Abstract
By inhibiting FtsZ ring formation near the cell ends, the MinC protein plays a critical role in proper positioning of the division apparatus in Escherichia coli. MinC activity requires that of MinD, and the MinE peptide provides topological specificity by suppressing MinC-MinD-mediated division inhibition specifically at the middle of the cell. We recently presented evidence that MinE not only accumulates in an FtsZ-independent ring structure at the cell’s middle but also imposes a unique dynamic localization pattern upon MinD in which the latter accumulates alternately in either one of the cell halves in what appears to be a rapidly oscillating membrane association-dissociation cycle. Here we show that functional green fluorescent protein-MinC displays a very similar oscillatory behavior which is dependent on both MinD and MinE and independent of FtsZ. The results support a model in which MinD recruits MinC to its site of action and in which FtsZ ring assembly at each of the cell ends is blocked in an intermittent and alternate fashion.
Cell division in prokaryotes is initiated by the localization of the tubulin-like GTPase FtsZ to the future division site. FtsZ assembles into a ring, and other proteins are then recruited to form the septal ring organelle which mediates cell envelope invagination (5, 23, 24, 34). Division of Escherichia coli normally occurs at the middle of the cell, but potential division sites (PDSs) are also present near each of the cell poles (1, 37). Restricting division to the cell’s midpoint requires the activity of the protein products of the min operon, MinC, MinD, and MinE (10). MinC can inhibit division at all PDSs by preventing formation of the FtsZ ring but normally requires the activity of the peripheral membrane ATPase MinD for this function (3, 8–10). How MinD stimulates MinC function is not clear, but the two proteins interact in two-hybrid assays, suggesting that they form a complex (18). Mutants lacking either MinC or MinD frequently divide near the cell ends, resulting in the production of small, chromosomeless minicells (9, 10, 12). MinE provides topological specificity to the system by suppressing MinCD division inhibition specifically at midcell, allowing FtsZ ring assembly at this site (10, 31, 32, 42).
We recently showed that a functional green fluorescent protein (Gfp)-tagged derivative of MinE accumulates in a ring at or near midcell. Formation of the MinE ring did not require MinC or FtsZ but depended on the presence of MinD (32). Conversely, functional Gfp-MinD was found to undergo a unique, dynamic, oscillatory localization cycle which is also independent of MinC and FtsZ but requires MinE (33). In the absence of MinE, Gfp-MinD accumulated along the periphery of the entire cell. In MinE+ cells, however, Gfp-MinD segregated to the periphery of one of the cell halves, where it dwelled for a short period of time (∼10 s), and then relocated to the opposite cell half, where it dwelled again, followed by relocation to its original position, and so on. A moderate increase in the Gfp-MinD/MinE ratio in the cell led to an increase in the average dwell time and the concomitant formation of polar septa, suggesting that a certain minimum oscillation frequency of MinD is required to efficiently suppress FtsZ ring formation at both poles (33).
These results support a model in which MinD stimulates division inhibition in one cell half at a time while MinE protects the midcell from MinC-MinD action (20, 33). This model is compatible with two possible mechanisms of MinC-MinD action, depending on the relative affinities of MinC for MinD on the one hand and for its presumed sites of action on the other. Thus, MinC may have a higher affinity for MinD and follow the latter from one cell half to the other. Alternatively, MinC may have a higher affinity for fixed cellular sites, such as cell poles, and become activated whenever the two proteins colocalize.
Supporting the first possibility, we here describe the dynamic properties of a functional Gfp-MinC fusion protein in live cells. Gfp-MinC oscillated from pole to pole in a membrane association-dissociation cycle that is similar to that of Gfp-MinD (33). Furthermore, segregation and oscillation of Gfp-MinC did not require FtsZ ring assembly but were dependent on both MinD and MinE. The results strongly support the notion that E. coli prevents polar division events from occurring by rapidly shuttling the MinC division inhibitor back and forth between the cell ends in a MinD-driven process, leaving only the middle of the cell competent to support septum formation.
MATERIALS AND METHODS
Strains, plasmids, and phages.
Strains PB103 (dadR trpE trpA tna) (11) and PB114 (PB103, ΔminCDE::aph) (10) have been described previously.
Plasmid pDR155 (bla+ lacIq+ Plac::minD), a pMLB1115 derivative lacking any minE sequences, was obtained in two steps. The 2,624-bp PacI/SacI fragment of pDB164 (10) was replaced with the 2,539-bp PacI/SacI fragment of pDR119 (33), yielding pDR150. This same PacI/SacI fragment was next released from pDR150 and used to replace the 3,547-bp PacI/SacI fragment of pDR112 (32).
