Abstract
Aspergillus fumigatus, a filamentous fungus producing bluish-green conidia, is an important opportunistic pathogen that primarily affects immunocompromised patients. Conidial pigmentation of A. fumigatus significantly influences its virulence in a murine model. In the present study, six genes, forming a gene cluster spanning 19 kb, were identified as involved in conidial pigment biosynthesis in A. fumigatus. Northern blot analyses showed the six genes to be developmentally regulated and expressed during conidiation. The gene products of alb1 (for “albino 1”), arp1 (for “aspergillus reddish-pink 1”), and arp2 have high similarity to polyketide synthases, scytalone dehydratases, and hydroxynaphthalene reductases, respectively, found in the dihydroxynaphthalene (DHN)-melanin pathway of brown and black fungi. The abr1 gene (for “aspergillus brown 1”) encodes a putative protein possessing two signatures of multicopper oxidases. The abr2 gene product has homology to the laccase encoded by the yA gene of Aspergillus nidulans. The function of ayg1 (for “aspergillus yellowish-green 1”) remains unknown. Involvement of the six genes in conidial pigmentation was confirmed by the altered conidial color phenotypes that resulted from disruption of each gene in A. fumigatus. The presence of a DHN-melanin pathway in A. fumigatus was supported by the accumulation of scytalone and flaviolin in the arp1 deletant, whereas only flaviolin was accumulated in the arp2 deletants. Scytalone and flaviolin are well-known signature metabolites of the DHN-melanin pathway. Based on DNA sequence similarity, gene disruption results, and biochemical analyses, we conclude that the 19-kb DNA fragment contains a six-gene cluster which is required for conidial pigment biosynthesis in A. fumigatus. However, the presence of abr1, abr2, and ayg1 in addition to alb1, arp1, and arp2 suggests that conidial pigment biosynthesis in A. fumigatus is more complex than the known DHN-melanin pathway.
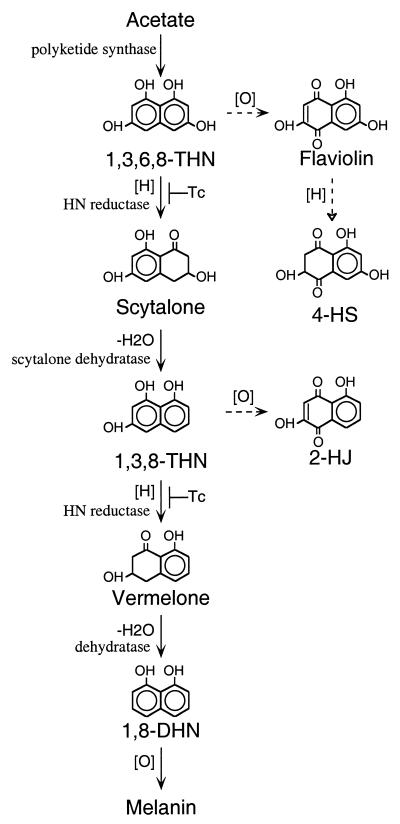
Many fungi produce melanins, which are dark brown and black pigments formed by oxidative polymerization of phenolic compounds. Melanin is known to contribute to the survival and longevity of fungal propagules (40). In addition, it is an important virulence factor for both plant- and animal-pathogenic fungi, e.g., Magnaporthe grisea, Colletotrichum lagenarium, Cryptococcus neoformans, Exophiala dermatitidis (Wangiella dermatitidis), and Aspergillus fumigatus (10, 14, 20, 21, 23, 29, 35, 37, 40). Pigment biosynthesis has been studied mainly in plant-pathogenic fungi, including Colletotrichum lagenarium, M. grisea, Verticillium dahliae, Cochliobolus miyabeanus, and Alternaria alternata (40). All these brown and black fungi synthesize their pigments through the dihydroxynaphthalene (DHN)-melanin pathway, in which hydroxynaphthalene (HN) and tetralone polyketides are the common intermediates (Fig. 1). DHN-melanin biosynthesis starts with polyketide synthase using acetate as a precursor (Fig. 1). An HN reductase then converts 1,3,6,8-tetrahydroxynaphthalene (1,3,6,8-THN) to scytalone (4, 40). Dehydration of scytalone forms 1,3,8-trihydroxynaphthalene (1,3,8-THN), which is converted to 1,8-DHN after an additional reduction and dehydration step. Finally, oxidative polymerization of 1,8-DHN gives the end product, DHN-melanin. These reduction steps are sensitive to tricyclazole, a fungicide that specifically inhibits HN reductase involved in DHN-melanin biosynthesis (40). To date, molecular genetic studies have identified three different genes involved in the DHN-melanin biosynthetic pathway in brown and black fungi. These genes encode polyketide synthases, HN reductases, and scytalone dehydratases, respectively (7, 14).
