Abstract
Arg-52 of the Escherichia coli melibiose carrier was replaced by Ser (R52S), Gln (R52Q), or Val (R52V). While the level of carrier in the membrane for each mutant remained similar to that for the wild type, analysis of melibiose transport showed an uncoupling of proton cotransport and a drastic reduction in Na+-coupled transport. Second-site revertants were selected on MacConkey plates containing melibiose, and substitutions were found at nine distinct locations in the carrier. Eight revertant substitutions were isolated from the R52S strain: Asp-19→Gly, Asp-55→Asn, Pro-60→Gln, Trp-116→Arg, Asn-244→Ser, Ser-247→Arg, Asn-248→Lys, and Ile-352→Val. Two revertants were also isolated from the R52V strain: Trp-116→Arg and Thr-338→Arg revertants. The R52Q strain yielded an Asp-55→Asn substitution and a first-site revertant, Lys-52 (R52K). The R52K strain had transport properties similar to those of the wild type. Analysis of melibiose accumulation showed that proton-driven accumulation was still defective in the second-site revertant strains, and only the Trp-116→Arg, Ser-247→Arg, and Asn-248→Lys revertants regained significant Na+-coupled accumulation. In general, downhill melibiose transport in the presence of Na+ was better in the revertant strains than in the parental mutants. Three revertant strains, Asp-19→Gly, Asp-55→Asn, and Thr-338→Arg strains, required a high Na+ concentration (100 mM) for maximal activity. Kinetic measurements showed that the N248K and W116R revertants lowered the Km for melibiose, while other revertants restored transport velocity. We suggest that the insertion of positive charges on membrane helices is compensating for the loss of Arg-52 and that helix II is close to helix IV and VII. We also suggest that Arg-52 is salt bridged to Asp-55 (helix II) and Asp-19 (helix I).
Bacterial secondary active transporters capture free energy from the movement of cations down their electrochemical gradient and use it to drive the transport of solutes such as sugars, amino acids, Krebs cycle intermediates, antibiotics, and inorganic ions across the cell membrane (27, 31, 33). The melibiose carrier (MelB) of Escherichia coli is a cation/substrate symporter which couples transport of Na+ and melibiose across the bacterial inner membrane (for reviews see references 22, 32, and 38). In addition to melibiose, MelB transports a variety of sugar substrates including α- and β-galactosides as well as some monosaccharides (46, 50). An interesting feature of MelB is its ability to couple sugar transport to three different cations, Na+, Li+, and H+, depending on the configuration of the transported sugar (44–46, 50). Sugar binding studies using membrane vesicles have shown that the presence of Na+ and Li+ ions increases the carrier’s affinity for galactosides and that the cations compete for a single binding site (6, 8).
The melB gene has been cloned (18) and sequenced (52). The primary amino acid sequence deduced from the gene sequence predicts a hydrophobic protein (70% apolar) with a molecular mass of 52 kDa (52). The results of hydropathy analysis and melB-phoA fusions have provided good evidence for a two-dimensional structure where the protein forms 12 α-helical transmembrane domains connected by hydrophilic loops (1, 34, 52).
E. coli MelB is a member of the galactoside-pentose-hexuronide family of bacterial transport proteins (32). The MelB subfamily consists of melibiose carriers from E. coli, Salmonella typhimurium, Klebsiella pneumoniae, and Enterobacter aerogenes. Although a high degree (78 to 85%) of amino acid identity exists among carriers in the MelB subfamily (30, 32), there are distinct differences in cation selectivity. For example, the MelB of E. coli couples H+, Na+, and Li+ to sugar transport, while the K. pneumoniae carrier couples either H+ or Li+, but not Na+ (16). The amino acid residues responsible for Na+ recognition were localized by constructing chimeras of the E. coli and K. pneumoniae melibiose carriers (15). Replacement of the first 81 amino acids of the K. pneumoniae carrier with those of the E. coli MelB was sufficient to allow the K. pneumoniae carrier to couple Na+ and sugar transport (15). Interestingly, a single-amino-acid substitution in helix II of K. pneumoniae, Ala-58→Asn, also resulted in Na+-coupled sugar transport (17).
Cation recognition in E. coli MelB has also been investigated by site-directed mutagenesis. Studies have focused primarily on acidic residues that reside on membrane-spanning helices in the amino-terminal portion of the carrier. Neutral amino acid substitutions for Asp-19 (helix I), Asp-55 (helix II), Asp-59 (helix II), and Asp-124 (helix IV) cause the loss of Na+-coupled sugar transport (32, 35, 36, 53). In these mutants, sugar binding is comparable to that of wild-type MelB in the absence of Na+, but this binding is no longer stimulated by Na+. Taken together, the results of these studies have led to a model in which Asp residues at positions 19, 55, 59, and 124 provide part of a network for the coordination of cations in E. coli MelB (22, 32, 36, 54).
The studies mentioned above show that the acidic amino acids on transmembrane helix II of the E. coli MelB, Asp-tt and Asp-59, are important for cation recognition. While the roles of these aspartates have been studied thoroughly, less is known about the positively charged residue Arg-52 in this helix. It has been reported that a substitution of Ala for Arg-52 leads to a 95% loss in carrier activity but that the remaining activity is still stimulated by Na+ and Li+ (54). In the present study, we further investigate the role of Arg-52. We use site-directed mutagenesis to substitute Gln, Val, and Ser for Arg-52 (R52Q, R52V, and R52S, respectively). We show that substitution of Arg-52 causes a dramatic loss of melibiose transport, with only a small amount of Na+-stimulated activity remaining. Subsequently, we use the Val-52, Ser-52, and Gln-52 mutant strains to isolate revertant strains which regain the ability to transport melibiose. Sequence analyses reveal that revertant mutations, with one exception, are found at locations other than position 52. The majority of these mutations result in substitution of amino acids located on transmembrane domains in both the amino and carboxyl halves of the protein. Our analysis of the melibiose transport properties in the strains with site-directed or second-site revertant mutations provide significant new information about the functional role of Arg-52. On the basis of our data, we suggest that specific transmembrane helices are close to one another in the three-dimensional structure of the protein. The data also suggest that Arg-52 is involved in an intrahelical salt bridge with Asp-55 and possibly in an interhelical salt bridge with Asp-19.
MATERIALS AND METHODS
Reagents.
Melibiose (O-α-d-galactopyranosyl-(1,6)-d-glucopyranose) was purchased from Sigma. [3H]melibiose was a generous gift from Gérard Leblanc of the Départment de Biologie Commissariat à l’Energie Atomique, Villefranche-su-mer, France. 35S-labeled protein A was purchased from Amersham. [α-33P]dATP was from Andotek. Restriction enzymes and ligase were from Pharmacia Biotech. Bacteriological media were from Difco. All other chemicals were reagent grade.
Bacterial strains and plasmids.
Plasmids used in this study are listed in Table 1. Plasmid DNA was isolated with the QIAprep Spin Miniprep Kit (Qiagen) and introduced into the appropriate bacterial strains by RbCl2 transformation. E. coli DW1 (lacI+ lacΔZY melΔAB) (50) and DW1/pSUMelA (lacI+ lacΔZY melA+ melΔB) (51) were used as host strains for plasmids expressing E. coli melB (GenBank accession no. K01991). The pTZ19U phagemid (Bio-Rad) was used for site-directed mutagenesis of melB. The plasmid pKKMB (2) that contains the gene for the melibiose carrier inserted into the vector pKK223-3 (Pharmacia Biotech) was used for the expression of melB. The plasmid pSUmelA (51) was used to express melA (α-galactosidase) (GenBank accession no. X04894) which allows cells to ferment melibiose.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant genotype | Source or reference |
|---|---|---|
| pTZ19U | lacO+P+Z′ Ampr | Bio-Rad |
| pSUmelA | melA+melΔB Cmr | 51 |
| pKK223-3 | Ampr | Pharmacia Biotech |
| pKKMB | melΔ(A)B+ Ampr | 2 |
| pR52S | melΔ(A)B+(Arg-52→Ser) Ampr | This study |
| pR52V | melΔ(A)B+(Arg-52→Val) Ampr | This study |
| pR52Q | melΔ(A)B+(Arg-52→Gln) Ampr | This study |
| pR52Ka | melΔ(A)B+(Arg-52→Lys) Ampr | This study |
| pR52S/D19G | melΔ(A)B+(Arg-52→Ser, Asp-19→Gly) Ampr | This study |
| pR52S/D55N | melΔ(A)B+(Arg-52→Ser, Asp-55→Asn) Ampr | This study |
| pR52S/P60Q | melΔ(A)B+(Arg-52→Ser, Pro-60→Gln) Ampr | This study |
| pR52S/W116R | melΔ(A)B+(Arg-52→Ser, Trp-116→Arg) Ampr | This study |
| pR52S/N244S | melΔ(A)B+(Arg-52→Ser, Asn-244→Ser) Ampr | This study |
| pR52S/S247R | melΔ(A)B+(Arg-52→Ser, Ser-247→Arg) Ampr | This study |
| pR52S/N248K | melΔ(A)B+(Arg-52→Ser, Asn-248→Lys) Ampr | This study |
| pR52S/I352V | melΔ(A)B+(Arg-52→Ser, Ile-352→Val) Ampr | This study |
| pR52V/W116Rb | melΔ(A)B+(Arg-52→Val, Trp-116→Arg) Ampr | This study |
| pR52V/T338R | melΔ(A)B+(Arg-52→Val, Thr-338→Arg) Ampr | This study |
| pR52Q/D55Nb | melΔ(A)B+(Arg-52→Gln, Asp-55→Asn) Ampr | This study |
pR52K (Arg-52→Lys) is a first-site revertant isolated from the pR52Q (Arg-52→Gln) site-directed mutant.