Plasmid pDR175 (aadA cI857 λpR::gfpmut2-minC) is a pGB2 (pSC101) derivative which encodes a 52.5-kDa fusion protein in which the complete Gfpmut2 peptide (7) is joined by the linker peptide ASMTGGQQMGRIP to residues 5 to 232 of MinC. The plasmid was constructed in several steps. Insertion of the 1,857-bp BamHI/EcoRI fragment of pDB124 (10) into the multicloning sequence of pET21c (Novagen) yielded pDB299. The 1,999-bp BglII/EcoRI fragment of pDB299 was next inserted into the multicloning sequence of pMLB1115 (10). Replacement of the 76-bp XbaI/BamHI fragment of the resulting plasmid (pCH2) with the 771-bp XbaI/BamHI fragment of pDR107c (33) gave rise to pDR115, and replacement of the 2,492-bp ClaI/NsiI fragment of this plasmid with the 1,495-bp ClaI/NsiI fragment of pDB171 (10) resulted in pDR121. Finally, pDR175 was obtained by replacement of the 1,096-bp XbaI/SalI fragment of pDB346 (32) with the 1,614-bp XbaI/SalI fragment of pDR121.
The phages λDB156 (imm21 bla+ lacIq+ Plac::minE), λDB175 (imm21 bla+ lacIq+ Plac::minDE), λDR122 (imm21 bla+ lacIq+ Plac::gfp-minDE), and λDR144 (imm21 bla+ lacIq+ Plac::sfiA) have been described previously (10, 32, 33). Phage λDR155 (imm21 bla+ lacIq+ Plac::minD) was obtained by crossing pDR155 with λNT5 (10).
Microscopy and other methods.
Cells were grown at either 30 or 37°C in M9 minimal salts medium supplemented with tryptophan (50 μg/ml), Casamino Acids (0.2%), maltose (0.2%), and isopropyl-β-d-thiogalactopyranoside (IPTG; as indicated) to an optical density (600 nm) of approximately 0.5. Doubling times ranged from 175 (30°C) to 90 (37°C) min.
For fluorescence and differential interference contrast microscopy, cells were immediately applied to a microscope slide and imaged as described previously (33). For phase-contrast micrographs, cells were chemically fixed (10) and viewed as described before (33). The positions of at least 149 septa were determined to calculate the percentage of polar septa. Western analyses were performed essentially as described previously (12).
RESULTS
Gfp-MinC is functional.
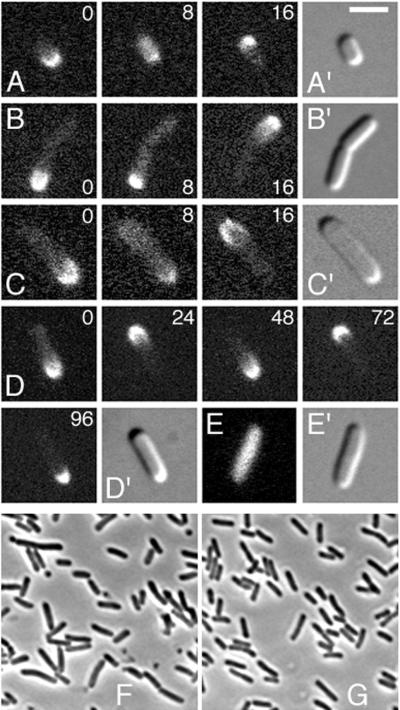
To study the location of MinC in living cells, we constructed pDR175 (λpR::gfp-minC cI857), a low-copy-number plasmid encoding a Gfp-MinC fusion in which the first four amino acids of MinC are replaced by a bright variant of Gfp (Gfpmut2 [7]). Transcription of gfp-minC from this plasmid is under control of the λpR promoter and a temperature-sensitive allele of the λ repressor such that expression of the fusion increases with increasing temperature (Fig. 1). The functionality of Gfp-MinC was studied by testing the ability of pDR175 to correct the minicell phenotype (Min−) of strain PB114(λDB175) [ΔminCDE(Plac::minDE)]. This strain lacks the chromosomal minCDE operon but is lysogenic for a phage which carries the minD and minE genes downstream from the lac promoter. When grown in the absence of IPTG (MinDE−), cells of strain PB114(λDB175)/pDR175 showed the Min− phenotype regardless of the temperature (Table 1). At 30 and 37°C, respectively, 45 and 55% of all septa were misplaced at a pole.
FIG. 1.
