Abstract
In the bacterium Escherichia coli, oxidized pyrimidines are removed by two DNA glycosylases, endonuclease III and endonuclease VIII (endo VIII), encoded by the nth and nei genes, respectively. Double mutants lacking both of these activities exhibit a high spontaneous mutation frequency, and here we show that all of the mutations observed in the double mutants were G:C→A:T transitions; no thymine mutations were found. These findings are in agreement with the preponderance of C→T transitions in the oxidative and spontaneous mutational databases. The major oxidized purine lesion in DNA, 7,8-dihydro-8-oxoguanine (8-oxoG), is processed by two DNA glycosylases, formamidopyrimidine DNA glycosylase (Fpg), which removes 8-oxoG opposite C, and MutY DNA glycosylase, which removes misincorporated A opposite 8-oxoG. The high spontaneous mutation frequency previously observed in fpg mutY double mutants was significantly enhanced by the addition of the nei mutation, suggesting an overlap in the substrate specificities between endo VIII and Fpg/MutY. When the mutational specificity was examined, all of the mutations observed were G:C→T:A transversions, indicating that in the absence of Fpg and MutY, endo VIII serves as a backup activity to remove 8-oxoG. This was confirmed by showing that, indeed, endo VIII can recognize 8-oxoG in vitro.
The spontaneous mutational burden results from a combination of replication errors and endogenous DNA damages. Replication errors are repaired by postreplication mismatch repair (for a review, see reference 39), while endogenous damages are primarily repaired by base excision repair (for reviews, see references 59 and 66). Both of these repair processes are highly conserved across species at the functional level, and in many cases the proteins involved are conserved at the level of amino acid sequence (reviewed in references 30, 43, and 58). This has resulted in Escherichia coli and the yeast Saccharomyces cerevisiae, both genetically tractable organisms, becoming paradigms for understanding the cellular roles of the enzymes involved.
The data thus far suggest that endogenous DNA damages play at least as great a role, if not a greater role, in the production of spontaneous mutations as do replication errors. Endogenous damages include spontaneous depurinations, deaminations, alkylations, and oxidative lesions produced during normal aerobic metabolism (32). It has become increasingly clear that oxidative DNA damages play a major role in spontaneous mutagenesis. A significant number of oxidized purines and pyrimidines are readily bypassed by DNA polymerases and can mispair with a noncognate base (for reviews, see references 19 and 61). Of particular note is DNA guanine, which is frequently oxidized to 7,8-dihydro-8-oxoguanine (8-oxoG), which, if not repaired, can be bypassed by DNA polymerases and pair with its cognate C as well as noncognate A (53), leading to G→T transversions (6, 40, 41, 67). E. coli has evolved a complicated strategy to avoid mutations from this commonly oxidized base (for a review, see reference 35). Formamidopyrimidine DNA glycosylase, Fpg (also called MutM), removes 8-oxoG paired with C in DNA (7, 55), while the MutY protein removes A opposite 8-oxoG (3, 33, 56) resulting from A mispairing with unrepaired 8-oxoG during replication. Finally, the MutT protein, an 8-oxodGTPase, removes oxidized dGTPs from the nucleotide pool, preventing their misincorporation opposite adenine (2). E. coli mutants defective in Fpg or MutY, and double mutants lacking both proteins, exhibit higher-than-wild-type spontaneous mutation frequencies, about 5- to 15-fold, 10- to 30-fold, and 200- to 800-fold, respectively, depending on the assay system used (34, 36, 42). Mutants lacking the MutT protein also exhibit high spontaneous mutation frequencies, of about 1,000-fold (1, 2).
The role of oxidized pyrimidines has been less well studied; however, both spontaneous and oxidative mutational spectra are replete with C→T transitions resulting from oxidized cytosines (reviewed in reference 61), and the major oxidized DNA cytosine products, uracil glycol, 5-hydroxycytosine, and 5-hydroxyuracil, are mutagenic (16, 29). Oxidized pyrimidines are repaired in E. coli by endonuclease III (endo III) and endonuclease VIII (endo VIII) (for reviews, see references 58 and 59), DNA glycosylases encoded by the nth and nei genes, respectively. Mutants defective in endo III exhibit a small mutator phenotype (25, 63), while mutants defective in endo VIII exhibit no mutator phenotype (25, 51). Double mutants lacking both proteins, however, exhibit a higher-than-wild-type (about 20-fold) spontaneous mutation frequency (25, 51).
In this paper we show that double mutants lacking both pyrimidine-specific endonucleases show increases only in spontaneous G:C→A:T transitions, which is in keeping with their substrate specificities. Surprisingly, triple mutants lacking Fpg, MutY, and endo VIII and quadruple mutants lacking all four DNA glycosylases exhibit significant synergistic effects, suggesting an overlap in the substrate specificities of the “pyrimidine-specific” and “purine-specific” enzymes. Since an increase only in G:C→T:A transversions was observed in both the triple and quadruple mutants, and since endo VIII can incise substrates containing 8-oxoG in vitro, this synergy appears to be due to the recognition and removal of 8-oxoG by endo VIII.