The pR52V/W116R and pR52Q/D55N strains show second-site revertant substitutions that were also obtained from the K52S parental strain. These two revertants were not studied in detail.
Site-directed mutagenesis.
The pKKMB plasmid was digested with EcoRI/HindIII to produce a 1,500-bp melB fragment. This fragment was ligated to the EcoRI/HindIII sites of the phagemid vector pTZ19U such that the antisense strand of the melB gene was incorporated into the plus strand (excreted strand) of the phagemid. Site-directed mutagenesis was performed by using the Muta-Gene phagemid kit (Bio-Rad) according to the manufacturer’s protocol. Mutations were introduced in the melB gene by using the following primers: for R52S, 5′-CTGGTGGCG(TCT)ATCTGGGATGCTATTAAC-3′; for R52V, 5′-CTGGTGGCG(GTA)ATCTGGGATGCTATTAAC-3′; and for R52Q, 5′-GGTGGCG(CAA)ATCTGGGATG-3′). These primers are complementary to the antisense strand of the melB gene except for mismatches (indicated in parentheses) that altered the desired codon. Mutations were verified by DNA sequencing (Amplicycle Sequencing Kit; Perkin-Elmer) of the melB coding sequence. Following mutagenesis, the pTZ19U-melB phagemid was digested with EcoRI/HindIII to remove the melB DNA fragment, and it was subsequently ligated to the EcoRI/HindIII sites of the expression vector pKK223-3.
Screen for second-site revertants.
E. coli DW1/pSUmelA cells with the melB R52Q, R52S, and R52V substitutions were used to screen for cells exhibiting a transport-positive phenotype. Cells grew initially as white colonies on MacConkey agar containing 0.4% melibiose (Difco). After 5 to 8 days of incubation at 37°C, small red isolates were picked and restreaked on the same medium to purify the colonies. Plasmid DNA isolated from red revertant colonies was used to transform DW1/pSUmelA to verify that the plasmid carrying the melB gene was responsible for the red phenotype on MacConkey medium containing melibiose. In order to eliminate the possibility of a cell containing two different types of melB plasmids (one with the original site-directed mutation and one with the original mutation plus a second-site mutation), DNA was diluted 1/10,000 and used to transform DW1/pSUmelA. In some cases, both white and red transformants were observed. A red clone was picked and used for analysis. Base substitutions were identified by sequencing the entire melB gene using primers at approximately 200-bp intervals.
Melibiose transport assays.
E. coli DW1 was used as the host strain for melibiose accumulation assays (15). Cells were grown to mid-log phase in Luria-Bertani (LB) medium containing ampicillin (100 μg/ml), harvested, and washed twice in a buffer containing 0.1 M morpholinepropanesulfonic acid (MOPS)-Tris (pH 7.0) and 0.5 mM MgSO4. The washed cells were resuspended in the same buffer to a density of approximately 3 × 109 cells/ml, and allowed to equilibrate to room temperature for 15 min. Transport was initiated by the addition of [3H]melibiose (0.2 mM; 0.5 μCi/ml) in the absence or presence of NaCl (10 or 100 mM) or LiCl (10 mM). A 0.2-ml aliquot was taken at various time points (0.5 to 15 min) and filtered rapidly through 0.65-μm-pore-size cellulose nitrate filters (Sartorius). To remove any remaining external sugar solution, filtered cells were washed with 5 to 10 ml of buffer. Filters were dissolved in 4 ml of Liquiscint (National Diagnostics) and counted. The volume of intracellular water was estimated to be 0.4 μl/6 × 108 cells (42) for calculations of sugar accumulation. Accumulation values are reported as the ratios of intracellular sugar concentration to extracellular sugar concentration.
E. coli DW1/pSUmelA was used as the host strain for downhill melibiose transport assays. Cells were grown to mid-log phase in LB medium containing ampicillin (100 mg/ml) and chloramphenicol (30 μg/ml). Cells were prepared as described in the previous paragraph, and transport assays were initiated by the addition of [3H]melibiose (0.8 mM; 0.5 μCi/ml) in the absence or presence of NaCl (10 or 100 mM) or LiCl (10 mM). Transport assays were performed as described in the previous paragraph. An estimate of 0.1 mg of protein/6 × 108 cells was used for calculations of 3[H]melibiose transport. Transport values are reported as nanomoles of [3H]melibiose per milligram of total cell protein.
Km and Vmax measurements.
The E. coli DW1/pSUmelA strain was used for the measurement of apparent Km and Vmax values for downhill transport of melibiose. Downhill transport was first tested at a variety of NaCl concentrations (0 to 200 mM) to determine which concentration gave maximal stimulation for each MelB derivative. In most cases, no significant stimulation was found above 10 mM NaCl. However, the R52S D19G, R52S D55N, and R52V T338R strains required 100 mM Na+ for maximal activity. Cells were prepared as described above for the downhill transport assay, and the initial rate (within 30 s) of melibiose transport was measured at six sugar concentrations (0.1 to 3.3 mM) to estimate the Km for melibiose in each strain. Six melibiose concentrations were chosen which bracketed the initial estimate, and measurements were repeated at least two more times. Kinetic values were determined by using a Lineweaver-Burk double-reciprocal plot.
Melibiose-induced Na+ transport.
Sodium uptake was measured as described previously (11). E. coli DW1 cells expressing wild-type, mutant, or revertant MelB carriers were grown to mid-log phase in LB medium containing ampicillin (100 mg/ml). Cells were harvested, washed twice, and resuspended in a solution containing 0.1 M MOPS-TMAH (pH 7.0) and 0.5 mM MgSO4. For the assay, an aliquot of cells was diluted to 2.5 mg of protein/ml with 0.1 M Tricine-TMAH (pH 8.0), and NaCl was added to a final concentration of 50 μM. Cells (6 ml) were placed in a closed plastic vial with holes in the lid to accommodate a ISE21Na sodium electrode (Radiometer), a REF201 reference electrode (Radiometer), a gas-tight syringe (Hamilton), and a tube for the introduction of argon. Cells were placed into anaerobic conditions by adding argon for 30 min, followed by the addition of melibiose to a concentration of 5 mM. Changes in sodium levels in the extracellular medium were monitored with a chart recorder (Linear Instruments). The sensitivity of the system was tested with a known amount of NaCl during each experiment. Melibiose and NaCl were added without introducing oxygen.
Melibiose-induced H+ transport.
The measurement of proton uptake was performed by the method of West (47) as modified by Wilson et al. (49). Briefly, E. coli DW1 cells expressing wild-type, mutant, or revertant MelB carriers were grown to mid-log phase in LB medium containing 100 mg of ampicillin per ml. Cells were washed twice and resuspended to a density of approximately 3.5 mg of protein/ml in an unbuffered 120 mM KCl solution. Cells (2.5 ml) were placed in a closed plastic vial with a lid containing apertures for the introduction of argon, a PHC4406 pH electrode (Radiometer) and a gas-tight syringe (Hamilton). Potassium thiocyanate (30 mM) was added, and cells were placed into anaerobic conditions by adding argon for 30 min. The addition of melibiose (anaerobically) (final concentration of 10 mM) was used to initiate proton uptake. Changes in the pH of the extracellular medium were monitored with a PHM64 pH meter (Radiometer) and recorded with a chart recorder (Linear Instruments) such that a 0.1-unit pH change caused a 25-cm deflection in the chart recording. Calibrations were performed with a known amount of HCl while maintaining the anaerobic conditions.
