Abstract
Yersinia enterocolitica is well equipped for siderophore piracy, encompassing the utilization of siderophores such as ferrioxamine, ferrichrome, and ferrienterochelin. In this study, we report on the molecular and functional characterization of the Yersinia fep-fes gene cluster orthologous to the Escherichia coli ferrienterochelin transport genes (fepA, fepDGC, and fepB) and the esterase gene fes. In vitro transcription-translation analysis identified polypeptides of 30 and 35 kDa encoded by fepC and fes, respectively. A frameshift mutation within the fepA gene led to expression of a truncated polypeptide of 40 kDa. The fepD, fepG, and fes genes of Y. enterocolitica were shown to complement corresponding E. coli mutants. Insertional mutagenesis of fepD or fes genes abrogates enterochelin-supported growth of Y. enterocolitica on iron-chelated media. In contrast to E. coli, the fep-fes gene cluster in Y. enterocolitica consists solely of genes required for uptake and utilization of enterochelin (fep) and not of enterochelin synthesis genes such as entF. By Southern hybridization, fepDGC and fes sequences could be detected in Y. enterocolitica biotypes IB, IA, and II but not in biotype IV strains, Yersinia pestis, and Yersinia pseudotuberculosis strains. According to sequence alignment data and the coherent structure of the Yersinia fep-fes gene cluster, we suggest early genetic divergence of ferrienterochelin uptake determinants among species of the family Enterobacteriaceae.
Acquisition of iron is essential for growth and survival of microorganisms. However, in neutral aqueous systems under aerobic conditions ferric iron (Fe3+) forms insoluble oxyhydroxide compounds, resulting in extremely low concentrations of free Fe3+. Similar conditions are met by microorganisms colonizing or invading vertebrate hosts where iron is tightly bound to host proteins such as transferrin, lactoferrin, or heme-containing proteins. For survival and multiplication in such an iron-restricted environment, facultative anaerobic microorganisms have developed high-affinity ferric iron uptake systems. Many cope with iron-deficient growth conditions by releasing small iron-chelating molecules termed siderophores, which subsequently can be taken up as ferric siderophores by specific transport systems (16, 33).
A large number of structurally diverse siderophores, which can be divided into three distinct major chemical classes, catecholates, hydroxamates, and heterocyclic compounds (e.g., pyochelin and yersiniabactin) (32), have been described.
Among the members of the family Enterobacteriaceae, the prototype of the catecholate siderophores, enterochelin (enterobactin), is widely distributed. Genes for enterochelin biosynthesis (ent), transport (fep), and the release of iron (ferric enterochelin esterase [fes]) are clustered. This enterochelin locus is about 20 kb in length and found in the genomes of Escherichia coli, Salmonella enterica, and Shigella species (15, 49, 58). The hydroxamate siderophore aerobactin is distributed with lower frequency among these three enterobacteria.
In contrast, pathogenic Yersinia species (Yersinia pestis, Yersinia pseudotuberculosis, and Yersinia enterocolitica) do not produce catecholate or hydroxamate siderophores (5, 37, 42). However, they are endowed with siderophore uptake systems for catecholate siderophores (enterochelin) and hydroxamate siderophores (e.g., ferrioxamine and ferrichrome) (37). Moreover, the mouse-virulent (high-pathogenic) Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica biogroup 1B strains are able to produce and utilize the unique heterocyclic siderophore yersiniabactin (Ybt) (6, 10, 19, 23, 24, 34, 38, 39). The yersiniabactin biosynthesis and transport genes are clustered within a 45-kb region of the genome referred to as a high-pathogenicity island (6, 17, 18, 21, 34, 38). In addition to the Ybt system, pathogenic yersiniae carry a gene cluster involved in the uptake of heme-containing compounds (56).
So far, little is known about catecholate siderophore uptake in yersiniae. A putative outer membrane receptor for ferric enterochelin of about 90 kDa has been detected by monoclonal antibodies raised against E. coli ferric enterochelin receptor FepA (45). Moreover, a 60-kDa outer membrane protein has been identified as the receptor for a catechol-cephalosporin antibiotic (CccA) (5). In E. coli, catecholate siderophores are transported through the cytoplasmic membrane by means of an ATP binding cassette (ABC) transporter system (FepDGC) (12, 13). In the cytoplasm, ferric enterochelin is degraded to ferrous iron (Fe2+) and 2,3-dihydroxybenzoyl serine derivatives by the esterase Fes. According to this observation, functional genes orthologous to fes and fepDGC may be present for the utilization of catecholate siderophores in yersiniae.
In order to identify genes involved in enterochelin uptake in yersiniae, we screened a genomic library of Y. enterocolitica serotype O8 for complementation of an E. coli fes mutant. We were able to identify a Yersinia gene cluster consisting of a set of genes (fepB, fepDGC, and fes) that reveal high identity to the corresponding genes of the enterochelin gene cluster of E. coli. In contrast to E. coli, the enterochelin locus of yersiniae does not carry genes involved in enterochelin biosynthesis (ent).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. E. coli strains, except for the strains harboring the pGP1-2 plasmid, were grown in Luria broth (LB) or on LB agar plates at 37°C. Yersinia strains and E. coli strains harboring the pGP1-2 plasmid were cultivated at 28°C in the same medium (3). Blood agar plates were used for conjugation experiments. Antibiotics, when required, were included in the culture media at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 30 μg/ml; and tetracycline, 15 μg/ml. Siderophore production was demonstrated on siderophore indicator colorimetric chromeazurol S (CAS) agar (50). For iron-deficient growth, strains were grown in NBD medium (nutrient broth plus 5 g of NaCl per liter and 200 μM 2,2′-dipyridyl). Siderophore feeding assays were performed as described elsewhere (24, 37, 43). With the E. coli aroB mutant strain AN272, the addition of 2,3-dihydroxybenzoic acid (2,3-DHBA; Sigma Chemical Co., St. Louis, Mo.) to a final concentration of 10 μg/ml was required.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Y. enterocolitica | ||