FIG. 1.
Biosynthetic pathway of melanin in brown and black fungi. The solid arrows indicate the main melanin pathway, and the dashed arrows indicate branching pathways from the melanin pathway. The hollow arrowhead indicates that the conversion requires more than one step. The reduction steps are indicated by [H] and the oxidation steps are indicated by [O]. Dehydration steps are labeled −H2O. A reduction step which can be inhibited by the fungicide tricyclazole is indicated with Tc.
Among fungi pathogenic to humans, pigment biosynthesis has been studied in the white yeast C. neoformans, the black yeast E. dermatitidis, and the filamentous fungus A. fumigatus. C. neoformans uses a laccase encoded by the CNLAC1 gene to produce DOPA-melanin in the presence of diphenolic compounds (27), while E. dermatitidis produces DHN-melanin constitutively (19). However, the genes involved in the biosynthesis of DHN-melanin in E. dermatitidis have yet to be identified. The only other human-pathogenic fungus in which pigment biosynthesis has been studied is A. fumigatus (34, 35). This ubiquitous opportunistic pathogen produces conidia bearing bluish green pigment. A. fumigatus is a saprophyte that propagates via highly dispersible conidia. The airborne conidia may be inhaled and cause a life-threatening disease, invasive aspergillosis, in patients with prolonged neutropenia (19, 38). A. fumigatus is thought to synthesize its conidial pigment by using a pathway similar to that of DHN-melanin because of its observed sensitivity to tricyclazole (34, 41). However, the biosynthesis of this unique bluish green pigment has not been studied in detail. A reddish pink conidial mutant, RP3, was restored to produce bluish green conidia by a cosmid from the A. fumigatus genomic library through a complementation screening. This cosmid was rescued from the complemented strain and designated pG1-1 (34). To date, two genes, alb1 (for “albino 1”) and arp1 (for “aspergillus reddish pink 1”), involved in conidial pigment biosynthesis in A. fumigatus have been mapped on this cosmid (34, 35). The arp1 gene encodes a putative scytalone dehydratase based on its amino acid sequence similarity to the scytalone dehydratases identified from plant-pathogenic fungi (34). Disruption of arp1 results in the production of reddish pink conidia. The disruption also significantly increases the ability of the human complement component C3 to bind to these conidia (34). Deposition of C3 on conidia is an important step for efficient phagocytosis of inhaled conidia. The alb1 gene product has high similarity to polyketide synthases from various fungi (35). Disruption of the alb1 gene results in an albino conidial phenotype and significantly reduced virulence in a murine model (35). Thus, conidial pigment biosynthesis in A. fumigatus appears to be an important virulence factor in the establishment of infection.
The three genes involved in the DHN-melanin biosynthetic pathway identified in brown and black fungi are dispersed in the genome of Colletotrichum lagenarium, while they are organized as a cluster in Alternaria alternata (14). Here, we report the identification of a six-gene cluster involved in conidial pigmentation in A. fumigatus. Biochemical analysis of the scytalone dehydratase and HN reductase gene deletants indicates that A. fumigatus utilizes a pathway similar to the DHN-melanin pathway for biosynthesis of its conidial pigment.
MATERIALS AND METHODS
Strains and media.
A. fumigatus B-5233, which produces bluish green conidia, was isolated from a patient with a fatal invasive aspergillosis. Strain B-5233/RGD4-2 is an arp1 gene deletant of B-5233 (34). Strains B-5233/RGD3-1 and B-5233/RGD10-1 are the arp2 gene deletants of B-5233. Both the arp1 and arp2 gene deletants produce reddish pink conidia.
