Abstract
Lipid particles of the yeast Saccharomyces cerevisiae were isolated at high purity, and their proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Major lipid particle proteins were identified by mass spectrometric analysis, and the corresponding open reading frames (ORFs) were deduced. In silicio analysis revealed that all lipid particle proteins contain several hydrophobic domains but none or only few (hypothetical) transmembrane spanning regions. All lipid particle proteins identified by function so far, such as Erg1p, Erg6p, and Erg7p (ergosterol biosynthesis) and Faa1p, Faa4p, and Fat1p (fatty acid metabolism), are involved in lipid metabolism. Based on sequence homology, another group of three lipid particle proteins may be involved in lipid degradation. To examine whether lipid particle proteins of unknown function are also involved in lipid synthesis, mutants with deletions of the respective ORFs were constructed and subjected to systematic lipid analysis. Deletion of YDL193w resulted in a lethal phenotype which could not be suppressed by supplementation with ergosterol or fatty acids. Other deletion mutants were viable under standard conditions. Strains with YBR177c, YMR313c, and YKL140w deleted exhibited phospholipid and/or neutral lipid patterns that were different from the wild-type strain and thus may be further candidate ORFs involved in yeast lipid metabolism.
All types of eukaryotic cells, such as plants (13, 20, 39), mammals (6), and yeasts (8), contain intracellular lipid particles. These particles consist of a highly hydrophobic core formed from neutral lipids (triacylglycerols and steryl esters) surrounded by a phospholipid monolayer in which only a few proteins are embedded. The structure of this cellular compartment is reminiscent of lipoproteins in mammals (24).
Principal studies of lipid particles from the yeast Saccharomyces cerevisiae were carried out by Clausen et al. (8). Triacylglycerols and steryl esters were identified as the main components (approximately 50% each), and it was suggested that lipid particles function as a storage for components needed for membrane formation. Under sterol depletion, steryl esters of lipid particles are mobilized, and sterols set free through this process are incorporated into cellular membranes (27). Under conditions of fatty acid deficiency, fatty acyl moieties of triacylglycerols and steryl esters can be incorporated into phospholipids (9).
The protein pattern of yeast lipid particles is rather simple. Biochemical studies led to the identification of some of these proteins. One of the major proteins of yeast lipid particles is sterol-Δ24-methyltransferase (Erg6p) (27). Localization studies of this protein revealed a 700- to 800-fold enrichment in lipid particles over the homogenate (27). Subsequently, another major lipid particle protein was identified as squalene epoxidase (Erg1p) (28). It was demonstrated that Erg1p is not exclusively localized to lipid particles but is also present in the endoplasmic reticulum, thus pointing to a relationship between these two compartments. Similar observations were made with Slc1p, a 1-acylglycerol-3-phosphate acyltransferase involved in the biosynthesis of phosphatidic acid (3, 34). Slc1p was identified as a component of lipid particles by two-dimensional electrophoresis and functional analysis by using an slc1 deletion strain. In addition, a glycerol-3-phosphate acyltransferase activity catalyzed by the hypothetical Gat1p was detected in lipid particles (3, 8, 43). (The nomenclature GAT1 is used in this paper for the gene encoding the putative major glycerol-3-phosphate acyltransferase of the yeast. The gene is not identical to GAT1 [also referred to as NIL1; ORF YFL021w] listed in the Proteome Database, encoding a zinc finger transcription factor that plays a supplemental role to Gln3p, which activates genes needed to use nonpreferred nitrogen sources.) The gene encoding this protein and the polypeptide itself, however, have not yet been identified.
In the present paper, we report the identification of the major yeast lipid particle proteins by systematic mass spectrometric analysis. This strategy allowed us to classify some proteins of known function as lipid particle components and to identify additional unassigned open reading frames (ORFs) which code for novel lipid particle proteins. Phenotypic analysis of strains with deletions of the respective ORFs is described, and common features of lipid particle proteins are discussed.
MATERIALS AND METHODS
Strains and culture conditions.
The haploid wild-type yeast strains S. cerevisiae X2180-1A (MATa SUC2 mal gal2 CUP1) and FY1679 (MATa ura3-52 trp1Δ63 leu2Δ1 hisΔ200) and the diploid wild-type strain FY1679 (MATa/α ura3-52/ura3-52 leu2Δ1/LEU2 his3Δ200/HIS3 trp1Δ63/TRP1 GAL2/GAL2) were used throughout this study.
Cells were grown aerobically in 2-liter Erlenmeyer flasks to the late logarithmic phase at 30°C in YPD medium (1% yeast extract [Oxoid], 2% peptone [Oxoid], 2% glucose [Merck, Darmstadt, Germany]). Five hundred milliliters of culture medium were inoculated with 0.3 ml of a preculture grown aerobically for 48 h. Growth was monitored by measuring the optical density at 600 nm.
Construction of deletion strains.
A dominant resistance marker module, kanMX4, containing the coding sequence of the Kanr gene of the Escherichia coli transposon Tn903 on vector pFA6a (38) was used to replace yeast ORFs. The Kanr gene encodes an aminoglycoside phosphotransferase activity (35) which renders S. cerevisiae resistant to the drug geneticin (G418) (21). A replacement strategy making use of short flanking homology regions to the target locus was used to construct deletion cassettes by PCR (38, 39). Deletion cassettes contained the ATG codon of the ORF to be deleted, the kanMX4 gene, and the stop codon of the ORF, thus eliminating the entire target ORF. Adjacent ORFs were not affected by this deletion procedure. All deletions were made in the FY1679 strain background.
