Abstract
DNA arrays of the entire set of Escherichia coli genes were used to measure the genomic expression patterns of cells growing in late logarithmic phase on minimal glucose medium and on Luria broth containing glucose. Ratios of the transcript levels for all 4,290 E. coli protein-encoding genes (cds) were obtained, and analysis of the expression ratio data indicated that the physiological state of the cells under the two growth conditions could be ascertained. The cells in the rich medium grew faster, and expression of the majority of the translation apparatus genes was significantly elevated under this growth condition, consistent with known patterns of growth rate-dependent regulation and increased rate of protein synthesis in rapidly growing cells. The cells grown on minimal medium showed significantly elevated expression of many genes involved in biosynthesis of building blocks, most notably the amino acid biosynthetic pathways. Nearly half of the known RpoS-dependent genes were expressed at significantly higher levels in minimal medium than in rich medium, and rpoS expression was similarly elevated. The role of RpoS regulation in these logarithmic phase cells was suggested by the functions of the RpoS dependent genes that were induced. The hallmark features of E. coli cells growing on glucose minimal medium appeared to be the formation and excretion of acetate, metabolism of the acetate, and protection of the cells from acid stress. A hypothesis invoking RpoS and UspA (universal stress protein, also significantly elevated in minimal glucose medium) as playing a role in coordinating these various aspects and consequences of glucose and acetate metabolism was generated. This experiment demonstrates that genomic expression assays can be applied in a meaningful way to the study of whole-bacterial-cell physiology for the generation of hypotheses and as a guide for more detailed studies of particular genes of interest.
The field of microbial physiology was launched in 1958 with the fundamental discovery that the macromolecule composition of the bacterial cell changes with the growth rate (58). Faster-growing cells contain proportionally more stable RNAs—rRNA and tRNA. The reason for this increased abundance of stable RNA is simple: in order to grow faster, bacteria must synthesize protein faster. The growth rate of the bacterial cell increases in proportion to the quality of the growth medium (although not necessarily in proportion to its exact composition), and this increase in growth rate is accomplished by an increase in the number of ribosomes and the concentrations of translation accessory factors (8). It is now understood that the seven Escherichia coli rRNA operons are under the control of growth rate-dependent promoters and that expression of the ribosomal proteins, translation factors, and the transcription apparatus are all tied to the cellular concentration of rRNA (8, 27, 35). The rate of transcription initiation of the growth rate-dependent rrn promoters is physiologically connected to the metabolic state of the cell by the concentration of nucleoside triphosphates—efficient transcription initiation from these promoters requires a high concentration of the initiating nucleotide (22). The presence of high-quality nutrients in the growth medium results in high intracellular nucleoside triphosphate concentrations; hence, this model unifies the idea that the quality of the growth medium dictates the growth rate of the cell.
Growth rate-dependent changes in cell composition are realized at the level of gene expression; for example, transcript levels corresponding to the protein components of the protein synthesis apparatus change in proportion to the growth rate as the rates of transcription or mRNA turnover are modulated (27, 35). Other changes in cellular physiology can be more subtle, such as redirection of intermediary metabolism in response to changes in growth medium composition or the flow of carbon and electrons that is coupled to ATP generation, although many of these adjustments in metabolism are accompanied by changes in the concentrations of metabolic enzymes and electron transport chain components (40, 41, 56, 63, 64). The expression of numerous other genes is affected by environmental stresses (9, 17, 26, 29, 48, 60, 69, 71). Almost all aspects of microbial physiology, including the myriad adjustments made by the cell in response to changes in the environment, have been cataloged by the scientific community in the form of the book Escherichia coli and Salmonella: Cellular and Molecular Biology. Since the publication of this compendium, the sequence of the E. coli genome has been completed and the way that we look at gene expression is forever changed (6). The genome sequence provides the tools necessary to take a global view of E. coli physiology.
Genomic expression assays provide an unprecedented ability not only to look at a single aspect of physiology but also to see how a particular gene, regulon, or modulon interacts with every other aspect of physiology. Genomewide methods have been developed for a number of uses, including drug discovery (43), measurement of gene copy number (50), discovery of disease-related genes in humans (18, 28), gene mapping (12), and gene expression: in humans (73), in yeast (13, 19, 31, 37, 65), and in Arabadopsis (59).
From the E. coli MG1655 genome sequence (6), 4,290 open reading frame (ORF)-specific primer pairs were designed for PCR amplification of all E. coli ORFs, and this set of 4,290 PCR-amplified, ORF-specific DNA fragments was used to develop DNA arrays for gene expression profiling (54). A similar set of ORF-specific DNA fragments was used to generate commercially available DNA macroarrays (12 by 24 cm) on nylon membranes (Sigma-GenoSys Biotechnologies, Inc., Woodland, Tex.). The advantage of the commercial arrays is that they can be used with equipment found in typical molecular biology laboratories. For these utilitarian investigations of bacterial physiology to be successful, it will be necessary to determine if DNA macroarrays can reveal differences in gene expression across the genome. Here we report on the expression profiles of E. coli under two very different growth conditions, and from the data we provide insights into growth rate-dependent gene expression, global regulation of biosynthetic regulons, and stress responses that appear to be involved in growth on minimal glucose medium.
MATERIALS AND METHODS
Growth conditions.
E. coli MG1655 cultures were grown in 50-ml batch cultures in 250-ml Erlenmeyer flasks at 37°C with aeration by gyrotary shaking (300 rpm). The culture media used were M63 minimal medium (57) containing 0.2% glucose and a rich medium, Luria broth (39) containing 0.2% glucose. Growth was monitored spectrophotometrically at 600 nm on a Spectronic 601 (Milton Roy). Cells were harvested in late logarithmic growth phase (absorbance at 600 nm = 0.6) from cultures that had been inoculated at low density and had maintained a constant growth rate for at least 10 generations.
Handling of RNA.
The ability to isolate pure, intact mRNA is critical to the success of genomic expression assays. Cells in growing cultures were pipetted directly into boiling lysis buffer. The lysed cells were extracted twice with phenol (pH 5.0) at 60°C and then with phenol-chloroform (66). The RNA was precipitated with isopropanol, redissolved in water, treated with DNase I, and applied to an RNeasy column. The purified RNA was redissolved in water and stored at −70°C in 2 volumes of ethanol.
Probe synthesis.
Hybridization probes were generated by standard cDNA synthesis. The protocol supplied by the manufacturer of the DNA arrays was suitable for achieving >70% incorporation of the 33P-labeled nucleotide. Since it is not possible to purify bacterial mRNA from total RNA (i.e., by purification of polyadenylated mRNA as in eukaryotes), the labeling protocol takes into account the presence of rRNA and tRNA, which constitute 85% of the total RNA. The C-terminal primer set (4,290 ORF-specific C-terminal primers [Sigma-GenoSys Biotechnologies, Inc.]) was used to generate the hybridization probe in a standard first-strand cDNA synthesis. Briefly, 1 μg of RNA was mixed with dATP, dGTP, and dTTP (final concentrations, 0.33 mM each), and cDNA-labeling primers (Sigma-GenoSys), in a volume of 25 μl of first-strand buffer, heated to 90°C for 2 min and cooled to 42°C in 20 min. Then 200 U of Superscript II, 10 U of RNase inhibitor, and 20 μCi of [α-32P]dCTP (2,000 to 3,000 Ci/mmol) were added, bringing the total volume to 30 μl, and the cDNA synthesis reaction mixture was incubated at 42°C for 2 h. Unincorporated nucleotides were removed by gel filtration through a G-50 Sephadex column (57).
Hybridization.
The DNA arrays (Panorama E. coli gene arrays) used in the hybridization experiments were produced by Sigma-GenoSys Biotechnologies, Inc. Each DNA array consists of a 12- by 24-cm positively charged nylon membrane on which 10 ng each of all 4,290 PCR-amplified ORF-specific DNA fragments are robotically printed in duplicate. The hybridization and washing steps were carried out as described by the manufacturer. Briefly, the blots were prehybridized in hybridization solution (5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 2% sodium dodecyl sulfate [SDS], 1× Denhardt’s reagent, 100 μg of sheared salmon sperm DNA per ml) at 65°C for 1 h in a 30- by 3.5-cm roller bottle in a hybridization oven. The entire cDNA probe, generated as described above, was added to 3 ml of hybridization buffer, and the blot was hybridized with this solution for 15 h at 65°C. The blots were washed with buffer (0.5× SSPE, 0.2% SDS) three times for 5 min each at room temperature and three times for 30 min each at 65°C. The blots were then wrapped in clear plastic food wrap and exposed to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.) for 48 h. For each of the data sets used in this study, the same blot was consecutively hybridized, stripped, and rehybridized (this can be done up to four times). The blots were stripped at 100°C with 1% SDS in Tris-EDTA buffer as specified by the manufacturer.
Data analysis.