| WA-314 | Clinical isolate harboring the virulence plasmid pYVO8; Ybt+ | 23 |
| WA-314 fes | fes mutant derived from WA-314; Ybt+ | This study |
| WA-C | Plasmidless derivative of strain WA-314; Ybt+ | 23 |
| WA-Str | Streptomycin-resistant derivative of WA-C; Ybt+ | This study |
| Y1852 | Derivative of WA-C; fur Ybt+ | 24 |
| WA-fepD587::Tn552kan | Derivative of WA-C; Tn552kan insertion in fepD gene; Ybt+ Smr | This study |
| WA-fes625::Tn552kan | Derivative of WA-C; Tn552kan insertion in fes gene; Ybt+ Smr | This study |
| Y-108-P | Y. enterocolitica serotype O3; clinical isolate; Ybt− | 23 |
| Y-NF | Y. enterocolitica serotype O5; clinical isolate; Ybt− | 23 |
| Y-5,27 | Y. enterocolitica serotype O5.27; clinical isolate; Ybt− | 23 |
| Y-96-P | Y. enterocolitica serotype O9; clinical isolate; Ybt− | 23 |
| Y-O13 | Y. enterocolitica serotype O13; clinical isolate; Ybt+ | 23 |
| Y-O20 | Y. enterocolitica serotype O20; clinical isolate; Ybt+ | 23 |
| Y. pseudotuberculosis | ||
| Y-P-I | Clinical isolate; serotype 1; Ybt+ | 23 |
| Y-P-II | Clinical isolate; serotype 2; Ybt− | 23 |
| Y-P-III | Clinical isolate; serotype 3; Ybt− | 20 |
| Y. pestis | ||
| KUMA | Pgm+ pYV− pPCP1 Ybt+ | 34 |
| EV 76 | Pgm− pYV+ pPCP1 Ybt− | 34 |
| Other Yersinia spp. | ||
| Y. frederiksenii | 23 | |
| Y. intermedia | 23 | |
| Y. kristensenii | 23 | |
| Escherichia coli | ||
| DH5α | endA1 hsdR17 (rK−mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | 22 |
| HB101 | Δ(gpt-proA)62 leuB6 thi-1 lacY1 hsdSB20 recA rps λ20(Strr) ara-14 galK2 xyl-5 mtl-1 supE44 mcrBB | 9 |
| WM1576 | thr leu lacY thi supE hsdR fhuA trxA (pGP1-2) | 57 |
| AN272 | aroB fes proA argE pheA tyr trp | 29 |
| AB1515.718 | purE42 proC14 leu-6 trpE38 thi-1 fhuA23 lacY1 (fepD::Tn5) | 12 |
| AB1515.764 | purE42 proC14 leu-6 trpE38 thi-1 fhuA23 lacY1 (fepG::Tn5) | 12 |
| AB1515.199 | purE42 proC14 leu-6 trpE38 thi-1 fhuA23 lacY1 (fepC::Tn5) | 12 |
| H1717 | aroB fhuF::λplac Mu | 55 |
| S17-1 λpir | pir+ tra+ Smr | 52 |
| Plasmids | ||
| pACYC184 | Cloning vector; Tcr Cmr | 11 |
| pBluescript KS(+) | Cloning vector; Apr | Stratagene |
| pT7-5 | Vector for T7 protein expression; Apr | 57 |
| pGP1-2 | Plasmid encoding T7 RNA polymerase; Kmr | 57 |
| pKAS-32 | Suicide vector; Apr | 53 |
| pUC19 | Cloning vector; Apr | |
| pAL101 | pUC19 carrying Tn552kan transposon (1.1-kb SpeI fragment); Kmr Apr | 30 |
| pSI10 | 16-kb Sau3AI WA-C chromosomal DNA in pACYC184; Cmr | This study |
| pB-1 and -2b | 6.0-kb BglII fragment of pSI10 in pBluescript KS(+); fes fepA* | This study |
| pH-1 and -2b | 7.5-kb HindIII fragment of pSI10 in pBluescript KS(+); fepDGC fes | This study |
| pH-1C | 3.5-kb ClaI/HindIII fragment of pH-1; fepDGC | This study |
| pHB3 | 3.3-kb BglII/HindIII fragment of pH-1; fepDGC | This study |
| pHB4 | 4.0-kb BglII/HindIII fragment of pH-1; fes | This study |
| pH-1fes::Tn552kan | pH-1 with Tn552kan insertion in fes gene; Kmr Apr | This study |
| pH-1fepD::Tn552kan | pH-1 with Tn552kan insertion in fepD gene; Kmr Apr | This study |
| pKASfes::Tn552kan | 5.3-kb EcoRI/KpnI fragment of pH-1fes::Tn552kan in pKAS-32; Apr | This study |
| pKASfepD::Tn552kan | 3.3-kb BclI fragment of pH-1fepD::Tn552kan in pKAS-32; Apr | This study |
Yersinia strains producing the siderophore yersiniabactin have been designated Ybt+, while those defective in yersiniabactin biosynthesis are Ybt−.
The inserts are cloned in both orientations.
Recombinant DNA methodology.
DNA was isolated, digested with restriction endonucleases, and ligated by standard methods (3, 48) according to the recommendations of the manufacturers (Boehringer Mannheim Biochemicals, Pharmacia LKB, and New England BioLabs Ltd.). DNA fragments less than 10 kb were recovered from agarose gels with the GeneClean II kit (Bio 101, Inc., La Jolla, Calif.). E. coli DH5α was transformed by the CaCl2 method (48), and Y. enterocolitica strains were transformed by electroporation (gene pulse apparatus; Bio-Rad Laboratories, Munich, Germany) according to the manufacturer’s instructions. To generate the genomic library of Y. enterocolitica WA-C, genomic DNA was isolated by the sodium dodecyl sulfate-proteinase K method (35) and partially digested with endonuclease Sau3AI. After electrophoretic separation, DNA fragments of 6 to 20 kb were isolated by electroelution, purified further by phenol and chloroform extraction, and ligated into the BamHI site of the pACYC184 vector (11). For hybridization, the restriction enzyme-digested genomic and plasmid DNA fragments were resolved through 0.8% agarose gels, and DNA was transferred to Zeta-Probe BT blotting membranes (Bio-Rad Laboratories) according to the method of Southern (54). After prehybridization at 68°C for 2 h and addition of heat-denatured probe, blots were incubated overnight at 68°C in the absence of formamide. The detection was performed with the ECF Random-Prime labelling and detection system (Amersham Pharmacia Biotech, Freiburg, Germany) according to the manufacturer’s instructions.
Protein analyses.
Protein expression was determined according to the procedure of Tabor and Richardson (57). For this, the 7.5-kb HindIII fragment as well as the 6.0-kb BglII fragment of the pSI10 plasmid was cloned in both orientations into the pBluescript KS(+) vector, generating the recombinant plasmids pH-1/pH-2 and pB-1/pB-2, respectively (Table 1 and Fig. 1). These plasmids were transformed into E. coli WM1576 carrying pGP1-2, a T7 RNA polymerase-encoding plasmid. Proteins were radiolabelled with 10 μCi of [35S]methionine (ICN Biomedicals GmbH, Eschwege, Germany), cells were treated at 100°C for 10 min in sample buffer (60 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate, 5% 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.001% bromophenol blue), and proteins were separated by electrophoresis on polyacrylamide gels (3, 28). Dried gels were exposed to Kodak Bio Max MR film at room temperature.
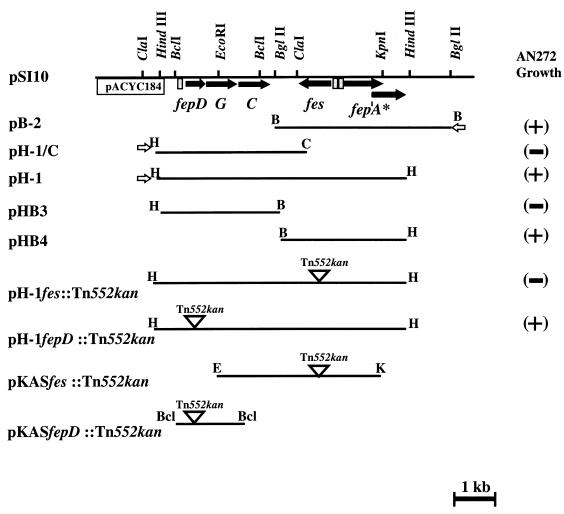
FIG. 1.