MATERIALS AND METHODS
Bacterial strains.
E. coli BW35 (KL16 [wild type]), BW402 (KL16 nth-1::kan), BW540 (KL16 nfo-1::kan), and BW9101 (KL16 Δxth-pncA) were generously provided by Bernard Weiss, Department of Pathology, Emory University, and have been described previously (10, 64, 65). SW2-8 (KL16 Δnei::cm) was described earlier (25). SW2-F (KL16 Δfpg::amp) is described below. Strain CSH11 mutY::mini-tet and strains CC101 to CC106 (11) were generously provided by Jeffrey Miller, Molecular Biology Institute and Department of Microbiology and Molecular Genetics, University of California, Los Angeles.
Construction of multiple mutants defective in DNA glycosylases specific for oxidative DNA damages.
CSH11 mutY::mini-tet was used to create SW2-Y (KL16 mutY::mini-tet) via P1vir transduction (38). Construction of the Δfpg::amp null mutant was as follows: the genomic DNA fragments surrounding fpg (726 bp upstream and 1,766 bp downstream) were amplified by PCR (with Pfu DNA polymerase [Stratagene]) and cloned into the phagemid pBC (Stratagene). A cassette carrying the ampicillin resistance gene was PCR amplified from the phagemid pBluescript II (Stratagene) and cloned in place of fpg in the above-mentioned construct. This resulted in a 950-bp deletion of the entire fpg gene as well as 20 bp from the 3′ end of rpmG, a gene which codes for the ribosomal protein L33 (31). Mutations in this gene show no phenotypic change and are virtually normal with respect to growth rate (8). The construct was used in a linear transformation into strain BW853 (recD::mini-Tn10) (provided by B. Weiss), as previously described (50). The resulting BW853 Δfpg::amp mutant was verified first via PCR and second by lack of incision of an 8-oxoG-containing substrate (data not shown). All further multiple-mutant strains used in this study were created via P1vir transduction (38) of the desired disruptions into the appropriate KL16 background strains.
DNA damages.
Oligonucleotides containing either 8-oxoG (Glen Research, Sterling, Va.) or thymine glycol (Tg) (see reference 18) were synthesized in the 54-mer sequence 5′ ATTCCAGACTGTCAATAACACGGXGGACCAGTCGATCCTGGGCTGCAGGAATTC 3′, where X represents the damage. The oligonucleotides were 5′ labeled with [γ-32P]ATP (NEN) by T4 polynucleotide kinase (New England Biolabs) and annealed to the appropriate complementary strands.
Enzymes and enzyme assays.
endo VIII, endo III, and Fpg proteins were overexpressed in E. coli and purified as described previously (25). All DNA cleavage reaction mixtures contained 5 fmol of substrate and were incubated at 37°C for 10 min. The reaction mixtures contained either 1 to 2 μg of crude cell extract in 10 mM Tris-HCl (pH 8.0) plus 50 mM NaCl and 10 mM EDTA or 1, 10, 100, or 1,000 pmol of purified enzyme in 10 mM Tris-HCl (pH 8.0) plus 50 mM NaCl. The reactions were stopped by the addition of formamide, and the products were separated by 8 M urea–polyacrylamide gel electrophoresis.
Determination of spontaneous mutation frequencies.
Overnight cultures of the appropriate CC101 to CC106 strains were plated directly onto M9 minimal medium (52) containing 2 mM MgSO4, 100 μm CaCl2, and either lactose or dextrose. The plates were incubated at 37°C for 48 h, and the Lac+ revertant fraction was determined. We have previously described calculation of the spontaneous mutation frequency to rifampin resistance (25). Mutation data were analyzed by one-way analysis of variance, with the natural logarithm of mutation frequency. Dunnett’s procedure was used to adjust for multiple comparisons with the fpg mutY genotype.
RESULTS
endo III and endo VIII repair endogenous premutagenic oxidized cytosine lesions.
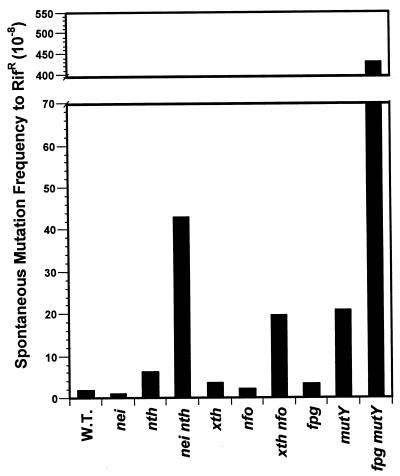
We and others have previously shown that E. coli mutants lacking both endo III and endo VIII (nth nei) are hypersensitive to the lethal effects of ionizing radiation (25) and hydrogen peroxide (51, 60), indicating that these enzymes are responsible for removing potentially lethal oxidative lesions in DNA. This is in contrast to mutants lacking Fpg protein, which are not hypersensitive to the lethal effects of ionizing radiation or hydrogen peroxide (5, 60). nth nei double mutants also exhibit increased spontaneous mutation frequency, as determined by both forward mutation and reversion assays (24). Table 1 and Fig. 1 show the spontaneous mutation frequencies to rifampin resistance of wild-type E. coli and single and double mutants lacking the oxidized pyrimidine-specific DNA glycosylases endo III and endo VIII, the oxidized purine-specific DNA glycosylases Fpg and MutY, and, for comparison, the apurinic (AP) site-specific AP endonucleases exonuclease III (xth) and endonuclease IV (endo IV) (nfo). As expected, synergy was observed in the double mutants lacking both pyrimidine-specific DNA glycosylases, both purine-specific DNA glycosylases, and both AP endonucleases because of the overlaps in the substrate specificities of these pairs of enzymes. Furthermore, the spontaneous mutation frequency observed in the nth nei double mutants was about twice that observed in mutants lacking both AP endonucleases (Table 1 and Fig. 1), suggesting that the oxidized pyrimidines recognized by endo III and endo VIII have a greater mutagenic potential than the AP sites recognized by the AP endonucleases. However, the mutation frequency observed in the absence of both pyrimidine-specific endonucleases was significantly lower (about 10-fold) than that observed in fpg mutY double mutants defective in the processing of 8-oxoG.