The exposed PhosphorImager screens were scanned with a pixel size of 100 μm (10,000 dots/cm2) on a STORM 840 PhosphorImager (Molecular Dynamics). The resulting TIFF image files were analyzed by determining the pixel density (intensity) for each spot in the array by using ImageQuant (version 5.0) software (Molecular Dynamics). A grid of individual ellipses corresponding to each of the DNA spots on the blots was laid down on the image to designate each spot to be quantified. Background was subtracted automatically by the software by using the local median background subtraction method. The intensities for each spot were exported from ImageQuant into a Microsoft Excel spreadsheet. Each ORF-specific spot was present in duplicate, and the intensities were averaged for analysis. Each averaged spot intensity was expressed as a percentage of the total of intensities of all the spots on the DNA array, which allowed direct comparison of the two conditions by normalizing with regard to the specific activity of the probes used. The correlation coefficients of the percent intensities determined individually for the duplicate spots on a single blot ranged from 0.986 to 0.999, and the standard deviations for the log ratios of intensities of the duplicate spots (determined as described below) ranged from 0.073 to 0.095 for four different hybridizations, thus providing a measure of reproducibility.
Two growth conditions were compared by determining the ratio of the corresponding averaged percent intensities of each pair of ORF-specific spots on the two blots. These ratios represent the relative transcript levels of each E. coli ORF under the two growth conditions. Ratios were calculated such that the log of the absolute value of the expression ratio was positive for percent intensities that were higher under the first condition and negative for percent intensities that were higher under the second condition. Also taken into account in the calculation were situations where the percent intensities for both conditions fell below a threshold value equal to the background, that is, when the gene was not expressed at detectable levels under either condition; in this case, the calculated log expression ratio was zero. A threshold value, equal to the background, was used to calculate ratios where a gene was not expressed at detectable levels under one of the growth conditions. A statistical analysis of the log expression ratios of all 4,290 genes in the minimal glucose versus gluconate experiment indicated a standard deviation from the mean (0.000) of 0.180. There is 95.5% confidence that any expression ratio is significant if the value of the log expression ratio is greater than 2 standard deviations (0.360) from the mean. Thus, a log expression ratio of 0.400 (2.5-fold) was considered to indicate significantly higher expression (99% confidence of each tail) in the analyses, and this value is shown graphically in Fig. 3 to 6. The experiment presented here, comparing the expression profile of cells grown on minimal versus rich medium, was repeated, and qualitatively similar data were obtained (data not shown). The blot-to-blot reproducibility of DNA macroarray hybridization data has been addressed in detail elsewhere (54).
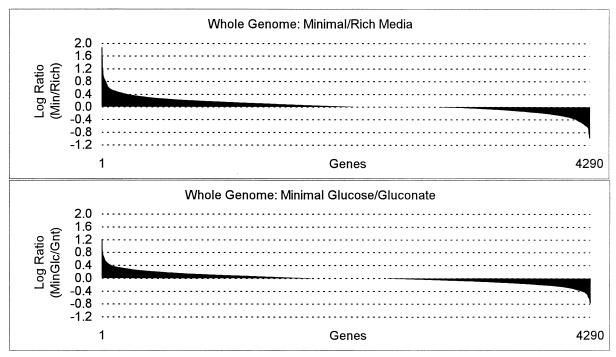
FIG. 3.
The log expression ratios of all E. coli genes were plotted for minimal glucose versus Luria broth plus glucose (top) and for minimal glucose versus minimal gluconate (bottom). The entire data sets were sorted in Excel spreadsheets by the log expression ratio values, and a bar chart was generated by the software, with individual genes plotted on the x axis and the log expression ratios plotted on the y axis. Genes more highly expressed under the first condition are positive, and genes more highly expressed under the second condition are negative. The horizontal divisions (dashed lines) represent 99% confidence levels, such that any gene with a value extending beyond the first horizontal division in either direction is significantly expressed at a higher level under that condition.
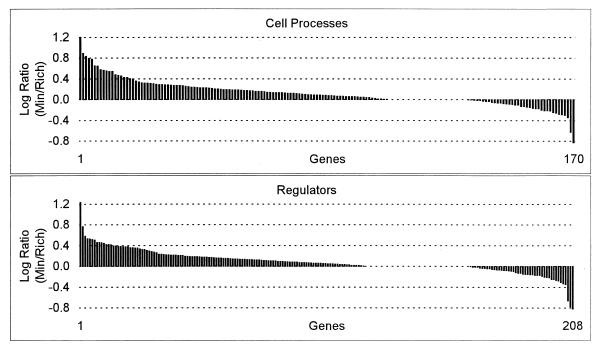
FIG. 6.
Log expression ratios of cell process genes and regulatory genes sorted by value for the minimal glucose versus Luria broth plus glucose experiment.
Functional groups.
Two schemes for functional grouping of genes have been applied to the expression data generated in these experiments. The first scheme assigns genes to groups in accordance with their cellular function, as described previously (6). The second scheme of functional assignments is that of Riley (55), version M54, submitted by Plunkett et al. (19a), as it appears on the E. coli K-12 MG1655 complete genome at the National Center for Biotechnology Information (43a).
Internet access to data.
An Internet accessible version of the expression data and details of the protocols has been created (49a). The data can also be accessed from a database (19a).
Chemicals.
SuperScript II, an RNase H− reverse transcriptase used for cDNA synthesis, was purchased from Gibco BRL (Bethesda, Md.). RNase inhibitor and DNase I were also purchased from Gibco BRL. PCR grade deoxyribonucleoside triphosphates were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.). RNeasy columns were purchased from Qiagen, Inc. (Valencia, Calif.). [α-33P]dCTP (2,000 to 3,000 Ci/mmol) was purchased from New England Nuclear (Wilmington, Del.). Biochemicals were purchased from Sigma (St. Louis, Mo.).
RESULTS AND DISCUSSION
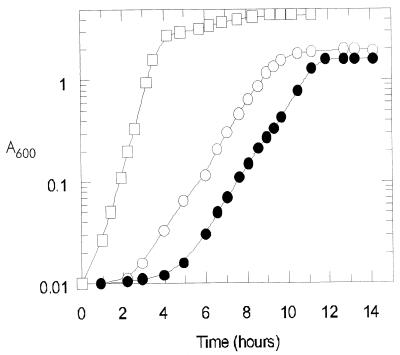
The genomic expression profiles of E. coli MG1655 growing on rich and on minimal culture media (Fig. 1) were determined. The rich medium (Luria broth) contained amino acids as the nitrogen source, a number of other preformed building blocks of macromolecule synthesis (e.g., nucleosides and vitamins, etc., provided by tryptone and yeast extract), and also glucose as a carbon and energy source. The minimal medium contained glucose as the sole carbon and energy source and ammonia as the nitrogen source. In glucose minimal medium, the carbon backbone of the glucose molecule was rearranged through the biosynthetic pathways to generate each of the building blocks de novo. In addition to having fundamentally different metabolisms, the two cultures grew at significantly different rates: G = 25 min on the rich medium and G = 57 min on minimal glucose medium. As a control, data are provided for a culture growing on minimal gluconate medium (G = 60 min).
FIG. 1.
Growth of E. coli MG1655 on Luria broth plus glucose (open squares), minimal glucose medium (open circles), and minimal gluconate medium (solid circles). Cells were harvested for genomic expression analysis at an absorbance at 600 nm (A600) of 0.6.
Whole-genome perspective.
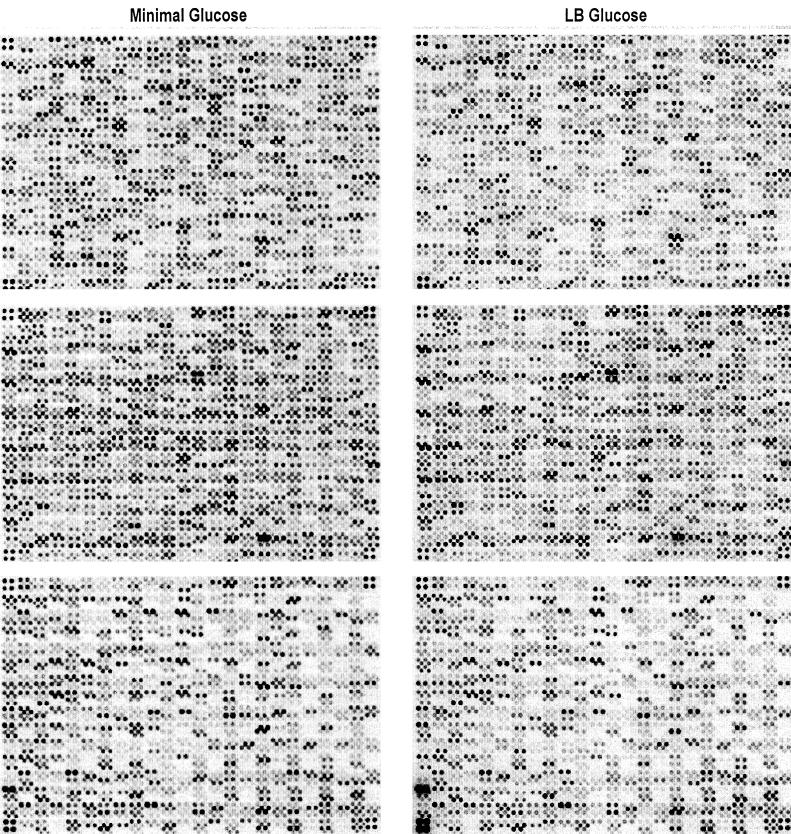
RNA isolated from the cultures in Fig. 1 were used to generate the probes used for hybridization of the DNA arrays shown in Fig. 2, and the data were quantified as described in Materials and Methods. Calculation of the log expression ratios of corresponding spots allowed pairwise comparisons of the relative transcript levels for each of the 4,290 E. coli protein-encoding genes under the different growth conditions. The log expression ratios indicate whether gene expression is higher under one condition or the other or remains unchanged. The results are summarized in Table 1 and presented in chart form in Fig. 3 to 6. It is important to keep in mind that in vivo transcript levels are dynamically balanced by the rates of transcription initiation and transcript turnover. Thus, the data presented here as expression ratios reflect the relative transcript levels for individual genes without providing any indication of the mechanism of regulation. Furthermore, some individual expression ratios may be in error, due to technical problems, including cross-hybridization, PCR failures, misapplied DNA spots on the arrays, or scatter in the data (see reference 54 for a more comprehensive review of the technical aspects of using E. coli DNA arrays). A few of the ratios are in conflict with published results, and it is possible that other ratios will not be validated in subsequent experiments. Thus, these data should not be taken as specific evidence for gene regulation and should be independently verified. Nevertheless, the general trends of the data are substantially clear and will be of value for generating experimental leads.
