Abstract
Inverse PCR was used to amplify major cold shock protein (MCSP) gene families from a diverse range of bacteria, including the psychrotolerant Yersinia enterocolitica, which was found to have two almost identical MCSP coding regions (cspA1 and cspA2) located approximately 300 bp apart. This tandem gene duplication was also found in Y. pestis, Y. pseudotuberculosis, and Y. ruckeri but not in other bacteria. Analysis of the transcriptional regulation of this MCSP gene in Y. enterocolitica, performed by using both reverse transcriptase-PCR and Northern blot assays, showed there to be two cold-inducible mRNA templates arising from this locus: a monocistronic template of approximately 450 bp (cspA1) and a bicistronic template of approximately 900 bp (cspA1/A2). The former may be due to a secondary structure between cspA1 and cspA2 causing either 3′ degradation protection of cspA1 or, more probably, partial termination after cspA1. Primer extension experiments identified a putative transcriptional start site (+1) which is flanked by a cold-box motif and promoter elements (−10 and −35) similar to those found in Escherichia coli cold-inducible MCSP genes. At 30°C, the level of both mRNA molecules was negligible; however, upon a temperature downshift to 10°C, transcription of the bicistronic mRNA was both substantial (300-fold increase) and immediate, with transcription of the monocistronic mRNA being approximately 10-fold less (30-fold increase) and significantly slower. The ratio of bicistronic to monocistronic mRNA changed with time after cold shock and was higher when cells were shocked to a lower temperature. High-resolution, two-dimensional protein gel electrophoresis showed that synthesis of the corresponding proteins, both CspA1 and CspA2, was apparent after only 10 min of cold shock from 30°C to 10°C. The data demonstrate an extraordinary capacity of the psychrotolerant Y. enterocolitica to produce major cold shock proteins upon cold shock.
The genus Yersinia contains 11 species of gram-negative facultative rods, including three pathogens of humans, Yersinia enterocolitica, Yersinia pseudotuberculosis, and Yersinia pestis, and the salmonid fish pathogen, Yersinia ruckeri (20). Y. enterocolitica causes gastroenteritis in humans and animals through its ingestion in contaminated food or water (34). It is the ability of Y. enterocolitica to grow at temperatures as low as −5°C (2, 35) that has mainly resulted in cases of bacterial septicemia from the transfusion of stored refrigerated blood products (31). On the other hand, Y. enterocolitica can grow at temperatures as high as 42°C (34).
The molecular basis of cold tolerance and the roles played by cold-inducible proteins are still poorly understood. The major focus of recent work has been on mesophilic bacteria such as Escherichia coli and Bacillus subtilis (reviewed in references 15 and 42). Initial work showed that when an exponential-phase culture of E. coli is shifted from 37 to 10°C, a novel set of at least 13 proteins is induced (24). Identification of a number of these cold shock proteins has revealed several polypeptides that are involved in transcription and translation. This has led to the suggestion that the cold shock response is an adaptive mechanism facilitating protein synthesis at low temperature (25) and, as reported recently, at early exponential-phase growth (4). In contrast to the relatively minor level of induction (2- to 10-fold) observed for most cold shock proteins during a temperature downshift in E. coli, the induction of a novel protein (initially designated as F10.6) was found to be considerably higher (24). This 70-amino-acid polypeptide was shown to be induced 200-fold following a shift from 37 to 10°C (24) and was subsequently termed the major cold shock protein (MCSP) or CspA (10). MCSPs are extremely widespread in eubacteria (reference 8 and this study) and belong to the most conserved group of nucleic acid-binding proteins yet defined in nature: the cold shock domain (CSD) proteins (40). They are characterized by the ability to preferentially bind to single-stranded nucleic acid sequences containing an ATTGG/CCAAT motif (16, 28, 36, 37, 40). In both prokaryotic and eukaryotic organisms, this ability has been shown to be due to two RNA-binding motifs, RNP-1 (KGFGF) and a partial RNP-2 (VFVH) (14, 40). Although a number of roles have been proposed for MCSPs, their most likely function is as molecular chaperones involved with the unfolding of mRNA secondary structures formed at low temperature (17, 22, 23).
E. coli, B. subtilis, Bacillus cereus, and Pseudomonas fragi have all been shown to have families of MCSP homologues, ranging from three in B. subtilis to nine in E. coli (16, 28, 29, 42). These proteins are all around 70 amino acids. Comparison of the four cold-inducible MCSPs of E. coli, CspA, CspB, CspG, and CspI, shows these proteins to have considerable homology (30, 39, 42). Moreover, alignment of the 5′ untranslated mRNA pertaining to these four MCSPs shows this homology to extend beyond each coding region, with the leading 160 bp of cspB and cspG having 70% or greater identity (30, 39). These four MCSPs of E. coli are differentially regulated at low temperature (7, 39).
To our knowledge, no information on the cold shock response of the psychrotolerant bacterium Y. enterocolitica is available. In this study, we show that Y. enterocolitica has an MCSP gene duplication that is induced by cold shock to give both monocistronic and bicistronic mRNAs. Furthermore, we demonstrate that both MCSPs are translated and that transcription and translation upon cold shock are extremely rapid.
MATERIALS AND METHODS
Inverse PCR amplification of multiple MCSP gene sequences from gram-positive and gram-negative bacteria.