Restriction map of pSI10 and various subclones used for protein expression and testing for complementation of growth of E. coli AN272. (+), plasmids which promote AN272 growth; (−), no growth stimulation. Arrows indicate the direction of transcription of genes included in the enterochelin uptake locus. Open inverted triangles represent sites of Tn552kan insertion. Open arrows represent the location of the T7 promoter. Grey shaded boxes upstream of fepDCG, fes, and fepA* (frameshifted fepA) represent potential Fur boxes.
Nucleotide sequencing.
In order to facilitate sequencing of the fepA-, fepDGC-, and fes-homologous genes of Y. enterocolitica O8 strain WA-C, a series of subclones of the plasmids pH-1 and pH-2, containing progressively smaller portions of the original inserts, were obtained by combined exonuclease III-S1 nuclease treatment (nested deletion kit; Amersham). The generated sets of smaller fragments were purified with anion-exchange resin columns (Qiagen, Hilden, Germany), and the nucleotide sequences of both strands were determined by the TaqDyeDideoxy terminator method with the ABI model 373A DNA sequencer (Applied Biosystems, Weiterstadt, Germany). Sequence analysis was performed with ANALYSIS software (version 1.2.0; Applied Biosystems), MacDNASIS software (version 2.0; Hitachi Software Engineering Co., Tokyo, Japan), and DNAMAN sequence analysis software (Lynnon BioSoft, Vandrevil, Quebec, Canada). The nucleotide sequences were compared to those in SWISSPROT, Pir, and GenPept at the National Center for Biotechnology Information by using the program blastX and to GenBank and EMBL by using the program blastN (1).
Transposon (Tn552kan) mutagenesis of fes and fepDGC genes.
The pH-1 plasmid covering the fepD and fes genes (Fig. 1 and 2) was used as a target for in vitro transposon mutagenesis. Tn552kan carrying a kanamycin cassette (kindly provided by Tom Griffin, Yale University) was inserted into these genes, thereby generating plasmids pH-1fepD::Tn552kan and pH-1fes::Tn552kan, respectively (30, 44). The insertion of Tn552kan was verified by PCR and sequencing. Subcloning of DNA fragments carrying Tn552kan targeted fepD and fes into the suicide vector pKAS-32, which gave rise to plasmids pKASfepD::Tn552kan and pKASfes::Tn552kan, respectively (53). E. coli S17-1 λpir was used for propagation of all suicide vector constructs and as a donor for introduction of these constructs into Y. enterocolitica O8 strain WA-Str. Chromosomal DNA of mutants was routinely tested by Southern hybridization with suitable DNA probes to confirm insertional inactivation of genes of interest.
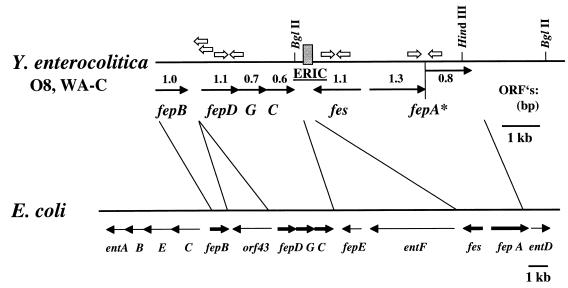
FIG. 2.
Comparison of the enterochelin gene cluster of Y. enterocolitica O8 strain WA-C to the E. coli gene cluster encoding proteins involved in biosynthesis and transport of enterochelin. Open arrows indicate the locations of primers used for RT-PCR and LA-PCR cloning procedures. The shaded box between fes and fepC represents the location of the ERIC sequence in Y. enterocolitica.
PCR analyses.
Genomic DNA (50 pg) or 1 μl of cells (104 to 105 CFU) of the Yersinia strains listed in Table 1 was used as a template in PCRs along with oligonucleotides (Metabion, Munich, Germany; Roth, Karlsruhe, Germany) as described below. The amplification mixtures consisted of either Taq DNA polymerase (ABI/Perkin-Elmer, Weiterstadt, Germany) or Pfu DNA polymerase (Stratagene, Heidelberg, Germany), 200 μM deoxynucleoside triphosphates, 1.5 μM MgCl2, and 0.4 μM primers. All PCRs were carried out in a GeneAmp PCR System 9700 thermal cycler (Perkin-Elmer). Products from these reactions were resolved by agarose gel electrophoresis. The initial denaturation step (94°C, 5 min) was followed by 30 cycles of denaturation (94°C, 1 min), annealing (annealing temperature [Tm], 1 min), and extension (72°C, 1 min) with one final extension step (72°C, 8 min). Sequences of the forward primers (FP) and reverse primers (RP) used for PCRs, the sizes of the amplified fragments (S), and the annealing temperatures (Tm) were as follows: (i) fepD.for (FP), 5′-GTG TGA TTG CCT TAC TAT TG-3′; fepD.rev (RP), 5′-CGG TCA TCC TTT TAT TAC GG-3′ (S, 397 bp; Tm, 55°C); (ii) fes.for (FP), 5′-GCC GGC AGG CAC AGC GTA AT-3′; fes.rev (RP), 5′-GGC CAA CCC ACC CAA AAC TT-3′ (S, 562 bp; Tm, 58°C); (iii) fepA.for (FP), 5′-TAC GCC AAA ATA CCT TAC GAT-3′; fepA.rev (RP), 5′-TGT AAA TAC ACC CCC ACC TGA-3′; (S, 438 bp; Tm, 56°C).
In order to determine the DNA region upstream of fepDGC (Fig. 2), the LA-PCR in vitro cloning technique (Takara Shuzo Co., Ltd., Kioto, Japan) was used. In brief, DNA linkers were ligated to chromosomal DNA of strain WA-C cleaved with EcoRI. Following ligation, a nested PCR was performed with primers derived from the linker together with primers derived from the fepD gene (S1.fepD, 5′-GGC GGG CTT CAT AGT GCG GTC ATC CTT TTA-3′; S2.fepD, 5′-GCC CGC CGC CCT GAG TTC CTA CCC AAT ACA-3′). The nucleotide sequence of a resulting 2-kb PCR fragment covering Yersinia fepB was determined.
Isolation of total RNA and RT-PCR.
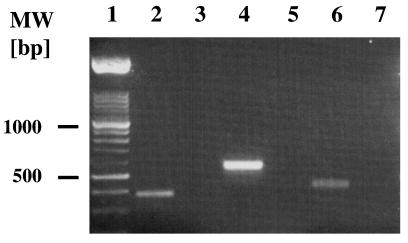
Y. enterocolitica WA-C was grown in LB medium at 26°C to an optical density at 600 nm (OD600) of 1.0. After centrifugation of 1 ml of the culture, the pellet was treated with the RNA Easy kit (Promega, Heidelberg, Germany) according to the manufacturer’s instructions. The RNA was dissolved in 50 μl of RNase-free water and stored at −70°C. For grown bacteria, several independent reverse transcriptase PCRs (RT-PCRs) were performed. As an initial step, contaminating DNA was digested by incubation with 2 U of RNase-free DNase (Promega) for 30 min at 48°C, DNase was heat inactivated at 70°C for 10 min, and the RNA concentration was determined spectrophotometrically. Ten nanograms of the total RNA was subsequently subjected to RT-PCR with primers listed above. For RT-PCR, the Access RT-PCR system (Promega) was used as recommended by the manufacturer. Controls consisting of reaction mixtures with RNA preparations without cDNA synthesis steps were tested with each primer pair. Products were analyzed by loading 20 μl of the PCR mixture into adjacent wells of a 1% agarose gel (Fig. 3).