FIG. 2.
DNA arrays of the entire set of E. coli genes hybridized with probes generated from RNA extracted from cells growing in late logarithmic phase on minimal glucose medium (left) and on Luria broth (LB) containing glucose (right).
TABLE 1.
Expression ratios of functional groups
| Functional group | No. of genesa
|
||||
|---|---|---|---|---|---|
| Total | Minimal glucose vs Luria broth plus glucose
|
Minimal glucose vs minimal gluconate
|
|||
| Higher on minimal | Higher on LB | Higher on glucose | Higher on gluconate | ||
| Whole genome | 4,290 | 225 | 119 | 80 | 82 |
| Amino acid biosynthesis | 97 | 22 | 0 | 3 | 0 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 106 | 9 | 1 | 0 | 2 |
| Carbon compound catabolism | 124 | 3 | 0 | 1 | 2 |
| Cell processes | 170 | 19 | 2 | 5 | 1 |
| Cell structure | 85 | 2 | 0 | 8 | 0 |
| Central intermediary metabolism | 149 | 15 | 1 | 4 | 5 |
| DNA replication, repair, restriction/modification | 105 | 1 | 0 | 1 | 1 |
| Energy metabolism | 136 | 14 | 5 | 4 | 3 |
| Fatty acid and phospholipid metabolism | 41 | 2 | 7 | 0 | 0 |
| Hypothetical, unclassified, unknown | 1,428 | 43 | 26 | 10 | 30 |
| Nucleotide biosynthesis and metabolism | 66 | 6 | 5 | 0 | 3 |
| Phage, transposon, or plasmid | 91 | 5 | 1 | 0 | 9 |
| Putative cell structure | 43 | 1 | 0 | 2 | 0 |
| Putative enzymes | 453 | 12 | 8 | 7 | 4 |
| Putative factors | 67 | 3 | 0 | 3 | 0 |
| Putative membrane proteins | 54 | 4 | 0 | 1 | 0 |
| Putative regulatory proteins | 167 | 11 | 0 | 4 | 1 |
| Putative transport proteins | 291 | 14 | 3 | 2 | 8 |
| Regulatory function | 208 | 14 | 3 | 6 | 3 |
| Transcription, RNA processing, and degradation | 28 | 0 | 1 | 0 | 1 |
| Translation and posttranslational modification | 128 | 0 | 53 | 6 | 1 |
| Transport and binding proteins | 254 | 24 | 2 | 13 | 8 |
Number of genes showing significant (99% confidence) log expression ratios (≥±0.400).
Expression levels of the majority of genes did not differ significantly (log ratio ≥ 0.4) between growth conditions. This was particularly true for the comparison of the cultures grown on minimal glucose versus minimal gluconate media; 80 genes (1.9%) were expressed at significantly higher levels on glucose, and 82 genes were expressed at significantly higher levels on gluconate (Table 1; Fig. 3). Thus, the overall similarity of these two growth conditions, being identical in basal medium composition, aeration, pH, and temperature and differing only in the nature of the carbon source, was reflected in their gene expression profiles. The comparison of genomic expression patterns of cells grown on minimal versus rich media was more revealing: 225 genes (5.2%) were expressed at significantly higher levels on minimal glucose, and 119 genes (2.8%) were expressed at significantly higher levels on rich medium (Fig. 3). A larger number of genes (3,496 versus 3,284 genes) had expression intensities above the background value on minimal glucose compared to rich medium (data not shown). By these measures, the cells growing on glucose minimal medium expressed more genes than did cells growing on rich medium. The nature of these differences in global gene expression was examined in detail, as described below.
Translation apparatus.
The culture containing rich medium plus glucose grew more than twice as fast as did the cultures on minimal media (Fig. 1). It is known that faster-growing cells synthesize protein faster and contain more ribosomes (27, 35). There are 128 known genes encoding the enzymes, factors, and structural components that make up the translation apparatus. Of these 128 genes of the translation apparatus, 53 (41.4%) were expressed at significantly higher levels in the cells growing on rich medium and none of them were expressed at significantly higher levels on the minimal medium. Of the 53 translation genes that were expressed at higher levels on rich medium, 42 encoded ribosomal proteins. These data are charted in Fig. 4 and can be compared to the data for the cultures on minimal glucose versus gluconate medium, which had nearly identical growth rates and showed very few significant differences in expression of the translation genes. A comparison of the general pattern of expression of the translation genes (Fig. 4) to that of the entire E. coli gene set (Fig. 3) further illustrates the dramatic increase in production of the translation apparatus in the faster-growing cells.
FIG. 4.
Log expression ratios of the translation apparatus genes sorted by value. The set of all translation apparatus genes is shown in the top two panels for the minimal glucose versus minimal gluconate and minimal glucose versus Luria broth plus glucose experiments (see the legend to Fig. 3). The bottom three panels show the results of the minimal glucose versus Luria broth plus glucose experiment for functionally grouped subsets of the translation apparatus genes.
(i) tRNA synthetase genes.
There are 37 known genes encoding the tRNA synthetases and other enzymes involved in tRNA modification. While none of the expression ratios of the tRNA synthetase genes varied significantly, it is clear from the chart in Fig. 4 that the transcript levels for these genes followed the same general trend as the complete set of translation genes. This result is consistent with the notion that synthesis of the tRNA synthetases is coupled to the synthesis of other ribosomal components (27).
(ii) Translation factors.
There are 17 known genes that encode factors involved in translation and ribosome modification, including the initiation and elongation factors, and 7 of these genes were expressed at significantly higher levels on rich medium (Fig. 4; Table 2). This result is generally consistent with the coupled synthesis of translation factors and ribosome components (27). The expression ratio of infB was significantly higher on rich medium. The regulation of infB, which is downstream of and cotranscribed with the transcription factor gene nusA, is complex and is thought to be the result of autoregulation of the extent of readthrough at upstream terminators by NusA (27). The expression ratio of infB was 1.8-fold higher than that of nusA (data not shown). The expression ratios of the translation elongation factor genes tsf, tufB, tufA, and fusA were all significantly higher, in that order, on rich medium, which is consistent with their coordinate regulation with the ribosomal protein genes (27). The growth rate-dependent regulation of tsf, tufA, and fusA, all of which are located in ribosomal protein operons, is the result of mRNA destabilization in slowly growing cells (27). Interestingly, regulation of tufB appears to be at least partially dependent upon Fis (68), and the fis gene had one of the highest expression ratios on rich medium, as described in more detail below. A fifth elongation factor encoded by efp has been shown to be essential in E. coli for protein synthesis and viability, although the details of efp regulation have not been published (2). The results of this study indicate that efp was expressed at a significantly higher level (log ratio = −0.425) in the faster-growing cells on rich medium, paralleling the expression of the other elongation factors.
TABLE 2.