Immunodetection of melibiose carrier in bacterial cells.
The amount of melibiose carrier present in each strain was determined as previously described (26). In summary, a known quantity of cells was lysed with NaOH-sodium dodecyl sulfate and neutralized on nitrocellulose filters. Filters were incubated with bovine serum albumin to block nonspecific binding, followed by incubation with a polyclonal antibody, anti-MBct10 (4), directed against the carboxyl-terminal 10 amino acids of the protein. 35S-protein A (Amersham) was used to label the bound antibody, and the amount of label was quantified by liquid scintillation counting. To correct for nonspecific adsorption, values obtained for the strain DW1/pKK223-3 (melB) were used as a background control in each experiment. Values for the mutants are presented as percentages of wild-type protein levels.
RESULTS
Arg-52 mutagenesis and revertant isolation.
In order to better define the role of Arg-52 in substrate translocation, we replaced this residue with Gln (R52Q), Ser (R52S), or Val (R52V). Derivative carriers were expressed from the pKK223-3 vector in the α-galactosidase-positive strain, E. coli DW1/pSUmelA. Cells were plated on MacConkey indicator plates containing melibiose as the sole fermentable carbon source. On this medium, the wild-type strain forms dark red colonies as the result of sugar transport via MelB and subsequent fermentation. All Arg-52 mutants displayed a white phenotype, suggesting that replacement of Arg-52 with a neutral residue causes disruption of melibiose uptake.
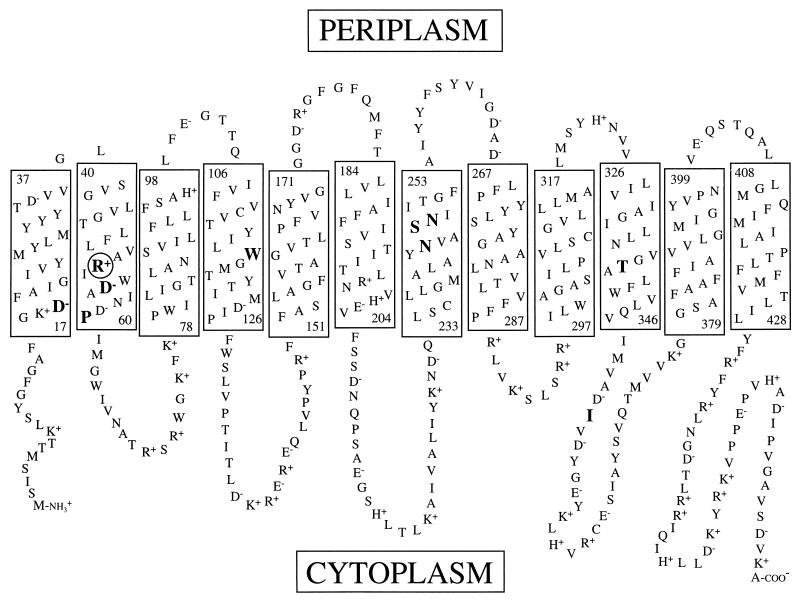
The white phenotype on MacConkey agar containing melibiose allows for a convenient screen for melibiose transport-competent revertants. Incubation of MelB mutants on melibiose-containing MacConkey agar for a prolonged period can produce spontaneous mutants which grow as red areas within the background of white cells, indicating melibiose fermentation. We streaked the R52S, R52Q, and R52V strains on MacConkey indicator plates containing 0.4% melibiose (11 mM) and incubated them at 37°C. After the strains were allowed to grow overnight, we observed only white colonies, but after 5 to 7 days, small red areas appeared within the white colonies. These red cells were purified by restreaking on the same type of medium until a uniform colony phenotype was observed, and then plasmid DNA was extracted and used to transform E. coli DW1/pSUmelA. To ensure that the mutation responsible for the red phenotype was carried on the melB-bearing plasmid, only those samples that retained a red phenotype after transformation were saved for further analysis. DNA sequence analysis revealed that in addition to the introduced mutations at codon 52, each revertant had an additional single-base substitution causing a missense mutation within the coding region of the melB gene. We identified three types of second-site revertants from the R52S, R52Q, and R52V mutants: (i) replacement of neutral residues with basic residues (gain of positive charge), (ii) replacement of acidic residues with neutral residues (loss of negative charge), and (iii) replacement of neutral amino acids with other neutral residues (Table 2). Amino acid substitutions were found at residues residing on transmembrane helices with one exception, I352V, which was located in a cytoplasmic loop between helices X and XI (Fig. 1 and Table 2). The R52Q strain also yielded a first-site substitution in which Gln-52 was replaced with Lys.
TABLE 2.
Phenotype and protein expression
| Mutant or revertant type | Strain | Mutation or substitutiona | Location in secondary modelb | Phenotype on melibiose MacConkeyc | Protein expression (%)d |
|---|---|---|---|---|---|
| melB control | White | ||||
| Wild-type | Red | 100 | |||
| Site-directed mutante | R52S | Arg-52→Ser | Helix II | White | 114 ± 15 |
| R52V | Arg-52→Val | Helix II | White | 101 ± 19 | |
| R52Q | Arg-52→Gln | Helix II | White | 112 ± 18 | |
| First-site revertantf | R52K | Arg-52→Lys | Helix II | Red | 85 ± 12 |
| Second-site revertant | R52S W116R | Trp-116 to Arg | Helix IV | Red | 43 ± 8 |
| R52S S247R | Ser-247 to Arg | Helix VII | Red | 105 ± 25 | |
| R52S N248K | Asn-248 to Lys | Helix VII | Red | 75 ± 18 | |
| R52S D19G | Asp-19 to Gly | Helix I | Pink | 12 ± 3 | |
| R52S D55N | Asp-55 to Asn | Helix II | Red | 92 ± 25 | |
| R52S P60Q | Pro-60 to Gln | Helix II | Red | 73 ± 3 | |
| R52S N244S | Asn-244 to Ser | Helix VII | Red | 72 ± 8 | |
| pR52S I352V | Ile-352 to Val | Loop X-XI | Red | 111 ± 21 | |
| R52V T338R | Thr-338 to Arg | Helix X | Red | 37 ± 4 |
For the second-site revertants, only the positions and identities of second-site revertant amino acid substitutions within the melibiose carrier are shown.
Position of the mutant or revertant amino acid residue within the secondary topological model for the melibiose carrier (Fig. 1).
MacConkey agar is a rich medium lacking a fermentable carbon source. Melibiose was added to the agar as the sole fermentable carbon source. The fermentation of melibiose on this medium causes cells to form red colonies. Cells not able to ferment melibiose form white colonies. The concentration of melibiose was 15 mM (0.5%).
Expression levels for melibiose carriers are reported as percentages of the expression found for the wild-type carrier. The values are averages of at least two separate measurements ± standard errors.
Strains in which Arg-52 has been replaced with Ser, Gln, or Val by site-directed mutagenesis. These strains were subsequently used for the isolation of revertants that regained melibiose transport activity.
First-site revertant isolated from the R52Q mutant strain.
FIG. 1.
Toplogical model of the melibiose carrier. Rectangles represent transmembrane domains. The numbers at the top and bottom of each rectangle indicate the first and last residue of each helix. The circled residue in helix II was subjected to site-directed mutagenesis in this study. Large bold residues indicate the positions of isolated second-site revertants. The model is based on that described by Pourcher et al. (34).
Determination of relative levels of carrier protein in mutant and revertant strains.
When constructing mutations in membrane proteins, it is always a possibility that the introduced amino acid change will disrupt proper membrane insertion and/or stability of the protein. To determine the relative amount of carrier protein present in the membrane of each mutant on revertant strain, we performed immunoblotting using a polyclonal antibody directed against the C-terminal 10 amino acids of the carrier. The amount of melibiose carrier protein expressed in each strain is presented as a percentage of that value (Table 2). The R52S, R52Q, and R52V strains produced protein levels similar to that of the wild type. Generally, the revertant strains showed about 70%, or higher, of wild-type protein levels. Two strains, R52S W116R and R52V T338R strains, had reduced, but significant amounts of protein at 43 and 37%, respectively. Only one revertant, R52S D19G strain, had significantly reduced protein levels, showing only 12% of the wild-type value.
Melibiose transport assays.