Genomic DNA was purified (1) from a wide range of both gram-positive and gram-negative bacteria, including the psychrotolerant Bacillus weihenstephanensis (WSBC10201) and the mesophilic B. cereus (WSBC10028), Enterococcus faecalis (NCTC 775), E. coli (W3110), Klebsiella pneumoniae (NCTC 9633), Listeria monocytogenes (ATCC 23074), Micrococcus luteus (NCTC 2665), Proteus vulgaris (NCTC 4175), Pseudomonas aeruginosa (PAO1), Salmonella typhimurium (LT2), Staphylococcus aureus (RN4220), and Y. enterocolitica (NCTC 10460). Approximately 1 μg of each DNA was dissolved in 50 μl of water, which was then subdivided into six 8-μl aliquots. Five aliquots were cut individually with the restriction enzymes AluI, HhaI, HpaII, MboI, and RsaI (Roche Diagnostics GmbH, Mannheim, Germany), while the sixth aliquot of uncut DNA acted as a control. After heat inactivation of the restriction enzyme, the cut DNAs were self-ligated (1 U of T4 DNA ligase; Promega) for 4 h at 16°C.
Using a compilation of MCSP DNA sequences gained from more than 30 species of bacteria (8), three degenerate oligonucleotide primers were designed (two pairs) with which to perform inverse PCR. The sequences of these primers are as follows: CSPIF1, 5′ [AG][AG]I GA[CT] GTI TTC GT[AT] CA[CT] TT[CT] I[GC]I GC 3′; CSPIF2, 5′ GGI T[AT]C AAA [AT]CI [CT]T[AG] CAI GAA GG[CT] CA 3′; and CSPIR1, 5′ [GC][GC][AT] [GT]I[AG] AT[AG] AAI CC[AG] AAI CCT TTI TC 3′ (bracketed nucleotides show the degeneracies used, and I represents inosine). PCR was performed with a Techne Progene automated thermocycler with 0.2-ml thin-walled PCR tubes. Reactions were carried out in 50-μl volumes containing 5 μl of 10× PCR buffer (supplied with Taq DNA polymerase; Eurogentec), 2 mM MgCl2, 100 pmol of each oligonucleotide primer, 0.2 mM each deoxynucleotide triphosphate (dATP, dCTP, dGTP, dTTP), 1 U of Taq DNA polymerase (Eurogentec), and 1 μl of a ligation mixture or uncut DNA. PCR conditions were as follows: 2 min at 95°C, followed by 35 cycles at 95°C for 15 s, 50°C for 2 min, and 72°C for 3 min, with a final extension at 72°C for 5 min. Amplified products were analyzed on a 1.5% agarose gel (NuSieve; FMC BioProducts). Reactions containing PCR products were then run on a low-melting-point agarose gel (SeaPlaque GTG; FMC BioProducts), and DNA fragments were liquid nitrogen band extracted by using a freeze-thaw procedure (32). Direct sequencing of PCR products was performed with an ABI 373A sequencer (Perkin-Elmer Applied Biosystems) with CSPIR1 and either CSPIF1 or CSPIF2 (depending on which oligonucleotide was used to perform PCR) as sequencing primers. DNA sequences gained from the latter procedure were then used to design specific PCR primers to amplify complete MCSP genes, including their missing central regions.
Preparation of RNA from Y. enterocolitica.
A 350-ml culture of Y. enterocolitica was grown at 30°C to an optical density at 600 nm of 0.5. This was then cold shocked to 10°C in an ice bath. Ten-milliliter samples were taken before (control) and shortly after cold shock (2 min) and then at 10, 20, 30, 45, 60, 90, and 120 min after shock. The cells were centrifuged, and the pellet was frozen in liquid N2. Total RNA was isolated from the pellets with guanidine-phenol buffer as described before (18).
Northern blot analysis of cold-induced Y. enterocolitica MCSP mRNA.
Northern blotting, with 20 μg of total RNA, was carried out as described previously (27) with the following changes. The RNA was blotted with a vacuum blotter at 70 mbar for 1 h. The membrane was then air dried at 37°C for 20 min before it was cross-linked with 0.3 J/cm2. Immediately after cross-linking, the membrane was prehybridized. The blot was washed at 30°C, if not indicated otherwise, as follows: two times for 5 min each in 2× SSPE (1× SSPE is 0.15 M NaCl, 100 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 20 min at 55°C in 0.2× SSPE, 20 min in blocking buffer, followed by 1 h of conjugation with anti-digoxigenin (DIG) AP (Roche Diagnostics), two times for 5 min each in blocking buffer, two times for 5 min each in 1× phosphate-buffered saline (plus 0.5% sodium dodecyl sulfate), two times for 5 min each in assay buffer, and finally transferred to the substrate solution (250 μM CDP-Star; Tropix) for 5 min at room temperature. The blot was exposed to Curix HC1.000G film (Agfa Gevart, Köln, Germany) for 3 to 15 min. Hybridization solution was made with DIG-labelled oligonucleotide (YeA1-DIG, 5′ GCC ACA ATA CTG TTT TGC CAC AAT ATG T 3′) or DIG-labelled PCR products amplified with the primers Melmack (5′ GCT GCT GGC ACG TAG TTA 3′) and Alex (5′ ACT GGG ACT GAG ACC GG 3′) in accordance with the Boehringer Mannheim manual.