Identification of Gfp-MinC with MinC-specific antiserum. Immunoblot showing Gfp-MinC (52.5 kDa; upper arrowhead) and native MinC (24.8 kDa; lower arrowhead) as detected with MinC-specific antiserum. Cells were grown to an optical density (600 nm) of 0.5 at 30°C (lane 6) or 37°C (lanes 1 to 5 and 7) either in the absence (lane 7) or presence (lanes 1 to 6) of 50 μM IPTG, and whole-cell extracts were prepared. Lanes 1 and 2 contain 44 μg of total protein from strain PB114 (ΔminCDE) and PB103 (WT), respectively. Lanes 3 to 7 contain 44 μg (lanes 3, 6, and 7), 8.8 μg (lane 4), or 4.4 μg (lane 5) of protein from strain PB114(λDB175)/pDR175 [ΔminCDE(Plac::minDE)/λpR::gfp-minC]. Samples in lanes 4 and 5 were mixed with appropriate amounts of PB114 extract such that each lane contained 44 μg of total protein.
TABLE 1.
Biological activity, cellular distribution, and oscillation parameters of Gfp-MinCa
| Expt | Strain | Genotype | Temp (°C) | −IPTG
|
+IPTG
|
Location and oscillation parametersb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phenotypec | Distributiond | Phenotype | Distribution | Dwell (range) | Shift (range) | Cycle | ||||
| 1 | PB114/pDR175 | ΔminCDE/λpR::gfp-minC | 37 | Min− | C | Min− | C | |||
| 2 | PB114(λDB156)/pDR175 | ΔminCDE(Plac::minE)/λpR::gfp-minC | 37 | Min− | C | Min− | C | |||
| 3 | PB114(λDR155)/pDR175 | ΔminCDE(Plac::minD)/λpR::gfp-minC | 37 | Min− | C | Sep− | M | |||
| 4 | PB114(λDB175)/pDR175 | ΔminCDE(Plac::minDE)/λpR::gfp-minC | 30 | Min− | NA | Min− | NA | |||
| 5 | PB114(λDB175)/pDR175 | ΔminCDE(Plac::minDE)/λpR::gfp-minC | 37 | Min− | C | WT | O | 11 (6–21) | 10 (7–16) | 42 |
| 6 | PB103/pDR175 | WT/λpR::gfp-minC | 37 | WT | O | WT | O | 11 (7–16) | 11 (8–18) | 44 |
| 7 | PB103(λDR122) | WT/(Plac::gfp-minDE) | 37 | WT | NA | WT | O | 11 (6–17) | 11 (8–17) | 44 |
Cells were grown at the indicated temperature in the absence or presence of 37 μM (experiment 7) or 50 μM (experiment 1 to 6) IPTG. To determine the division phenotype, cells were chemically fixed and observed by phase-contast microscopy.
To determine the location and oscillation parameters of Gfp-MinC (experiment 1 to 6) or Gfp-MinD (experiment 7), cells grown in the presence of IPTG at 37°C were immediately observed by fluorescence microscopy. The dwell period was defined as the period in which the location of the fluorescent fusion appeared static, and the shift period was defined as the period between two consecutive dwell periods. One complete oscillation cycle equals two dwell plus two shift periods. Values are given in seconds and represent the average of at least 69 events in 20 individual cells. Ranges are given in parentheses.
WT, less than 1% of total septa were polar; Min−, minicell producing, more than 14% of septa were polar; Sep−, septation deficient, no division septa formed.
O, segregating and oscillating; M, present along entire membrane, no net movement obvious; C, cytoplasmic, no obvious specific localization; NA, not applicable.
When grown in the presence of 50 μM IPTG (MinDE+), the phenotype of PB114(λDB175)/pDR175 depended on the incubation temperature. At 30°C, cells were still clearly Min− (Fig. 2F; Table 1), although the percentage of polar septa (15%) was reduced significantly when compared to that of cells grown in the absence of IPTG. As judged from immunoblot analyses with MinC-specific antiserum, Gfp-MinC was expressed at a low but detectable level at this temperature (Fig. 1, lane 6), providing a likely explanation for the partial suppression of the Min− phenotype under these conditions. When cells were grown at 37°C, the level of Gfp-MinC increased to a fewfold that of native MinC in the wild-type (WT) strain, PB103 (Fig. 1, lanes 2 to 5). Importantly, cells displayed a normal division phenotype under these conditions (Fig. 2G; Table 1), with less than 1% (2 of 215) of septa being polar. In addition to full-length fusion, cell extracts also contained some smaller species that reacted with anti-MinC antiserum (Fig. 1, lanes 3 to 7). Although these likely represented incomplete translation and/or breakdown products of Gfp-MinC, it cannot be ruled out that they actively contributed to suppression of the Min− phenotype. With this caveat in mind, we conclude that the full-length Gfp-MinC fusion retained a level of biological activity close to that of the native protein.
FIG. 2.