TABLE 1.
Spontaneous mutation frequencies in E. coli mutants lacking oxidative base excision repair enzymes
| Genotypea | Spontaneous mutation frequency to Rifr (10−8)
|
Approx mutation frequency relative to:
|
||||
|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg ± SD | Wild type | fpg mutY | |
| Wild type* | 2.2 | 1.6 | 2.1 | 2.0 ± 0.3 | 1 | |
| nei* | 0.8 | 0.5 | 2.0 | 1.1 ± 0.8 | <1 | |
| nth* | 6.0 | 8.9 | 4.2 | 6.4 ± 2.4 | 3 | |
| fpg | 2.1 | 6.5 | 2.3 | 3.6 ± 2.5 | 2 | |
| mutY | 22 | 24 | 17 | 21 ± 3.6 | 11 | |
| xth | 2.5 | 4.3 | 4.3 | 3.7 ± 1.0 | 2 | |
| nfo | 2.5 | 1.2 | 3.3 | 2.3 ± 1.1 | 1 | |
| nei nth* | 33 | 70 | 28 | 44 ± 23 | 22 | |
| nei fpg | 1.4 | 3.0 | 0.7 | 1.7 ± 1.2 | <1 | |
| nei mutY | 22 | 18 | 25 | 22 ± 3.5 | 11 | |
| nth fpg | 6.0 | 8.6 | 17 | 10 ± 5.7 | 5 | |
| nth mutY | 58 | 53 | 38 | 50 ± 10 | 25 | |
| fpg mutY | 360 | 460 | 480 | 430 ± 64 | 220 | |
| xth nfo | 25 | 23 | 12 | 20 ± 7.0 | 10 | |
| nei nth fpg | 29 | 48 | 57 | 45 ± 14 | 22 | |
| nei nth mutY | 31 | 31 | 37 | 33 ± 3.5 | 17 | |
| fpg mutY | 360 | 460 | 480 | 430 ± 64 | 1 | |
| nei fpg mutY | 820 | 1,500 | 1,400 | 1,200 ± 370 | 3 | |
| nth fpg mutY | 520 | 530 | 740 | 600 ± 120 | 1.4 | |
| nei nth fpg mutY | 650 | 750 | 1,400 | 930 ± 410 | 2.2 | |
*, previously reported by Jiang et al. (24).
FIG. 1.
Spontaneous forward mutation frequencies to rifampin resistance in E. coli base excision repair mutants. Mid-log-phase cultures were plated on Luria-Bertani medium with or without 100 μg of rifampin per ml and incubated for 15 h at 37°C. Mutants per 108 cells are shown. The data represent the average of the three experiments summarized in Table 1. W.T., wild type.
The strains constructed by Cupples and Miller (12) were then used to examine the specificity of spontaneous mutagenesis in the nth nei mutant background. Table 2 shows that all of the spontaneous mutations observed in the nth nei double mutants were G:C→A:T transitions, suggesting that oxidized cytosines were the primary premutagenic lesions responsible, which is in keeping with the substrate specificity of the enzymes. As has been previously observed (34), double mutants lacking Fpg and MutY proteins exhibit an extremely high frequency of G:C→T:A transversions (Table 3), which is in keeping with their substrate specificities for processing 8-oxoG.
TABLE 2.
Specificity of spontaneous reversions formed in a double nth nei mutant lacking endo III and endo VIII
| Strain | Reversion event | Lac+ revertants/109 cells (avg ± SD)
|
Approx fold increase | |
|---|---|---|---|---|
| Wild type | nth nei | |||
| CC101 | A:T→C:G | 1.1 ± 0.7 | 0.3 ± 0.1 | <1 |
| CC102 | G:C→A:T | 0.8 ± 0.3 | 34 ± 4.9 | 43 |
| CC103 | G:C→C:G | 1.8 ± 0.8 | 0.6 ± 0.4 | <1 |
| CC104 | G:C→T:A | 1.8 ± 1.6 | 2.8 ± 0.8 | 1.5 |
| CC105 | A:T→T:A | 1.2 ± 0.7 | 2.4 ± 1.4 | 2 |
| CC106 | A:T→G:C | 0.5 ± 0.3 | 0.1 ± 0.1 | <1 |
TABLE 3.