Aspergillus minimal medium contained 1% glucose, 10 mM NaNO3, and trace elements (13). Malt extract medium contained 2% glucose, 2% malt extract, and 0.1% peptone. All cultures were grown at 37°C. Asparagine-sucrose agar (ASA) medium, which is identical to the alkaline medium (TM medium) previously described (43), was used to culture A. fumigatus for the detection of flaviolin and 2-hydroxyjuglone (2-HJ) accumulation (35). Modified ASA medium, which was used for tricyclazole inhibition and scytalone feeding experiments, contained 30 μg of tricyclazole per ml, 1 mM scytalone, or both, as previously described (35).
Preparation and analysis of nucleic acids.
Total DNA and RNA were isolated from Aspergillus cultures as described previously (32). Cultures grown in Aspergillus minimal medium were harvested for total RNA preparation at 0 and 14 h after induction of conidiation, as described by Parta et al. (25). DNA sequencing was done with a Sequenase version 2.0 kit (U.S. Biochemical, Cleveland, Ohio) and an ABI automatic DNA sequencing system (Perkin-Elmer, Foster City, Calif.). Purified DNA fragments were recovered with the GeneClean II kit (Bio 101, Vista, Calif.). DNA cloning and Southern blot analyses were done by standard methods (30). Hybond-N nylon membranes (Amersham, Arlington Heights, Ill.) were used for blot analysis. DNA probes were labeled with [α-32P]dCTP (Amersham) by using a Prime It kit (Stratagene, La Jolla, Calif.). The Genetics Computer Group program Gap (9), with the default parameters, was used to analyze the similarity and identity between proteins.
Plasmids.
Cosmid pG1-1, a cosmid containing the vector pCosHX and a 42.5-kb genomic DNA of A. fumigatus, was obtained via plasmid rescue from a complemented conidial color mutant, RP3/G1-1 (34).
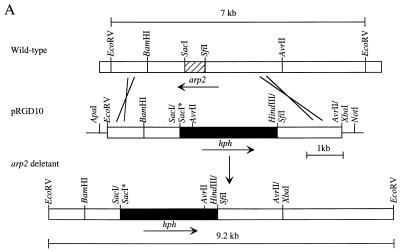
To disrupt the arp2 gene, pRGD10 (see Fig. 3A) was constructed as follows. pBC KS+ (Stratagene) was digested with SacI and SacII and then self-ligated to eliminate the SacI site, producing pBCKS58. The 11-kb HindIII DNA fragment of pG1-1 was cloned into the HindIII site of pCosHX to produce pGH10 (34). The 7-kb EcoRV DNA fragment from pGH10 was cloned into the SmaI-EcoRV sites of pBCKS58. The resulting construct was digested with AvrII and XbaI, polished with T4 DNA polymerase, and self-ligated to yield pRGAS40. The 0.6-kb SacI-SfiI DNA fragment of pRGAS40 was then replaced with the 2.8-kb SacI-HindIII (blunt-ended) DNA fragment of pAN7-1 containing the hygromycin B phosphotransferase gene (hph) (28) to give the gene disruption construct pRGD10 (see Fig. 3A). The disruption construct pRGD3 was identical to pRGD10, except that the hph gene was inserted in the opposite orientation. A similar strategy was used to disrupt abr1, abr2, and ayg1 in B-5233, by using disruption constructs pRGD24, pRGD17, and pRGD15S1, respectively. All of the gene deletants were confirmed by Southern blot analysis (data not shown).
FIG. 3.