Genes of the translation apparatus showing significant expression ratios
| Gene | Gene product | Log ratio (minimal/rich medium)a |
|---|---|---|
| rplY | 50S ribosomal subunit protein L25 | −0.405 |
| ileS | Isoleucine-tRNA synthetase | −0.414 |
| rpsU | 30S ribosomal subunit protein S21 | −0.416 |
| rpmH | 50S ribosomal subunit protein L34 | −0.419 |
| efp | Elongation factor P | −0.425 |
| fusA | GTP-binding protein chain elongation factor G | −0.435 |
| tufA | Protein chain elongation factor Tu | −0.441 |
| slyD | FKBP-type peptidylprolyl cis-trans isomerase | −0.450 |
| rplL | 50S ribosomal subunit protein L7/L12 | −0.452 |
| rpsO | 30S ribosomal subunit protein S15 | −0.461 |
| rpmA | 50S ribosomal subunit protein L27 | −0.462 |
| infB | Protein chain initiation factor 2 | −0.463 |
| rpmD | 50S ribosomal subunit protein L30 | −0.466 |
| rpmE | 50S ribosomal subunit protein L31 | −0.471 |
| rpsN | 30S ribosomal subunit protein S14 | −0.471 |
| rplD | 50S ribosomal subunit protein L4, regulates S10 operon | −0.475 |
| prfB | Peptide chain release factor 2 | −0.481 |
| rpsD | 30S ribosomal subunit protein S4 | −0.483 |
| rplF | 50S ribosomal subunit protein L6 | −0.494 |
| rpsG | 30S ribosomal subunit protein S7, initiates assembly | −0.499 |
| ppiA | Peptidylprolyl cis-trans isomerase A | −0.501 |
| rpsI | 30S ribosomal subunit protein S9 | −0.503 |
| rpsK | 30S ribosomal subunit protein S11 | −0.504 |
| rplK | 50S ribosomal subunit protein L11 | −0.514 |
| rpmB | 50S ribosomal subunit protein L28 | −0.518 |
| rpmC | 50S ribosomal subunit protein L29 | −0.522 |
| rplA | 50S ribosomal subunit protein L1, regulates L1 and L11 | −0.529 |
| rpsL | 30S ribosomal subunit protein S12 | −0.530 |
| rpsB | 30S ribosomal subunit protein S2 | −0.540 |
| rplS | 50S ribosomal subunit protein L19 | −0.548 |
| rplN | 50S ribosomal subunit protein L14 | −0.553 |
| rplC | 50S ribosomal subunit protein L3 | −0.555 |
| rplP | 50S ribosomal subunit protein L16 | −0.570 |
| rplU | 50S ribosomal subunit protein L21 | −0.575 |
| rplQ | 50S ribosomal subunit protein L17 | −0.577 |
| rplV | 50S ribosomal subunit protein L22 | −0.580 |
| rpsT | 30S ribosomal subunit protein S20 | −0.587 |
| rplE | 50S ribosomal subunit protein L5 | −0.587 |
| rplR | 50S ribosomal subunit protein L18 | −0.596 |
| rplM | 50S ribosomal subunit protein L13 | −0.603 |
| rplI | 50S ribosomal subunit protein L9 | −0.607 |
| prmA | Methylase for 50S ribosomal subunit protein L11 | −0.615 |
| rpsA | 30S ribosomal subunit protein S1 | −0.620 |
| rplW | 50S ribosomal subunit protein L23 | −0.627 |
| rpsS | 30S ribosomal subunit protein S19 | −0.634 |
| rplB | 50S ribosomal subunit protein L2 | −0.636 |
| rpsR | 30S ribosomal subunit protein S18 | −0.637 |
| rpsJ | 30S ribosomal subunit protein S10 | −0.644 |
| rpsE | 30S ribosomal subunit protein S5 | −0.646 |
| rplJ | 50S ribosomal subunit protein L10 | −0.676 |
| tufB | Protein chain elongation factor Tu | −0.688 |
| rplX | 50S ribosomal subunit protein L24 | −0.875 |
| tsf | Protein chain elongation factor Ts | −0.990 |
Log expression ratios of measured transcript levels determined for the two cultures. The log expression ratio is positive for genes that were more highly expressed on minimal glucose medium and negative for genes that were more highly expressed on Luria broth plus glucose.
(iii) Ribosomal proteins.
Of the 55 genes encoding the ribosomal proteins, 42 were expressed at significantly higher levels in the more rapidly growing cells in rich medium (Fig. 4; Table 2). This result is consistent with the paradigm of growth rate-dependent regulation of ribosome number (35). Although the ribosomal S10 operon is at least partially regulated at the transcriptional level, it is generally accepted that regulation of the 21 ribosomal protein operons is not at the level of transcription initiation (23, 35). Rather, the regulation of ribosomal protein synthesis involves a combination of translational control and transcriptional control at the level of mRNA stability. In general, growth conditions which lead to a decreased rate of ribosome synthesis result in an excess of ribosomal proteins, with certain ones serving as autoregulators by binding to their transcript and decreasing the translation rate of the mRNA, thus leading to destabilization of the transcript (35). While not all of the ribosomal protein operons have been studied at this level of detail, the experiment presented here indicates that most of the operons are regulated in such a way that their transcript levels are higher in faster growing cells. Clearly, these data demonstrate that any regulatory mechanism that contributes to the dynamic control of a particular mRNA concentration, whether it be the rate of transcription or the rate of turnover, can be visualized in genomic expression assays. The global regulation and coordination of ribosome number and components of the translation apparatus was the most obvious result of this experiment.
Nitrogen metabolism.
The minimal medium used in this study contained ammonia as the nitrogen source and the rich medium contained amino acids as the nitrogen source. In general, cells growing on minimal medium are limited for amino acids while cells growing on rich medium are limited for nucleotides (47, 52, 76). These differences were reflected in the transcript levels of the genes involved in nitrogen assimilation and biosynthesis of amino acids. The genes involved in assimilation of ammonia as the nitrogen source were expressed at significantly higher levels on minimal medium, including gdhA, which encodes glutamate dehydrogenase, and gltD, which encodes a subunit of glutamate synthase (Table 3). While it is known that gdhA is transcriptionally regulated by ammonia, next to nothing is known about the mechanism (53). The gltBD operon is subject to complex regulation by certain amino acids and in a concentration-dependent fashion by leucine-responsive protein (Lrp) (20); thus, the high induction ratio of gltBD on minimal medium (0.329 for gltB; 0.889 for gltD) can be explained by amino acid repression in rich medium and a high induction ratio of Lrp on minimal medium (see below). Conversely, glnA, which encodes glutamine synthase and is induced by nitrogen limitation (as indicated by a low ratio of intracellular glutamine to α-ketoglutarate), had the highest (although not significantly so) expression ratio (−0.316) of any of the amino acid biosynthetic genes in rich medium (52). In summary, the genes involved in ammonia assimilation were induced for growth on minimal medium where ammonia was the nitrogen source.
TABLE 3.
Genes of nitrogen metabolism and biosynthesis showing significant expression ratios
| Functional group | Gene | Gene Product | Log ratio (minimal/rich medium) |
|---|---|---|---|
| Amino acids | ilvC | Ketol-acid reductoisomerase | 0.977 |
| leuD | Isopropylmalate isomerase subunit | 0.951 | |
| tyrA | Chorismate mutase-T and prephenate dehydrogenase | 0.934 | |
| gltD | Glutamate synthase, small subunit | 0.889 | |
| aroF | 3-Deoxy-d-arabinoheptulosonate-7-phosphate synthase | 0.847 | |
| leuA | 2-Isopropylmalate synthase | 0.809 | |
| leuB | 3-Isopropylmalate dehydrogenase | 0.756 | |
| serA | d-3-Phosphoglycerate dehydrogenase | 0.633 | |
| ygjG | Probable ornithine aminotransferase | 0.623 | |
| leuC | 3-Isopropylmalate isomerase (dehydratase) subunit | 0.566 | |
| trpB | Tryptophan synthase, beta protein | 0.563 | |
| glyA | Serine hydroxymethyltransferase | 0.541 | |
| gdhA | NADP-specific glutamate dehydrogenase | 0.519 | |
| trpC | N-(5-Phosphoribosyl)anthranilate isomerase and indole-3-glycerolphosphate synthetase | 0.518 | |
| ilvG1 | Acetolactate synthase II, large subunit, interrupted | 0.513 | |
| trpE | Anthranilate synthase component I | 0.511 | |
| metE | Tetrahydropteroyltriglutamate methyltransferase | 0.499 | |
| cysK | Cysteine synthase A, O-acetylserine sulfhydrolase A | 0.497 | |
| aspC | Aspartate aminotransferase | 0.482 | |
| trpA | Tryptophan synthase, alpha protein | 0.481 | |
| argA | N-Acetylglutamate synthase | 0.444 | |
| pheA | Chorismate mutase P and prephenate dehydratase | 0.439 | |
| serC | 3-Phosphoserine aminotransferase | 0.401 | |
| Vitamins and cofactors | nrdH | Glutaredoxin-like protein; hydrogen donor | 0.638 |
| hemC | Porphobilinogen deaminase | 0.552 | |
| entE | 2,3-Dihydroxybenzoate-AMP ligase | 0.543 | |
| grxB | Glutaredoxin 2 | 0.506 | |
| gst | Glutathionine S-transferase | 0.501 | |
| folE | GTP cyclohydrolase I | 0.489 | |
| ggt | Gamma-glutamyltranspeptidase | 0.477 | |
| entF | ATP-dependent serine activating enzyme | 0.459 | |
| entB | 2,3-Dihydro-2,3-dihydroxybenzoate synthetase, isochorismatase | 0.451 | |
| entC | Isochorismate hydroxymutase 2 enterochelin biosynthesis | 0.428 | |
| thiH | Thiamine biosynthesis | −0.473 | |
| Nucleotides | pyrI | Aspartate carbamoyltransferase, regulatory subunit | 0.628 |
| pyrB | Aspartate carbamoyltransferase, catalytic subunit | 0.464 | |
| prsA | Phosphoribosylpyrophosphate synthetase | −0.472 | |
| ndk | Nucleoside diphosphate kinase | −0.484 | |
| pfs | ORF, hypothetical protein | −0.515 | |
| gmk | Guanylate kinase | −0.646 | |
| upp | Uracil phosphoribosyltransferase | −0.646 | |
| Fatty acid and phospholipid metabolism | cfa | Cyclopropane fatty acyl phospholipid synthase | 0.618 |
| fadA | 3-ketoacyl-CoA thiolase | 0.494 | |
| fabA | β-Hydroxydecanoyl thioester dehydrase | −0.413 | |
| fabD | Malonyl-CoA-[acyl-carrier-protein] transacylase | −0.437 | |
| fabI | Enoyl-[acyl carrier protein] reductase | −0.468 | |
| accC | Acetyl-CoA carboxylase, biotin carboxylase subunit | −0.557 | |
| fabH | 3-Oxoacyl-[acyl carrier protein] synthase III | −0.571 | |
| fabF | 3-Oxoacyl-[acyl carrier protein] synthase II | −0.576 | |
| fabZ | (3R)-Hydroxymyristol acyl carrier protein dehydratase | −0.608 |
Biosynthesis of amino acids.