Specificity of spontaneous G:C→T:A reversions formed in multiple mutants lacking DNA glycosylases specific for oxidized purines and pyrimidines
| Genotype | Lac+ revertants per 109 cells
|
||||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg ± SD | Relative to fpg mutY (approx) | |
| Wild type (CC104) | 1.6 | 0.3 | 3.5 | 1.8 ± 1.6 | N/Aa |
| fpg mutY | 5,900 | 4,000 | 5,400 | 5,100 ± 990 | 1 |
| fpg mutY nei | 7,100 | 8,100 | 13,000 | 9,400 ± 3,200 | 1.8 |
| fpg mutY nth | 6,400 | 6,800 | 7,900 | 7,000 ± 780 | 1.4 |
| fpg mutY nei nth | 6,800 | 9,200 | 8,000 | 8,000 ± 1,200 | 1.6 |
Not applicable.
Effects of multiple mutants lacking DNA glycosylases specific for oxidized DNA bases on spontaneous mutagenesis.
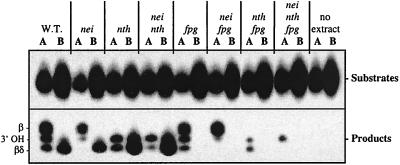
All mutant combinations for the oxidized base-specific DNA glycosylases were constructed as described in Materials and Methods. To confirm that the putative mutant combinations were actually deficient in their respective encoded proteins, mutant extracts were examined for the ability to recognize the appropriate substrates. Figure 2 shows the activity of cell extracts from various mutant combinations on substrates containing either Tg (lanes A), a major substrate for endo III and endo VIII, or 8-oxoG (lanes B), the primary substrate for Fpg. (The chemistry of these reactions is reviewed in references 9 and 13.) The lyase activity of endo III, in a β-elimination reaction, leaves an α,β-unsaturated aldehyde attached to the 3′ side of the nick (β-product), while the lyase activities of endo VIII and Fpg, in a double elimination, leave a phosphate attached to the 3′ side of the nick (β,δ-product). The AP endonucleases hydrolyze the DNA backbone at sites of base loss, leaving an OH group attached to the 3′ side of the nick (62). The AP endonucleases can also remove the β and β,δ products produced by endo III and endo VIII and Fpg, resulting in the 3′ OH product (14).
FIG. 2.
Oxidized pyrimidine- and purine-specific DNA glycosylase activities in crude cell extracts of E. coli mutants. Lanes A, Tg:A 54-mer; lanes B, 8-oxoG:C 54-mer. Both damages are at position 24 from the radiolabeled 5′ end. Reaction mixtures included 1 to 2 μg of crude cell extract, 5 fmol of substrate, and 10 mM EDTA and were incubated at 37°C for 10 min. See Materials and Methods for additional reaction conditions. W.T., wild type.
Figure 2 shows that in the wild-type extract with the Tg substrate (lane A), endo III β-elimination product, endo VIII δ-elimination product, and 3′ OH hydrolysis product produced by endo IV activity on the β- and β,δ-elimination products were all found. (Exonuclease III activity would not be observed in these extracts, since EDTA was present [49].) In the lane containing the 8-oxoG substrate (lane B) and wild-type extract, the δ-elimination product produced by Fpg lyase and some hydrolysis products were found. Examination of the nei and nth mutant extracts in the lanes containing Tg substrate shows the δ-elimination product to be missing from the nei mutant extract and the β-elimination product to be missing from the nth mutant extract; the nei nth double mutant extract was totally devoid of the β-product, although a small amount of δ-product was present. We take this δ-product to be the result of Fpg action on the Tg substrate, since we have previously shown that the enzymatic efficiency of Fpg on a Tg-containing substrate is only about ninefold lower than that of endo III (20). In the four extracts containing the fpg mutation, no δ-product was observed with the 8-oxoG substrate (lanes B); in the nei fpg mutant extract, the δ-product from the Tg substrate was missing, while in the nth fpg mutant extract, no β-product with the Tg substrate was seen (lanes A). Finally, and as expected, in the triple mutant, no lyase products were found with either substrate; only a small amount of hydrolysis product was observed with the Tg-containing substrate, perhaps due to endo IV action on degraded Tg (urea) (28). When extracts containing the mutY mutation were examined, the results were the same as those shown in Fig. 2 (data not shown), since MutY removes A from the unlabeled strand when it is paired with 8-oxoG and its activity would not be detected in this assay. Taken together, the results observed with the various mutant extracts show that the putative genes were indeed disrupted; furthermore, the data are consistent with the known activities of endo III, endo VIII, and Fpg.