FIG. 3.
Detection of the transcription of fepD-, fes-, and fepA-homologous genes of Y. enterocolitica by RT-PCR. Lane 1, DNA molecular size (MW) markers; lanes 2 and 3, fepD; lanes 4 and 5, fes; lanes 6 and 7, fepA-homologous gene. PCR mixtures in lanes 2, 4, and 6 contained RT, whereas mixtures in lanes 3, 5, and 7 lacked RT and served as negative controls.
FURTA.
The Fur titration assay (FURTA) was performed as described previously (55). In brief, plasmid pSI10 was double digested with the restriction enzymes HindIII and BglII. A 3.3-kb HindIII/BglII fragment, containing the putative Fur box of the fepDGC operon, and a 4.0-kb HindIII/BglII fragment, containing the Fur boxes of the fepA and fes genes, were subcloned in pBluescript KS(+) vector, resulting in plasmids pHB3 and pHB4, respectively (Fig. 1). These plasmids, along with the pBluescript KS(+) vector as negative control, were transformed into E. coli H1717. Transformants were plated on MacConkey agar and evaluated for Lac+ phenotype (55).
Isolation of enterochelin and enterochelin feeding bioassay.
Enterochelin was isolated as described by Langman et al. (29). Briefly, supernatant from the enterochelin-hyperproducing E. coli strain AN311 was acidified with H2SO4 and extracted with ethyl acetate. After washing with sodium phosphate buffer in order to remove enterochelin by-products such as 2,3-dihydroxybenzoyl serine, the enterochelin-containing ethyl acetate fraction was concentrated by rotary evaporation (crude enterochelin).
The strains to be tested were grown in NB medium (8 g of nutrient broth and 5 g of NaCl per 1 liter of distilled water) to an OD600 of 0.5. Thirty microliters of the bacteria was seeded in 10 ml of 0.6% H2O top agar on 1% NB agar, both containing the iron chelator 2,2′-dipyridyl at a concentration of 200 μM (24). Filter disks impregnated with 10 μl of a methanolic solution of enterochelin were used. The crude enterochelin solution was adjusted to an OD578 of 0.1 (37). The diameters of the zone of enhanced bacterial growth around the filter paper were determined after 24 h of culture at 26°C (Yersinia) and 37°C (E. coli).
Nucleotide sequence accession number.
The nucleotide sequences of the fes and fepDGC genes have been deposited in the GenBank database and assigned the accession no. U41370 and AF082879, respectively.
RESULTS
Identification of a Yersinia fes homologue by genetic complementation.
To identify a ferric enterochelin siderophore uptake system of Y. enterocolitica, a genomic library derived from Y. enterocolitica O8 strain WA-C was introduced into E. coli AN272 (fes aroB), a mutant defective in the synthesis of the enterochelin esterase Fes. The recombinant clones were selected for fes complementation on NBD medium containing DHBA. The addition of 2,3-DHBA was required for enterochelin biosynthesis of AN272. Ten representative colonies were chosen for plasmid extraction. All of these clones carried a unique plasmid designated pSI10 (Fig. 1). By using different combinations of restriction enzymes, a physical map of pSI10 was determined (Fig. 1). A 7.5-kb HindIII fragment of pSI10 was shown to restore the fes mutation of E. coli AN272.
Sequence analysis of the Y. enterocolitica O8 fes gene.
For further characterization, the 7.5-kb HindIII fragment was subcloned into the vector pBluescript KS(+) in both orientations and designated pH-1 and pH-2. Sequencing of the entire 7.5-kb HindIII fragment revealed a single open reading frame (ORF) of 1,059 bp, showing 54% identity to the fes gene of E. coli (Fig. 2). Upstream of this ORF, a putative promoter region comprising a sequence with 68% identity to the E. coli Fur-binding consensus sequence (FBS) was found (Table 2) (14). A putative Shine-Dalgarno sequence is located upstream of the initiating methionine codon. Moreover, an imperfect inverted repeat is located beyond the translational stop codon and may serve as a transcriptional terminator. The deduced amino acid sequence, encoded by the 1,059-bp ORF, was analyzed for homologous proteins in the Swiss-Prot database and was found to be highly homologous to the E. coli Fes sequence with 40% identity and 56% similarity over a stretch of 293 amino acids as well as 37% identity and 57% similarity to Fes of Erwinia chrysanthemi. The putative Yersinia Fes consists of 353 amino acids and has a calculated size of 39,837 Da.
TABLE 2.
Comparison of the Fur boxes from fes, fepDGC, and fepA of Y. enterocolitica O8 strain WA-C to the E. coli Fur box consensus sequence (FBS)
| Fur box | Sequencea | % Identity |
|---|---|---|
| Yersinia fes | 5′ GCAAATAATAATACTTCTC3′ | 68.4 |
| Yersinia fepA | 5′ GATAATAATAAATCTGGGT 3′ | 57.9 |
| Yersinia fepDGC | 5′ AATAATGATAATCAAAACT 3′ | 73.7 |
| Fur box consensus sequence | 5′ GATAATGATAATCATTATC 3′ |
Nucleotides identical to those in the Fur box consensus sequence are indicated by bold type.
A fepA homologue carrying a frameshift mutation.
Upstream of fes, two overlapping ORFs with a total size of 2,175 bp, exhibiting sequence identity to the 5′ and 3′ halves of the E. coli enterochelin receptor gene fepA, respectively, were identified. Upstream of the first ORF, a potential promoter was found, overlapped by a sequence revealing identity to the E. coli FBS (Table 2). From this, we presume a single-frameshift deletion within the initially intact fepA gene. The putative frameshift deletion in the fepA-homologous gene was confirmed by sequencing of PCR products of chromosomal DNA of Y. enterocolitica O8 strain WA-C.
In E. coli, the enterochelin gene cluster is terminated by the entD gene that is located immediately downstream of fepA (Fig. 2). The entD gene encodes an enzyme involved in the biosynthesis of enterochelin. In E. coli, the entD gene contains a high frequency of rare codons and is downstream of two repetitive extragenic palindromic sequences, suggesting chromosomal rearrangements in this region. In Y. enterocolitica, however, no entD-homologous gene is found at the corresponding position downstream of fepA.
Enterochelin transport proteins are encoded downstream of fes.