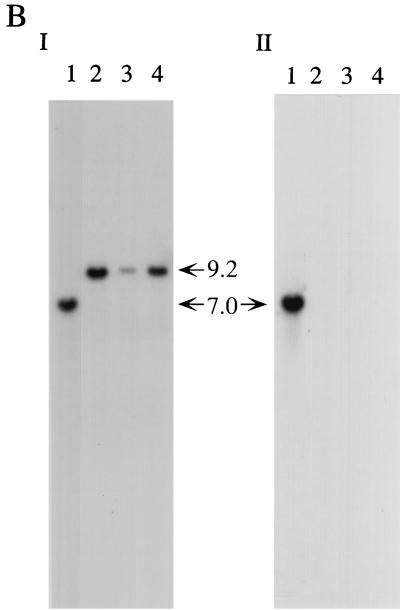
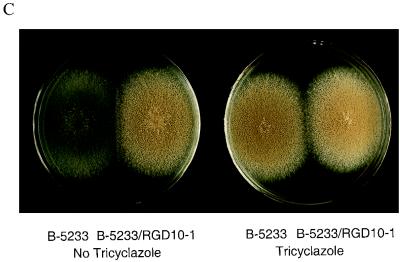
arp2 gene disruption in B-5233. (A) Diagram of a double-crossover event via homologous recombination. Open boxes represent DNA from A. fumigatus. Solid boxes represent the hph gene. Asterisks indicate SacI sites that were destroyed during vector construction. (B) Southern blot analysis of B-5233 and the arp2 deletants. Total DNA was digested with EcoRV. The blot was analyzed with pRGD10 (I) or a 0.6-kb SacI-SfiI DNA fragment (hatched box in Fig. 3A) as a probe (II). Lanes: 1, B-5233; 2, B-5233/RGD10-1; 3, 5233/RGD10-2; 4, 5233/RGD10-3. The sizes of hybridized DNA fragments are indicated by arrows. (C) Conidial color phenotype of B-5233 and B-5233/RGD10-1. Cultures were grown on Aspergillus minimal medium for 3 days with (right) or without (left) tricyclazole.
Transformation of A. fumigatus.
Protoplasts of A. fumigatus were prepared with mureinase (Amersham) as previously described (34) and transformed by the standard polyethylene glycol transformation method (44). Transformants were selected on Aspergillus minimal medium supplemented with 200 μg of hygromycin B per ml.
Tricyclazole inhibition assay.
A. fumigatus strains were point inoculated on agar media consisting of malt extract, malt extract plus 1% ethanol, or malt extract plus 1% ethanol and tricyclazole (8 μg/ml). Conidiated cultures were photographed after 3 days of incubation at 37°C.
TLC analysis.
Technical-grade tricyclazole (5-methyl-1,2,4-triazole[3,4-b] benzothiazole) was obtained from Eli Lilly Research Laboratories (Greenfield, Ind.). Flaviolin and 2-HJ were synthesized as previously described (3, 42). Scytalone and 4-hydroxyscytalone were purified from the Brm-1 mutant strain of Verticillium dahliae (3).
ASA medium was inoculated with 5 × 105 conidia and incubated at 24°C for 8 days as previously described (35). Then culture extracts were analyzed for the presence of flaviolin, 2-HJ, scytalone, and 4-hydroxyscytalone (4-HS) by ethyl acetate extraction and thin-layer chromatography (TLC) procedures as described previously (35, 41).
Nucleotide sequence accession numbers.
The sequence data have been submitted to the GenBank database under accession no. AF025541 (alb1), U95042 (arp1), AF099736 (arp2), AF116901 (abr1), AF116902 (ayg1), and AF104823 (abr2).
RESULTS
Cloning and disruption of arp2.