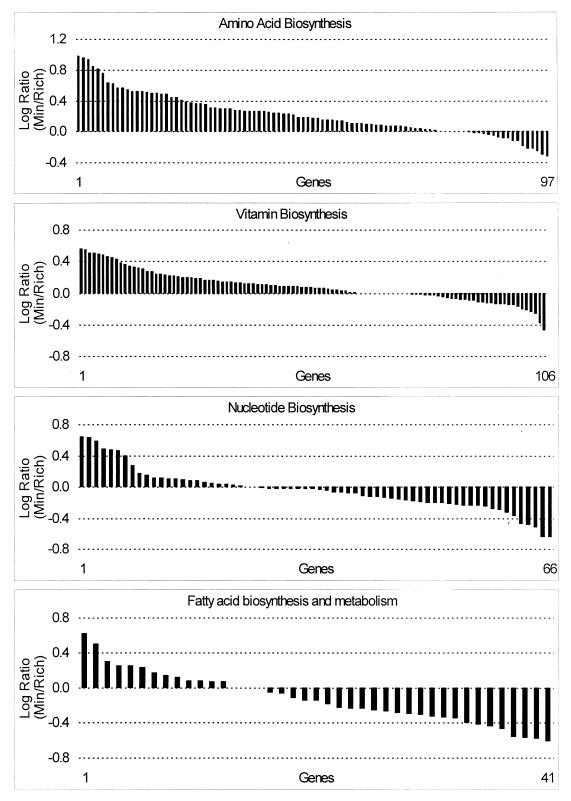
The overall expression pattern of the genes encoding the enzymes of amino acid biosynthesis indicated that these were generally induced for growth on minimal medium (Fig. 5; Table 3). The argA gene, which encodes N-acetylglutamate synthase, the first enzyme of the pathway, and also ygjG, a probable ornithine aminotransferase, were expressed at significantly higher levels on minimal medium. Expression of the genes of the branched-chain amino acid biosynthetic pathways (67)was significantly elevated in minimal medium. The first gene of the ilvGMEDA operon, which encodes the enzymes of isoleucine and valine synthesis, was expressed at significantly higher levels on minimal medium. Interestingly, the monocistronic gene ilvC, which is derepressed exclusively by valine, had a log expression ratio of 0.977 on minimal medium, the highest of any of the amino acid biosynthesis genes. The leucine biosynthetic genes, encoded by the leuABCD operon, were all expressed at significantly higher levels on minimal medium. The high expression ratios of the leucine and valine biosynthetic genes are consistent with the relatively high abundance of these two amino acids (third and fourth most abundant, respectively) in E. coli cells (45). The genes encoding the first enzymes of the four branches of the aromatic amino acid biosynthetic pathways were all significantly elevated in cells grown on minimal medium (51). The first step of the “common pathway” of chorismate synthesis, encoded by aroF, and the first step of tyrosine biosynthesis, encoded by tyrA, form an operon, in that order, and had log expression ratios of 0.847 and 0.934, respectively. The pheA gene was significantly elevated, as were four of the five genes of the trpEDCBA operon; the trpD transcript level was high in both minimal and rich media. The gene encoding the first step in serine biosynthesis, serA, and the gene which codes for the enzyme that forms glycine from serine, glyA, were expressed at significantly higher levels on minimal medium. The cysK gene, which encodes cysteine synthase A, was expressed at significantly higher levels on minimal medium, while cysM, the gene encoding cysteine synthase B, was expressed at slightly higher levels on rich medium. The cysE product, serine transacetylase, forms a multifunctional complex with the cysK product, and the relative expression ratios of cysK and cysE (0.497 versus −0.024) are consistent with the cysE product being much less abundant in the enzyme complex (36). The uniquely MetR-regulated methionine synthase gene, metE, was expressed at a significantly higher level on minimal medium, in contrast to the remaining MetJ-regulated genes of methionine biosynthesis, which did not vary significantly (25). The cobalamin-dependent methionine synthase encoded by metH was not expressed on minimal or rich media (data not shown). Overall, 8 of the 22 amino acid biosynthesis genes which were significantly elevated on minimal medium corresponded to the first step in the biosynthetic pathway. Thus, significant elevation of the first step in the amino acid biosynthetic pathways in cells grown on minimal medium was a recurring regulatory theme, consistent with the roles of these steps in controlling the flow of precursor metabolites out of the central pathways and into biosynthesis. Increased expression of the amino acid biosynthetic genes on minimal medium was indicative of the need to generate these building blocks from the sole carbon source, glucose.
FIG. 5.
Log expression ratios of biosynthetic genes were sorted by value for the minimal glucose versus Luria broth plus glucose experiment and are grouped by related pathways.
Biosynthesis of vitamins, cofactors, prosthetic groups and carriers.
Expression of the 106 genes involved in biosynthesis of vitamins, cofactors, prosthetic groups, and carriers followed the same trend as the genes of amino acid biosynthesis, although the expression ratios were generally not so large (Fig. 5; Table 3). Among the genes expressed at significantly higher levels on minimal medium were hemC, involved in porphyrin biosynthesis; the first three genes of the entCEBA operon and entF, involved in enterobactin biosynthesis; grxB, encoding glutaredoxin 2; gst, encoding glutathione S-transferase; folE, encoding the first step in tetrahydrofolate biosynthesis; and ggt, involved in glutathione biosynthesis. Only one gene, thiH, had an expression ratio that was significantly higher on rich medium, but this was in contrast to the remainder of the thi genes, which in general were expressed at modestly higher levels on minimal medium. Given that the vitamins and cofactors synthesized by the pathways in this functional group are needed in very small amounts, so small that they are rarely included in chemical composition tables (45), it is not surprising that most of these genes did not have significant expression ratios. Nevertheless, the general trend of higher expression in minimal medium is again indicative of the need to generate these building blocks de novo from glucose.
Nucleotide biosynthesis.
While expression of the genes involved in biosynthesis of amino acids, vitamins, enzyme cofactors, and prosthetic groups, etc., was generally elevated on minimal glucose medium, expression of the genes involved in nucleotide salvage and biosynthesis was more evenly divided between the two growth conditions (Fig. 5; Table 3). The pyrBI genes, which form an operon encoding the first step of pyrimidine biosynthesis, were expressed at significantly higher levels on minimal medium, perhaps reflecting the presence of uridine in the rich medium, which would tend to repress these genes (47). There are three enzymes involved in conversion of ribonucleotides to deoxyribonucleotides (33, 47), but of the three corresponding genetic loci only the nrdHIEF operon was expressed at significantly higher levels on minimal glucose medium. In previous studies, hyperinduction of nrdEF by hydroxyurea was measured, but this was the first experiment comparing nrdEF transcript levels under normal growth conditions (33), and this was the first indication that the genes encoding the NrdEF accessory proteins, NrdH and NrdI (34), are coregulated with nrdEF. There were five genes that were expressed at significantly higher levels in rich medium: the prsA gene, which encodes an enzyme that forms the first precursor of purine biosynthesis, and upp, gmk, pfs, and ndk, all of which encode enzymes involved in nucleotide salvage or interconversion, consistent with the availability of nucleotides in the rich medium.
Fatty acid biosynthesis and degradation.
The cfa gene, which encodes an enzyme responsible for postsynthetic formation of cyclopropane fatty acids from unsaturated fatty acids, had a significantly high expression ratio on minimal medium (Table 3; also see Table 6). Since cfa is transcribed from an RpoS-dependent promoter (16), this result is consistent with elevated expression of rpoS on minimal medium (see below). All of the genes of the fad regulon (13) of fatty acid degradation (except for fadA [possibly an erroneous result]) were significantly elevated on rich medium, including fadB, which is in the fadAB operon, and fadD, which together encode the fatty acid oxidation multienzyme complex (Fig. 5). Also significantly elevated on rich medium were fadR, the repressor of the fad genes, and fadL, which encodes a long-chain fatty acid transporter. These results tend to indicate that the cells grown on rich medium were exposed to exogenous long-chain fatty acids, leading to induction of the fad regulon (14). Interestingly, the ato genes, which are involved in degradation of four-carbon fatty acids, were modestly elevated on minimal medium, and the sensor of the two-component regulator of these genes, encoded by atoS, was significantly elevated on minimal medium. These results suggest that the cells grown on minimal glucose medium were exposed to acetoacetate, which is the inducer of the ato genes (14). E. coli is not known to form acetoacetate from glucose, and it is possible that some closely related compound such as acetolactate, which is formed by E. coli, serves as an inducer of the ato genes (46, 67).
TABLE 6.