Primer extension.
Primer extension was conducted as described previously (26) with the following minor changes: 5 μg of total RNA (shock at 10°C for 10 min) was incubated with 4 μl of the primer YeA1R2(+1) (2 pmol; 5′ GCC ACA ATA CTG TTT TGC CAC 3′) and 2.6 μl of H2O for 1 min at 94°C and then 10 min at 50°C. All subsequent steps were conducted at 50°C. A sequencing reaction for a ladder was carried out with different plasmids and primers in accordance with the manual of the Sequenase V2.0 DNA sequencing kit (Amersham Pharmacia Biotech, Freiburg, Germany).
RT-PCR of cold-induced Y. enterocolitica bicistronic cspA1/A2 mRNA.
Reverse transcriptase (RT)-PCR was carried out with the primer YeRTR (5′ GGC TAT CAC CTT CAT CGC 3′) for MCSP mRNA as described for primer extension but without using labelled dATP. PCR was conducted by using the primers YeRTF (5′ CAT CGG TTT GGA CAC CAG AC 3′) and YeRTR, with the following parameters: 95°C for 10 s, 50°C for 15 s, and 72°C for 20 s for 20 cycles.
Southern blot analysis of Y. enterocolitica DNA.
Ten micrograms of DNA was cut with the restriction enzymes SspI, EcoRV, EcoRI. Fragments were run on a 1.2% Tris-acetate-EDTA agarose gel and blotted on a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech). Hybridization and washing were carried out as described for Northern blotting above.
Prediction of secondary mRNA structure.
The secondary structure of the bicistronic cspA1/A2 mRNA from Y. enterocolitica was obtained by using the folding software program MFOLD (38, 43, 43a).
Two-dimensional polyacrylamide gel electrophoresis.
For the preparation of protein samples, shocked cells were centrifuged and the pellets were resuspended in solubilization buffer as described previously (12). Cell lysis was performed by a single passage through a French press (SLM Aminco Inc., Rochester, N.Y.) cell at 20,000 lb/in2. The cell extract was then centrifuged for 45 min (15,400 × g at 4°C). Two-dimensional gel electrophoresis of the supernatant was performed as described before (11-13a) by using immobilized pH gradient (IPG) recipes (pH 4 to 7 and pH 5 to 6) described previously (33). For analytical purposes, samples of approximately 70 μg, for microseparation ca. 700 μg, of protein per gel were used. Proteins were resolved by isoelectric focusing with Pharmacia’s DryStrip kit. The protein solution was applied at the anodic side of the IPG gel strips. The sample was run into the gel at low voltages (gradient pH 4 to 7, 150, 300, and 600 V, each for 3 h; gradient pH 5 to 6, 150, 300, and 600 V, each for 5 h). Isoelectric focusing was done for 12 h at 3,500 V (gradient pH 4 to 7) and for 12 h at 1,500 V and 12 h at 3,500 V (gradient pH 5 to 6). The IPG gel strips either were used immediately for the second-dimension run or were stored at −80°C. The IPG gel strips were equilibrated two times as described previously (12). In the second dimension, self-casted sodium dodecyl sulfate-pore gradient gels on plastic backing (12) with a gel size of 190 by 250 by 0.5 mm3 were run for 0.75 h at 300 V and 4.5 h at 600 V on a Multiphor electrophoresis unit (Pharmacia). The gels were stained with silver (3), or the proteins were blotted onto polyvinylidene difluoride membranes (Millipore) with Tris-borate transfer buffer (12) and stained with Coomassie brilliant blue R-250 (Serva). Selected protein spots either were directly applied to an automated protein sequencer to obtain N-terminal amino acid sequences or were digested with trypsin and the mass of the peptide fragments was analyzed with an automated mass spectrometer (matrix-assisted laser desorption ionization [MALDI]; Toplab, Martinsried, Germany).
Nucleotide sequence accession numbers.
Complete sequences of MCSPs were deposited in the GenBank database and include E. coli cspB-cspF (accession no. AF003590) and cspH (accession no. AF003591), M. luteus cspA (accession no. AF019905), P. aeruginosa cspA (accession no. U82822), S. aureus cspB and cspC (accession no. AF003592 and AF003593), S. typhimurium cspH (accession no. AF006035), and Y. enterocolitica cspA1/A2 (accession no. U82821).
RESULTS
A cspA gene duplication in Y. enterocolitica.
Inverse PCR of bacterial genomic DNA, performed with the degenerate MCSP oligonucleotide pairs CSPIR1-CSPIF1 and CSPIR1-CSPIF2, resulted in the amplification of at least one MCSP sequence from all bacteria tested (see Materials and Methods for the list of strains). In the majority of cases, control PCRs containing uncut genomic DNA did not give amplified products. Exceptions to this were E. coli and Y. enterocolitica, which were found to give products of between 400 and 500 bp. Sequencing of the PCR products from E. coli and Y. enterocolitica confirmed these DNAs to each contain two MCSP sequences, E. coli having divergent MCSP coding regions (corresponding to cspB-cspF and cspG-cspH, respectively) and Y. enterocolitica having a tandem repeat (Fig. 1).