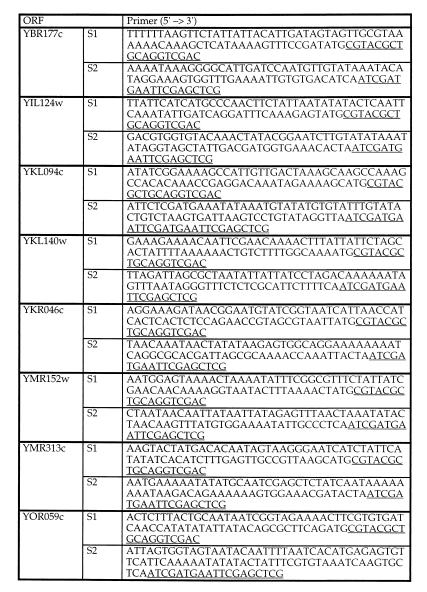
To generate marker DNA flanked by short homology regions, a pair of oligonucleotide primers with 70 nucleotides homologous to the target locus at the 5′ end followed by 18 to 19 nucleotides homologous to pFA6a-kanMX4 were used (Fig. 1). A 1.65-kb PCR fragment was generated with Pwo polymerase (Boehringer, Mannheim, Germany) by using approximately 50 ng of gel-purified NotI-digested pFA6a-kanMX4 plasmid or 150 ng of plasmid as a template in a standard PCR mixture. This mixture contained the PCR buffer [10 mM Tris-Cl− (pH 8.85), 25 mM KCl, 5 mM (NH4)2SO4] with 2 mM MgSO4, 0.2 mM (each) deoxynucleoside triphosphates, and a 1 μM concentration of primers in a total volume of 25 μl. After a denaturation step for 5 min at 94°C, fragments were amplified for 10 cycles of 30 s at 94°C, 30 s at 54°C, and 105 s at 72°C and for 20 cycles of 30 s at 94°C, 30 s at 65°C, and 105 s at 72°C, followed by a final elongation step for 12 min at 72°C. PCR fragments were ethanol precipitated and 400 to 700 ng was used for transformation.
FIG. 1.
Deletion of eight ORFs encoding yeast lipid particle proteins. Primers were used for the deletion of ORFs by the short flanking homology method. The underlined sequences are homologous to the kanMX4 gene. S1, sequence 1; S2, sequence 2.
Diploid FY1679 yeast cells were transformed by using the high-efficiency lithium acetate transformation protocol (14). Transformed cells were grown in YPD at 30°C overnight and then spread on YPD plates containing 200 mg of G418 (Calbiochem, La Jolla, Calif.) per liter. After incubation for 2 to 3 days, large colonies were transferred to fresh YPD-G418 plates. Only those clones that yielded colonies were considered as positive transformants and further checked for correct integration of the respective deletion cassette.
Correct replacement of the respective ORFs by the kanMX4 module in G418-resistant transformants was verified by analytical PCR with Dynazyme polymerase and whole yeast cell extracts (17). In brief, oligonucleotides were designed to bind outside the target locus, within the target locus, and within the marker module (38, 39). In diploid yeast transformants, the correct integration of the marker resulted in the appearance of one PCR fragment characteristic for the wild-type allele and one fragment characteristic for the mutated allele.
Diploid yeast transformants were sporulated in liquid medium containing 0.3% potassium acetate and 0.02% raffinose for 3 to 5 days at room temperature. Tetrad dissection was performed on YPD plates. At least nine tetrads were dissected for each ORF and incubated at 30°C for 2 to 3 days prior to phenotypic analyses.
Isolation of lipid particle proteins.
Highly purified yeast lipid particles with an enrichment factor of 700 to 800 for triacylglycerols, steryl esters, and Erg6p over the homogenate were prepared from cells grown to the late logarithmic phase as described by Leber et al. (27). Prior to protein analysis, the lipid particle fraction was delipidated. Nonpolar lipids were extracted with 2 volumes of diethyl ether. The organic phase was withdrawn, residual diethyl ether was removed under a stream of nitrogen, and proteins were precipitated from the aqueous phase with trichloroacetic acid at a final concentration of 10%. The protein pellet was solubilized in 0.1% sodium dodecyl sulfate (SDS)–0.1% NaOH. The protein was quantified by the method of Lowry et al. (30) with bovine serum albumin as a standard.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by the method of Laemmli (26). Samples were dissociated at 37°C because treatment at higher temperatures resulted in hydrolysis of several lipid particle proteins. Western blot analysis was carried out as described by Haid and Suissa (15), and immunoreactive proteins were detected by enzyme-linked immunosorbent assay with rabbit antisera as the first antibody and goat anti-rabbit immunoglobulin G linked to peroxidase or phosphatase as the second antibody.
Identification of lipid particle proteins.
Protein bands stained with Coomassie brilliant blue were excised from the SDS polyacrylamide gel and stored at −20°C. Proteins in gel slices were digested with trypsin as described by Shevchenco et al. (37). In brief, gel slices were washed with 50 mM NH4HCO3–acetonitrile (1:1 [vol/vol]) followed by dehydration with acetonitrile and drying by vacuum centrifugation. Then proteins were reduced with 200 μl of 10 mM dithiothreitol–50 mM NH4HCO3 at 56°C and alkylated in 200 μl of 55 mM iodoacetamide–50 mM NH4HCO3 for 15 min. The gel pieces were washed several times in 50 mM NH4HCO3, dehydrated with acetonitrile, and dried by vacuum centrifugation. Then proteins were digested overnight with trypsin (modified trypsin; Promega) at 37°C. The resulting peptides were extracted with 25 mM NH4HCO3–aceto-nitrile (1:1 [vol/vol]) and subsequently with 25 mM NH4HCO3–5% formic acid. Combined extracts were dried by vacuum centrifugation.
Mass spectrometric analysis of generated peptide mixtures was performed by Protana A/S, Odense, Denmark, by using a nanoelectrospray system coupled to a Finnigan LCQ ion trap mass spectrometer (Finnigan, San Jose, Calif.). Peptide mixtures were purified on purification capillaries with POROS R2 perfusion chromatography material. Peptides were eluted in 50% methanol–5% formic acid directly into the nanospray capillary by centrifugation.
Proteins were identified by querying a nonredundant sequence database containing more than 300,000 entries with partial amino acid sequences (peptide sequence tags) deduced from mass spectra. The search software was PepSea, version 1.0 (Protana A/S).