Genes regulated by RpoS showing significant expression ratios
| Functional group | Gene | Gene product | Log ratio (minimal/rich medium) |
|---|---|---|---|
| Catabolism | galK | Galactokinase | 0.519 |
| poxB | Pyruvate oxidase | 0.535 | |
| Cell processes | bolA | Possible regulator of murein genes | 0.564 |
| cbpA | Curved DNA binding protein | 0.540 | |
| fic | Induced in stationary phase, affects cell division | 0.654 | |
| glgS | Glycogen biosynthesis | 0.794 | |
| glgC | Glucose-1-phosphate adenylyltransferase | 0.649 | |
| glgB | 1,4-α-Glucan branching enzyme | 0.430 | |
| katE | Catalase | 0.893 | |
| osmC | Osmotically inducible protein | 1.204 | |
| osmY | Hyperosmotically inducible periplasmic protein | 0.580 | |
| otsB | Trehalose-6-phosphate phophatase | 0.777 | |
| otsA | Trehalose-6-phosphate synthase | 0.556 | |
| Energy metabolism | frdD | Fumarate reductase, anaerobic | 0.401 |
| glpD | sn-Glycerol-3-phosphate dehydrogenase | 0.616 | |
| FA and PL metabolism | cfa | Cyclopropane fatty acyl phospholipid synthase | 0.618 |
| Hypothetical | hdeA | ORF, hypothetical protein | 1.872 |
| hdeB | ORF, hypothetical protein | 1.705 | |
| Regulatory function | dps | Global regulator, starvation | 1.402 |
| rpoS | Sigma S (sigma 38) factor | 0.766 | |
| wrbA | trp repressor binding protein | 0.420 |
Expression of the genes of fatty acid biosynthesis was generally elevated on rich medium, and, with the exception of fabB and fabG, all of the fab genes were significantly elevated (Fig. 5; Table 3). The relative expression ratios of the genes in the fabHDG-acpP-fabF operon corresponded very closely to measurements of transcript levels by Northern analysis (77). In addition, accA, which encodes a component of acetyl coenzyme A (acetyl-CoA) carboxylase, was elevated on rich medium. The transcription rate of accC is growth rate dependent; the rate is higher in faster-growing cells (16). With the exception of FadR activation of fabA, less is known about the regulation of the fab genes (16). The data presented here, indicating that the fab genes were generally expressed at higher levels on rich medium, suggest that regulation of the phospholipid biosynthesis genes could be growth rate dependent (Fig. 5). This is a reasonable hypothesis, given that faster-growing cells must make membrane components more rapidly. However, the genomic expression data do not prove this hypothesis, and it is also possible that regulation of the fab genes is mediated by a signal molecule(s) in the rich medium. Further research in this area will help to clarify the global regulation of phospholipid biosynthesis.
Carbon and energy metabolism.
The cells grown on rich medium showed nothing remarkable with respect to the expression pattern of genes involved in carbon and energy metabolism. Of the 409 genes of carbon catabolism, central metabolism, and energy metabolism, only 8 were expressed at significantly higher levels on rich medium (Table 4). These included nuoM and nuoN of the large operon encoding NADH dehydrogenase I and cyoA of the operon encoding cytochrome oxidase c (24), suggesting that aerobic respiration was elevated under this growth condition.
TABLE 4.
Genes of carbon and energy metabolism showing significant expression ratios
| Functional group | Gene | Gene product | Log ratio (minimal/rich medium) |
|---|---|---|---|
| Catabolism | amyA | Cytoplasmic alpha-amylase | 0.679 |
| nanA | N-Acetylneuraminate lyase | 0.581 | |
| poxB | Pyruvate oxidase | 0.535 | |
| galK | Galactokinase | 0.519 | |
| ptr | Protease III | −0.629 | |
| clpP | ATP-dependent proteolytic subunit | −0.689 | |
| Central metabolism | gadA | Glutamate decarboxylase isozyme | 1.569 |
| gadB | Glutamate decarboxylase isozyme | 1.497 | |
| aceA | Isocitrate lyase | 0.928 | |
| gltD | Glutamate synthase, small subunit | 0.889 | |
| aceB | Malate synthase A | 0.871 | |
| gltA | Citrate synthase | 0.746 | |
| gpmA | Phosphoglyceromutase 1 | 0.724 | |
| mdh | Malate dehydrogenase | 0.587 | |
| rpiB | Ribose 5-phosphate isomerase B | 0.580 | |
| phnJ | Phosphonate metabolism | 0.538 | |
| icdA | Isocitrate dehydrogenase | 0.506 | |
| zwf | Glucose-6-phosphate dehydrogenase | 0.503 | |
| nrdE | Ribonucleoside-diphosphate reductase 2, alpha subunit | 0.489 | |
| nrdF | Ribonucleoside-diphosphate reductase 2, beta chain | 0.478 | |
| tpiA | Triosephosphate isomerase | 0.442 | |
| talA | Transaldolase A | 0.439 | |
| pfkB | 6-Phosphofructokinase II | 0.423 | |
| speE | Spermidine synthase | −0.531 | |
| Energy metabolism | nrfC | Formate-dependent nitrite reductase | 0.805 |
| dld | d-Lactate dehydrogenase | 0.726 | |
| nrfA | Periplasmic cytochrome c(552) | 0.617 | |
| glpD | sn-Glycerol-3-phosphate dehydrogenase (aerobic) | 0.616 | |
| nrfB | Formate-dependent nitrite reductase | 0.600 | |
| qor | Quinone oxidoreductase | 0.497 | |
| ppc | Phosphoenolpyruvate carboxylase | 0.477 | |
| atpG | Membrane-bound ATP synthase, F1 sector, gamma subunit | 0.451 | |
| nuoJ | NADH dehydrogenase I chain J | 0.419 | |
| dsbE | Disulfide oxidoreductase | 0.418 | |
| hyfB | Hydrogenase 4 membrane subunit | 0.415 | |
| frdD | Fumarate reductase | 0.401 | |
| fdnI | Formate dehydrogenase N, cytochrome b556 gamma subunit | 0.400 | |
| nuoN | NADH dehydrogenase I chain N | −0.412 | |
| ackA | Acetate kinase | −0.446 | |
| cyoA | Cytochrome o ubiquinol oxidase subunit II | −0.461 | |
| nuoM | NADH dehydrogenase I chain M | −0.537 | |
| fdoG | Formate dehydrogenase O, major subunit | −0.967 |
Cells grown on minimal glucose medium expressed 31 of the 409 of the carbon and energy metabolism genes at significantly higher levels. These included genes involved in d-lactate utilization (dld), acetate formation (poxB), regulation of poxB expression (rpoS), acetate utilization (aceA, aceB, gltA, icd, and mdh), and coupling of glucose and acetate cometabolism (uspA) (Tables 4 and 5). The elevated expression of these genes implicates metabolism of acetate and d-lactate as being perhaps the prominent feature of glucose metabolism in minimal medium. Under this growth condition, cells first consume glucose, which causes repression of the glyoxylate bypass and tricarboxylic acid cycle (15). Simultaneously, the cells excrete acetate and lesser amounts of d-lactate as overflow metabolites (46). As glucose is consumed and acetate accumulates, cells switch smoothly to cometabolism of glucose and acetate (1, 4, 14). This switch involves induction of the tricarboxylic acid cycle and glyoxylate bypass enzymes required to provide energy and to replenish intermediates used for amino acid biosynthesis (14).
TABLE 5.
Genes involved in cell processes and global regulation showing significant expression ratios
| Functional group | Gene | Gene product | Log ratio (minimal/rich medium) |
|---|---|---|---|
| Cell processes | osmC | Osmotically inducible protein | 1.204 |
| katE | Catalase | 0.893 | |
| msyB | Acidic protein suppresses mutants lacking function of protein export | 0.837 | |
| glgS | Glycogen biosynthesis | 0.794 | |
| otsB | Trehalose-6-phosphate phophatase | 0.777 | |
| fic | Induced in stationary phase, affects cell division | 0.654 | |
| glgC | Glucose-1-phosphate adenylyltransferase | 0.649 | |
| osmY | Hyperosmotically inducible periplasmic protein | 0.580 | |
| bolA | Possible regulator of murein genes | 0.564 | |
| otsA | Trehalose-6-phosphate synthase | 0.556 | |
| cspD | Cold shock protein | 0.540 | |
| cbpA | Curved DNA binding protein | 0.540 | |
| envY | Envelope protein, thermoregulation of porin biosynthesis | 0.477 | |
| uspA | Universal stress protein | 0.469 | |
| motA | Proton conductor component of motor | 0.457 | |
| glgB | 1,4-α-Glucan branching enzyme | 0.430 | |
| sodC | Superoxide dismutase precursor (Cu-Zn) | 0.430 | |
| pbpG | Penicillin binding protein 7 | 0.406 | |
| acrF | Integral transmembrane protein | 0.400 | |
| sodB | Superoxide dismutase, iron | −0.904 | |
| Cell structure | nlpD | Lipoprotein | 0.629 |
| slp | Outer membrane protein induced after carbon starvation | 0.549 | |
| Regulatory function | dps | Global regulator, starvation conditions | 1.402 |
| rpoS | RNA polymerase, sigma S (sigma38) factor | 0.766 | |
| atoS | Sensor protein AtoS for response regulator AtoC | 0.580 | |
| rseA | Sigma E factor, negative regulatory protein | 0.538 | |
| arsR | Transcriptional repressor ars operon | 0.530 | |
| molR1 | Molybdate metabolism regulator | 0.521 | |
| lrp | Regulator for leucine (or lrp) regulon | 0.511 | |
| srlR | Regulator for gut (srl), glucitol operon | 0.464 | |
| tar | Methyl-accepting chemotaxis protein II | 0.463 | |
| phoU | Negative regulator for pho regulon | 0.457 | |
| narP | Nitrate/nitrite response regulator (sensor NarQ) | 0.444 | |
| rpoE | Sigma E factor; heat shock and oxidative stress | 0.425 | |
| wrbA | trp repressor binding protein | 0.420 | |
| rcsB | Positive regulator for colanic capsule biosynthesis (sensor, RcsC) | 0.416 | |
| fadR | Negative regulator for fad regulon, positive activator of fabA | −0.662 | |
| cspA | Cold shock protein, transcriptional activator of hns | −0.776 | |
| fis | Site-specific DNA inversion stimulation factor; DNA binding protein | −0.818 |
Evidence has been published which suggests that pyruvate oxidase (PoxB) forms acetate from pyruvate during the transition from exponential growth to stationary phase: poxB expression requires RpoS and thus is elevated during transition phase (11). That cells grown in glucose minimal medium exhibited elevated poxB levels supports the contention that acetate was formed via pyruvate oxidation. The elevated expression of rpoS during late logarithmic growth (Table 5) also argues that RpoS may play a crucial role in regulating acetate metabolism.