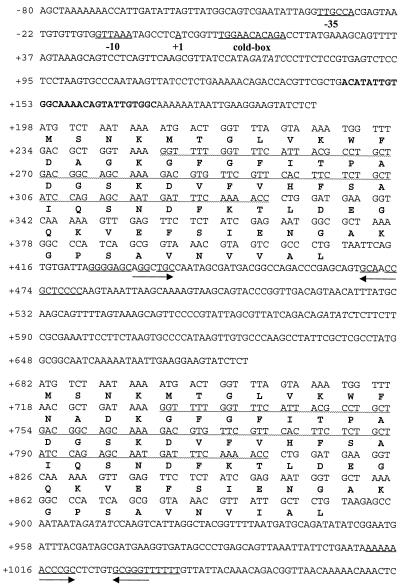
FIG. 1.
Nucleotide sequence of cspA1/cspA2 from Y. enterocolitica. cspA1 (upper protein sequence) and cspA2 (lower protein sequence) differ only in the 13th and 15th amino acids and in the third amino acid of the C terminus. Putative transcriptional start site (+1), putative promoter regions (−10, −35), and the cold-box are indicated. Sequence in boldface type shows the position of the A1 probe (YeA1-Dig). The underlined sequence indicated by upper facing arrows shows the putative transcriptional termination structure of the monocistronic mRNA. The underlined sequence indicated by lower facing arrows shows the transcriptional termination structure of the polycistronic mRNA. The sequence underlined by a wavy line in cspA1 can fold in an antiparallel direction to the same region of cspA2, forming an extensive secondary structure. EcoRV restriction sites, which cut at positions +73, +579, and +909, are indicated by italics.
Comparison of the two MCSP coding regions from Y. enterocolitica, designated cspA1 and cspA2, shows these sequences to be almost identical (96.7%), with their deduced protein sequences only differing in amino acids 13 and 15 (CspA1, DAG; CspA2, NAD) and a single amino acid at their C termini (CspA1, VVAL; CspA2, VIAL). Furthermore, there is a large degree of homology between the 5′ untranslated regions of these two genes, with the first 175 bp upstream of cspA1 and the corresponding 154 bp upstream of cspA2 showing approximately 70% identity. Primer extension experiments identified the 5′ end of the cspA1/A2 mRNA as lying 197 bp upstream of the first coding region (cspA1, ATG) (data not shown). This is probably the transcriptional start site +1 because it is in the same position as that in E. coli and is flanked by a cold-box motif and promoter elements (−10 and −35) similar to those found in E. coli MCSP genes (7, 10, 21).
Interestingly, the coding regions of the Y. enterocolitica cspA1/A2 mRNA can fold and hybridize to each other, resulting in an extensive secondary structure. This secondary structure begins with the 17th codon in cspA1 (GGU), which can bind in an antisense direction to the 44th codon of cspA2 (ACC) and vice versa (Fig. 1).
Transcription of Y. enterocolitica MCSP gene duplication.
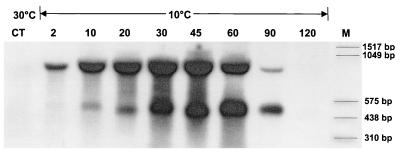
Northern blot analysis of Y. enterocolitica mRNA was conducted at both 30°C and after cold shock to 10°C, hybridizing with the oligonucleotide A1 probe specific to the 5′ untranslated region of cspA1. This analysis showed that two cold-inducible mRNA templates are produced from this MCSP sequence (Fig. 2): a monocistronic template of approximately 450 bp (cspA1) and a bicistronic template of approximately 900 bp (cspA1/A2). At 30°C, the level of the bicistronic mRNA was low, with the level of monocistronic mRNA almost negligible. However, upon a temperature downshift to 10°C, transcription was both substantial and immediate.
FIG. 2.
Northern blots of Y. enterocolitica cold shocked to 10°C. The signals were gained by a probe binding to the 5′ untranslated region of cspA1 (YeA1-Dig). Time after cold shock is given in minutes. The upper signal is approximately 900 bp and represents the bicistronic mRNA. The lower signal is approximately 450 bp and represents the monocistronic mRNA. CT, control.
To confirm that the bicistronic transcript contained the complete coding regions of both cspA1 and cspA2, RT-PCR was performed with mRNA taken at 30°C and at 10°C after 30 min by using an oligonucleotide back primer downstream of the cspA2 coding region. After 20 PCR cycles, an intense product of approximately 900 bp was obtained from the cold-shocked sample, with virtually no product obtained from the control sample (data not shown). The former product was sequenced and contained both cspA1 and cspA2 gene sequences (Fig. 1).
To ensure that the A1 probe used in the Northern blots specifically hybridized to cspA1/A2, a Southern blot analysis of Y. enterocolitica genomic DNA was conducted. This procedure confirmed that the A1 probe hybridized to only one region of this bacterium’s DNA (data not shown). Furthermore, by including an EcoRV digest of this DNA (a restriction enzyme known to cut before cspA1, between cspA1/A2 and after cspA2 [see Fig. 1], to give 456-bp and 330-bp DNA fragments, respectively), it can be confirmed that both signals visible on the Northern blots are mRNAs flanked by cspA1 and not a second MCSP gene sequence.