Dynamic properties of functional Gfp-MinC in live cells. Phase-contrast (F and G), fluorescence (A to E), and differential interference contrast (A′ to E′) micrographs showing properties of Gfp-MinC. Cells were grown at 30°C (F) or 37°C (A to E and G) either in the absence (E) or presence (A to D and G) of 50 μM IPTG. (A to D) Time-lapse images showing segregation and oscillation of Gfp-MinC in the presence of MinD and MinE in strains PB114(λDB175)/pDR175 [ΔminCDE(Plac::minDE)/λpR::gfp-minC] (A, B, and D) and PB103(λDR122) (WT/λpR::gfp-minC) (C). Times are indicated in seconds. (E) Random distribution of Gfp-MinC in the absence of MinD and MinE in strain PB114(λDB175)/pDR175. (F and G) Correction of the minicell phenotype (Min−) of strain PB114(λDB175)/pDR175 by Gfp-MinC. Bar, 2 (A to E) or 5 (F and G) μm.
Segregation and oscillation of Gfp-MinC.
Microscopic inspection of normally dividing cells of PB114(λDB175)/pDR175, grown at 37°C (Gfp-MinC+) in the presence of 50 μM IPTG (MinDE+), showed a dynamic distribution of Gfp-MinC fluorescence (Fig. 2A, B, and D) that was remarkably similar to that observed for Gfp-MinD (33). Compared to Gfp-MinD (33), the Gfp-MinC fusion produced a significantly weaker fluorescent signal, resulting in a relatively low signal/background ratio. Nevertheless, Gfp-MinC could be readily observed to segregate to one of the cell halves, where it accumulated at the periphery of the pole and a variable portion of the adjacent cylindrical portion of the cell. The protein dwelled for a short period of time and then moved to take up a similar position in the opposite half of the cell, where it dwelled again before returning to its original position, and so on. Figure 2D shows a typical cell in which Gfp-MinC relocated four times in a span of 96 s. We observed up to 15 relocation events per cell within a span of 305 s, before the signal became too weak to be detected. Segregation and oscillation of Gfp-MinC could be observed in virtually all cells in the population, including very small (Fig. 2A) and constricting (Fig. 2B) cells. As previously noticed with Gfp-MinD (33), the fluorescent signal at the cell’s periphery weakened as it appeared to increase in the cytoplasm during each shift period (Fig. 2A to C, 8 s), suggesting that a portion or all of Gfp-MinC dissociated from the membrane before relocation to the opposite cell half.
Identical behavior of Gfp-MinC was observed when the fusion was expressed in WT cells (Fig. 2C). Cells of strain PB103/pDR175 (WT/λpR::gfp-minC) divided normally at 37°C (Table 1), indicating that expression of Gfp-MinC at this level did not noticeably interfere with the activities of the native Min proteins.
The average oscillation cycle of Gfp-MinC, consisting of two dwell plus two shift periods, was ∼43 s in both PB114(λDB175)/pDR175 and PB103/pDR175 cells. Significantly, this oscillation rate was virtually identical to that of Gfp-MinD in cells of strain PB103(λDR122) [WT/(Plac::gfp-minDE)] grown under similar conditions (Table 1).
These results showed that the behavior of Gfp-MinC in normally dividing cells is remarkably similar to that previously observed for Gfp-MinD (33) and suggested a role for native MinD in the oscillatory behavior of Gfp-MinC in the present experiments.
Segregation and oscillation of Gfp-MinC require both MinD and MinE.
We previously showed that MinE, but not MinC, is required for segregation and oscillation of Gfp-MinD. However, Gfp-MinD localized evenly along the entire periphery of both MinE− MinC+ and MinE− MinC− cells, indicating that MinD associates with the membrane, even in the absence of MinE and MinC (33).
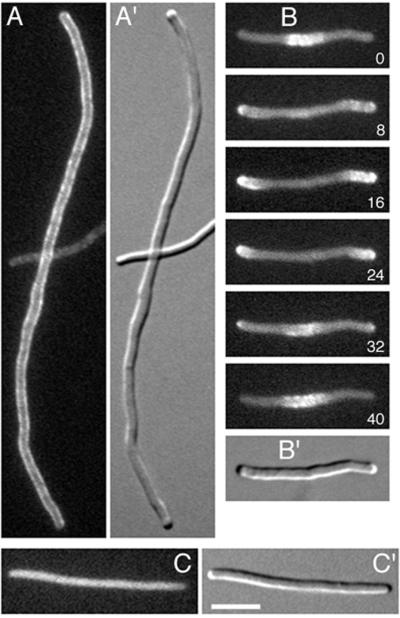
To study the dependency of Gfp-MinC localization on MinD and MinE, pDR175 was introduced into strain PB114(λDR155) [ΔminCDE(Plac::minD)] and cells were grown at 37°C (Gfp-MinC+) in the absence or presence of IPTG. In the presence of 50 μM IPTG (MinD+ MinE−), cells formed long nonseptate filaments, confirming that Gfp-MinC acted as a potent division inhibitor. Furthermore, the fusion failed to show the typical segregation and oscillation behavior seen in normally dividing cells but, instead, appeared evenly distributed along the periphery of the entire filament (Fig. 3A; Table 1). These results suggested that MinD was responsible for recruiting Gfp-MinC to the membrane in these filaments. Support for this idea came from the observation of PB114(λDR155)/pDR175 cells that had been grown in the absence of IPTG (MinD− MinE−). As expected, these cells did not form filaments but showed the Min− phenotype. In addition, Gfp-MinC was present diffusely throughout the cytoplasm, with no obvious preference for the membrane or any other specific location (Table 1). The same cytoplasmic distribution of Gfp-MinC was also observed in strain PB114(λDB175)/pDR175 after growth in the absence of IPTG (Fig. 2E; Table 1) as well in the nonlysogenic strain PB114/pDR175 (Table 1). Expression of minE had no effect on the random distribution of Gfp-MinC in MinD− cells. Thus, cells of strain PB114(λDB156)/pDR175 [ΔminCDE(Plac::minE)/λpR::gfp-minC], which is lysogenic for a phage carrying minE downstream of the lac promoter, showed a random distribution of fluorescence both in the absence (MinD− MinE−) and presence (MinD− MinE+) of IPTG (Table 1).