Mutants lacking uspA exhibit diauxic growth on minimal glucose medium. This behavior probably occurs because of a failure to assimilate acetate until glucose becomes completely exhausted (49). In the wild-type cells examined here, expression of uspA was significantly elevated during growth on glucose minimal medium (Table 5), supporting the argument that UspA somehow plays a critical role in coupling of glucose and acetate cometabolism.
In summary, the evidence presented here provides some insight into the global control of carbon flow in cells growing on glucose in minimal medium. The data argue that acetate overflow metabolism is an important aspect of growth on glucose as the sole carbon and energy source, RpoS may play a role in regulating carbon metabolism genes in late-logarithmic-phase cells, and the universal stress protein, UspA, may coordinate glucose and acetate cometabolism.
Cellular processes and global regulators.
Growth on minimal medium with glucose as the sole carbon and energy source places a burden on the cell to synthesize its amino acids de novo or starve. Thus, cells growing on minimal glucose medium are partially starved for amino acids, certainly a stressful situation and potentially having dramatic consequences on global gene regulation, elevating transcript levels of stress-inducible genes, and invoking the stringent response (8, 9, 29, 32). Several of the genes known to be regulated by the stringent-response signal molecule, ppGpp, were found to be differentially regulated on minimal and rich media (Fig. 6; Table 5). Most notable of these genes was rpoS, encoding the stationary-phase sigma factor, which was significantly elevated on minimal medium. In fact it appeared that RpoS-dependent gene expression was a prominent feature of the genomic expression pattern of cells grown on minimal medium (Table 6). It is not clear from these genomic expression assays if the elevated level of the rpoS transcript was the result of regulation by ppGpp, although this would be consistent with the positive correlation between ppGpp concentration and RpoS levels (9), because it was also found that expression of nlpD (which encodes a lipoprotein and is operonic with rpoS) was significantly elevated on minimal medium (Table 2). Thus, these data do not distinguish between the possibilities that the higher level of rpoS transcription was driven by the nlpD promoter or by the rpoS promoters located within the upstream nlpD gene (29, 38). Production of RpoS is also subject to complex posttranscriptional and translational regulation, and therefore it cannot be presumed that rpoS transcript levels are correlated with RpoS activity (29). However, the number of RpoS-inducible genes that were observed to be expressed at significantly elevated levels on minimal medium (21 of them) argues strongly in this case that the rpoS transcript level correlated with RpoS function. Interestingly, of the 21 RpoS-dependent genes which were significantly elevated on glucose minimal medium, more than half are known to be involved in the physiological changes that highlight entry into stationary phase (32). However, the cells used in these experiments were in late logarithmic growth phase, still in steady-state growth. Although most studies have focused on the role of RpoS in preparing cells for entry into stationary phase, it has been suggested that RpoS may play a role in logarithmic phase as well (29), and the results presented here support this idea. Since this question is likely to receive further attention, a time course study of genomic expression in cells growing on minimal glucose medium in batch culture would be invaluable.
Some of the regulatory genes had significant expression ratios that were consistent with the elevated transcript levels of their target genes, such as fadR, which was expressed at significantly higher levels on rich medium (as noted above), and lrp, which was expressed at significantly higher levels on minimal medium, correlating well with the elevated expression of several genes of the amino acid biosynthetic pathways (48). Several other regulatory genes showing significant expression ratios in this experiment are pleiotropic, and their roles under the growth conditions reported here are not as well understood (Table 5). Among the regulatory genes that were more highly expressed on rich medium are cspA, which encodes a cold shock transcription factor, and fis, which encodes a factor involved in site-specific recombination and pleiotropic transcriptional regulation (Table 5). Recent evidence indicates that cspA is expressed in cells that have not been subjected to cold stress and that its expression is higher in early logarithmic growth phase (7), a pattern of regulation that is remarkably similar to that of fis (3, 21, 70), which is also known to be more highly expressed in rich medium (35). Significantly higher in cells grown on minimal medium was dps, which encodes a DNA binding protein induced by starvation (42).
What these general DNA binding proteins, Dps and Fis, together with HN-S, seem to have in common is their involvement in growth rate-dependent regulation of gene expression, and it is nearly impossible to discuss these regulators without mentioning RpoS, which either regulates expression of or is regulated by these other factors (8, 9, 29, 32, 35). Together with RpoS, HN-S-dependent gene expression was prominent in the genomic transcription pattern of cells grown on minimal medium, and in fact the four genes with the highest expression ratios on minimal medium, hdeA, hdeB, gadA, and gadB (dps was fifth highest [Tables 4 and 6]) are known to be regulated by HN-S (5, 74, 75). Interestingly, hns expression was similar in minimal and rich media, suggesting either that expression of the gad and hde genes was not regulated by HN-S under these conditions or that hns expression (or HN-S function) is subject to posttranscriptional regulation by a mechanism which has yet to be described. In fact, expression of gadB and hdeAB in Shigella flexneri (72) and also gadA and gadB in E. coli (10) is RpoS dependent.
The apparent connection between these HN-S-regulated genes is their involvement in acid resistance (10). The unlinked genes, gadA and gadB, encoding homologous glutamate decarboxylases (62), are thought to be induced during fermentation as a result of acid stress (5, 61, 72). The gadB gene appears to form an operon with xasA (gadC) in E. coli (10) and is known to be cotranscribed with xasA (gadC) in S. flexneri (72); gadC mutants of E. coli are acid sensitive (30). Not surprisingly, xasA (gadC) had a significant log expression ratio on minimal medium (0.580 [data not shown]). Clustered together with gadA and hdeAB are several other genes that showed significantly higher expression ratios on minimal medium, i.e., hdeD, yhiE, and yhiX (log expression ratios of 0.872, 0.852, and 1.096, respectively [data not shown]). The functions of these five genes are all unknown, but it has been shown that hdeAB mutants of S. flexneri are acid sensitive (72). The yhiX gene, which encodes an AraC-like protein, is a likely candidate for regulation of the gad and hde genes, given its position downstream of gadA and its high expression ratio on minimal medium. Furthermore, alignment of the gadA, gadB, and hdeD-hdeAB regulatory regions (200 bp upstream of start codons) revealed a 19-bp sequence which is perfectly conserved in gadA and gadB and of which 15 bp are conserved in all three (data not shown). In summary, the results suggest that these HN-S/RpoS-dependent genes comprise a system for acid tolerance. It is interesting to speculate that RpoS plays a role in these logarithmic-phase cells of coordinating induction of the acid tolerance genes, together with the genes of organic acid metabolism, under conditions of glucose overflow metabolite formation.
Conclusion.
In the single experiment presented here, the hallmark features of growth on minimal and rich media were revealed. Across the genome, we observed differences in the expression of functionally grouped genes that paralleled the physiology of these two growth conditions. Cells grown in rich medium with a good carbon and energy source, glucose, grew rapidly, turning off the pathways of biosynthesis and elevating the expression of the genes involved in macromolecule synthesis, most prominently protein synthesis. Cells in minimal medium faced the need to synthesize all of their building blocks from a single carbon and energy source, again glucose, and this burden was reflected not just in the turning on of biosynthetic pathways but also in the elevated expression of regulators of cell processes and regulons involved in stress tolerance. The most prominent features of growth on glucose minimal medium were the formation of overflow metabolites, in particular acetate, and protection of the cell from the stress of living in a self-formed acidic environment. All of these aspects of physiology were revealed not by painstaking and careful analysis in the laboratory of each system but, rather, by deduction from the genomic expression patterns of cells grown under these two rather different conditions. These deductions would not have been possible were it not for countless microbial physiology experiments published over the past 50 years (44). On the other hand, from the one simple experiment reported here, the tremendous potential of functional genomics is obvious. As a result of this experiment, several genes were added to functional groups on the basis of coregulation with similar and related genes. Also, several testable hypotheses were generated, in particular those involving the flow of carbon to acetate, coupling of glucose and acetate cometabolism, and acid resistance, the importance of which has been previously pointed to in enteric bacteria (4).