Comparison of mono- and bicistronic mRNA synthesis.
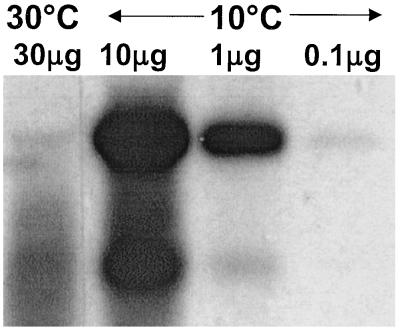
To compare the levels of monocistronic and bicistronic mRNA transcribed from this MCSP gene sequence before and after cold shock, quantitative Northern blot analysis was performed. mRNA was isolated at 30°C and compared to a dilution range of mRNA taken at 10°C after 30 min. This showed that at this time point the levels of bicistronic and monocistronic mRNA were induced 300- and 30-fold, respectively (Fig. 3). However, at 30°C the level of monocistronic mRNA was approximately fourfold higher than that of the bicistronic mRNA. When cold shocked to 10°C, a high level of bicistronic mRNA was observed at 30 min, whereas a high level of monocistronic mRNA was not recorded until 60 min, after which time both transcripts diminished. Neither transcript was visible 120 min after the initial temperature downshift (Fig. 2).
FIG. 3.
Northern blot of a dilution series of cold-shocked Y. enterocolitica mRNA (10°C, 30 min) detected by using the A1-probe. The relative amounts of bi- and monocistronic mRNA from cspA1/A2 were determined with the software Image Master 1D Elite (Amersham Pharmacia Biotech). The bicistronic mRNA shows a 300-fold increase; the monocistronic one shows a 30-fold increase.
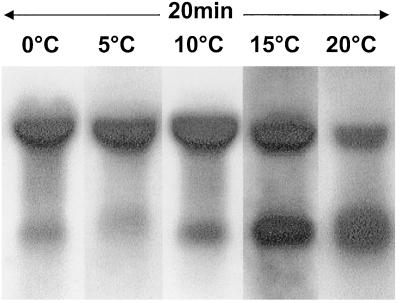
Comparison of the two mRNA transcripts shows that the ratio of bicistronic and monocistronic mRNA increases with decreasing temperature (Fig. 4). At 0 and 5°C, bicistronic mRNA is present at a high level, with relatively little monocistronic mRNA appearing. In contrast, at 15°C, similar levels of bi- and monocistronic mRNA are present, while at 20°C, the level of monocistronic mRNA seems to be higher.
FIG. 4.
Northern blots of cold-shocked Y. enterocolitica mRNA obtained at different temperatures with the A1 probe. Each cold shock lasted for 20 min. Changes in the proportion of bicistronic and monocistronic mRNA are most obvious at 15 and 20°C.
Cold-induced synthesis of Y. enterocolitica CspA1 and CspA2.
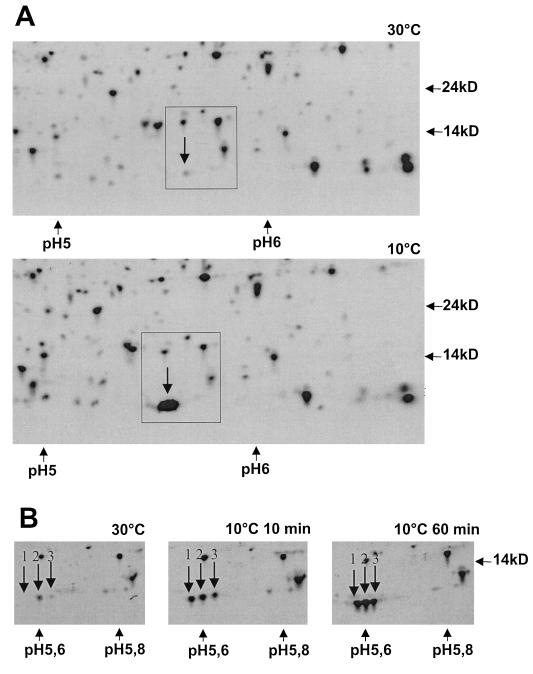
Protein extracts gained from cultures of Y. enterocolitica that were grown at 30°C and then cold shocked at 10°C for increasing periods of time were analyzed by two-dimensional gel electrophoresis. Initial studies using conventional gel gradients between pH 4 and 7 identified MCSPs around 7 kDa (Fig. 5A). However, it was not possible to determine how many separate MCSPs this spot contained. To increase resolution, protein extracts were focused in a narrow immobilized pH gradient between pH 5 and 6, over 18 cm, which to our knowledge has not been used before. As can be seen in Fig. 5B, the protein spot of interest actually consists of at least three proteins (marked 1, 2, and 3 on Fig. 5B). The N-terminal sequences of each of these spots was determined. Since it was unclear which of the spots 2 or 3 contains CspA2 (or another unknown MCSP), a peptide mass fingerprint (MALDI) was executed with each of these spots. The data are summarized in Table 1. The mixture of Csps in spot 2 is most probably due to carryover.
FIG. 5.