In silicio sequence analysis.
Information about proteins with known and hypothetical functions was retrieved from the Yeast Protein Database (36a), the Saccharomyces Genome Database (37a), and the Munich Information Center for Protein Sequences (33a). Hydrophobicity analyses were performed according to Kyte and Doolittle (25), and homology searches were done by using BLAST Search (2).
Lipid analysis.
Lipids of whole yeast cells were extracted after cell disruption by the procedure of Folch et al. (11). Individual phospholipids were separated by two-dimensional thin-layer chromatography on silica gel 60 plates (Merck) by using chloroform–methanol–25% NH3 (65:35:5 [vol/vol/vol]) as the first developing solvent and chloroform-acetone-methanol-acetic acid-water (50:20:10:10:5 [vol/vol/vol/vol/vol]) as the second developing solvent. Phospholipids were visualized on thin-layer chromatography plates by staining with iodine vapor, scraped off the plate, and quantified by the method of Broekhuyse (5).
For the analysis of neutral lipids, extracts were applied to silica gel 60 plates with the aid of a sample applicator (Linomat IV; CAMAG, Muttenz, Switzerland), and chromatograms were developed in an ascending manner by using the solvent system light petroleum-diethyl ether-acetic acid (25:25:1 [vol/vol/vol]) for the first third of the total distance. Then, the plates were dried briefly and further developed to the top of the plate by using the solvent system light petroleum-diethyl ether (49:1 [vol/vol]). Quantification of ergosterol and ergosteryl esters was carried out by densitometric scanning at 275 nm with ergosterol as a standard. Triacylglycerols were visualized by postchromatographic staining with a chromatogram immersion device (CAMAG). Plates were dipped for 6 s into a developing reagent consisting of 0.63 g of MnCl2 · 4H2O, 60 ml of water, 60 ml of methanol, and 4 ml of concentrated sulfuric acid, briefly dried, and heated at 100°C for 30 min. Quantification of acylglycerols was carried out by densitometric scanning at 400 nm with triolein (NuCheck, Inc., Elysian, Maine) as a standard.
Individual sterols were analyzed after alkaline hydrolysis (29) of the lipid extract by gas liquid chromatography (GLC). GLC was performed on a Hewlett-Packard 5890 equipped with a flame ionization detector operated at 320°C, using a capillary column (Hewlett-Packard 5; 30 m by 0.32 mm by 0.25 μm film thickness). After a 1-min hold at 150°C, the temperature was increased to 310°C at 10°C/min. The final temperature was held for 10 min. Nitrogen was used as the carrier gas, and 1-μl aliquots of samples were injected onto the column. Relative retention times of sterols were similar, as described previously (36, 41).
Fatty acids were also analyzed by GLC. Lipids extracted as described above were subjected to methanolysis with BF3-methanol (14%) and converted to methyl esters (33). Fatty acid methyl esters were separated by GLC by using the same equipment as described above. A temperature program of 2 min at 150°C, increasing 10°C/min to 300°C, and 5 min at 300°C was used. Fatty acids were identified by comparing them to commercial fatty acid methyl ester standards (NuCheck).
RESULTS
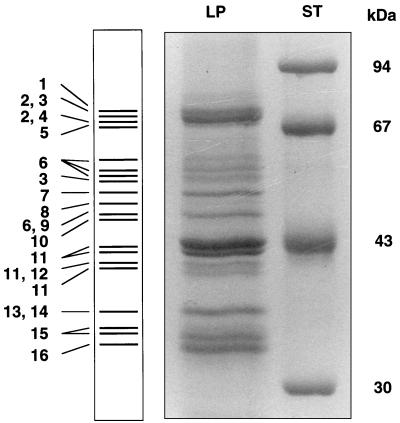
To identify proteins of yeast lipid particles, polypeptides of a highly enriched lipid particle fraction were separated by SDS-PAGE (Fig. 2), reisolated from the gel, and subjected to mass spectrometric analysis as described in Materials and Methods. This strategy led to the identification of the major yeast lipid particle proteins as summarized in Table 1. In some of the bands excised from the gel (Fig. 2), more than one protein was detected, namely Fat1p and Faa4p (the amount of Faa4p was greater than Fat1p), Erg7p and Fat1p (the amount of Erg7p was greater than Fat1p), Tgl1p and the YOR059c gene product (approximately equal amounts), Erg6p and the YDL193w gene product (equal amounts), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Yju3p (the amount of Yju3p was greater than GAPDH). Since the mass spectrometric method employed did not allow exact quantification of proteins identified in the same band, the molar ratio could be only roughly estimated. Some proteins or their polypeptide fragments were identified in several bands, namely Fat1p, Tgl1p, Erg6p, and the gene product of YIL124w (Fig. 2). The reason for this finding is most likely degradation of proteins during the isolation procedure. Alternatively, different posttranslationally modified forms of proteins might be present on the lipid particle surface, although such modifications could not be demonstrated by the methods used for this study.
FIG. 2.
Protein pattern of lipid particles. Lipid particle proteins (LP) were separated by SDS-PAGE, reisolated from the gel, and subjected to mass spectrometric analysis as described in Materials and Methods. ST, standard proteins. The numbering of the bands is the same as that shown in Tables 1 and 2.
TABLE 1.