Colony phenotype on MacConkey plates containing melibiose (Table 2) provides only a qualitative measure of carrier function. To obtain a more quantitative assessment of transporter activity, we measured downhill melibiose transport using whole cells under aerobic conditions, with an external concentration of 0.8 mM [3H]melibiose and a variety of cationic conditions (Table 3). In this experiment, sugar flows into the cell via the melibiose carrier and is rapidly cleaved by α-galactosidase so that the external sugar concentration always remains higher than the intracellular concentration. In the wild-type carrier, maximal stimulation of melibiose transport has been found to require 10 mM Na+. Our data show that the R52S, R52Q, and R52V parental strains had extremely low melibiose transport activity with 10 mM Na+ (8, 5, and 0% of the wild-type level, respectively) and that R52V transport was best with 100 mM Na+. In addition, H+-coupled melibiose transport activity was lost for the R52V and R52S mutants and severely impaired in the R52Q mutant. Interestingly, the R52K first-site revertant, isolated from the R52Q mutant, had downhill transport values that exceeded the wild-type values under all of the conditions tested.
TABLE 3.
Downhill melibiose transport
| melB strain | Amt of melibiose transporteda in the presence of:
|
|||
|---|---|---|---|---|
| H+ | 10 mM Na+ | 100 mM Na+ | 10 mM Li+ | |
| Wild-type | 35 ± 9 | 149 ± 40 | 42 ± 2 | 87 ± 4 |
| R52K | 50 ± 5 | 206 ± 17 | 98 ± 5 | 148 ± 11 |
| R52Q | 4 ± 1 | 8 ± 1 | 4 ± 1 | 2 ± 1 |
| R52V | 0 ± 0 | 0.1 ± 0.1 | 4 ± 1 | 0.9 ± 0.3 |
| R52S | 0.2 ± 0.1 | 12 ± 4 | 8 ± 1 | 4 ± 0.3 |
| R52S W116R | 6 ± 0.7 | 99 ± 7 | 93 ± 9 | 68 ± 7 |
| R52S S247R | 2 ± 2 | 25 ± 11 | 13 ± 1 | 22 ± 2 |
| R52S N248K | 2 ± 0.5 | 60 ± 19 | 31 ± 1 | 81 ± 16 |
| R52V T338R | 1 ± 1 | 3 ± 0.4 | 10 ± 1 | 6 ± 1 |
| R52S D19G | 0.8 ± 0.4 | 2 ± 1 | 8 ± 1 | 4 ± 0.7 |
| R52S D55N | 2 ± 0.4 | 23 ± 1 | 65 ± 2 | 9 ± 0.7 |
| R52S P60Q | 0.9 ± 0.1 | 15 ± 7 | 6 ± 2 | 7 ± 0.6 |
| R52S N244S | 1 ± 0.3 | 41 ± 3 | 19 ± 7 | 18 ± 0.9 |
| R52S I352V | 4 ± 3 | 40 ± 16 | 33 ± 1 | 20 ± 0.9 |
The concentration of radiolabeled melibiose was 0.8 mM, and values given represent transport at 10 min. The values are given in nanomoles of [3H]melibiose per milligram of protein. Transport values were calculated, assuming 0.1 mg of protein for every 6 × 108 cells. The data are averages of at least two separate experiments with duplicate determinations in each experiment ± standard errors.
Most of the second-site revertants isolated from the R52S and R52V mutants regained downhill melibiose transport function in the presence of Na+ and Li+. Two revertants, the R52S D19G and R52S P60Q revertants, showed no recovery or marginal recovery of activity, respectively. In contrast, H+-coupled transport remained defective for the revertant strains. Only the R52S I352V and R52S W116R revertants showed limited H+-coupled melibiose transport activity. Among revertants where a positively charged residue had been added, the R52S W116R and R52S N248K revertants had good downhill transport with 10 mM Na+ (66 and 40% of wild-type, respectively). However, in the R52S S247R revertant, transport activity was only slightly better than that of the R52S parental strain. The R52V T338R revertant required 100 mM Na+ for optimal activity, but transport was still low. In general, Li+-coupled transport in these revertants follows the same trend as in the wild type, being slightly lower than optimal Na+-coupled transport. One exception was the R52S N248K revertant where 10 mM Li+ gave the highest downhill transport activity.
For those revertants where a negatively charged residue was removed, R52S D19G and R52S D55N revertants, 100 mM Na+ was required for optimal activity. The R52S D55N revertant had fairly good transport activity under these conditions (44% of the wild-type level). The R52S D19G revertant did not recover transport activity that exceeded that of the R52S revertant. While this appears to contradict the pink phenotype on melibiose-containing MacConkey medium, it should be noted that revertants were selected in the presence of 12 mM melibiose compared to 0.8 mM melibiose used in the in vitro transport assay. Apparently the long incubation time (16 h) and relatively high melibiose concentration on the MacConkey medium were sufficient to allow the R52S D19G revertant to produce pink colonies.
For those strains where a neutral residue was substituted with another neutral residue, 10 mM Na+ stimulated the highest melibiose uptake. Both the R52S N244S and R52S I352V revertants had significantly increased transport activity (28 and 27% of the wild-type level, respectively) compared to the R52S parental strain. However, transport in the R52S P60Q revertant was only slightly better than that of the R52S strain.
To further detail the transport properties of mutant and revertant strains, we measured apparent Km and Vmax values for downhill melibiose transport (Table 4). We determined initial transport rates at a variety of sodium concentrations (0 to 200 mM), and the concentration which stimulated the highest initial transport rate was used for kinetic measurements (data not shown). For all strains where kinetic measurements were possible, with the exception of the R52S D55N revertant, raising the Na+ concentration above 10 mM did not significantly increase transport rates and this concentration was used. The requirement for 100 mM Na+ in the R52S D55N revertant was not unexpected, as the Asp-55 residue has been shown to be critical for Na+-stimulated sugar transport. The wild-type carrier had a Km of 0.22 mM and a Vmax of 71 nmol of melibiose/min/mg of protein (Table 4). The R52K strain had a normal Km and a Vmax similar to that of the wild type. In contrast, the R52S mutant had both an increased Km and a decreased Vmax compared to those of the wild-type strain. Transport activity in the R52Q and R52V mutants was too low for determination of Km and Vmax. In two revertants, the R52S W116R and R52S N248K revertants, the apparent affinity for melibiose was better than that of the R52S parent, suggesting that the insertion of a positive charge in these revertants was compensating for the loss of Arg-52. Although the other revertants assayed had high Km values, they all had an increased Vmax values compared to that of the R52S parental strain. Strikingly, the Vmax for the R52S D55N strain was almost threefold higher than that of the wild type, although this revertant required a higher Na+ concentration.
TABLE 4.
Kinetic analysis of sodium-coupled downhill melibiose transporta
| Strain | Mean apparent Km (mM) ± SE | Mean apparent Vmax (nmol of melibiose/min/mg of protein) ± SE |
|---|---|---|
| Wild-type | 0.22 ± 0.06 | 71 ± 14 |
| R52K | 0.24 ± 0.05 | 153 ± 55 |
| R52S | 0.75 ± 0.06 | 18 ± 3 |
| R52S W116R | 0.63 ± 0.02 | 64 ± 10 |
| R52S S247R | 2.7 ± 0.3 | 50 ± 3 |
| R52S N248K | 0.24 ± 0.06 | 39 ± 8 |
| R52S D55N | 7.4 ± 1.7 | 209 ± 57 |
| R52S P60Q | 6.5 ± 1.0 | 40 ± 3 |
| R52S N244S | 2.4 ± 0.48 | 72 ± 14 |
| R52S I352V | 1.2 ± 0.08 | 48 ± 10 |
Transport of melibiose after 30 s was measured at six different concentrations which bracketed an initial estimate of the Km for each strain. The values for Km and Vmax are the averages of at least two separate experiments with three datum points in each experiment. Transport was done in the presence of 10 mM Na+ for all strains except for the R52S D55N strain for which 100 mM Na+ was required.
Melibiose accumulation.
The coupling of cation and sugar transport allows the melibiose carrier to accumulate sugar against a concentration gradient at the expense of the cation gradient. In the wild-type MelB, the preferred cation for transport is Na+, but H+ or Li+ can also be utilized, depending on the conformation of the sugar substrate. Measurement of melibiose accumulation (uphill transport) in the presence of cations allows an assessment of the coupling between cation and sugar transport. E. coli DW1 (α-galactosidase negative) was used for melibiose accumulation assays. We measured accumulation of melibiose at an external concentration of 0.2 mM for a period of 15 min. We show that in the presence of 10 mM Na+, the wild-type carrier was able to accumulate melibiose 153-fold (Table 5), while only 51-fold accumulation was found when the Na+ concentration was increased to 100 mM. The inhibitory effect of Na+ above 10 mM is a characteristic of the melibiose carrier that has not been explained. In the presence of 10 mM Li+, the wild-type carrier accumulated 87-fold. In the absence of added cation (proton-coupled transport), the carrier was able to accumulate melibiose only sixfold.
