Cloning and transcription regulation of the Ferric Uptake Regulatory gene of Campylobacter jejuni TGH9011. Gene. 1995;164:25–31. doi: 10.1016/0378-1119(95)00477-n. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christman M F, Storz G, Ames B N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci USA. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coy M, Neilands J B. Structural dynamics and functional domains of the Fur protein. Biochemistry. 1991;30:8201–8210. doi: 10.1021/bi00247a016. [DOI] [PubMed] [Google Scholar]
- 12.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballa A, Helmann J D. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant K A, Park S F. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology. 1995;141:1369–1376. doi: 10.1099/13500872-141-6-1369. [DOI] [PubMed] [Google Scholar]
- 16.Ketley J M. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143:5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 17.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maciver I, Hansen E J. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect Immun. 1996;64:4618–4629. doi: 10.1128/iai.64.11.4618-4629.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S F. The use of the hipO gene, encoding benzoylglycine amidohydrolase, as a reporter of gene expression in Campylobacter coli. In: Lastovica A J, Newell D G, Lastovica E E, editors. Campylobacter, Helicobacter & related organisms. Cape Town, South Africa: Institute of Child Health, Red Cross Children’s Hospital; 1998. p. 528. [Google Scholar]
- 20.Patzer S I, Hantke K. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 21.Pesci E C, Cottle D L, Pickett C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purdy D, Park S F. Cloning, nucleotide sequence and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology. 1994;140:1203–1208. doi: 10.1099/13500872-140-5-1203. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning, a laboratory manual. 2nd ed. Cold Spring Harbor, New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Sanger Centre. posting date. Campylobacter jejuni page. [Online.] http://www.sanger.ac.uk/Projects/C_jejuni/. [29 July 1999, last date accessed.] 19 July 1999. [Google Scholar]
- 25.Stojiljkovic I, Hantke K. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur) Mol Gen Genet. 1995;247:199–205. doi: 10.1007/BF00705650. [DOI] [PubMed] [Google Scholar]
- 26.Storz G, Tartaglia L A, Ames B N. Transcriptional regulator of oxidative stress-inducible genes—direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 27.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 28.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 29.van Vliet, A. H. M., J. D. Rock, L. N. Madeleine, and J. M. Ketley. Submitted for publication.
- 30.van Vliet A H M, Wood A C, Henderson J, Wooldridge K G, Ketley J M. Genetic manipulation of enteric Campylobacter species. Methods Microbiol. 1998;27:407–419. [Google Scholar]
- 31.van Vliet A H M, Wooldridge K G, Ketley J M. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang J Y, Mushegian A, Lory S, Jin S G. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]