Identification of genes encoding lipid particle proteinsa
| No. | Gene or ORF designation | Function [hypothetical]b | MW | pI | ORF | Reference |
|---|---|---|---|---|---|---|
| 1 | FAA1 | Long-chain fatty acid CoA ligase (for degradation or incorporation in PL) | 77,866 | 7.70 | YOR317c | 23 |
| 2 | FAT1 | Very long-chain fatty acid activator; fatty acid transport protein (exogenous long-chain fatty acids) | 71,700 | 8.40 | YBR041c | 10, 40 |
| 3 | FAA4 | Long-chain fatty acid CoA ligase (for degradation or incorporation into PL) | 77,267 | 6.33 | YMR246w | 22 |
| 4 | ERG7 | Lanosterol synthase | 83,318 | 6.30 | YHR072w | 4 |
| 5 | YMR313c | Unknown | 73,612 | 8.74 | YMR313c | |
| 6 | TGL1 | [Triacylglycerol lipase] | 62,979 | 6.60 | YKL140w | 1 |
| 7 | ERG1 | Squalene epoxidase | 55,120 | 6.27 | YGR175c | 19, 28 |
| 8 | YBR177c | [UFP007 family (not characterized); weak homologue to probable membrane receptor] | 51,255 | 7.84 | YBR177c | |
| 9 | YOR059c | [Lipase serine active site] | 51,114 | 9.20 | YOR059c | |
| 10 | YIM1 | [Homologue to mitochondrial inner membrane protease] | 41,651 | 8.24 | YMR152w | |
| 11 | ERG6 | Sterol-Δ24-methyltransferase | 43,421 | 5.59 | YML008c | 27 |
| 12 | YDL193w | [Weak homologue to Ca2+ channel protein] | 42,555 | 6.73 | YDL193w | |
| 13 | TDH1 | GAPDH | 35,619 | 8.41 | YJL052w | 32 |
| TDH2 | 35,715 | YJR009c | ||||
| TDH3 | 35,615 | YGR192c | ||||
| 14 | YJU3 | [Lysophospholipase] | 35,549 | 8.55 | YKL094w | 12 |
| 15 | YIM4 | [Oxidoreductase (short-chain dehydrogenase or reductase family)] | 32,822 | 9.34 | YIL124w | |
| 16 | YKR0456 | Unknown | 31,246 | 8.85 | YKR046c | |
| SLC1c | 1-acylglycerol-3-phosphate acyltransferase | 33,903 | 9.76 | YDL052c | 3, 34 |
MW, molecular weight; pI, isoelectric point; PL, phospholipid.
Functions in brackets are hypothetical.
The gene product of SLC1 was not detected during the mass spectrometric approach described in this work but was characterized earlier by biochemical and molecular biological means.
Lipid particles contain enzymes involved in lipid metabolism.
A number of lipid particle proteins identified in this study had been previously characterized by function, although in some cases the precise subcellular localization of the respective protein was questionable. Among these components, there are several enzymes involved in ergosterol biosynthesis (Erg1p, Erg6p, and Erg7p) and fatty acid activation (Faa1p, Faa4p, and Fat1p). It has to be mentioned, however, that the occurrence of all these proteins is most likely not restricted to lipid particles. For example, Erg1p and Erg6p were also found in the endoplasmic reticulum (28). Also, Fat1p was originally reported to be a fatty acid transporter (10) and assumed to be associated with the plasma membrane. Recent results (40), however, have demonstrated that Fat1p has very-long-chain fatty acid acyl coenzyme A (CoA) synthetase activity and may be a component of peroxisomes. Unspecific coisolation of proteins with lipid particles is unlikely because (i) the protein pattern of lipid particles can be obtained in a highly reproducible way and (ii) proteins of other subcellular compartments are not randomly associated with lipid particles. One exception may be GAPDH, whose association with lipid particles (Table 1 and Fig. 2) came as a surprise. Three different forms of GAPDH are known (32). They are encoded by the ORFs YJL052w, YJR009c, and YGR192c (Table 1). Mass spectrometry did not allow us to distinguish between these three forms, because differences in the amino acid sequence of the isoforms are only minor. The amount of GAPDH in lipid particles was very low, and Western blot analysis with monospecific antibodies failed to detect the protein in this fraction (data not shown). Thus, it is likely that the presence of GAPDH, which is a cytosolic enzyme, is due to unspecific interaction with the surface of lipid particles. A tendency of GAPDH to associate with other subcellular membranes in a rather unspecific way has been reported before (42).
Characterization of mutants with deletions of ORFs encoding lipid particle proteins with unknown functions.
The fact that almost all lipid particle proteins of known function are enzymes of lipid biosynthetic pathways led us to speculate that the other hypothetical proteins of lipid particles might also be involved in lipid metabolism. To obtain some general information about these uncharacterized proteins, strains with deletions of the respective ORFs were constructed and subjected to phenotypic analysis. Lack of the hypothetical proteins resulted in only one case in lethality (YDL193w), whereas all other mutants were viable under standard conditions (Table 2). The YDL193w deletion strain could not be rescued by supplementation with ergosterol or long-chain fatty acids under aerobic or anaerobic conditions (data not shown). Thus, involvement of this gene in sterol or fatty acid biosynthesis appears unlikely. Since the YDL193w gene product shows weak homology to Ca2+ channel proteins in different organisms (Table 1), complementation of the deletion defect by addition of various ions to the culture medium was tested. Supplementation of the mutant with neither Ca2+ nor other divalent or monovalent cations restored growth. The other deletion strains were viable and grew like the wild type on YPD medium at 15, 30, and 37°C. The only exception was the mutant with a deletion of YBR177c, which exhibited a slight temperature sensitivity at 37°C but grew better than the wild type at 15°C.
TABLE 2.