FIG. 3.
Gfp-MinC localization in filaments. Fluorescence (A to C) and differential interference contrast (A′ to C′) images showing the distribution of Gfp-MinC in cells in which FtsZ ring assembly is blocked. Cells were grown at 37°C with 50 μM IPTG. (A) Gfp-MinC localization in MinE− filaments of strain PB114(λDR155)/pDR175 [ΔminCDE(Plac::minD)/λpR::gfp-minC]. (B) Time-lapse images of Gfp-MinC localization in a SfiA-induced filament of strain PB103(λDR144)/pDR175 [WT(Plac::sfiA)/λpR::gfp-minC]. Times are indicated in seconds. (C) Random distribution of Gfp-MinC in a SfiA-induced filament of strain PB114(λDR144)/pDR175 [ΔminCDE(Plac::sfiA)/λpR::gfp-minC]. Bar, 5 μm.
We conclude that MinD recruits MinC to the membrane and that both MinD and MinE are required for the oscillatory behavior of Gfp-MinC seen in normally dividing cells.
Segregation and oscillation of Gfp-MinC do not require FtsZ ring assembly.
SfiA (SulA) is a MinCDE-independent division inhibitor which is normally not expressed but is induced as part of the SOS response to DNA damage. The protein binds FtsZ directly (15, 18), inhibits polymer formation in vitro (26, 38), and prevents FtsZ ring assembly in vivo (3). In contrast, SfiA does not interfere with MinE ring formation, and SfiA-induced filaments contain multiple MinE ring structures (32). To examine the behavior of Gfp-MinC under conditions where FtsZ ring assembly is prevented, we constructed strain PB103(λDR144)/pDR175 [WT(Plac::sfiA)/λpR::gfp-minC], in which sfiA is under control of the lac promoter. When grown in the presence of 50 μM IPTG, cells formed nonseptate filaments. Similar to what we observed with Gfp-MinD in FtsZ− filaments (33), Gfp-MinC accumulated in membrane-associated segments throughout the length of these filaments. A time lapse series of a short filament containing three such segments is shown in Fig. 3B. At time zero (t = 0), Gfp-MinC is present in the central segment but absent from both polar segments. Eight seconds later, the fusion is present throughout the filament, with some accumulation at the membrane at the two cell poles. At t = 16 s, the fusion is located exclusively at the membrane in the two polar segments. Between t = 24 s and t = 32 s, the fusion is in transition from the polar segments to the central segment, with decreased accumulation at the polar segments and increased accumulation in the central segment. At t = 40 s, the cycle is complete, and the fusion has returned to the membrane of the central segment. Just as observed with Gfp-MinD in FtsZ− filaments (33), longer filaments contained an increasing number of segments and Gfp-MinC appeared to move from the membrane of one segment to the membrane of neighboring segments (data not shown). As in WT cells (see above), segregation and oscillation of Gfp-MinC in SfiAinduced filaments were dependent on the presence of MinD and MinE, and the fusion was evenly distributed throughout the cytoplasm of SfiA-induced filaments of strain PB114 (λDR144)/pDR175 [ΔminCDE(Plac::sfiA)/λpR::gfp-minC] (Fig. 3C).
We conclude that assembly of FtsZ rings is not required for segregation and oscillation of Gfp-MinC.