TABLE 5.
Steady-state melibiose accumulation
| melB strain | Melibiose accumulation valuea in the presence of:
|
|||
|---|---|---|---|---|
| H+ | 10 mM Na+ | 100 mM Na+ | 10 mM Li+ | |
| Wild-type | 6 ± 1.4 | 153 ± 21 | 51 ± 5 | 87 ± 4 |
| R52K | 4 ± 1 | 127 ± 14 | 97 ± 9 | 115 ± 6 |
| R52Q | 0.4 ± 0.1 | 5 ± 1 | 2 ± 0.2 | 5 ± 1 |
| R52V | 0.2 ± 0.1 | 0.8 ± 0.2 | 1 ± 0.4 | 0.2 ± 0.2 |
| R52S | 0.3 ± 0.1 | 9 ± 1 | 1 ± 0.2 | 2 ± 0.2 |
| R52S W116R | 1 ± 0.4 | 23 ± 4 | 41 ± 5 | 10 ± 2 |
| R52S S247R | 0.6 ± 0.1 | 24 ± 5 | 8 ± 0.5 | 39 ± 6 |
| R52S N248K | 1 ± 0.8 | 69 ± 5 | 28 ± 1 | 66 ± 9 |
| R52V T338R | 0.7 ± 0.2 | 3 ± 0.5 | 10 ± 1 | 4 ± 0.7 |
| R52S D19G | 0.2 ± 0.3 | 2 ± 0.6 | 5 ± 0.6 | 2 ± 0.1 |
| R52S D55N | 0.8 ± 0.2 | 6 ± 1 | 19 ± 2 | 1 ± 0.2 |
| R52S P60Q | 0.5 ± 0.2 | 4 ± 0.3 | 1 ± 0.1 | 2 ± 0.4 |
| R52S N244S | 0.5 ± 0.2 | 8 ± 2 | 2 ± 0.1 | 4 ± 0.8 |
| R52S I352V | 0.4 ± 0.2 | 7 ± 1 | 1 ± 0.1 | 5 ± 0.5 |
The concentration of radiolabeled melibiose in transport assays was 0.2 mM, and values given represent transport at 15 min. Each value is given as the ratio of intracellular melibiose concentration to the extracellular melibiose concentration. Transport values were calculated assuming 0.4 μl of intracellular water for every 109 cells. The data are averages of at least two separate experiments with duplicate determinations in each experiment ± standard errors.
The replacement of Arg-52 with a polar or nonpolar amino acid dramatically reduced the carrier’s ability to accumulate melibiose. The R52V strain was unable to support uphill transport under any of the conditions tested. The R52S and R52Q strains accumulated melibiose to a small but significant extent in the presence of 10 mM NaCl (nine- and fivefold, respectively). The R52K revertant showed accumulation similar to that of the wild type in the presence of protons and 10 mM Na+ and better accumulation than the wild type in the presence of 100 mM Na+ and 10 mM Li+.
In the case of the second-site revertants, we observed no recovery of proton-coupled melibiose accumulation. However, we could show uphill transport activity for revertant strains in the presence of Na+ and Li+. For revertants of the R52S mutant where a positively charged residue was added (the R52S W116R, R52S N248K, and R52S S247R revertants), 10 mM Na+ or Li+ stimulated good accumulation (Table 5). The R52S N248K revertant showed the highest accumulation at 69-fold. Interestingly, different cationic conditions stimulated optimal accumulation for the R52S W116R and R52S N247R revertants. The R52S W116R strain was stimulated maximally by 100 mM Na+, while the R52S S247R strain gave the best uptake with 10 mM Li+. The R52V T338R revertant also required 100 mM Na+ for optimal stimulation of uphill transport.
In the revertants where a negative charge was removed (the R52S D19G and R52S D55N revertants), 100 mM Na+ was needed for optimal melibiose accumulation. The R52S D55N strain accumulated 19-fold, while the R52S D19G strain accumulated only 5-fold. The three revertants which had neutral residues substituted for other neutral residues (the R52S P60Q, R52S N244S, and R52S I352V revertants) had little or no accumulation activity above that of the R52S parental mutant under all conditions tested.
Melibiose-stimulated cation transport.
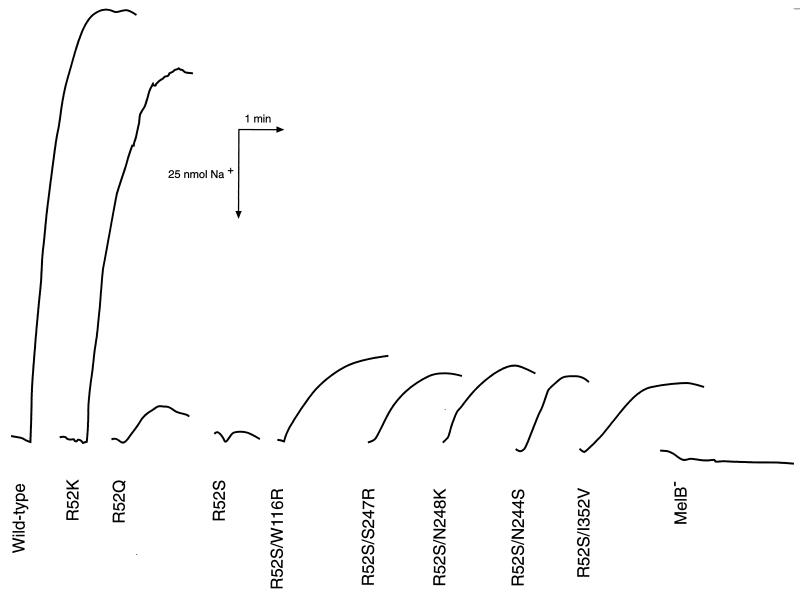
In addition to uphill transport assays, the coupling of cation and sugar influx can be tested by assaying cation transport directly. The coupled transport of cation and sugar causes a measurable decrease in the extracellular concentration of cation (H+ or Na+) which can be detected with pH- or Na+-sensitive electrodes. In order to assay the Na+-coupled transport reaction, we tested E. coli DW1 cells expressing mutant or revertant MelB carriers for melibiose-stimulated Na+ transport. When the wild-type melibiose carrier was exposed to 5 mM melibiose, a large, rapid, inwardly directed movement of Na+ was indicated by an upward deflection of the chart recording (Fig. 2). In contrast, the R52S mutant had a small response that was on the border of detection for this assay, and the R52V mutant was indistinguishable from cells lacking a melibiose carrier (not shown). The R52Q mutant gave a small deflection, indicating Na+ influx, and the R52K revertant was able to efficiently transport Na+, producing a deflection similar to that found for the wild type. Five revertants showed partial activity for Na+-coupled transport under the conditions tested. All of the revertants which reside on transmembrane helix VII (R52S N244S, R52S S247R, and R52S N248K), the R52S W116R revertant of helix IV, and the R52S I352V revertant in loop X-XI had similar sodium uptake (approximately 25% of wild type). The remaining revertants did not have demonstrable melibiose-stimulated Na+ influx.
FIG. 2.
Melibiose-stimulated Na+ transport. Melibiose was added to a final concentration of 5 mM, and the change in extracellular Na+ was monitored with a sodium-selective electrode and chart recorder. An upward deflection in the chart recording indicates sugar-stimulated Na+ uptake into the cell.
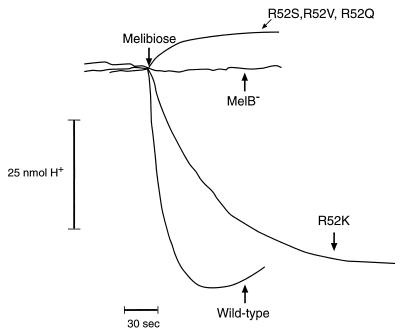
Using a pH electrode, we also measured proton coupling to sugar transport as an alkalinization of the external medium following the addition of melibiose to an anaerobic cell suspension. Upon the addition of melibiose, the wild-type carrier showed a rapid influx of protons, indicated by a downward deflection in the chart recording, due to the obligatory coupling of H+ and sugar during the transport reaction (Fig. 3). In contrast, the R52S, R52V, and R52Q strains showed no proton uptake. However, the R52K revertant had melibiose-stimulated proton uptake that was similar to that found for the wild-type carrier. In agreement with the in vitro transport data, none of the isolated second-site revertant strains showed proton uptake in these experiments.
FIG. 3.