Characterization of gene products present in yeast lipid particles
| No. | Gene or ORF designation | Gene product
|
Deletion strain phenotype | |
|---|---|---|---|---|
| TM | Hydrophobicity domain ≥2a | |||
| 1 | FAA1 | 1 | 2 (middle and C terminal) | Viable |
| 2 | FAT1 | 3 | 2 (N terminal and middle) | Viable |
| 3 | FAA4 | 0 | 2 (middle and C terminal) | Viable |
| 4 | ERG7 | 0 | 1 (C terminal) | Sterol auxotroph |
| 5 | YMR313c | 0 | 1 (C terminal) | Viable |
| 6 | TGL1 | 1 | 1 (N terminal) | Viable |
| 7 | ERG1 | 2 | 3 (N and C terminal) | Sterol auxotroph |
| 8 | YBR177c | 0 | 1 (middle) | Viable (temperature sensitive) |
| 9 | YOR059c | 1 | 1 (middle) | Viable |
| 10 | YIM1 | 0 | 0 | Viable |
| 11 | ERG6 | 0 | 0 | Viable |
| 12 | YDL193w | 1 | 1 (middle) | Lethal |
| 13 | TDH1 | 0 | 0 | Viable |
| TDH2 | 0 | 0 | Viable | |
| TDH3 | 0 | 0 | Viable | |
| 14 | YJU3 | 0 | 0 | Viable |
| 15 | YIM4 | 0 | 2 (middle and C terminal) | Viable |
| 16 | YKR0456 | 1 | 0–1 | Viable |
| SLC1 | 1 | 2 (N terminal) | Viable | |
Hydrophobicity index from Kyte and Doolittle plots (25); the positions of the hydrophobic domains within the polypeptide are shown in parentheses.
Haploid strains with deletions of unassigned ORFs were subjected to systematic lipid analysis. In the mutant with a deletion of YBR177c, the phospholipid pattern was slightly but significantly different from that of the wild-type strain. In this mutant, cellular levels of phosphatidylinositol (PtdIns) and phosphatidic acid were increased at the expense of phosphatidylethanolamine and phosphatidyldimethylethanolamine (Table 3). The strain with a deletion of YMR313c contained significantly higher amounts of triacylglycerols and the strains with deletions of YBR177c and YKL140w had higher amounts of ergosteryl esters than the wild type (Table 4). The pattern of individual sterols (Table 5) was similar in all deletion strains, with the exception of the mutant having a deletion of YBR177c. This strain contained a smaller amount of fecosterol and a higher level of lanosterol than the wild type. Phospholipid, sterol, and neutral lipid compositions of the other deletion strains were like those of the wild type. The fatty acid composition of all deletion strains was similar to that of FY1679 (data not shown). In summary, these results suggest that the gene products of YBR177c, YMR313c, and YKL140w are candidates for involvement in lipid metabolism through secondary effects. Since we did not observe all-or-nothing effects, it is unlikely that major enzymes of lipid metabolism are directly affected by the respective mutations.
TABLE 3.
Phospholipid composition of deletion strains
| ORF deleted | % of total phospholipidsa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PtdCho | PtdDMEtn | PtdEtn | PtdIns | PtdSer | PA | CL | Lyso-PL | |
| None (FY1679) | 45.7 | 2.5 | 21.7 | 17.1 | 6.6 | 2.4 | 2.9 | 1.1 |
| YBR177c | 48.7 | 0.6 | 15.9 | 20.5 | 6.3 | 5.7 | 1.3 | 1.0 |
| YIL124w | 44.4 | 2.0 | 21.0 | 19.1 | 7.0 | 2.2 | 4.0 | <0.1 |
| YKL094w | 47.6 | 2.5 | 21.7 | 16.4 | 7.8 | 2.0 | 1.8 | 0.2 |
| YKL140w | 40.2 | 2.4 | 20.3 | 21.3 | 9.0 | 2.1 | 3.4 | 1.3 |
| YKR046c | 45.1 | 2.5 | 21.0 | 17.0 | 6.9 | 2.6 | 2.7 | 2.2 |
| YMR152w | 46.3 | 2.4 | 21.8 | 16.5 | 7.1 | 2.3 | 3.1 | 0.6 |
| YMR313c | 45.6 | 2.4 | 22.0 | 16.9 | 7.6 | 2.3 | 2.8 | 0.5 |
| YOR059c | 45.1 | 3.4 | 23.3 | 16.3 | 7.9 | 2.0 | 2.0 | 0.1 |
Mean values, with a standard deviation of ±10%, are from three independent experiments. PtdCho, phosphatidylcholine; PtdDMEtn, phosphatidyldimethylethanolamine; PtdEtn, phosphatidylethanolamine; PtdIns, phosphatidylinositol; PtdSer, phosphatidylserine; PA, phosphatidic acid; CL, cardiolipin; Lyso-PL, lysophospholipids.
TABLE 4.
Neutral lipid composition of deletion strains
| ORF deleted | Ratio (mg/mg) ofa:
|
||
|---|---|---|---|
| Ergosterol/phospholipid | Ergosteryl esters/phospholipid | Triacylglycerols/phospholipid | |
| None (FY1679) | 0.23 | 0.10 | 0.13 |
| YBR177c | 0.23 | 0.17 | 0.12 |
| YIL124w | 0.20 | 0.10 | 0.16 |
| YKL094w | 0.26 | 0.12 | 0.10 |
| YKL140w | 0.23 | 0.26 | 0.16 |
| YKR046c | 0.26 | 0.10 | 0.09 |
| YMR152w | 0.25 | 0.11 | 0.09 |
| YMR313c | 0.21 | 0.11 | 0.33 |
| YOR059c | 0.18 | 0.10 | 0.10 |
Mean values, with a standard deviation of ±10%, are from three independent experiments.
TABLE 5.
Sterol composition of deletion strains
| ORF deleted | % of total sterolsa
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Squalene | Zymosterol | Fecosterol | Ergosta-5,7,4(28)-trienol | Episterol | Lanosterol | 4,4-Dimethylsterol | Ergosterol | |
| None (FY1679) | 6.7 | 7.1 | 6.0 | 8.2 | 6.3 | 7.8 | 1.6 | 56.3 |
| YBR177c | 7.0 | 8.0 | 2.2 | 8.2 | 6.5 | 10.2 | 1.0 | 56.9 |
| YIL124w | 5.9 | 8.3 | 5.5 | 9.7 | 4.4 | 6.9 | 1.3 | 57.7 |
| YKL094w | 6.1 | 8.1 | 5.9 | 8.0 | 6.0 | 7.4 | 1.9 | 56.6 |
| YKR046c | 7.1 | 7.1 | 5.4 | 8.1 | 5.7 | 7.3 | 2.0 | 57.4 |
| YKL140w | 6.3 | 7.5 | 6.8 | 4.5 | 8.1 | 9.2 | 1.6 | 56.0 |
| YMR152w | 6.3 | 7.5 | 5.8 | 8.7 | 5.9 | 7.8 | 1.7 | 56.3 |
| YMR313c | 8.2 | 8.4 | 5.5 | 8.4 | 5.0 | 8.3 | 2.0 | 54.3 |
| YOR059c | 5.0 | 7.6 | 6.0 | 8.9 | 5.6 | 8.0 | 1.8 | 57.2 |
Mean values, with a standard deviation of ±3%, are from three independent experiments.