Downstream of fes, in an opposite orientation a set of three ORFs (Fig. 1 and 2) forming a putative Fur-regulated operon as indicated by a preceding FBS was identified (Table 2). The nucleotide sequences of these three ORFs were found to be homologous to the E. coli genes fepD, -G, and -C, which encode proteins involved in ferric enterochelin transport (Fig. 2). Accordingly, the deduced amino acid sequence of the polypeptides encoded by the Yersinia homologous fepDGC genes revealed striking similarity to the set of integral membrane proteins involved in iron transport through high-affinity periplasmic transport systems (Yersinia-E. coli FepD, 51% identity and 73% similarity over 315 amino acids; Yersinia-E. coli FepG, 65% identity and 78% similarity over 246 amino acids). Hydropathy profiles of the predicted proteins Yersinia FepD and Yersinia FepG indicate that these are extremely hydrophobic proteins with eight and six predicted transmembrane helices, respectively (31). The deduced amino acid sequence of the Yersinia FepC homologue shows a strong identity to highly conserved regions of peripheral membrane ATP-binding proteins. A consensus sequence has been determined for both the amino- and the carboxy-terminal regions of nucleotide-binding proteins (51), with the predicted Yersinia FepC sequence containing 11 of 12 amino acids of the first region and 10 of 11 of the second region, including the completely conserved GKS (Walker motif A) and DEP (Walker motif B) amino acid sequence motifs (25, 59). The presence of a consensus nucleotide-binding site suggests FepC-homologous function in Yersinia. In order to obtain sequence information on the region upstream of fepDGC which is not located on the recombinant plasmid pSI10, the LA-PCR cloning method was used. Thus, upstream of fepDGC we found an ORF homologous to fepB. The deduced putative polypeptide has 59% identity and 75% similarity with the periplasmic binding protein FepB of E. coli (Fig. 2). In contrast to the enterochelin cluster of E. coli, no orf43-homologous gene could be detected in Y. enterocolitica (Fig. 2).
An ERIC sequence is located within the fep-fes gene cluster of Y. enterocolitica O8.
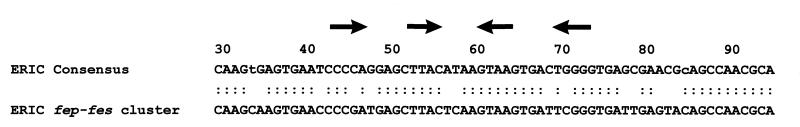
Sequencing of the DNA region between Yersinia fepC and fes revealed a 126-bp nucleotide sequence with extensive (81%) identity to the enterobacterial repetitive intergenic consensus (ERIC) sequence (27) (Fig. 2 and 4). It has 84% identity to another ERIC sequence of Y. enterocolitica O8 strain WA-314 found upstream of the ybtA gene of the yersiniabactin siderophore gene cluster (41). As is common for ERIC sequences, this element is located in an intergenic region and shares a core inverted repeat (Fig. 4) which could potentially form a stem-loop structure. The sequence does not resemble any known insertion sequence or transposable element. Interestingly, compared to the enterochelin gene cluster of E. coli, the ERIC sequence in the Y. enterocolitica chromosome is located at the position where orthologous entF and fepE genes are expected (Fig. 2). Thus, the presence of an ERIC sequence at this position may suggest that the biosynthesis genes have been deleted in Yersinia.
FIG. 4.
Comparison of the ERIC sequence from the Y. enterocolitica fes-fepDGC DNA region with the consensus (27). The core inverted repeats found in all ERIC sequences are indicated by the arrows.
Identification of polypeptides encoded by the fes-fep gene cluster of Y. enterocolitica O8.
The transcription of fepDGC as well as of Yersinia fes and fepA was confirmed by RT-PCR. Amplification products of expected sizes were obtained in those samples containing RT whereas samples without RT revealed no PCR product (Fig. 3). This indicated that all three genes were transcribed in Y. enterocolitica O8 strain WA-C.
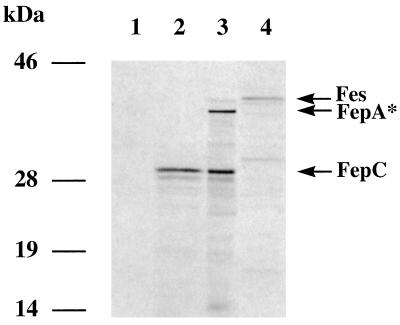
To identify polypeptides encoded by the Yersinia fes-fep gene cluster, the T7-based expression system of Tabor and Richardson was used (57). Appropriate restriction subfragments of the pSI10 plasmid were subcloned downstream of the T7 promoter of pBluescript KS(+), resulting in plasmids pH-1, pH-1/C, and pB-2, respectively (Fig. 1). For [35S]methionine labelling, these plasmids were introduced into E. coli WM1576 carrying the gene for T7 RNA polymerase. Expression of the fepDGC operon (pH-1C plasmid) resulted in a 30-kDa polypeptide. The molecular weight of this polypeptide corresponds closely to that of FepC of E. coli (51). It was not possible to detect FepD and FepG by T7 expression. This has already been reported for E. coli FepD and FepG and is probably due to the hydrophobic character of these proteins (51). The expression of genes from the entire 7.5-kb HindIII fragment (pH-1) reveals an additional polypeptide of about 35 kDa. This size corresponds closely to the predicted truncated polypeptide of the Yersinia FepA (Fig. 1 and 5). According to the orientation of T7 transcription, the fes gene product was not expressed by using pH-1 and pH-1C. Therefore, plasmid pB-2 was subjected to T7-based expression, revealing synthesis of a protein with an estimated molecular mass of 40 kDa, which is similar to the size of the known E. coli Fes protein (Fig. 2).
FIG. 5.
T7 protein expression assay with E. coli WM1576 harboring pGP1-2 and other plasmids. Shown are autoradiograms of plasmid-encoded proteins labelled with 35S-amino acids. Lane 1, pACYC184; lane 2, pH-1/C; lane 3, pH-1; lane 4, pB-2. Arrows indicate the presumed expression products: Fes, truncated FepA*, and FepC.
Functional analysis of the Y. enterocolitica fes-fep gene cluster.
To determine the role of the Y. enterocolitica fes-fep gene cluster in utilization of catecholate siderophores, we investigated the ability of Yersinia fes-fep genes to complement orthologous E. coli mutants. For this purpose, Yersinia fes and fepD genes of plasmid pH-1 were inactivated by Tn552kan insertion, resulting in pH-1fes::Tn552kan and pH-1fepD::Tn552kan plasmids. pH-1fepD::Tn552kan and pH-1 were introduced into E. coli fepD and fepG mutant strains (AB1515.718 and AB1515.764, respectively). The pH-1 and pH-1fes::Tn552kan plasmids were transferred into E. coli fes mutant strain AN272. All transformants were tested for enterochelin-supported growth on NBD agar plates supplemented with 100 μM 2,2′-dipyridyl (Table 3). The pH-1 plasmid conferred enterochelin-supported growth on all the E. coli mutants. By introduction of plasmids carrying insertionally inactivated Yersinia fes or fepD (pH-1fes::Tn552kan or pH-1fepD::Tn552kan), it was not possible to complement the corresponding E. coli mutant. Interestingly, under higher iron-chelating conditions (200 μM 2,2′-dipyridyl) only E. coli AN272 fes carrying pH-1 showed enterochelin-supported growth. Obviously, complementation of corresponding E. coli mutants with orthologous fepD and fepG was less efficient than that with orthologous Yersinia fes. Presumably, Yersinia FepD and FepG are not optimal partners for the corresponding E. coli Fep proteins to form a highly efficient enterochelin transport system.
TABLE 3.