Two-dimensional gels from Y. enterocolitica. Molecular weights and pH values are indicated. MCSPs are indicated by downward arrows. (A) Windows of the broad-range gels showing proteins from pH 4 to 9 at 30°C and after a shock of 60 min to 10°C; (B) windows of the narrow-range gels from pH 5 to 6 of the control (30°C) and shock experiments to 10°C for 10 min and 60 min.
TABLE 1.
Summary of results from N-terminal sequencing and MALDI
| Spot no. | N-terminal sequence | Peptide massesa | Corresponding peptide fragment sequencesa | MCSP contained in spot |
|---|---|---|---|---|
| 1 | SNKMTGLVKWFDAGKG | NDb | B, A1 | |
| 2 | SNKMTGLVKWFNADKG | 723.3 | 11-KWFDAGKG-16 (CspB or CspA1) | A1, A2, B |
| MSNKMTGLVKWFNADKG | 780.3 | 11-KWFNADKG-16 (CspA2) | ||
| 926.5 | 61-KGPSAVNVVAL-70 (CspA1) | |||
| 940.5 | 61-KGPSAVNVIAL-70 (CspA2) | |||
| 3 | SNKMTGLVKWFNADKG | 780.3 | 11-KWFNADKG-16 (CspA2) | A2 |
| 940.5 | 61-KGPSAVNVIAL-70 (CspA2) |
Peptide masses and fragments from MALDI are examples showing fragments at the relevant amino acid signatures which are specific for CspA1, CspA2, and CspB (6), respectively.
ND, not determined.
MCSP tandem gene duplications in other species of Yersinia.
To determine whether the MCSP gene duplication found in Y. enterocolitica was present in other species of Yersinia, three additional members of this genera were investigated. Y. pseudotuberculosis (biotype III; environmental isolate from H. Wolf-Watz, University of Umea, Umea, Sweden) and Y. ruckeri (NCIMB 1316) were both probed by PCR, using the oligonucleotide primers CSPIR1 and CSPIF2, and in both cases gave a 450-bp product (similar in size to that gained from Y. enterocolitica). Sequencing of these DNA fragments (data not shown) confirmed the flanking regions of each product to be MCSP sequences homologous to Y. enterocolitica cspA1/A2.
Analysis of the Y. pestis database (Pathogen Sequencing Group, Sanger Centre, Cambridge, United Kingdom) (37a) confirmed an MCSP gene duplication to also be present in this Yersinia species. Due to a number of unknown bases being present in the latter sequence, this entire region of DNA was PCR amplified and resequenced (Y. pestis DNA was a gift from R. Titball, Defence Evaluation Research Agency, Porton Down, United Kingdom). Like those of Y. enterocolitica, the sequences of CspA1 and CspA2 of Y. pestis show only minor changes. Furthermore, both of these proteins are 94% identical to their comparative homologues in Y. enterocolitica. Alignment of the DNA sequences of both of these MCSP gene duplications, Y. enterocolitica cspA1/A2 with Y. pestis cspA1/A2, shows that there is considerable homology (85% identity) throughout these entire sequences (approximately 950 bp), including the 5′ untranslated region (data not shown).
DISCUSSION
Only Yersinia species carry a tandem cspA gene duplication.
Recently, it has been shown that Lactococcus lactis has two sets of MCSP genes, cspA-cspB and cspC-cspD, both being separated by approximately 300 bp. Comparison of these two MCSP loci showed them to be homologous, with 79% identity over 800 bp (41). Similarly, in this study we amplified two MCSP loci from E. coli that are located at 35 and 22 min on the chromosome. Each of these loci again contains two MCSP genes, cspB-cspF and cspG-cspH, that are separated by approximately 300 bp and show high levels of homology (more than 70% identity over 750 bp). We found no evidence to suggest that S. typhimurium has the cspBF-cspGH duplication found in E. coli, although subsequent work has demonstrated these sequences to be present in Shigella species (data not shown). The high levels of homology shown between these genes (loci) give strong evidence that they have arisen by sequence duplication.
In contrast to Y. enterocolitica, all of the above MCSP genes of L. lactis and E. coli are transcribed divergently and monocistronically. In each of the latter bacteria, it is the single MCSP locus that has been duplicated and not tandem genes, as in the case of Y. enterocolitica, Y. pestis, Y. pseudotuberculosis, and Y. ruckeri. Furthermore, although families of MCSPs have been identified in a wide range of bacteria (16, 17, 28, 29, 42), Y. enterocolitica is the first bacterium in which bicistronic MCSP mRNA has been found. This finding is especially intriguing in this particular bacterium due to its exceptional ability to grow at low temperature.
Differential appearance of mono- and bicistronic cspA1/A2 mRNA.
It has been shown that cspA mRNA is constitutively transcribed in E. coli but that, at elevated temperatures, this mRNA is rapidly degraded and may not be translated. Following cold shock, cspA mRNA is specifically and temporally stabilized (9). These findings could help to explain the immediate and substantial quantities of bicistronic mRNA produced by Y. enterocolitica upon cold shock from 30 to 10°C (Fig. 2). A sequence that is characteristic of a conventional transcription termination site can be found downstream of the cspA2 coding region (Fig. 1). This site, which is approximately 900 bp downstream of the +1, is most probably where the bicistronic messenger terminates.