Most lipid particle proteins lack transmembrane spanning domains.
Computer analysis of functionally characterized and uncharacterized yeast lipid particle proteins did not unveil obvious common motifs that could be regarded as targeting sequences of these polypeptides to lipid particles. The observation, however, that most lipid particle proteins of yeast lack transmembrane spanning (TM) domains or contain only one TM domain (Table 2) deserves our attention. The only exceptions are Fat1p and Erg1p, with two or three putative TM segments. Preferential association of proteins lacking TM domains with lipid particles is most likely because the surface membrane of lipid particles is a phospholipid monolayer (27). Thus, proteins containing hydrophobic stretches different from TM domains may preferentially interact with the surface of lipid particles.
DISCUSSION
Mass spectrometric analysis was used to identify major proteins of lipid particles of the yeast S. cerevisiae. This approach supported previous findings concerning the localization of Erg6p and Erg1p to lipid particles (27, 28). In addition, several proteins with known function could be attributed to the lipid particle fraction during this study, namely Erg7p, Faa1p, Faa4p, and Fat1p (4, 10, 22, 23, 40). Furthermore, some novel gene products encoded by unassigned ORFs were identified as lipid particle components.
What is the physiological role of lipid particle proteins? Since several enzymes of ergosterol synthesis are located on lipid particles, it is tempting to speculate that these proteins actively participate in cellular sterol formation. This is very likely for Erg6p (27) and Erg7p (4a), which are enzymatically active in lipid particle preparations. Similarly, a glycerol-3-phosphate acyltransferase encoded by the unidentified GAT gene and the 1-acylglycerol-3-phosphate acyltransferase Slc1p, which contribute to phosphatidic acid biosynthesis, were previously identified as lipid particle components (3, 7, 43). It has to be mentioned, however, that Slc1p escaped detection by the mass spectrometric approach most likely due to its low abundance. None of the unassigned ORFs could be identified as GAT since lipid particles of all deletion strains tested contained wild-type levels of glycerol-3-phosphate acyltransferase activity (2a).
In contrast to the above-mentioned enzymes, it was shown that squalene epoxidase (Erg1p) of isolated lipid particles is not enzymatically active in vitro, whereas Erg1p present in the endoplasmic reticulum fraction exhibits enzymatic activity (28). It was argued that a component, probably a reductase, that is present in the endoplasmic reticulum but absent from lipid particles may be the missing cofactor. Interaction of the endoplasmic reticulum with lipid particles may activate Erg1p of the latter compartment.
The presence of enzymatically inactive proteins on the surface of lipid particles, as described for Erg1p, may also be interpreted as a regulatory phenomenon. If Erg1p of lipid particles does not contribute to ergosterol synthesis in vivo, this protein might be put on hold on the surface of this compartment for a situation which requires enhancement of lipid biosynthesis. Under these conditions, enzymes could be immediately mobilized from lipid particles and translocated to their site of activation, e.g., the endoplasmic reticulum, thus providing lipids within a short time without new polypeptide synthesis. The idea of depositing proteins in lipid particles during formation of this compartment has been previously advocated by Lum and Wright (31) when studying overexpression of 3-hydroxy-3-methylglutaryl CoA reductase in Schizosaccharomyces pombe. This enzyme, which accumulated first in so-called karmellae, was deposited in lipid particles upon degradation of the former organelle.
The function of several gene products located on yeast lipid particles remains to be demonstrated. With one exception, YDL193w, a deletion of ORFs encoding lipid particle proteins affected neither cell viability nor the formation of lipid particles in a significant way. All these deletion strains contain lipid particles of normal size and physical properties as shown by microscopic inspection and isolation of the respective fractions (44). We can only speculate at present that some proteins located on the surface of lipid particles may be involved in the deposition of triacylglycerols and/or steryl esters in or mobilization of these lipids from this compartment. The gene product of YMR313c may be a candidate for such a function because the strain having a deletion of this ORF accumulates triacylglycerols to some extent (Table 4).
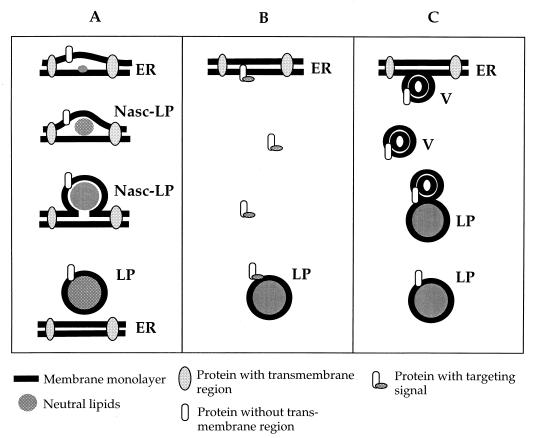
An intriguing question concerns targeting and transfer of proteins to lipid particles. Through localization studies of Erg1p (28) and of Slc1p and Gat1p (3), it was demonstrated that lipid particles and the endoplasmic reticulum share a certain set of proteins and thus appear to be related compartments. At present, we can only speculate about the relationship of these two subcellular fractions. The fact that (i) all lipid particle proteins characterized so far are involved in lipid metabolism and (ii) most lipid particle proteins contain either none or only a small number of TM domains may serve as the basis for the following hypothesis (Fig. 3A). Several enzymes involved in lipid synthesis may be located in specific domains of the endoplasmic reticulum. Clustering of these enzymes might cause local accumulation of newly formed lipids, especially those which are unable to integrate into a phospholipid bilayer, namely triacylglycerols and steryl esters. These neutral lipids may form microdroplets (preforms of lipid particles) between the two leaflets of the endoplasmic reticulum membrane bilayer which bud off after reaching a certain size. The presence of steryl ester synthases Are1p and Are2p in the endoplasmic reticulum (45) is in line with this model. Recent findings from our laboratory (3a) suggest that the endoplasmic reticulum is also the major site of triacylglycerol synthesis, indicating that both neutral lipid species of lipid particles are formed in the same compartment. It has to be noted, however, that triacylglycerol synthase activity has previously been attributed to the lipid particle fraction by Christiansen (7). The assay used by this author, however, did not allow for distinguishing between acylation of the substrate diacylglycerol and transacylation reactions.