DISCUSSION
Recently, we showed that functional Gfp-MinD rapidly oscillates between the two cell halves in a membrane association-dissociation cycle which is dependent on MinE but does not require MinC or FtsZ (33). Here we showed that biologically active Gfp-MinC undergoes a very similar localization cycle in which the protein oscillates between cell ends at about the same rate as that observed for Gfp-MinD. Like Gfp-MinD (33), segregation and oscillation of Gfp-MinC required the presence of MinE and occurred independently of FtsZ rings. However, whereas Gfp-MinD localizes independently of MinC, the localization of Gfp-MinC appeared to be directly dictated by MinD. Thus, like MinD itself (33), Gfp-MinC preferentially associated along the entire periphery of MinE− filaments, but the fusion failed to accumulate at any specific location in the absence of MinD regardless of the presence of MinE.
Combined with the knowledge that MinC and MinD show a strong interaction in two-hybrid assays (18), these observations support a model for MinC action which is summarized in Fig. 4. In the absence of MinD and MinE, MinC has no intrinsic affinity for any special site and is present throughout the cell (Fig. 4A). In MinE− cells, MinD associates with the cytoplasmic membrane and recruits MinC. The even distribution of MinC-MinD along the membrane results in a block of FtsZ ring assembly at all PDSs and the formation of nonseptate filaments (Fig. 4B). In WT cells, MinE accumulates in a ring at midcell and stimulates the dynamic oscillatory behavior of MinD. MinC follows along with MinD and either remains associated with MinD throughout oscillation or disengages during disassembly of MinD from the membrane, only to reengage when MinD assembles at the membrane in the opposite half. In either case, MinC actively interferes with FtsZ ring assembly at only one of the cell poles at a time (Fig. 4C).
FIG. 4.
Model for MinCDE action in preventing aberrant septation events. Symbols: ●, MinC;  , MinD;
, MinD;  , the MinE ring. PDSs are represented by either a minus (blocked by MinC-MinD) or a plus (not blocked, available for FtsZ ring assembly) sign. (A) In the absence of MinD and MinE, MinC localizes nonspecifically to the cytoplasm and has no effect on septal or FtsZ ring formation. (B) In the presence of MinD, and absence of MinE, MinC associates with MinD along the entire membrane, preventing FtsZ ring formation at all PDSs. (C) In WT cells, MinC co-oscillates with MinD from one side of the MinE ring to the other, actively interfering with FtsZ ring assembly at each cell end in a sequential and rapidly repeating fashion. (D) In cells lacking FtsZ rings, multiple MinE rings define three or more cell segments. As in WT cells, MinC co-oscillates with MinD between the segments flanking each MinE ring. Note that although the figure suggests the presence of a limited number of regularly spaced PDSs in each cell, the proposed mechanism of MinCDE action does not depend on the exact number or nature of the PDSs and is equally tenable whether potential sites for FtsZ ring assembly are (co)determined by positioning of the nucleoids (41) or any other mechanism.
, the MinE ring. PDSs are represented by either a minus (blocked by MinC-MinD) or a plus (not blocked, available for FtsZ ring assembly) sign. (A) In the absence of MinD and MinE, MinC localizes nonspecifically to the cytoplasm and has no effect on septal or FtsZ ring formation. (B) In the presence of MinD, and absence of MinE, MinC associates with MinD along the entire membrane, preventing FtsZ ring formation at all PDSs. (C) In WT cells, MinC co-oscillates with MinD from one side of the MinE ring to the other, actively interfering with FtsZ ring assembly at each cell end in a sequential and rapidly repeating fashion. (D) In cells lacking FtsZ rings, multiple MinE rings define three or more cell segments. As in WT cells, MinC co-oscillates with MinD between the segments flanking each MinE ring. Note that although the figure suggests the presence of a limited number of regularly spaced PDSs in each cell, the proposed mechanism of MinCDE action does not depend on the exact number or nature of the PDSs and is equally tenable whether potential sites for FtsZ ring assembly are (co)determined by positioning of the nucleoids (41) or any other mechanism.
Given the evidence for a direct interaction between MinC and MinD, the formal possibility that the two proteins segregate to opposite cell halves and oscillate out of phase by half a cycle is far less likely. So far, attempts to exclude this possibility by observing cells in which the two proteins are tagged with different color Gfp derivatives have failed, due to insufficient signal intensities of the nongreen varieties. However, in MinE+ cells in which Gfp-MinC and Gfp-MinD are expressed simultaneously, we have observed that fluorescence still clearly segregates to only one cell end at a time, indicating that the two proteins indeed co-oscillate in the same direction (data not shown).
It is also noteworthy that the average oscillation rate of Gfp-MinC measured in WT cells (∼40 s/cycle) was very close to that of Gfp-MinD in cells in which gfp-minD was coexpressed with minE to maintain a normal MinD-to-MinE ratio (33). This finding not only further supports the idea that the behavior of Gfp-MinC directly reflects that of native MinD in the present experiments but also suggests that oscillation parameters measured with the Gfp-tagged proteins fairly accurately reflect those of native MinC-MinD.