With shock at temperatures not quite as low, monocistronic mRNA is produced predominantly (Fig. 4); this may be sufficient to overcome the impediment caused by this mild cold shock. The most significant difference between the appearance of these two transcripts at 10°C is that bicistronic mRNA is immediately available for translation at 10°C, whereas monocistronic mRNA does not significantly emerge until 30 min later (Fig. 2).
The ratio of monocistronic to bicistronic mRNA increases with the duration of the cold shock (Fig. 2) and decreasing shock temperature (Fig. 4). This change of the ratio of monocistronic to bicistronic mRNA could be reflective of transcription termination downstream of cspA1. Although a classical termination structure does not appear to be present between cspA1 and cspA2, sequence homology which indicates the formation of a large loop structure (Fig. 1) exists; this structure may act as a temperature-dependent second termination site. It cannot be excluded, on the other hand, that this loop structure acts as 3′ degradation protection. Investigations of the rpsO operon in E. coli showed that a secondary structure (t1) is located between rspO and pnp, which can act either as a terminator or as 3′ degradation protection. A second terminator (t2) is found downstream of pnp. Therefore, rpsO is sometimes transcribed monocistronically and sometimes bicistronically together with pnp (5, 19). Further investigations of the cspA1/A2 tandem in Yersinia will show whether a similar mechanism occurs in this bacterium.
Both CspA1 and CspA2 are synthesized.
In case the bicistronic mRNA indeed contributes to an elevated cold shock response, it should be fully translated; i.e., CspA2 should be present after cold shock. By electrophoresing proteins on two-dimensional gels with an extremely narrow pH range between 5 and 6 (Fig. 5B), it is possible to separate cold-inducible MCSPs. At least three cold-inducible MCSPs could be found, CspA1, CspA2, and CspB (see Results). Surprisingly, CspA1 and CspA2 separate at the narrow-range two-dimensional gel, although their sequences differ by only three amino acids (see Results). We conclude that CspA2 is synthesized and contributes to the cold shock response of Y. enterocolitica.
Does Y. enterocolitica have a higher translational capacity for MCSP than E. coli?
Studies in E. coli have shown that cspA mRNA is maximally induced 30-fold following a temperature downshift from 37 to 15°C and that this peak induction occurs after approximately 60 min at this lower temperature (9). These data correspond well with the induction of monocistronic cspA1 mRNA in Y. enterocolitica (Fig. 2). In contrast, a high level of bicistronic cspA1/A2 mRNA in Y. enterocolitica occurs after only 30 min and is induced at least 300-fold, 10-fold more than the monocistronic transcript (Fig. 2 and Fig. 3) of Y. enterocolitica and E. coli. If it is true that the basal levels of the monocistronic MCSP mRNAs are similar in both bacteria at non-cold shock temperatures, Y. enterocolitica would have a much greater translational capacity than E. coli upon cold shock with respect to these single MCSP sequences (loci).
Additionally, the fact that this transcript contains two copies of the coding region, which are both translated, means that Y. enterocolitica is able to synthesize this protein more effectively than bacteria that have monocistronic MCSP mRNAs only, such as E. coli, assuming that translation levels of these two mRNAs are comparable. Indeed, synthesis of CspA1 and CspA2 is clearly seen only 10 min after cold shock. However, in order to finally conclude that the rapid accumulation of these MCSPs is essential for Y. enterocolitica’s superior ability to adapt to low temperature, a knockout mutant of cspA2 needs to be constructed.
ACKNOWLEDGMENTS
K. Neuhaus and K. P. Francis contributed equally to this work.
The majority of the gene sequences reported in this manuscript were discovered by Kevin P. Francis in the laboratory of Gordon S. A. B. Stewart at the University of Nottingham (ROPA grant 42/CEL 04626). We thank Günther Boguth and Christian Obermaier for support and technical assistance in preparing the two-dimensional gels.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. 1:2.4.1. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 2.Bergann T, Kleemann J, Sohr D. Model investigations into psychrotrophic growth of Yersinia enterocolitica. J Vet Med Ser B. 1995;42:523–531. [PubMed] [Google Scholar]
- 3.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 4.Brandi A, Spurio R, Gualerzi C O, Pon C L. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 1999;18:1653–1659. doi: 10.1093/emboj/18.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coburn G A, Mackie G A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog Nucleic Acids Res. 1999;62:55–109. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson J H. Yersinia enterocolitica cold shock protein CspB (cspB) gene, complete cds. EMBL/DDBJ accession no. AF070484. 1998. [Google Scholar]
- 7.Etchegaray J-P, Jones P G, Inouye M. Differential thermoregulation of two highly homologous cold shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–178. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 8.Francis K P, Stewart G S A B. Detection and speciation of bacteria through PCR using universal major cold-shock protein primer oligomers. J Ind Microbiol Biotechnol. 1997;19:286–293. doi: 10.1038/sj.jim.2900463. [DOI] [PubMed] [Google Scholar]
- 9.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein J, Pollitt N S, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görg A. Two-dimensional electrophoresis. Nature. 1991;349:545–546. [Google Scholar]