Complementation of E. coli mutants of ferric enterochelin esterase (AN272 fes) and ferric enterochelin permease (AB1515.718 fepD and AB1515.764 fepG)a
| Plasmid used for complementation | Complementation of mutant at concn (μM) of 2,2′-dipyridyl
|
||
|---|---|---|---|
| AN272 fes, 200 | AB1515.718 fepD and AB1515.764 fepG
|
||
| 100 | 200 | ||
| pACYC184 | φ | φ | φ |
| pBluescript KS(+) | φ | φ | φ |
| pSI10 | + | + | φ |
| pH-1 | + | + | φ |
| pH-1fes::Tn552kan | φ | ND | ND |
| pH-1fepD::Tn552kan | ND | φ | φ |
Lack of complementation (φ) or positive complementation (+) by the bioassay as described in Materials and Methods. ND, not determined.
According to this assumption, we expected that coexpression of Yersinia fepDGC and E. coli fepDGC in E. coli should result in mixed FepDGC transport complexes with impaired ferric enterochelin utilization and consequently in enterochelin hyperproduction. On the other hand, it is also conceivable that the periplasmic catecholate siderophore binding protein FepB of E. coli does not interact properly with the Yersinia FepDGC complex. In fact, E. coli DH5α harboring plasmid pH-1 produced much larger haloes on CAS agar (CAS++ phenotype) than the parental strain (CAS+ phenotype), indicating enhanced enterochelin production. Progressively truncated subfragments of the 7.5-kb HindIII fragment of pH-1 were obtained by exonuclease treatment. By subcloning these subfragments in DH5α, we found that the CAS++ phenotype was dependent on the integrity solely of the Yersinia fepDGC operon. Thus, a Fur capture phenomenon caused by introduction of Yersinia Fur boxes on the recombinant plasmid into E. coli DH5α could largely be excluded.
However, the deduced Fur boxes located on the Y. enterocolitica fes-fep gene cluster are functional in E. coli. This could be demonstrated for plasmid pHB3 (carrying promoters of fes and fepA) and for plasmid pHB4 (fepDGC promoter) by using the FURTA (reference 55 and data not shown).
Construction of Yersinia mutants defective in fepD and fes.
To determine the role of fes and fepDGC in catecholate siderophore uptake in Y. enterocolitica, we constructed fes and fepD mutants by allelic exchange with the suicide plasmids pKASfes::Tn552kan and pKASfepD::Tn552kan (Fig. 1). The resulting mutants WA-fes625::Tn552kan and WA-fepD587::Tn552kan together with the parental strain WA-C were tested for enterochelin utilization by a bioassay as described elsewhere (37). In contrast to strain WA-C, the growth of the fepD and fes mutant strains was not supported by applying enterochelin-soaked filter disks. However, enterochelin-supported growth became evident after introduction of plasmid pH-1 into the mutant strains. Thus, the Yersinia fes-fep gene cluster is involved in ferric enterochelin siderophore transport and utilization.
Distribution of the Y. enterocolitica fes-fep gene cluster among different Yersinia species.
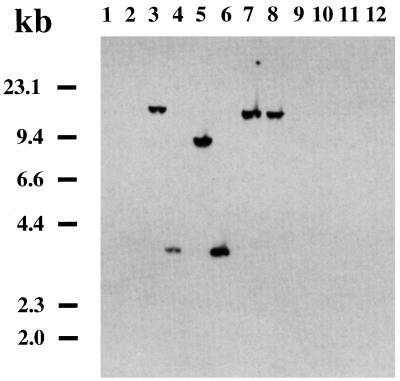
In order to investigate the distribution of the fes gene among different serovars of Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis, as well as among nonpathogenic Yersinia species (Yersinia frederiksenii, Yersinia intermedia, and Yersinia kristensenii), Southern hybridizations were performed with a PCR-derived fes gene probe. Hybridization of ClaI-digested genomic DNA (Fig. 6) showed that the Yersinia fes gene is detectable in all Y. enterocolitica serovars pathogenic to humans except serovar O3 (14 strains of serovar O3 were tested [47]). However, the fes gene is absent in Y. pseudotuberculosis serovars 1, 2, and 3; Y. pestis KUMA; and the nonpathogenic Yersinia species. In Y. enterocolitica O8, the fes probe reacted with a fragment of about 9 kb; in serovars O5, O13, and O20, a hybridizing band of approximately 18 kb was detected. Serovars O5.27 and O9 revealed a hybridizing band of about 3 kb. Corresponding results were observed for ClaI-digested genomic DNA with a probe derived from Yersinia fepD and fepA genes, indicating that fes-positive strains were also positive for fepD and fepA (data not shown). The conservation of the fes gene in different Y. enterocolitica serovars was determined by sequencing of PCR fragments covering the fes gene. Identity of 98.6 to 100% was found for the Yersinia fes gene. However, the base pair exchanges observed within the fes genes have no impact on changes of the predicted amino acid sequence.
FIG. 6.
Southern blotting of ClaI-digested chromosomal DNA of different Y. enterocolitica serovars and E. coli DH5α. The preparation was probed with a labelled PCR product generated from the Yersinia fes gene. Lane 1, E. coli DH5α; lane 2, Y. enterocolitica O3; lane 3, O5; lane 4, O5.27; lane 5, O8; lane 6, O9; lane 7, O13; lane 8, O20; lane 9, Y. pseudotuberculosis I; lane 10, Y. pseudotuberculosis II; lane 11, Y. pseudotuberculosis III; lane 12, Y. pestis pgm+.
As shown above, the fepA gene of Y. enterocolitica O8 appears to be interrupted by a frameshift. To investigate the presence of the frameshifted fepA gene among other Y. enterocolitica serovars, suitable primers were designed to amplify the central part of fepA by PCR. Only Y. enterocolitica O20, O5, and O13 yielded abundant PCR products which were subsequently sequenced. The serotype O20 strain carries an identical frameshift deletion of the fepA homologue. Interestingly, sequencing of the corresponding gene in Y. enterocolitica serotype O5 and O13 strains revealed a 31-bp insertion of DNA within this region leading to a single ORF. However, the significance of the fepA frameshift mutation remains to be clarified.
DISCUSSION
The objective of this study was to characterize the ferric enterochelin uptake and utilization of Y. enterocolitica serotype O8. For this purpose, we used an E. coli fes mutant as the recipient of a Yersinia genomic library and selected for fes complementation. We were able to isolate an ∼7.5-kb fragment of genomic DNA from Y. enterocolitica O8 composed of five genes arranged in three distinct transcriptional units (Fig. 1 and 2). The first unit consists of functional genes highly homologous to the E. coli fepDGC genes. This operon encodes three polypeptides which collectively resemble a cytoplasmic membrane transport system for ferric enterochelin belonging to the superfamily of ABC transporters (12, 25, 51). FepC has signature ATP-binding motifs (25, 59), while FepD and FepG are endowed with characteristics of integral membrane permeases (51). By the LA-PCR cloning technique, a gene homologous to E. coli fepB could be detected immediately upstream of fepDGC. The E. coli FepB represents a periplasmic binding protein involved in enterochelin uptake. The second transcriptional unit consists of a single gene that has high homology with the E. coli fes gene. It encodes an esterase which is involved in the release of iron from ferric enterochelin. The third transcriptional unit consists of two overlapping ORFs homologous to fepA of E. coli, indicating that a frameshift mutation might have taken place in fepA of Y. enterocolitica O8. In line with this observation, T7 polymerase expression of the third transcriptional unit leads to a truncated polypeptide of 35 kDa instead of about 83 kDa. However, a nonfunctional FepA protein does not exclude ferricatecholate siderophore uptake as has been shown for E. coli and S. enterica. For these bacteria, it is known that besides FepA other outer membrane receptor proteins such as Fiu, Cir, and IroN can support catecholate siderophore transport through the outer membrane (4, 36). The catecholate-like receptor CccA of Y. enterocolitica O8 (5) and two further iron-repressible outer membrane proteins of 75 and 90 kDa which might be involved in ferric enterochelin uptake have been described (45). In spite of this ambiguity concerning ferricatecholate siderophore transport through the outer membrane, we have shown that both Yersinia fepDGC and fes are functional in corresponding E. coli mutants. Moreover, inactivation of fepDGC or fes in Y. enterocolitica results in abrogation of ferric enterochelin uptake. In summary, Y. enterocolitica is endowed with the required genes for ferric enterochelin uptake (fepDGC and fepB) and utilization (fes).