FIG. 3.
Hypothetical models describing targeting and transfer of proteins to lipid particles. (A) Budding of a neutral lipid-rich domain from the endoplasmic reticulum is accompanied by association of proteins with the phospholipid monolayer of newly formed lipid particles. According to this model, polypeptides without TM regions would preferentially associate with lipid particles. Nasc-LP, nascent lipid particles. (B) Association of proteins with preexisting lipid particles though a targeting signal on the polypeptide. (C) Transport of proteins to preexisting lipid particles by vesicle (V) flow. LP, lipid particles; ER, endoplasmic reticulum.
Fluorescence microscopic evidence obtained recently in our laboratory (23a) demonstrated the appearance of small, newly formed lipid particles in proximity to the endoplasmic reticulum, thus supporting the model presented in Fig. 3A. According to the molecular shape concept (18), PtdIns-rich domains in the endoplasmic reticulum might facilitate the budding process. This hypothesis is in line with the finding that PtdIns comprises approximately 30% of total lipid particle phospholipids (27), whereas PtdIns is only a minor component among endoplasmic reticulum bulk phospholipids. During the budding process, newly formed lipid particles may be enwrapped by an endoplasmic reticulum-derived phospholipid monolayer, which indeed forms the surface membrane of lipid particles (27). Proteins with none or only a low number of TM domains initially present in the endoplasmic reticulum may remain associated with the phospholipid monolayer of lipid particles during the budding process, whereas proteins with typical TM regions may be largely excluded.
Although the above-mentioned hypothesis of lipid particle biosynthesis is consistent with experimental evidence obtained during our studies and compatible with the theory of oil body formation in plants (16), alternative possibilities of lipid particle formation should be considered. As an example, proteins could be directed to the surface of preformed lipid particles by a targeting signal (Fig. 3B). Although no such typical motifs were found in lipid particle proteins, signals based on conformational properties that escaped our attention may be important in that respect. As a further possible mechanism for assembly of proteins to lipid particles, transport of proteins through vesicle flux might be considered (Fig. 3C). Experimental evidence for such a mechanism, however, is also missing. The protein encoded by YBR177c, which is slightly homologous to a probable human membrane receptor (HPS1) (Table 1), might be regarded as a candidate for facilitating such a vesicle docking process. The fact that the mutant with a deletion of YBR177c contains lipid particles with a slightly different protein pattern than the wild type could be an argument for this hypothesis. A detailed analysis of this mutant and characterization of the YBR177c gene product will be required to address this question.
ACKNOWLEDGMENTS
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (projects 11491 to G.D. and F706 to S.D.K.), EUROFAN project BIO4-CT97-2294, and the Austrian Ministry of Science and Transportation.
K.A. and D.Z. contributed equally to this work and should be considered co-first authors.
REFERENCES
- 1.Abraham P R, Mulder A, Van’t Riet J, Planta R J, Raue H A. Molecular cloning and physical analysis of an 8.2 kb segment of chromosome XI of Saccharomyces cerevisiae reveals five tightly linked genes. Yeast. 1992;8:227–238. doi: 10.1002/yea.320080309. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Lipman D J. Protein database searches for multiple alignments. Proc Natl Acad Sci USA. 1990;87:5509–5513. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Athenstaedt, K. Unpublished observation.
- 3.Athenstaedt K, Daum G. Biosynthesis of phosphatidic acid in lipid particles and the endoplasmic reticulum of the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:7611–7616. doi: 10.1128/jb.179.24.7611-7616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Athenstaedt, K., and D. Sorger. Unpublished data.
- 4.Balliano G, Viola F, Ceruti M, Cattel L. Characterization and partial purification of squalene-2,3-oxide cyclase from Saccharomyces cerevisiae. Arch Biochem Biophys. 1992;293:122–129. doi: 10.1016/0003-9861(92)90374-6. [DOI] [PubMed] [Google Scholar]
- 4a.Balliano, G. 1999. Personal communication.