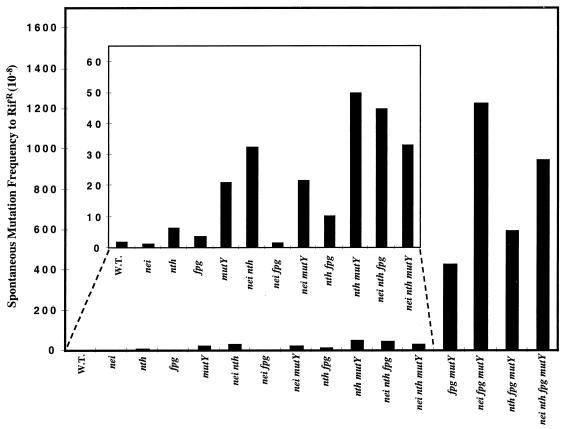
The spontaneous forward mutation frequency to rifampin resistance was then determined for all combinations of mutants, and these results are contained in Table 1 and depicted in Fig. 3. As can be seen, with the exception of nth mutY, combinations of nth or nei single or nth nei double mutants defective in the pyrimidine-specific DNA glycosylases together with a single mutation in either mutY or fpg resulted in additive or, in a few cases, possibly less than additive spontaneous mutation frequencies. We cannot explain the synergy observed with nth mutY, since we have not been able to demonstrate any overlap in substrate specificity between endo III and MutY. However, the introduction of either a nei single or both nth and nei mutations into an fpg mutY mutant background resulted in a statistically significant (P < 0.05) increase in spontaneous mutation frequency over fpg mutY double mutants, about 3-fold and 2.2-fold, respectively (Fig. 3). These results suggest an overlap in the processing of substrates for the glycosylases in the “pyrimidine- and purine-specific” pathways.
FIG. 3.
Spontaneous mutation frequencies observed in E. coli multiple mutants lacking oxidized pyrimidine- and purine-specific DNA glycosylases. Mid-log-phase cultures were plated on Luria-Bertani medium with and without 100 μg of rifampin per ml and incubated for 15 h at 37°C. Mutants per 108 cells are shown. The data represent the average of the three experiments summarized in Table 1. W.T., wild type.
In order to determine what this overlap in substrate specificity might be, the fpg mutY double mutant combination together with the single and double nth and nei mutant combinations was moved into the Cupples/Miller strains in order to determine specific base changes. To our surprise, and as shown in Table 3, increases in G:C→T:A transversions were observed in the nei fpg mutY triple and nth fei fpg mutY quadruple mutants, suggesting that endo VIII was capable of removing 8-oxoG in vivo. All potential reversions were examined in both the triple and quadruple mutants, and no increases other than G:C→T:A transversions were found (data not shown). Moreover, the increase in the number of G:C→T:A transversions in the nei fpg mutY triple mutant compared to the fpg mutY double mutant was statistically significant (P < 0.05). There was no significant increase in G:C→A:T transitions in the quadruple mutant over what was observed with the nth nei double mutant (data not shown), and the small number of G:C→A:T transitions found in the nth fpg mutY triple mutant (data not shown) is in agreement with the small mutator phenotype observed in nth single mutants (Table 1).
endo VIII can recognize and incise 8-oxoG lesions.
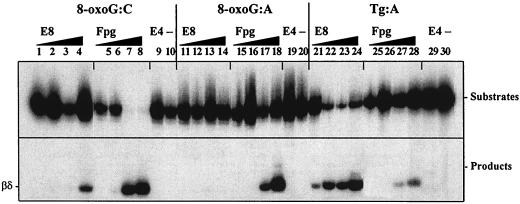
To confirm that endo VIII is indeed capable of incising substrates containing 8-oxoG, purified endo VIII was incubated with oligonucleotide substrates containing a single 8-oxoG lesion. As can be seen in Fig. 4, at high concentrations of endo VIII, a δ-elimination product was seen when 8-oxoG was positioned opposite C (lane 4) but not opposite A (lanes 11 to 14). Clearly, Tg was a much better substrate for endo VIII (compare lanes 1 through 4 to lanes 21 through 24), while 8-oxoG opposite C was a better substrate for Fpg (compare lanes 1 through 4 to lanes 5 through 8). At high concentrations, Fpg also incised 8-oxoG paired with A (lanes 17 and 18), as has been previously observed (37, 54). (The relative catalytic efficiency of Fpg on 8-oxoG:C versus 8-oxoG:A substrates is about 17-fold [54].) Tg is also a substrate for Fpg (lanes 27 and 28), as we have previously observed (20). Failure of endo IV to cleave the 8-oxoG oligonucleotide (lanes 9 and 10, 19 and 20, and 29 and 30) indicated that the substrate did not contain AP sites which would have confounded the results, since AP sites are the best substrates for endo VIII and Fpg (20). The same results were observed with substrates containing 8-oxoG paired with C in three other sequence contexts and in an additional sequence context where 8-oxoG was paired with A (data not shown).
FIG. 4.
Activity of purified endo VIII and Fpg on damaged DNA substrates. Damages are as specified and are at position 24 from the radiolabeled 5′ end. Enzyme concentrations for endo VIII (E8) and Fpg are 0.001, 0.01, 0.1, and 1.0 μM (increasing as depicted). endo IV (2 μM) was used as a control for the presence of AP sites. —, lanes contain no enzyme. Reaction mixtures contained 5 fmol of substrate and were incubated at 37°C for 10 min. See Materials and Methods for additional reaction conditions.