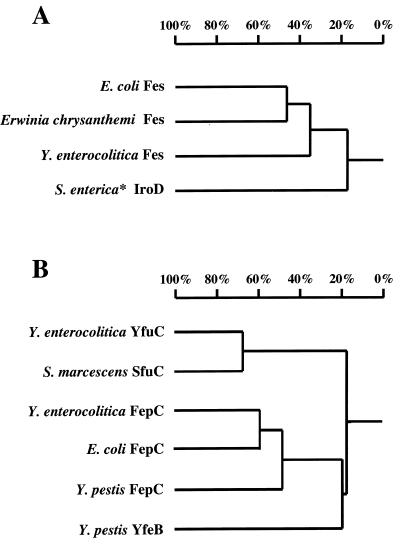
The high degree of amino acid sequence similarity between Y. enterocolitica and E. coli Fes as well as between FepDGC and FepB indicates a common evolutionary origin (Fig. 7). In addition, the overall G+C content of the Yersinia fes-fep gene cluster (49%) is close to the average G+C content of Y. enterocolitica (46 to 48%) and lower than that of the E. coli fes-fep gene cluster (54%). Taken together, these data are suggestive of an early divergent evolution of the fes-fep gene cluster within members of the family Enterobacteriaceae. However, comparison of the gene arrangement of the fes-fep gene cluster of E. coli with that of Y. enterocolitica reveals some striking differences (Fig. 2). The entF and fepE homologues of E. coli are missing within the 7.5-kb fes-fep gene cluster of Yersinia.
FIG. 7.
Homology tree determined by pairwise alignment of amino acid sequences by the method developed by Wilbur and Lipman (1, 60). (A) Homology tree of E. coli Fes, E. chrysanthemi Fes, Y. enterocolitica Fes, and IroD of S. enterica (∗, Salmonella enterica subsp. enterica serotype Typhi). (B) Homology tree for iron transport proteins belonging to the superfamily of ABC transporters: Y. enterocolitica YfuC, S. marcescens SfuC (2), Y. enterocolitica FepC, E. coli FepC, Y. pestis FepC, and Y. pestis YfeB (8).
To explain the absence of fepE in the fes-fep gene cluster of Y. enterocolitica, one has to consider the function of the fepE product in E. coli. FepE of E. coli had originally been thought to be involved in ferric enterochelin uptake. However, a recent study has demonstrated high sequence identity between fepE of E. coli and the cld gene of Shigella flexneri encoding the lipopolysaccharide chain length determinant Cld (26). Moreover, the G+C content of fepE (46%) is distinct from that of other fep genes in E. coli (55 to 61%), suggesting different origins. Thus, fepE might not be involved in ferric enterochelin uptake. It is therefore not surprising that Y. enterocolitica can utilize ferric enterochelin in spite of the absence of fepE in the fes-fep gene cluster. Beside entF and fepE, the entD gene is also missing in the enterochelin gene cluster of Yersinia. These genes might have been deleted because they are not necessary or of no advantage for Y. enterocolitica. On the other hand, it is unknown whether the E. coli enterochelin gene cluster (including the fes, fep, and ent genes) might have evolved by stepwise accumulation of coherent functional clusters (e.g., biosynthesis module and transport module) with subsequent rearrangements resulting in the functional mixed enterochelin cluster organization. Thus, as an alternative to the hypothesis of ent-fepE deletion in Yersinia, it is also conceivable that the fepA-fes-fepDGC cluster (including fepB upstream of fepDGC) is the conserved descendant of an ancestor of a catecholate siderophore transport or utilization module, with an insertion of entF and fepE in the E. coli enterochelin gene cluster at a later evolutionary stage. This question may be answered after analyzing the enterochelin gene cluster of other members of the family Enterobacteriaceae.
Furthermore, in this study we investigated the presence of the fes gene in pathogenic species of Yersinia by Southern hybridization and DNA sequencing. Surprisingly, the Yersinia fes probe hybridized exclusively with genomic DNA of different serotypes and biogroups of Y. enterocolitica. However, the fes-fep gene cluster was not detectable in Y. enterocolitica serotype O3 biogroup 4, Y. pestis, and Y. pseudotuberculosis as well as nonpathogenic Yersinia spp. (Y. frederiksenii, Y. intermedia, and Y. kristensenii). The fes gene sequences of different serotypes showed high identity (98.6 to 100%), indicating a common origin. The distribution of the fes-fep gene cluster is in good agreement with the results of the enterochelin feeding bioassay obtained for the corresponding strains.
Other iron uptake systems are known for Yersinia species. For Y. enterocolitica, the yfu gene (46) which has high similarity to the Sfu iron uptake system of Serratia marcescens has been described (40). Recently, an ABC transporter system for iron uptake (yfe cluster) in Y. pestis (8) that is also present in Y. pseudotuberculosis and Y. enterocolitica has been discovered. This Yfe system restored growth of an E. coli mutant deficient in enterochelin biosynthesis. However, it remains to be clarified how far the Yfe system is involved in ferric enterochelin uptake, since the Yfe system is involved in transport of other metal ions in addition to iron (7, 8). Moreover, putative proteins with similarity to E. coli FepDGC are deposited in the Y. pestis genome database (61), whereas no Fes-homologous polypeptide is present. In Y. pestis, the fepDGC-homologous genes are arranged in a single operon consisting of six ORFs. The deduced amino acid sequence of one ORF reveals similarity to the ferripyoverdin receptor and to the E. coli ferrioxamine B-ferricoprogen receptor but has only moderate similarity to the corresponding FepA protein of Y. enterocolitica described in this study. Of note, in Y. pestis as in Y. enterocolitica, no ent-orthologous genes encoding enterochelin biosynthesis enzymes are present in the neighborhood of the fepDGC-homologous gene locus. Figure 7 shows the homology trees determined by pairwise alignment of amino acid sequences of the different ferric enterochelin esterase orthologs (Fig. 7A) and the different ABC iron uptake transporter proteins identified in Yersinia species and other members of the family Enterobacteriaceae as well as in E. chrysanthemi (Fig. 7B) (1, 60).