ACKNOWLEDGMENTS
We thank John Cronan, Rick Gourse, Larry Reitzer, and Alan Wolfe for stimulating dialogue during the course of writing this paper.
This research was supported by grants to T.C. from the NSF (MCB-9723593) and to F.R.B. from the NIH (R01GM35682-12).
REFERENCES
- 1.Amarasingham C R, Davis B D. Regulation of α-ketoglutarate dehydrogenase formation in Escherichia coli. J Biol Chem. 1965;240:3664–3668. [PubMed] [Google Scholar]
- 2.Aoki H, Dekany K, Adams S L, Ganoza M C. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- 3.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender R A. Variations on a theme by Escherichia. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 4–9. [Google Scholar]
- 5.Blankenhorn D, Phillips J, Slonczewski J L. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J Bacteriol. 1999;181:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Brandi A, Spurio R, Gualerzi C O, Pon C L. Massive presence of the Escherichia coli “major cold-shock protein” CspA under non-stress conditions. EMBO J. 1999;18:1653–1659. doi: 10.1093/emboj/18.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bremer H, Denis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 9.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 10.Castanie-Cornet M P, Penfound T A, Smith D, Elliott J F, Foster J W. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y Y, Wang A Y, Cronan J E., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 12.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 13.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 14.Clark D P, Cronan J E. Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 343–357. [Google Scholar]
- 15.Cronan J E, LaPorte D. Tricarboxylic acid cycle and glyoxylate bypass. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 206–216. [Google Scholar]
- 16.Cronan J E, Rock C O. Biosynthesis of membrane lipids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 612–636. [Google Scholar]
- 17.Csonka L N, Epstein W. Osmoregulation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1210–1223. [Google Scholar]
- 18.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 19.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 19a.E. coli Genome Center. Functional assignment. [Online.] http://www.genetics.wisc.edu. [1 September 1999, last date accessed.] 22 January 1999. [Google Scholar]
- 20.Ernsting B R, Denninger J W, Blumenthal R M, Matthews R G. Regulation of the gltBDF operon of Escherichia coli: how is a leucine-insensitive operon regulated by the leucine-responsive regulatory protein? J Bacteriol. 1993;175:7160–7169. doi: 10.1128/jb.175.22.7160-7169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkel S E, Johnson R C. The Fis protein: it’s not just for DNA inversion anymore. Mol Microbiol. 1992;6:3257–3265. doi: 10.1111/j.1365-2958.1992.tb02193.x. [DOI] [PubMed] [Google Scholar]
- 22.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 23.Gausing K. Regulation of ribosome production in Escherichia coli: synthesis and stability of ribosomal RNA and of ribosomal protein messenger RNA at different growth rates. J Mol Biol. 1977;115:335–354. doi: 10.1016/0022-2836(77)90158-9. [DOI] [PubMed] [Google Scholar]
- 24.Gennis R B, Stewart V. Respiration. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 217–261. [Google Scholar]
- 25.Greene R C. Biosynthesis of methionine. In: Neidhardt F C, Curtiss III R, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 542–560. [Google Scholar]
- 26.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 27.Grunberg-Manago M. Regulation of the expression of aminoacyl-tRNA synthetases and translation factors. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1432–1457. [Google Scholar]
- 28.Heller R A, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley D E, Davis R W. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci USA. 1997;94:2150–2155. doi: 10.1073/pnas.94.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1497–1512. [Google Scholar]
- 30.Hersh B M, Farooq F T, Barstad D N, Blankenhorn D L, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 32.Huisman G W, Siegele D A, Zambrano M M, Kolter R. Morphological and physiological changes during stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1672–1682. [Google Scholar]
- 33.Jordan A, Aragall E, Gibert I, Barbe J. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol Microbiol. 1996;19:777–790. doi: 10.1046/j.1365-2958.1996.424950.x. [DOI] [PubMed] [Google Scholar]
- 34.Jordan A, Aslund F, Pontis E, Reichard P, Holmgren A. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- 35.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- 36.Kredich N M. Biosynthesis of cysteine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 442–457. [Google Scholar]
- 37.Lashkari D A, DeRisi J L, McCusker J H, Namath A F, Gentile C, Hwang S Y, Brown P O, Davis R W. Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci USA. 1997;94:13057–13062. doi: 10.1073/pnas.94.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loewen P C, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 39.Luria S E, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1526–1538. [Google Scholar]
- 41.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 42.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. doi: 10.1128/jb.179.16.5188-5194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, Bassett D E, Jr, Hartwell L H, Brown P O, Friend S H. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 43a.National Center for Biotechnology Information. revision date. Escherichia coli K-12 MG1655 complete genome. [Online.] http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/framik?db=Genome&gi=115. [2 September 1999, last date accessed.] 13 October 1998. [Google Scholar]
- 44.Neidhardt F C. The enteric bacterial cell and the age of bacteria. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1–3. [Google Scholar]
- 45.Neidhardt F C, Umbarger H E. Chemical composition of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 46.Neijssel O M, DeMattos M J T, Tempest D W. Growth yield and energy distribution. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1683–1692. [Google Scholar]
- 47.Neuhard J, Kelln R A. Biosynthesis and conversions of pyrimidines. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 580–599. [Google Scholar]
- 48.Newman E B, Lin R T, D’Ari R. The leucine/LRP regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1513–1525. [Google Scholar]
- 49.Nystrom T, Neidhardt F C. Isolation and properties of a mutant of Escherichia coli with an insertional inactivation of the uspA gene, which encodes a universal stress protein. J Bacteriol. 1993;175:3949–3956. doi: 10.1128/jb.175.13.3949-3956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.OU Bacterial Functional Genomics. Expression data. [Online.] http://www.ou.edu/cas/botany-micro/faculty/tconway/global.html. [2 September 1999, last date accessed.] 1 September 1999. [Google Scholar]
- 50.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo W L, Chen C, Zhai Y, Dairkee S H, Ljung B M, Gray J W, Albertson D G. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 51.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 458–484. [Google Scholar]
- 52.Reitzer L J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 391–407. [Google Scholar]
- 53.Riba L, Becerril B, Servin-Gonzalez L, Valle F, Bolivar F. Identification of a functional promoter for the Escherichia coli gdhA gene and its regulation. Gene. 1988;71:233–246. doi: 10.1016/0378-1119(88)90040-6. [DOI] [PubMed] [Google Scholar]
- 54.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 55.Riley M. Genes and proteins of Escherichia coli K-12. Nucleic Acids Res. 1998;26:54. doi: 10.1093/nar/26.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saier M H, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1325–1343. [Google Scholar]
- 57.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Schaechter E, Maaloe O, Kjeldgaard N O. Dependence on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 59.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 60.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1539–1549. [Google Scholar]
- 61.Small P L, Waterman S R. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 1998;6:214–216. doi: 10.1016/s0966-842x(98)01285-2. [DOI] [PubMed] [Google Scholar]
- 62.Smith D K, Kassam T, Singh B, Elliott J F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith M W, Neidhardt F C. Proteins induced by aerobiosis in Escherichia coli. J Bacteriol. 1983;154:344–350. doi: 10.1128/jb.154.1.344-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith M W, Neidhardt F C. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol. 1983;154:336–343. doi: 10.1128/jb.154.1.336-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong S, Porco A, Isturiz T, Conway T. Cloning and molecular genetic characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J Bacteriol. 1996;178:3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 442–457. [Google Scholar]
- 68.Verbeek H, Nilsson L, Bosch L. The mechanism of trans-activation of the Escherichia coli operon thrU(tufB) by the protein FIS. A model. Nucleic Acids Res. 1992;20:4077–4081. doi: 10.1093/nar/20.15.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker G C. The SOS response of Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- 70.Walker K A, Atkins C L, Osuna R. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol. 1999;181:1269–1280. doi: 10.1128/jb.181.4.1269-1280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wanner B L. Phosphorous assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 72.Waterman S R, Small P L. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 73.Welford S M, Gregg J, Chen E, Garrison D, Sorensen P H, Denny C T, Nelson S F. Detection of differentially expressed genes in primary tumor tissues using representational differences analysis coupled to microarray hybridization. Nucleic Acids Res. 1998;26:3059–3065. doi: 10.1093/nar/26.12.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshida T, Ueguchi C, Yamada H, Mizuno T. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in a hns deletion mutant. Mol Gen Genet. 1993;237:113–122. doi: 10.1007/BF00282791. [DOI] [PubMed] [Google Scholar]
- 75.Yoshida T, Yamashino T, Ueguchi C, Mizuno T. Expression of the Escherichia coli dimorphic glutamic acid decarboxylases is regulated by the nucleoid protein H-NS. Biosci Biotechnol Biochem. 1993;57:1568–1569. doi: 10.1271/bbb.57.1568. [DOI] [PubMed] [Google Scholar]
- 76.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 561–579. [Google Scholar]
- 77.Zhang Y, Cronan J E., Jr Polar allele duplication for transcriptional analysis of consecutive essential genes: application to a cluster of Escherichia coli fatty acid biosynthetic genes. J Bacteriol. 1996;178:3614–3620. doi: 10.1128/jb.178.12.3614-3620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]