DISCUSSION
The addition of the nei mutation to E. coli carrying an nth mutation conferred a significant increase in the spontaneous mutation frequency (about sevenfold). Interestingly, all of the mutations observed in the nei nth double mutant were C→T transitions (Table 2). This is consistent with the ability of both enzymes to recognize oxidized cytosines (26), with the observed high frequency of C→T transitions in the spontaneous and oxidative mutational spectra (reviewed in reference 61), and with the ability of the major oxidized cytosine products to pair with A (29, 44, 48). Interestingly, although both enzymes recognize oxidized thymines equally well (26), no T events were recorded. We have previously shown that ring saturation products of DNA thymine, such as dihydrothymine and Tg, pair with A (23, 24) and are not mutagenic (15, 21), although when Tg is present in a sequence context that does not block DNA polymerases it is slightly mutagenic (4). Thus, the biological results, showing no spontaneous T mutations, are in agreement with the in vitro studies and suggest that spontaneously oxidized DNA thymines, although frequently produced, are not important premutagenic lesions. In our hands (25) and as previously reported (63), the absence of endo III conferred a slight mutator effect, suggesting that not all lesions recognized by endo III are recognized and removed by endo VIII.
endo VIII also plays a role in the repair of 8-oxoG lesions, as evidenced by its synergy with Fpg and MutY (Fig. 2 and Table 3). Removal of 8-oxoG does not appear to be a primary role for endo VIII, nor does endo VIII appear to occupy a particular niche in the repair of 8-oxoG, since no G→T transversions were observed in the nei single mutant. It has been suggested (22) that a second enzyme in human cells that recognizes 8-oxoG may be involved in removing incorporated 8-oxodGMP residues in the nascent strand when inserted opposite adenine. This could occur when 8-oxodGTP escapes the action of MutT-like enzymes. This cannot be the role that endo VIII plays in E. coli since, if this were true, the presence of the nei mutation would result in A→C transversions which were not seen in any nei mutant combination either alone or in fpg mutY or nth mutant backgrounds. Also, we did not observe any endo VIII activity on 8-oxoG paired with A (Fig. 4) in two different sequence contexts, and the addition of the nei mutation to a mutT mutant background does not increase the spontaneous mutation frequency over that observed in the mutT mutant background alone (4a). So, it does not appear that endo VIII specifically acts on 8-oxoG after replication but rather removes 8-oxoG while scanning the DNA for oxidized pyrimidines in a mode similar to that of Fpg for oxidized purines. Interestingly, although MutY acts after replication to prevent mutations, it also does not seem to be nascent strand specific, since the presence of MutY is a mutator in a mutT- minus background (57).
It is interesting that the contribution of oxidized pyrimidines to the spontaneous mutation frequency is much lower than that of 8-oxoG, as evidenced by comparing the forward mutation frequency in the absence of endo III and endo VIII (4.4 × 10−7 [Table 1]) to that observed in mutants lacking Fpg, MutY, and endo VIII (12 × 10−6 [Table 1]). Since the oxidized cytosines, uracil glycol and 5-hydroxyuracil, always mispair (45, 47) and 5-hydroxycytosine mispairs to an extent similar to 8-oxoG (46, 47), it appears that 8-oxoG must be formed in DNA substantially more frequently than cytosine glycol, the unstable parent of the above-mentioned cytosine lesions. This would not be expected necessarily from simple hydroxyl radical chemistry and suggests that other reactive species, such as singlet oxygen, may play an important role in the formation of 8-oxoG DNA lesions.
It is important to note that in vitro, with damage-containing oligodeoxynucleotide substrates, the activity of endo VIII on 8-oxoG is rather poor compared to its activity on Tg (Fig. 4). In fact, no endo VIII activity on 8-oxoG was seen in extracts (Fig. 2) where the effective endo VIII concentration was much lower than used in the experiments represented by Fig. 4 with the purified enzyme. Since the 8-oxoG activity of endo VIII is functional in vivo, it is possible that, in vitro, we are lacking the complete subset of proteins required. There are other examples of broad cross-specificities among the DNA glycosylases; for example, the “purine-specific” Fpg also recognizes oxidized pyrimidines (17), and E. coli 3-methyladenine DNA glycosylase, AlkA, recognizes a broad range of substrates (66).
The role of endo VIII in the repair of 8-oxoG appears to be relatively less important than its role in the repair of oxidized cytosine products, since introduction of the nei mutation into the fpg mutY mutant background confers a two- to threefold enhancement of the spontaneous mutation frequency to rifampin resistance, while the addition of the nei mutation to an nth mutant background confers a sevenfold enhancement. It should be pointed out, however, that C→T transitions are represented more frequently than G→T transversions in the rpoB mutations that have been sequenced (27). When single base changes were scored, a greater enhancement of C→T transitions was also observed by the addition of the nei mutation to the nth mutant background, about 4.4-fold, compared to 1.8-fold when the nei mutation was added to the fpg mutY double-mutant background. Taken together, these data suggest that the major cellular role for endo VIII is to remove premutagenic oxidized cytosine residues and that, in addition, endo VIII can function as a backup enzyme for Fpg protein in removing 8-oxoG paired with C.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R37CA33657, awarded by the National Cancer Institute.