In summary, the results of this study are in line with the observation that Y. enterocolitica can utilize catecholate siderophores but is unable to produce enterochelin. These results also emphasize the role of fes- and fepDGC-orthologous genes for active transport across the cytoplasmic membrane as well as for utilization of diverse catecholate siderophores. Catecholate siderophore receptors in the outer membrane are frequently targets of bacteriocins or phages: for example, the enterochelin receptor FepA is a target for the bacteriocin colicin B. In the case of Yersinia, colicin B-producing members of the Enterobacteriaceae may have selected for FepA-negative Y. enterocolitica strains without abolishing ferric enterochelin uptake. Thus, the fep-fes gene cluster may contribute to better survival of yersiniae in ecological niches such as the gut lumen and the environment. Studies are under way to clarify this hypothesis.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to J.H. (HE 1297/8-1).
We thank T. Griffin for providing the Tn552kan in vitro transposon mutagenesis system and C. F. Earhart for providing E. coli AB1515.768, AB1515.718, and AB1515.199 and plasmid pGP111.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Angerer A, Gaisser S, Braun V. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggest a periplasmic-binding-protein-dependent iron transport mechanism. J Bacteriol. 1990;172:572–578. doi: 10.1128/jb.172.2.572-578.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 4.Baumler A J, Norris T L, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäumler A J, Koebnik R, Stojiljkovic I, Heesemann J, Braun V, Hantke K. Survey on newly characterized iron uptake systems of Yersinia enterocolitica. Zentbl Bakteriol Orig A. 1993;278:416–424. doi: 10.1016/s0934-8840(11)80858-3. [DOI] [PubMed] [Google Scholar]
- 6.Bearden S W, Fetherston J D, Perry R D. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect Immun. 1997;65:1659–1668. doi: 10.1128/iai.65.5.1659-1668.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bearden S W, Perry R D. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 8.Bearden S W, Staggs T M, Perry R D. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer H W, Roulland Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 10.Carniel E, Mazigh D, Mollaret H H. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chenault S S, Earhart C F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 13.Chenault S S, Earhart C F. Identification of hydrophobic proteins FepD and FepG of the Escherichia coli ferrienterobactin permease. J Gen Microbiol. 1992;138:2167–2171. doi: 10.1099/00221287-138-10-2167. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (Fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earhart C F. Ferrienterobactin transport in Escherichia coli. In: Winkelmann G, van der Helm D, Neilands J B, editors. Iron transport in microbes, plants and animals. Weinheim, Germany: VCH Verlagsgesellschaft mbH; 1987. pp. 67–84. [Google Scholar]
- 16.Earhart C F. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1075–1090. [Google Scholar]
- 17.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 18.Fetherston J D, Lillard J W, Jr, Perry R D. Analysis of the pesticin receptor from Yersinia pestis: role in iron-deficient growth and possible regulation by its siderophore. J Bacteriol. 1995;177:1824–1833. doi: 10.1128/jb.177.7.1824-1833.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fetherston J D, Schuetze P, Perry R D. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 20.Galyov E E, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensible virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 21.Guilvout I, Mercereau Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of enteropathogenic Yersinia species. FEMS Microbiol Lett. 1987;48:229–233. [Google Scholar]
- 24.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane protein of 65000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 25.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 26.Hong M, Payne S M. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol. 1997;24:779–791. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 27.Hulton C S, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Langman L, Young I G, Frost G E, Rosenberg H, Gibson F. Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. J Bacteriol. 1972;112:1142–1149. doi: 10.1128/jb.112.3.1142-1149.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leschziner A E, Griffin T J, Grindley N D. Tn552 transposase catalyzes concerted strand transfer in vitro. Proc Natl Acad Sci USA. 1998;95:7345–7350. doi: 10.1073/pnas.95.13.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohana Rao J K, Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986;869:197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- 32.Neilands J B. Iron absorption and transport in microorganisms. Annu Rev Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 33.Neilands J B. Siderophores of bacteria and fungi. Microbiol Sci. 1984;1:9–14. [PubMed] [Google Scholar]
- 34.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettis G S, McIntosh M A. Molecular characterization of the Escherichia coli enterobactin cistron entF and coupled expression of entF and the fes gene. J Bacteriol. 1987;169:4154–4162. doi: 10.1128/jb.169.9.4154-4162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabsch W, Voigt W, Reissbrodt R, Tsolis R M, Bäumler A J. Salmonella typhimurium IroN and FepA proteins mediate uptake of enterobactin but differ in their specificity for other siderophores. J Bacteriol. 1999;181:3610–3612. doi: 10.1128/jb.181.11.3610-3612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabsch W, Winkelmann G. The specificity of bacterial siderophore receptors probed by bioassays. Biol Met. 1991;4:244–250. doi: 10.1007/BF01141188. [DOI] [PubMed] [Google Scholar]
- 38.Rakin A, Heesemann J. Yersiniabactin/pesticin receptor: a component of an iron uptake system of highly pathogenic Yersinia. Contrib Microbiol Immunol. 1995;13:244–247. [PubMed] [Google Scholar]
- 39.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 40.Rakin, A., E. Saken, and J. Heesemann. 1999. Unpublished data.
- 41.Rakin, A., S. Schubert, C. Pelludat, D. Brem, and J. Heesemann. The high-pathogenicity island of Yersinia. In J. Hacker and J. B. Kaper (ed.), Pathogenicity islands, in press. ASM Press, Washington, D.C.
- 42.Reissbrodt R, Rabsch W. Further differentiation of Enterobacteriaceae by means of siderophore-pattern analysis. Zentbl Bakteriol Mikrobiol Hyg. 1988;268:306–317. doi: 10.1016/s0176-6724(88)80015-4. [DOI] [PubMed] [Google Scholar]
- 43.Reissbrodt R, Rabsch W, Chapeaurouge A, Jung G, Winkelmann G. Isolation and identification of ferrioxamine G and E in Hafnia alvei. Biol Met. 1990;3:54–60. [Google Scholar]
- 44.Rowland S J, Dyke K G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 45.Rutz J M, Abdullah T, Singh S P, Kalve V I, Klebba P E. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J Bacteriol. 1991;173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saken E. Direct submission to the databases (GenBank accession no. Z47200). 1995. (Unpublished data.) [Google Scholar]
- 47.Saken E, Roggenkamp A, Aleksic S, Heesemann J. Characterisation of pathogenic Yersinia enterocolitica serogroups by pulsed-field gel electrophoresis of genomic NotI restriction fragments. J Med Microbiol. 1994;41:329–338. doi: 10.1099/00222615-41-5-329. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schmitt M P, Payne S M. Genetics and regulation of enterobactin genes in Shigella flexneri. J Bacteriol. 1988;170:5579–5587. doi: 10.1128/jb.170.12.5579-5587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 51.Shea C M, McIntosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 52.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1988;1:784–785. [Google Scholar]
- 53.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 54.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 55.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. . (Erratum, 240:271.) [DOI] [PubMed] [Google Scholar]
- 56.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsolis R M, Baumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilbur W J, Lipman D J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci USA. 1983;80:726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yersinia pestis Genome Project website. [Online.] http://microbios1.mds.qmw.ac.uk/yersinia/. [30 August 1999, last date accessed.] 1999. [Google Scholar]