- 12.Görg A, Weiss W. High resolution two-dimensional electrophoresis of proteins using immobilized pH gradients. In: Celis J, editor. Cell biology: a laboratory handbook. 2nd ed. Vol. 4. 1998. pp. 386–397. [Online.] Academic Press, Inc., New York, N.Y. [Google Scholar]
- 13.Görg A, Postel W, Günther S. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 1988;9:531–546. doi: 10.1002/elps.1150090913. [DOI] [PubMed] [Google Scholar]
- 13a.Görg A. posting date. Two-dimensional electrophoresis of proteins using immobilized pH gradients. [Online.] http://www.weihenstephan.de/blm/deg. [9 June 1999, last date accessed.] 1999. [Google Scholar]
- 14.Graumann P, Marahiel M A. The major cold shock protein of Bacillus subtilis CspB binds with high affinity to the ATTGG- and CCAAT sequences in single stranded oligonucleotides. FEBS Lett. 1994;338:157–160. doi: 10.1016/0014-5793(94)80355-2. [DOI] [PubMed] [Google Scholar]
- 15.Graumann P, Marahiel M A. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166:293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- 16.Graumann P, Schröder K, Schmid R, Marahiel M A. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol. 1996;178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graumann P, Wendrich T M, Weber M H W, Schröder K, Marahiel M A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 18.Grsic S, Sauerteig S, Neuhaus K, Albrecht M, Rossiter J, Ludwig-Müller J. Physiological analysis of transgenic Arabidopsis thaliana plants expression on nitrilase isoform in sense or antisense direction. J Plant Physiol. 1998;153:446–456. [Google Scholar]
- 19.Hajnsdorf E, Régnier P. E. coli rpsO mRNA decay: RNase E processing at the beginning of the coding sequence stimulates poly(A)-dependent degradation of the mRNA. J Mol Biol. 1999;286:1033–1043. doi: 10.1006/jmbi.1999.2547. [DOI] [PubMed] [Google Scholar]
- 20.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T, editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 21.Jiang W, Fang J, Inouye M. The role of the 5′-end untranslated region of the mRNA for CspA, the major cold shock protein of Escherichia coli, in cold shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Hou Y, Inouye M. CspA, the major cold shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 23.Jones P G, Inouye M. The cold shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 24.Jones P G, van Bogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loessner M J, Scherer S. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J Bacteriol. 1995;177:6601–6609. doi: 10.1128/jb.177.22.6601-6609.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löw R, Rausch T. Sensitive, nonradioactive Northern blots using alkaline transfer of total RNA and PCR-amplified biotinylated probes. BioTechniques. 1994;17:1026–1028. [PubMed] [Google Scholar]
- 28.Mayr B, Kaplan T, Lechner S, Scherer S. Identification and purification of a family of dimeric major cold shock protein homologs from the psychrotolerant Bacillus cereus WSBC 10201. J Bacteriol. 1996;178:2916–2925. doi: 10.1128/jb.178.10.2916-2925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel V, Labadie J, Hebraud M. Effect of different temperature upshifts on protein synthesis by the psychrotrophic bacterium Pseudomonas fragi. Curr Microbiol. 1996;33:16–25. doi: 10.1007/s002849900067. [DOI] [PubMed] [Google Scholar]
- 30.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–2997. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumeister B, Körner K, Schwalbe B, Kubanek B. Fatal Yersinia enterocolitica septicemia after transfusion of red cells. Case report and review of the literature. Infusther Transfusmed. 1997;24:14–19. [Google Scholar]
- 32.Qian L, Wilkinson M. DNA fragment purification—removal of agarose 10 minutes after electrophoresis. BioTechniques. 1991;10:736. [PubMed] [Google Scholar]
- 33.Righetti P G. Immobilized pH gradients. Theory and methodology. Amsterdam, The Netherlands: Elsevier; 1990. [Google Scholar]
- 34.Roberts T A, Baird-Parker C A, Tompkin R B. Microorganisms in foods. London, United Kingdom: Blackie Academic & Professional; 1996. pp. 458–478. [Google Scholar]
- 35.Robins-Browne R M. Yersinia enterocolitica. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 192–215. [Google Scholar]
- 36.Schindelin H, Marahiel M A, Heinemann U. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold shock protein. Nature. 1993;364:164–168. doi: 10.1038/364164a0. [DOI] [PubMed] [Google Scholar]
- 37.Schnuchel A, Wiltschuk R, Czisch M, Herrier M, Willimsky G, Graumann P, Mahariel M A, Holak T A. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature. 1993;364:169–171. doi: 10.1038/364169a0. [DOI] [PubMed] [Google Scholar]
- 37a.The Sanger Centre. revision date. [Online.] http://www.sanger.ac.uk/Projects/Y_pestis/blast_server.shtml. [6 September 1999, last date accessed.] 16 July 1999. [Google Scholar]
- 38.Walter A E, Turner D H, Kim J, Lyttle M H, Mueller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N, Yamanaka Y, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolffe A P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 41.Wouters J A, Sanders J-W, Kok J, de Vos W M, Kuipers O P, Abee T. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology. 1998;144:2885–2893. doi: 10.1099/00221287-144-10-2885. [DOI] [PubMed] [Google Scholar]
- 42.Yamanaka K, Fang L, Inouye M. The cspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 43a.Zuker M. copyright date. [Online.] http://www.ibc.wustl.edu/∼zuker/rna/form1.cgi. [6 September 1999, last date accessed.] 1999. [Google Scholar]