Melibiose-stimulated proton transport. Melibiose was added to a final concentration of 10 mM, and the change in extracellular pH was monitored with a pH electrode and chart recorder. A downward deflection indicates alkalinization of the extracellular medium caused by sugar-stimulated proton uptake into the cell. The R52S, R52V, and R52Q mutant strains gave similar results in this experiment and are all represented by a single tracing.
DISCUSSION
The relationship between structure and function in membrane transport proteins often focuses on charged amino acids that reside on membrane-spanning α-helices. The presence of certain charged residues within the hydrophobic environment of a membrane domain has been found to influence substrate recognition. For example, in the lactose permease of E. coli, Glu-325 is critical for efficient proton coupling (9, 13), and recent studies suggest that Glu-126 and Arg-144 are required for substrate binding (12, 40). In addition, two distinct models of lactose permease function rely heavily on the participation of charged residues within membrane helices (13, 19). Charged residues in membrane domains have also been found to interact through interhelical ion pairs or salt bridges. Evidence for a salt bridge has been presented for bacteriorhodopsin (43), the voltage-gated Na+ channel (5), the H+ ATPase of Saccharomyces cerevisiae (41), CFTR (7), the lactose carrier of E. coli (21, 23, 24, 39), and a vesicular monoamine transporter (28). The location of interhelical salt bridges can help to define the three-dimensional structure of a membrane carrier by positioning membrane helices relative to one another. This type of information is useful for membrane proteins where attempts at crystallization have been unsatisfactory.
In the current study, we wanted to determine if Arg-52 is important for sugar transport by replacing Arg-52 with neutral amino acids and analyzing the transport properties of the mutant carriers. Arg-52 was targeted for two reasons, it resides on the same face of helix II as Asp-55 and Asp-59, both of which are important for Na+-stimulated sugar transport (35, 36, 53), and it is highly conserved within the galactoside-pentose-hexuronide protein family (32). In this study, we have shown that the replacement of Arg-52 with Ser, Gln, or Val leads to a dramatic reduction in melibiose transport activity without reducing the level of carrier protein in the membrane. These mutant carriers lacked proton-driven sugar accumulation and melibiose-stimulated proton uptake, while a small amount of Na+-stimulated melibiose transport activity and melibiose-stimulated Na+ uptake was detected for the R52S and R52Q strains. The R52V strain had only limited downhill transport that required high sodium (100 mM) and had no accumulation activity. A fourth substitution at position 52 (R52K) was isolated as a first-site revertant of the R52Q mutant where lysine was substituted for Arg-52. Characterization of this revertant showed that the R52K strain had transport similar to, and in some cases better than, the wild-type carrier. We conclude that while Arg-52 is not absolutely required for Na+-coupled transport, a positive charge is required at this position for coupling to the proton electrochemical gradient and for efficient cation-substrate cotransport. The reduction in transport activity in Arg-52 mutants may be attributed to several different problems. For example, Arg-52 could participate directly in cation and/or sugar binding. For sugar-binding proteins that have been crystallized (e.g., arabinose-binding protein [37]), arginine residues were found to make hydrogen-bonding contacts with sugar substrates. Alternatively, the positive charge might interact with nearby aspartate residues regulating the pKa of these side chains which participate in the coordination of cations. In the case of the melibiose carrier in which sodium binding increases carrier affinity for sugar substrates, disruption of the coordination site for cations could result in decreased transport activity.
The low transport activity of the R52S, R52V, and R52Q strains allowed for the isolation of second-site revertants that regained melibiose transport activity. MacConkey indicator plates with relatively high concentrations of melibiose (12 mM) and sodium (86 mM) were used for isolation of functional revertants. The goal of the revertant isolation was to determine if Arg-52 was salt bridged to residues of opposite charge (e.g., Asp-55 and/or Asp-59). A second-site revertant in which a negative charge had been neutralized would provide evidence for such an interaction. Interestingly, in addition to revertants in which negative charges were removed, two other types of revertants were identified. These included substitution of positively charged residues for neutral residues and substitution of neutral residues for neutral residues. In all, nine distinct substitution sites were found, eight of which reside on transmembrane domains.
Two second-site revertants isolated from the R52S mutant, Asp-19→Gly (helix I) and Asp-55→Asn (helix II), provide evidence for participation of Arg-52 in a salt bridge. In agreement with a role for Na+ coordination for these Asp residues, each revertant required high Na+ (100 mM) for the best activity. Comparison of the apparent Km for melibiose downhill transport in the R52S mutant and the R52S D55N revertant show that the affinity for melibiose is poor in the revertant. However, an increase in the velocity of melibiose transport (threefold higher than that for the wild type) for this revertant allowed melibiose uptake. We believe that our data strongly indicate that an intrahelical salt bridge exists between Arg-52 and Asp-55. Evidence for intrahelical salt bridges has also been found in the lactose carrier in which His-322 and Glu-325 interact (25) and in the Pi-linked hexose phosphate antiporter of E. coli (UhpT), where an intrahelical ion pair between Asp-388 and Lys-391 serves as a determinant of substrate selectivity (14). The transport activity in the R52S D19G revertant was too low for accurate measurement of apparent affinity for melibiose. This was most likely due to the low expression levels for this revertant. However, it is interesting that even with poor expression, this revertant was able to give a melibiose transport-positive phenotype (pink colonies on melibiose-containing MacConkey medium), suggesting that neutralizing Asp-19 restores transport activity in the R52S mutant by removing an uncompensated negative charge. It is possible that Arg-52 interacts with both Asp-19 and Asp-55. In the lactose carrier of E. coli, there is evidence that Lys-319 interacts with both Asp-240 and Glu-269 (24). In addition, a survey of salt bridges in proteins suggests that one third of all salt bridges are of a complex nature where more than two charged residues are joined (29). An alternative explanation is that Arg-52 pairs with each Asp residue at different phases of the transport cycle. Although this idea is highly speculative for the melibiose carrier, a molecular mechanism has been proposed for the lactose permease that is dependent on Lys-319 shifting between salt bridge interactions with Glu-325, Asp-240, and Glu-269 (19).
A second type of revertant isolated from the R52S and R52V strains involved replacing neutral residues in helices IV, VII, and X with Arg or Lys. Substitution of a positively charged residue emphasizes the importance of Arg-52 and a particularly strong selection for such a charge within a membrane-spanning domain. It is tempting to speculate that these revertant amino acid substitutions are compensating for the loss of Arg-52 by placing a positively charged side chain in the space vacated by this residue. If this idea were correct, it would suggest that helices IV, VII, and X are close to helix II in the three-dimensional structure of the protein. The notion that helices IV and VII are close to helix II is supported by the in vitro transport data where the R52S W116R, R52S S247R, and R52S N248K revertants are found to have the highest recovery of melibiose accumulation in the presence of Na+. These revertants were also among those in which we could demonstrate melibiose-induced Na+ transport. In addition, the R52S W116R revertant has an apparent Km for melibiose that is lower than that of the R52S mutant, and the R52S N248K revertant had an apparent Km for melibiose similar to that of the wild type. While the Km in the R52S S247R strain was high, the Vmax for melibiose transport was much better than that measured for the R52S parental strain. It is possible that helix VII is close to helix II so that a revertant at position 247 is able to provide partial compensation for the loss of Arg-52, while Asn-248 is in better register with Arg-52 so that insertion of a positive charge here will give substantial compensation for the loss of Arg-52. The idea that helix IV is close to helix II is also supported by previous studies where we used a D55S mutant to select for functional revertants, and a G117D substitution was isolated (48). It was concluded that the G117D revertant regained transport activity by compensating for the lost negative charge in D55S. We suggest that Trp-116 is close to Arg-52 in the three-dimensional structure of the protein. The final second-site revertant in this category was the R52V T338R revertant, in which a compensatory positive charge is inserted on helix X. Although this revertant had low transport activity, it was able to accumulate melibiose in the presence of high sodium.
The third type of revertant identified involved replacing uncharged residues with other uncharged residues. Two of these revertants, the R52S P60Q (helix II) and R52S I352V (loop X-XI) revertants, have substitutions that are adjacent to highly conserved aspartate residues (Asp-59 and Asp-351). It is interesting that an I352V mutant was also found in a screen for melibiose mutants with diminished recognition for methyl-β-d-thiogalactopyranoside (TMG) (3). In that study (3), it was suggested that this loop may be near the sugar binding site and that mutations here alter sugar recognition. It is possible that the R52S I352V revertant alters the local environment of the highly conserved Asp-351, altering sugar transport. The Pro-60 residue is adjacent to Asp-59, which is critical for Na+ binding. It is possible that the Pro-60→Gln substitution alters the structure of helix II, putting Asp-55 and/or Asp-59 in a better position to interact with Na+ to restore partial activation of melibiose transport. The last revertant in this class, R52S N244S (helix VII), had the effect of restoring melibiose transport velocity (Vmax) to the wild-type level (Table 4).