How MinC prevents FtsZ ring assembly is not known. Two-hybrid studies failed to show an interaction between the two proteins (18), and it is well possible that MinC interferes with the activity of another factor required for FtsZ ring assembly, such as a hypothetical membrane-associated molecule that might nucleate FtsZ polymerization. How MinD stimulates MinC activity is also not clear. The present results suggest one straightforward possibility, however; i.e., by recruiting MinC to the membrane, MinD may simply act to increase the local concentration of MinC to effective levels. Compatible with this idea is the fact that an increase in the cellular concentration of MinC to more than ∼30-fold its normal level is sufficient to cause division inhibition, even in the absence of MinD (12). Whether this idea is correct or not, the finding that Gfp-MinC associates with MinD at the cell’s periphery indicates that MinC-MinD-mediated division inhibition is a membrane-associated event, arguing against mechanisms whereby MinC directly modifies cytoplasmic FtsZ pools to a polymerization-incompetent state.
This work extends and supports our observations on the remarkable properties of the Min proteins in living E. coli cells (32, 33). One of the surprising implications is that MinC-MinD prevents aberrant FtsZ ring formation intermittently, at only one of the cell poles at a time. This mode of action appears not to have been conserved in the gram-positive rod Bacillus subtilis (14, 20, 25) and begs the question why such an oscillatory mechanism might have developed. MinD and MinE determine each other’s localization pattern (32) and, as suggested before (33), one attractive possibility is that oscillation of MinD between the cell segments on either side of a MinE ring is coupled to positioning of the ring. In this view, oscillation of MinD provides cells with a measuring device which allows the positioning of MinE at midcell, in the case of WT cells containing one ring, or at regularly spaced intervals, in the case of filaments containing multiple rings.
The idea that the Min system, in addition to preventing aberrant FtsZ ring assembly, may also function as a measuring device is supported by recent work by Yu and Margolin (41) in which they analyzed the placement of FtsZ rings in Min− mutants containing additional mutations that affect nucleoid partitioning (Par−). Placement of division septa in Min− mutants is typically not random but is restricted to a narrow area near each cell pole and regular positions between segregated nucleoids (2, 13, 21, 22, 37). One factor that is likely to contribute to this nonrandom pattern is a phenomenon called nucleoid occlusion which is based on the observation that septum formation appears to be inhibited in the close vicinity of the nucleoid(s) of certain DNA replication and topoisomerase mutants (19, 27, 28, 30, 36, 41). The mechanism of this inhibitory effect is not understood, although it is conceivable that the formation of septal rings at envelope sites directly surrounding a nucleoid would be sterically hindered by an abundance of membrane-associated transcription and translation complexes (29). In some models, positioning of the nucleoid is proposed to be the sole determining factor for positioning of the division apparatus (2, 39, 40), but this idea is refuted by a host of experimental data (references 4, 6, 16, 35, and 36 and references therein). For instance, mutants affected in nucleoid replication and segregation typically form filamentous cells which release chromosomeless cells from their ends. In several such mutants, septal placement is not random and the size distribution of the DNA-less rods is close to that of normal newly born cells, suggesting that cells can somehow measure a certain distance from the cell pole (4, 6, 17, 36).
Compelling evidence that the Min proteins are involved in defining this distance comes from the observation that the positioning of FtsZ rings becomes essentially random in the nucleoid-free segments of ΔminCDE parC double mutants (41). One of several interesting predictions of this work is that MinC-MinD in WT cells inhibits FtsZ ring assembly not only at the extreme cell ends but throughout most of the cell envelope except for a relatively narrow zone at the cell’s center defined by the MinE ring (41). Our observations on the distribution of Gfp-MinC and Gfp-MinD are compatible with this possibility insofar that, during dwell periods, the association of the two proteins with the membrane is clearly not restricted to the polar caps. This was especially obvious in the case of Gfp-MinD, which was frequently seen to cover the membrane from a pole to approximately the middle of the cell (33). In general, Gfp-MinC did not appear to extend as far toward midcell, although the protein was still observed to cover at least one-quarter of the membrane in most cells. Whether this was due to low signal intensities or whether factors, in addition to the location of MinD, further bias Gfp-MinC localization toward the cell ends is not clear. In any event, additional analyses of the behavior of the Min proteins in relation to nucleoid dynamics should prove valuable in testing the proposal by Yu and Margolin that the combination of MinCDE action and nucleoid occlusion may be sufficient to explain septal placement in E. coli (41).
Clearly, the dynamic behavior of MinC and MinD raises many new questions requiring additional experimentation. Further elucidation of the mechanisms underlying membrane assembly-disassembly of MinC-MinD and of the role(s) of the MinE ring is likely to contribute significantly to our understanding of the spatial organization of bacterial cells.