We are grateful to Pam Vacek for help with statistical analysis and to the Vermont Cancer Center for support of the Biometry Facility.
REFERENCES
- 1.Akiyama M, Horiuchi T, Sekiguchi M. Molecular cloning and nucleotide sequence of the mutT mutator of Escherichia coli that causes A:T to C:G transversion. Mol Gen Genet. 1987;206:9–16. doi: 10.1007/BF00326530. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama M, Maki H, Sekiguchi M, Horiuchi T. A specific role of MutT protein: to prevent dG · dA mispairing in DNA replication. Proc Natl Acad Sci USA. 1989;86:3949–3952. doi: 10.1073/pnas.86.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Au K G, Clark S, Miller J H, Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci USA. 1989;86:8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu A K, Loechler E L, Leadon S A, Essigmann J M. Genetic effects of thymine glycol: site-specific mutagenesis and molecular modeling studies. Proc Natl Acad Sci USA. 1989;86:7677–7681. doi: 10.1073/pnas.86.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Blaisdell, J. O., and S. S. Wallace. Unpublished observations.
- 5.Boiteux S, Huisman O. Isolation of a formamidopyrimidine-DNA glycosylase (fpg) mutant of Escherichia coli K12. Mol Gen Genet. 1989;215:300–305. doi: 10.1007/BF00339732. [DOI] [PubMed] [Google Scholar]
- 6.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—T and A—C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 7.Chung M H, Kasai H, Jones D S, Innoue H, Ishikawa H, Ohtsuka E, Nishimura S. An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat Res. 1991;254:1–12. doi: 10.1016/0921-8777(91)90035-n. [DOI] [PubMed] [Google Scholar]
- 8.Coleman S H, Maguire B A, Wild D G. Ribosome assembly in three strains of Escherichia coli with mutations in the rpmB,G operon. J Gen Microbiol. 1993;139:707–716. doi: 10.1099/00221287-139-4-707. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham R P. DNA glycosylases. Mutat Res. 1997;383:189–196. doi: 10.1016/s0921-8777(97)00008-6. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupples C G, Cabrera M, Cruz C, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cupples C G, Miller J H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David S S, Williams S D. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 14.Demple B, Johnson A, Fung D. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci USA. 1986;83:7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 16.Feig D I, Sowers L C, Loeb L A. Reverse chemical mutagenesis: identification of the mutagenic lesions resulting from reactive oxygen species-mediated damage to DNA. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatahet Z, Kow Y W, Purmal A A, Cunningham R P, Wallace S S. New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 18.Hatahet Z, Purmal A A, Wallace S S. A novel method for site specific introduction of single model oxidative DNA lesions into oligodeoxyribonucleotides. Nucleic Acids Res. 1993;21:1563–1568. doi: 10.1093/nar/21.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatahet Z, Wallace S. Translesion DNA synthesis. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. Vol. 1. 1998. pp. 229–262. : DNA repair in prokaryotes and lower eukaryotes. Humana Press, Inc., Totowa, N.J. [Google Scholar]
- 20.Hatahet, Z., and S. S. Wallace. Submitted for publication.
- 21.Hayes R C, Petrullo L A, Huang H M, Wallace S S, LeClerc J E. Oxidative damage in DNA. Lack of mutagenicity by thymine glycol lesions. J Mol Biol. 1988;201:239–246. doi: 10.1016/0022-2836(88)90135-0. [DOI] [PubMed] [Google Scholar]
- 22.Hazra T K, Izumi T, Maidt L, Floyd R A, Mitra S. The presence of two distinct 8-oxoguanine repair enzymes in human cells: their potential complementary roles in preventing mutation. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ide H, Petrullo L A, Hatahet Z, Wallace S S. Processing of DNA base damage by DNA polymerases. Dihydrothymine and β-ureidoisobutyric acid as models for instructive and noninstructive lesions. J Biol Chem. 1991;266:1469–1477. [PubMed] [Google Scholar]
- 24.Ide H, Wallace S S. Dihydrothymidine and thymidine glycol triphosphates as substrates for DNA polymerases: differential recognition of thymine C5-C6 bond saturation and sequence specificity of incorporation. Nucleic Acids Res. 1988;16:11339–11354. doi: 10.1093/nar/16.23.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang D, Hatahet Z, Melamede R J, Kow Y W, Wallace S S. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 27.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 28.Kow Y W, Wallace S S. Exonuclease III recognizes urea residues in oxidized DNA. Proc Natl Acad Sci USA. 1985;82:8354–8358. doi: 10.1073/pnas.82.24.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreutzer D A, Essigmann J M. Oxidized, deaminated cytosines are a source of C→T transitions in vivo. Proc Natl Acad Sci USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krokan H E, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325(Pt. 1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J S, An G, Friesen J D, Isono K. Cloning and the nucleotide sequence of the genes for Escherichia coli ribosomal proteins L28 (rpmB) and L33 (rpmG) Mol Gen Genet. 1981;184:218–223. doi: 10.1007/BF00272908. [DOI] [PubMed] [Google Scholar]