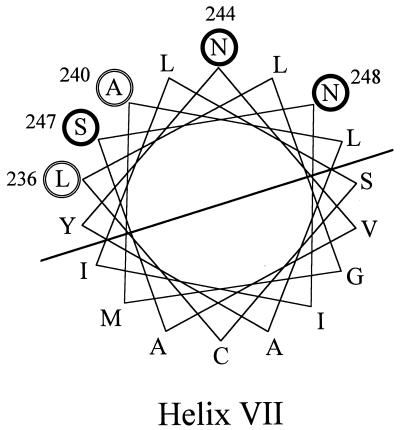
Interestingly, three of the isolated revertants reside on helix VII, tow of which introduced a positive charge. Two previous studies found substitution on helix VII. Mutants that are resistant to the inhibitory effect of a high Li+ concentration include Leu-236→Phe and Ala-240→Thr or Val mutants (20). These mutants also lost the capacity to couple H+ and sugar transport. The isolation of TMG-resistant mutants also identified Ala-240→Thr or Val substitutions (3). Thus, substitution of amino acids on helix VII modifies both sugar and cation transport properties in the melibiose carrier. A helical wheel model (Fig. 4) shows that substitutions found on helix VII define one side of that helix. We suggest that helix VII is important for sugar and cation transport and that it is close to helix II in the three-dimensional structure of the carrier.
FIG. 4.
Helix VII of the melibiose carrier. Residues substituted in this study (thick circles) and residues from previous studies that affect cation and/or sugar recognition (double circles) are indicated.
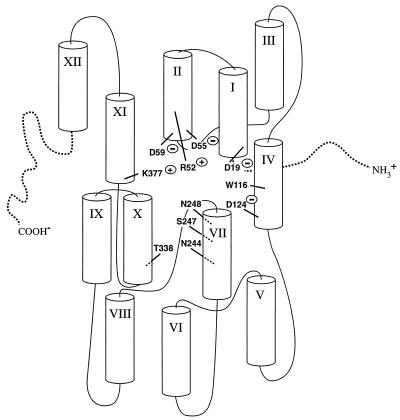
In the absence of direct physical evidence for the three-dimensional structure of the melibiose carrier, site-directed mutagenesis combined with revertant isolation can provide information concerning residues important for substrate recognition as well as the positioning of membrane helices relative to one another. Our characterization of substitutions for Arg-52 provide evidence that a positive charge at position 52 is important for efficient Na+-stimulated melibiose transport and is critical for H+-coupled melibiose transport. Isolation of functional revertants suggests that Arg-52 is salt bridged to Asp-55 and Asp-19. In addition, revertants with insertion of positive charges on transmembrane domains IV, VII, and X provide evidence for a helical packing arrangement that puts these transmembrane domains close to helix II in the three-dimensional structure of the protein. Figure 5 provides a hypothetical arrangement of helices in the melibiose carrier. This figure is meant to emphasize the positions of revertants of Arg-52 isolated in this study and of other charged residues important for carrier activity. Helix XI is included, because evidence has been presented to suggest that it is close to helix IV (51) and we have found evidence that Lys-377 (helix XI) is salt bridged to Asp-59 (helix II) (10). Information from this study will be used to target specific areas of the protein for biochemical studies in the cysteine-less melibiose carrier, including cysteine-scanning mutagenesis of helix IV and VII as well as engineering of double cysteine mutants for chemical cross-linking to verify the proximity of specific helices.
FIG. 5.
Hypothetical arrangement of the membrane helices in the melibiose carrier.
ACKNOWLEDGMENTS
This work was supported in part by NIH grant DK05736.
We thank Anupam B. Jena for technical assistance and Leena Karttunen for helpful suggestions during the preparation of this manuscript.
REFERENCES
- 1.Botfield M C, Naguchi K, Tsuchiya T, Wilson T H. Membrane topology of the melibiose carrier of Escherichia coli. J Biol Chem. 1992;267:1818–1822. [PubMed] [Google Scholar]
- 2.Botfield M C, Wilson T H. Carboxyl-terminal truncations of the melibiose carrier of Escherichia coli. J Biol Chem. 1989;264:11643–11648. [PubMed] [Google Scholar]
- 3.Botfield M C, Wilson T H. Mutations that simultaneously alter both sugar and cation specificity in the melibiose carrier of Escherichia coli. J Biol Chem. 1988;263:12909–12915. [PubMed] [Google Scholar]
- 4.Botfield M C, Wilson T H. Peptide-specific antibody for the melibiose carrier of Escherichia coli localizes the carboxyl terminus to the cytoplasmic face of the membrane. J Biol Chem. 1989;264:11649–11652. [PubMed] [Google Scholar]
- 5.Catterall W A. Structure and function of voltage-sensitive ion channels. Science. 1988;242:50–61. doi: 10.1126/science.2459775. [DOI] [PubMed] [Google Scholar]
- 6.Cohn D E, Kaback H R. Mechanism of the melibiose porter in membrane vesicles of Escherichia coli. Biochemistry. 1980;19:4237–4243. doi: 10.1021/bi00559a015. [DOI] [PubMed] [Google Scholar]
- 7.Cotten J F, Welsh M J. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR: evidence for disruption of a salt bridge. J Biol Chem. 1999;274:5429–5435. doi: 10.1074/jbc.274.9.5429. [DOI] [PubMed] [Google Scholar]
- 8.Damiano-Forano E, Bassilana M, Leblanc G. Sugar binding properties of the melibiose permease in Escherichia coli membrane vesicles. Effects of Na+ and H+ concentrations. J Biol Chem. 1986;261:6893–6899. [PubMed] [Google Scholar]
- 9.Franco P J, Brooker R J. Functional roles of Glu-269 and Glu-325 within the lactose permease of Escherichia coli. J Biol Chem. 1994;269:7379–7386. [PubMed] [Google Scholar]
- 10.Franco, P. J., A. B. Jena, and T. H. Wilson. 1999. Unpublished data.