ACKNOWLEDGMENTS
We thank Cynthia Hale for help in plasmid construction, technical advice, and comments on the manuscript.
This work was supported by NIH grant GM-57059, NSF Young Investigator Award MCB94-58197, and generous donations from The Elizabeth M. and William C. Treuhaft Fund, The Frank K. Griesinger Trust, Arline H. Garvin, James S. Blank, Charles E. Spahr, Alfred M. Taylor, and Theodore J. Castele (to P.A.J.D.). D.M.R. was supported by an NRSA Institutional Training Grant (T32GM08056) from the National Institutes of Health.
ADDENDUM IN PROOF
Marston and Errington recently showed that the cellular location of Bacillus subtilis MinC is also directly dictated by that of MinD (A. L. Marston and J. Errington, Mol. Microbiol. 33:84–96, 1999).
REFERENCES
- 1.Adler H I, Fisher W D, Cohen A, Hardigree A A. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci USA. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Åkerlund T, Bernander R, Nordström K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botello E, Nordström K. Effects of chromosome underreplication on cell division in Escherichia coli. J Bacteriol. 1998;180:6364–6374. doi: 10.1128/jb.180.23.6364-6374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 6.Cook W R, Rothfield L I. Nucleoid-independent identification of cell division sites in Escherichia coli. J Bacteriol. 1999;181:1900–1905. doi: 10.1128/jb.181.6.1900-1905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 8.de Boer P A J, Crossley R E, Hand A R, Rothfield L I. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer P A J, Crossley R E, Rothfield L I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 11.de Boer P A J, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer P A J, Crossley R E, Rothfield L I. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donachie W D, Begg K J. Division potential in Escherichia coli. J Bacteriol. 1996;178:5971–5976. doi: 10.1128/jb.178.20.5971-5976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards D H, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 15.Higashitani A, Higashitani N, Horiuchi K. A cell division inhibitor SulA of Escherichia coli directly interacts with FtsZ through GTP hydrolysis. Biochem Biophys Res Commun. 1995;209:198–204. doi: 10.1006/bbrc.1995.1489. [DOI] [PubMed] [Google Scholar]
- 16.Hill T M, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179:1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirota Y, Jacob F, Ryter A, Buttin G, Nakai T. On the process of cellular division in E. coli. I. Asymmetrical cell division and production of DNA-less bacteria. J Mol Biol. 1968;35:175–192. doi: 10.1016/s0022-2836(68)80046-4. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussain K, Begg K G, Salmond G P C, Donachie W D. ParD: a new gene coding for a protein required for chromosome partitioning and septum localization in Escherichia coli. Mol Microbiol. 1987;1:73–81. doi: 10.1111/j.1365-2958.1987.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs C, Shapiro L. Bacterial cell division: a moveable feast. Proc Natl Acad Sci USA. 1999;96:5891–5893. doi: 10.1073/pnas.96.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffé A, Boye E, D’Ari R. Rule governing the division pattern in Escherichia coli minB and wild-type filaments. J Bacteriol. 1990;172:3500–3502. doi: 10.1128/jb.172.6.3500-3502.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin P A, Shim J J, Grossman A D. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 24.Margolin W. A green light for the bacterial cytoskeleton. Trends Microbiol. 1998;6:233–238. doi: 10.1016/s0966-842x(98)01280-3. [DOI] [PubMed] [Google Scholar]
- 25.Marston A L, Thomaides H B, Edwards D H, Sharpe M E, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder E, El’Bouhali M, Pas E, Woldringh C L. The Escherichia coli minB mutation resembles gyrB in defective nucleoid segregation and decreased negative supercoiling of plasmids. Mol Gen Genet. 1990;221:87–93. doi: 10.1007/BF00280372. [DOI] [PubMed] [Google Scholar]
- 28.Mulder E, Woldringh C L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanninga N. Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:110–129. doi: 10.1128/mmbr.62.1.110-129.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orr E, Fairweather N F, Holland I B, Pritchard R H. Isolation and characterization of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177:103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- 31.Pichoff S, Vollrath B, Touriol C, Bouché J-P. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 32.Raskin D M, de Boer P A J. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 33.Raskin D M, de Boer P A J. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 35.Sharpe M E, Errington J. Postseptational chromosome partitioning in bacteria. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–503. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 37.Teather R M, Collins J F, Donachie W D. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woldringh C L, Mulder E, Huls P G, Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- 40.Woldringh C L, Mulder E, Valkenburg J A C, Wientjes F B, Zaritsky A, Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990;141:39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- 41.Yu X-C, Margolin W. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol. 1999;32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C-R, de Boer P A J, Rothfield L I. Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc Natl Acad Sci USA. 1995;92:4314–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]