- 32.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 33.Lu A L, Chang D Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988;54:805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- 34.Michaels M L, Cruz C, Grollman A P, Miller J H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaels M L, Miller J H. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michaels M L, Pham L, Cruz C, Miller J H. MutM, a protein that prevents G·C→T·A transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 1991;19:3629–3632. doi: 10.1093/nar/19.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michaels M L, Tchou J, Grollman A P, Miller J H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 38.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- 39.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 40.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G·C→T·A transversions in simian kidney cells. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman A P. Site-specific mutagenesis using a gapped duplex vector: a study of translesion synthesis past 8-oxodeoxyguanosine in E. coli. Mutat Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 42.Nghiem Y, Cabrera M, Cupples C G, Miller J H. The mutY gene: a mutator locus in Escherichia coli that generates G:C—T:A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikh S S, Mol C D, Tainer J A. Base excision repair enzyme family portrait: integrating the structure and chemistry of an entire DNA repair pathway. Structure. 1997;5:1543–1550. doi: 10.1016/s0969-2126(97)00303-1. [DOI] [PubMed] [Google Scholar]
- 44.Purmal A, Lampman G, Kow Y, Wallace S. The sequence context-dependent mispairing of 5-hydroxycytosine and 5-hydroxyuridine in vitro. Ann N Y Acad Sci. 1994;726:361–363. doi: 10.1111/j.1749-6632.1994.tb52852.x. [DOI] [PubMed] [Google Scholar]
- 45.Purmal A A, Bond J P, Lyons B A, Kow Y W, Wallace S S. Uracil glycol deoxynucleoside triphosphate is a better substrate for DNA polymerase I Klenow fragment than thymine glycol deoxynucleoside triphosphate. Biochemistry. 1998;37:330–338. doi: 10.1021/bi972153d. [DOI] [PubMed] [Google Scholar]
- 46.Purmal A A, Kow Y W, Wallace S S. 5-Hydroxypyrimidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res. 1994;22:3930–3935. doi: 10.1093/nar/22.19.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purmal A A, Kow Y W, Wallace S S. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purmal A A, Lampman G W, Bond J P, Hatahet Z, Wallace S S. Enzymatic processing of uracil glycol, a major oxidative product of DNA cytosine. J Biol Chem. 1998;273:10026–10035. doi: 10.1074/jbc.273.16.10026. [DOI] [PubMed] [Google Scholar]
- 49.Rogers S G, Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65:201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- 50.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J Bacteriol. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 53.Shibutani S, Takeshita M, Grollman A P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 54.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman A, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 55.Tchou J, Kasai H, Shibutani S, Chung M H, Laval J, Grollman A P, Nishimura S. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai-Wu J J, Liu H F, Lu A L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A-C and A-G mispairs. Proc Natl Acad Sci USA. 1992;89:8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vidmar J, Cupples C. MutY repair is mutagenic in mutT− strains of Escherichia coli. Can J Microbiol. 1993;39:892–894. doi: 10.1139/m93-133. [DOI] [PubMed] [Google Scholar]
- 58.Wallace S S. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150:S60–S79. [PubMed] [Google Scholar]
- 59.Wallace S S. Oxidative damage to DNA and its repair. In: Scandalios J, editor. Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 49–90. [Google Scholar]
- 60.Wallace S S, Harrison L, Jiang D, Blaisdell J O, Purmal A A, Hatahet Z. Processing and consequences of oxidative DNA base lesions. NATO ASI Ser Ser A. 1998;302:419–430. [Google Scholar]
- 61.Wang D, Kreutzer D A, Essigmann J M. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 62.Warner H R, Demple B F, Deutsch W A, Kane C M, Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci USA. 1980;77:4602–4606. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss B, Cunningham E, Chan E, Tsaneva I R. AP endonucleases of Escherichia coli. In: Friedberg E C, Hanawalt P, editors. Mechanisms of consequences of DNA damage processing. A. R. New York, N.Y: Liss; 1988. pp. 133–142. [Google Scholar]
- 64.Weiss B, Cunningham R P. Genetic mapping of nth, a gene affecting endonuclease III (thymine glycol-DNA glycosylase) in Escherichia coli K-12. J Bacteriol. 1985;162:607–610. doi: 10.1128/jb.162.2.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White B J, Hochhauser S J, Cintron N M, Weiss B. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J Bacteriol. 1976;126:1082–1088. doi: 10.1128/jb.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson D M, III, Engelward B P, Samson L. Prokaryotic base excision repair. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. Vol. 1. 1998. pp. 29–64. : DNA repair in prokaryotes and lower eukaryotes. Humana Press, Inc., Totowa, N.J. [Google Scholar]
- 67.Wood M L, Dizdaroglu M, Gajewski E, Essigmann J M. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]