- 11.Franco P J, Wilson T H. Alteration of Na+-coupled transport in site-directed mutants of melibiose carrier of Escherichia coli. Biochim Biophys Acta. 1996;1282:240–248. doi: 10.1016/0005-2736(96)00062-4. [DOI] [PubMed] [Google Scholar]
- 12.Frillingos S, Gonzalez A, Kaback H R. Cysteine-scanning mutagenesis of helix Iv and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 13.Frillingos S, Sahintoth M, Wu J H, Kaback H R. Cys-scanning mutagenesis: a novel approach to structure-function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 14.Hall J A, Fann M C, Maloney P C. Altered substrate selectivity in a mutant of an intrahelical salt bridge in UhpT, the sugar phosphate carrier of Escherichia coli. J Biol Chem. 1999;274:6148–6153. doi: 10.1074/jbc.274.10.6148. [DOI] [PubMed] [Google Scholar]
- 15.Hama H, Wilson T H. Cation-coupling in chimeric melibiose carriers derived from Escherichia coli and Klebsiella pneumoniae. The amino-terminal portion is crucial for Na+ recognition in melibiose transport. J Biol Chem. 1993;268:10060–10065. [PubMed] [Google Scholar]
- 16.Hama H, Wilson T H. Primary structure and characteristics of the melibiose carrier of Klebsiella pneumoniae. J Biol Chem. 1992;267:18371–18376. [PubMed] [Google Scholar]
- 17.Hama H, Wilson T H. Replacement of alanine 58 by asparagine enables the melibiose carrier of Klebsiella pneumoniae to couple sugar transport to Na+ J Biol Chem. 1994;269:1063–1067. [PubMed] [Google Scholar]
- 18.Hanatani M, Yazyu H, Shiota-Niiya S, Moriyama Y, Kanazawa H, Futai M, Tsuchiya T. Physical and genetic characterization of the melibiose operon and identification of the gene products in Escherichia coli. J Biol Chem. 1984;259:1807–1812. [PubMed] [Google Scholar]
- 19.Johnson J L, Brooker R J. A K319N/E325Q double mutant of the lactose permease cotransports H+ with lactose: implications for a proposed mechanism of H+ lactose symport. J Biol Chem. 1999;274:4074–4081. doi: 10.1074/jbc.274.7.4074. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami T, Akizawa Y, Ishikawa T, Shimamoto T, Tsuda M, Tsuchiya T. Amino acid substitutions and alteration in cation specificity in the melibiose carrier of Escherichia coli. J Biol Chem. 1988;263:14276–14280. [PubMed] [Google Scholar]
- 21.King S C, Hansen C L, Wilson T H. The interaction between aspartic acid 237 and lysine 358 in the lactose carrier of Escherichia coli. Biochim Biophys Acta. 1991;1062:177–186. doi: 10.1016/0005-2736(91)90390-t. [DOI] [PubMed] [Google Scholar]
- 22.Leblanc G, Pourcher T, Zani M-L. The melibiose permease of Escherichia coli: importance of the NH2-terminal domains for cation recognition by the Na+/sugar cotransporter. In: Reuss L, Russell J M, Jennings M L, editors. Molecular biology and function of carrier proteins. New York, N.Y: The Rockefeller University Press; 1993. pp. 213–227. [PubMed] [Google Scholar]
- 23.Lee J I, Hwang P P, Hansen C, Wilson T H. Possible salt bridges between transmembrane alpha-helices of the lactose carrier of Escherichia coli. J Biol Chem. 1992;267:20758–20764. [PubMed] [Google Scholar]
- 24.Lee J I, Hwang P P, Wilson T H. Lysine 319 interacts with both glutamic acid 269 and aspartic acid 240 in the lactose carrier of Escherichia coli. J Biol Chem. 1993;268:20007–20015. [PubMed] [Google Scholar]
- 25.Lee J I, Varela M F, Wilson T H. Physiological evidence for an interaction between Glu-325 and His-322 in the lactose carrier of Escherichia coli. Biochim Biophys Acta. 1996;1278:111–118. doi: 10.1016/0005-2736(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 26.Lolkema J S, Puttner I B, Kaback H R. Site-directed mutagenesis of Pro327 in the lac permease of Escherichia coli. Biochemistry. 1988;27:8307–8310. doi: 10.1021/bi00422a003. [DOI] [PubMed] [Google Scholar]
- 27.Maloney P C. Bacterial transporters. Curr Opin Cell Biol. 1994;6:571–582. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 28.Merickel A, Kaback H R, Edwards R H. Charged residues in transmembrane domains Ii and Xi of a vesicular monoamine transporter form a charge pair that promotes high affinity substrate recognition. J Biol Chem. 1997;272:5403–5408. doi: 10.1074/jbc.272.9.5403. [DOI] [PubMed] [Google Scholar]
- 29.Musafia B, Buchner V, Arad D. Complex salt bridges in proteins: statistical analysis of structure and function. J Mol Biol. 1995;254:761–770. doi: 10.1006/jmbi.1995.0653. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki N, Kuroda M, Shimamoto T, Tsuchiya T. Characteristics of the melibiose transporter and its primary structure in Enterobacter aerogenes. Biochim Biophys Acta. 1997;1326:83–91. doi: 10.1016/s0005-2736(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 31.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poolman B, Knol J, van der Does C, Henderson P J, Liang W J, Leblanc G, Pourcher T, Mus-Veteau I. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol Microbiol. 1996;19:911–922. doi: 10.1046/j.1365-2958.1996.397949.x. [DOI] [PubMed] [Google Scholar]
- 33.Poolman B, Konings W N. Secondary solute transport in bacteria. Biochim Biophys Acta. 1993;1183:5–39. doi: 10.1016/0005-2728(93)90003-x. [DOI] [PubMed] [Google Scholar]
- 34.Pourcher T, Bibi E, Kaback H R, Leblanc G. Membrane topology of the melibiose permease of Escherichia coli studied by melB-phoA fusion analysis. Biochemistry. 1996;35:4161–4168. doi: 10.1021/bi9527496. [DOI] [PubMed] [Google Scholar]
- 35.Pourcher T, Deckert M, Bassilana M, Leblanc G. Melibiose permease of Escherichia coli: mutation of aspartic acid 55 in putative helix II abolishes activation of sugar binding by Na+ ions. Biochem Biophys Res Commun. 1991;178:1176–1181. doi: 10.1016/0006-291x(91)91016-6. [DOI] [PubMed] [Google Scholar]
- 36.Pourcher T, Zani M L, Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. I. Effect on Na+-dependent transport and binding properties. J Biol Chem. 1993;268:3209–3215. [PubMed] [Google Scholar]
- 37.Quiocho F A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- 38.Reizer J, Reizer A, Saier M H. A functional superfamily of sodium/solute symporters. Biochim Biophys Acta Rev. 1994;1197:133–166. doi: 10.1016/0304-4157(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 39.Sahin-Toth M, Dunten R L, Gonzalez A, Kaback H R. Functional interactions between putative intramembrane charged residues in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10547–10551. doi: 10.1073/pnas.89.21.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahin-Toth M, le Coutre J, Kharabi D, le Maire G, Lee J C, Kaback H R. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38:813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 41.Sen Gupta S, DeWitt N D, Allen K E, Slayman C W. Evidence for a salt bridge between transmembrane segments 5 and 6 of the yeast plasma-membrane H+-ATPase. J Biol Chem. 1998;273:34328–34334. doi: 10.1074/jbc.273.51.34328. [DOI] [PubMed] [Google Scholar]
- 42.Shiota S, Yamane Y, Futai M, Tsuchiya T. Escherichia coli mutants possessing an Li+-resistant melibiose carrier. J Bacteriol. 1985;162:106–109. doi: 10.1128/jb.162.1.106-109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern L J, Khorana H G. Structure-function studies on bacteriorhodopsin. X. Individual substitutions of arginine residues by glutamine affect chromophore formation, photocycle, and proton translocation. J Biol Chem. 1989;264:14202–14208. [PubMed] [Google Scholar]
- 44.Tsuchiya T, Oho M, Shiota-Niiya S. Lithium ion-sugar cotransport via the melibiose transport system in Escherichia coli. Measurement of Li+ transport and specificity. J Biol Chem. 1983;258:12765–12767. [PubMed] [Google Scholar]
- 45.Tsuchiya T, Raven J, Wilson T H. Co-transport of Na+ and methyl-beta-D-thiogalactopyranoside mediated by the melibiose transport system of Escherichia coli. Biochem Biophys Res Commun. 1977;76:26–31. doi: 10.1016/0006-291x(77)91663-1. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchiya T, Wilson T H. Cation-sugar cotransport in the melibiose transport system of Escherichia coli. Membr Biochem. 1978;2:63–79. doi: 10.3109/09687687809063858. [DOI] [PubMed] [Google Scholar]
- 47.West I C. Lactose transport coupled to proton movements in Escherichia coli. Biochem Biophys Res Commun. 1970;41:655–661. doi: 10.1016/0006-291x(70)90063-x. [DOI] [PubMed] [Google Scholar]
- 48.Wilson D M, Hama H, Wilson T H. GLY113→ASP can restore activity to the ASP51→SER mutant in the melibiose carrier of Escherichia coli. Biochem Biophys Res Commun. 1995;209:242–249. doi: 10.1006/bbrc.1995.1495. [DOI] [PubMed] [Google Scholar]
- 49.Wilson D M, Tsuchiya T, Wilson T H. Methods for the study of the melibiose carrier of Escherichia coli. Methods Enzymol. 1986;125:377–387. doi: 10.1016/s0076-6879(86)25032-6. [DOI] [PubMed] [Google Scholar]
- 50.Wilson D M, Wilson T H. Cation specificity for sugar substrates of the melibiose carrier in Escherichia coli. Biochim Biophys Acta. 1987;904:191–200. doi: 10.1016/0005-2736(87)90368-3. [DOI] [PubMed] [Google Scholar]
- 51.Wilson T H, Wilson D M. Evidence for a close association between helix Iv and helix Xi in the melibiose carrier of Escherichia coli. Biochim Biophys Acta Biomembr. 1998;1374:77–82. doi: 10.1016/s0005-2736(98)00132-1. [DOI] [PubMed] [Google Scholar]
- 52.Yazyu H, Shiota-Niiya S, Shimamoto T, Kanazawa H, Futai M, Tsuchiya T. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1984;259:4320–4326. [PubMed] [Google Scholar]
- 53.Zani M L, Pourcher T, Leblanc G. Mutagenesis of acidic residues in putative membrane-spanning segments of the melibiose permease of Escherichia coli. II. Effect on cationic selectivity and coupling properties. J Biol Chem. 1993;268:3216–3221. [PubMed] [Google Scholar]
- 54.Zani M L, Pourcher T, Leblanc G. Mutation of polar and charged residues in the hydrophobic NH2-terminal domains of the melibiose permease of Escherichia coli. J Biol Chem. 1994;269:24883–24889. [PubMed] [Google Scholar]