- 5.Broekhuyse R M. Phospholipids in tissues of the eye. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim Biophys Acta. 1968;260:449–459. doi: 10.1016/0005-2760(68)90038-6. [DOI] [PubMed] [Google Scholar]
- 6.Christiansen K, Jensen P K. Membrane bound lipid particles from beef heart. Chemical composition and structure. Biochim Biophys Acta. 1972;260:449–459. doi: 10.1016/0005-2760(72)90060-4. [DOI] [PubMed] [Google Scholar]
- 7.Christiansen K. Triacylglycerol synthesis in lipid particles from baker’s yeast (Saccharomyces cerevisiae) Biochim Biophys Acta. 1978;530:78–90. doi: 10.1016/0005-2760(78)90128-5. [DOI] [PubMed] [Google Scholar]
- 8.Clausen M K, Christiansen K, Jensen P K, Behnke O. Isolation of lipid particles from bakers yeast. FEBS Lett. 1974;43:176–179. doi: 10.1016/0014-5793(74)80994-4. [DOI] [PubMed] [Google Scholar]
- 9.Daum G, Paltauf F. Triacylglycerols as fatty acid donors for membrane phospholipid biosynthesis in yeast. Monatsh Chem. 1980;111:355–363. [Google Scholar]
- 10.Faergeman N M, Di Rosso C C, Elberger A, Knudsen J, Black P N. Disruption of the Saccharomyces cerevisiae homologue to the murine fatty acid transport protein impares uptake and growth on long-chain fatty acids. J Biol Chem. 1997;272:8531–8538. doi: 10.1074/jbc.272.13.8531. [DOI] [PubMed] [Google Scholar]
- 11.Folch J, Lees M, Sloane-Stanley G H. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 12.Forrova H, Kolarov J, Ghislan M, Goffeau A. Sequence of the novel essential gene YJU2 and two flanking reading frames located within a 3.2 kb EcoRI fragment from chromosome X of Saccharomyces cerevisiae. Yeast. 1992;8:419–422. doi: 10.1002/yea.320080509. [DOI] [PubMed] [Google Scholar]
- 13.Gemmerich A R. Ultrastructural and enzymatic studies on the development of microbodies in germinating spores of the fern Anemia phyllitidis. Z Pflanzenphysiol. 1981;102:69–80. [Google Scholar]
- 14.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 15.Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 16.Huang A H C. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- 17.Huxley C, Green E D, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- 18.Israelachvili J N, Marcelja S, Horn R G. Physical principles of membrane organization. Q Rev Biophys. 1980;13:121–200. doi: 10.1017/s0033583500001645. [DOI] [PubMed] [Google Scholar]
- 19.Jandrositz A, Turnowsky F, Högenauer G. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene. 1991;107:155–160. doi: 10.1016/0378-1119(91)90310-8. [DOI] [PubMed] [Google Scholar]
- 20.Jayaram S, Bal A K. Oleosomes (lipid bodies) in nitrogen-fixing peanut nodules. Plant Cell Environ. 1991;14:195–203. [Google Scholar]
- 21.Jimenez A, Davies J. Expression of transposable antibiotic resistance elements in Saccharomyces. Nature. 1980;287:869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- 22.Johnson D R, Knoll L J, Levin D E, Gordon J I. Saccharomyces cerevisiae contains four fatty acid activating (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J Cell Biol. 1994;127:751–762. doi: 10.1083/jcb.127.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knoll L J, Johnson D R, Gordon J I. Biochemical studies of the three Saccharomyces cerevisiae acylCoA synthetases, Faa1p, Faa2p and Faa3p. J Biol Chem. 1994;269:16348–16356. [PubMed] [Google Scholar]
- 23a.Kohlwein, S. D. 1999. Unpublished results.
- 24.Kostner G M, Laggner P. Chemical and physical properties of lipoproteins. In: Fruchart J C, Sheperd J, editors. Clinical biochemistry—human plasma lipoproteins. Berlin, Germany: Walther de Gruyter; 1989. pp. 23–54. [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Leber R, Zinser E, Zellnig G, Paltauf F, Daum G. Characterization of lipid particles of the yeast Saccharomyces cerevisiae. Yeast. 1994;10:1421–1428. doi: 10.1002/yea.320101105. [DOI] [PubMed] [Google Scholar]
- 28.Leber R, Landl K, Zinser E, Ahorn H, Spök A, Kohlwein S D, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis T A, Rodriguez R J, Parks L W. Relationship between intracellular sterol content and sterol esterification and hydrolysis in Saccharomyces cerevisiae. Biochim Biophys Acta. 1987;921:205–212. doi: 10.1016/0005-2760(87)90020-8. [DOI] [PubMed] [Google Scholar]
- 30.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 31.Lum P L, Wright R. Degradation of HMG-CoA reductase-induced membranes in the fission yeast, Schizosaccharomyces pombe. J Cell Biol. 1995;131:81–94. doi: 10.1083/jcb.131.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAlister L, Holland J P. Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1985;260:15019–15027. [PubMed] [Google Scholar]
- 33.Morrison W R, Smith L M. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J Lipid Res. 1964;5:600–608. [PubMed] [Google Scholar]
- 33a.Munich Information Centre for Protein Sequences. revision date. [Online.] http://www.mips.biochem.mpg.de/. [May 1999, last date accessed.] 1999. [Google Scholar]
- 34.Nagiec M M, Wells G B, Lester R L, Dickson R C. A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- 35.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycine resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 36.Patterson G W. Relation between structure and retention time of sterols in gas chromatography. Anal Chem. 1971;43:1165–1170. [Google Scholar]
- 36a.Proteome Inc. copyright date. [Online.] http://quest7.proteome.com/YPDhome.html. [May 1999, last date accessed.] 1999. [Google Scholar]
- 37.Shevchenco A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 37a.Stanford University. revision date. [Online.] http://genome-www.stanford.edu/Saccharomyces/. [May 1999, last date accessed.] 1999. [Google Scholar]
- 38.Wach A, Brachat A, Pohlmann R, Phillipsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 39.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruption in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 40.Watkins P A, Lu J-F, Steinberg S J, Gould S J, Smith K D, Braiterman L T. Disruption of the Saccharomyces cerevisiae FAT1 gene decreases very long-chain fatty acyl-CoA synthetase activity and elevates intracellular very long-chain fatty acid concentrations. J Biol Chem. 1998;273:18210–18219. doi: 10.1074/jbc.273.29.18210. [DOI] [PubMed] [Google Scholar]
- 41.Xu S, Norton R A, Crumley F G, Nes W D. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow column liquid chromatography, thin-layer chromatography, gas liquid chromatography and high performance liquid chromatography. J Chromatogr. 1988;452:377–398. doi: 10.1016/s0021-9673(01)81462-x. [DOI] [PubMed] [Google Scholar]
- 42.Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]
- 43.Zinser E, Sperka-Gottlieb C D M, Fasch E-V, Kohlwein S D, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zweytick, D., and K. Athenstaedt. Unpublished observation.
- 45.Zweytick, D., et al. Submitted for publication.