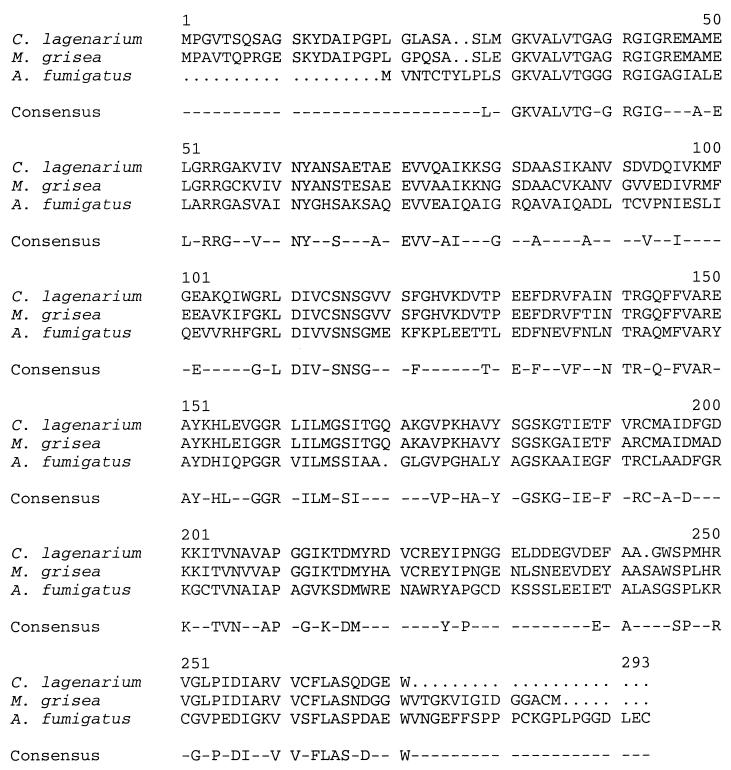
Previous analysis of the arp1 gene led to the discovery that the DNA adjacent to arp1 encodes a 1.1-kb transcript designed ORF2, which is detected only in the conidiating stage (34). Here, we sequenced the genomic and cDNA clones corresponding to the 1.1-kb transcript. The cDNA contains an 819-nucleotide open reading frame (ORF) which encodes a putative protein of 273 amino acids. No introns are present within orf2. A BLAST search with the deduced amino acid sequence revealed that the putative protein has 46% identity and 67% similarity to the HN reductase from Magnaporthe grisea (37) and 48% identity and 70% similarity to the HN reductase from Colletotrichum lagenarium (26) (Fig. 2).
FIG. 2.
Amino acid alignment of HN reductases. The amino acid sequence comparison between Arp2p and HN reductases of Magnaporthe grisea (GenBank NCBI ID 1127197) and Colletotrichum lagenarium (GenBank accession no. D86079) is shown (18). The alignment was performed with the Genetics Computer Group program Pileup, using the default parameters (9). Dots represent gaps which were introduced during alignment. Dashes indicate the nonconsensus residues among three proteins.
The sequence similarity of the putative protein to the HN reductases involved in the DHN-melanin biosynthesis suggested that this ORF is involved in pigment biosynthesis. Gene disruption was performed to substantiate the potential involvement of the gene in conidial pigmentation. A gene disruption construct of this ORF, pRGD10, was made by replacing a 0.6-kb coding region of this gene with a 2.8-kb DNA fragment containing the hph gene, which is used as a selection marker for transformation (Fig. 3A). Plasmid pRGD3 was a similar disruption construct, except that the hph gene was in the opposite orientation. Several transformants of the wild-type strain B-5233 with these plasmids produced reddish pink conidia. These transformants were analyzed by Southern blot hybridization to confirm the disruption of the gene. The pRGD10 DNA probe hybridized to a 7.0-kb DNA segment of B-5233 (Fig. 3B, panel I). In contrast, the transformants generated a 9.2-kb hybridizing signal which resulted from the loss of the 0.6-kb SacI-SfiI fragment from the gene and the gain of a 2.8-kb fragment from hph (Fig. 3B, panel I). These results indicate that pRGD10 was integrated into the homologous locus in these transformants. Deletion of the wild-type copy of the gene in these transformants was confirmed by using the 0.6-kb SacI-SfiI DNA fragment as a probe. There was no detectable signal in these transformants, in contrast to the 7.0-kb signal in B-5233 (Fig. 3B, panel II). Thus, we concluded that the reddish pink conidial color phenotype resulted from the replacement of the wild-type copy with a disrupted copy by homologous recombination through a double crossover. A similar hybridization pattern was also observed in the reddish pink conidial color transformants which received pRGD3 (data not shown). Since disruption of this gene results in mutants producing reddish pink conidia, we designated the gene arp2 (for “aspergillus reddish pink 2”).
The conidial color of both the arp1 and arp2 deletants is reddish pink. However, the color of the arp2 deletants appeared darker than that of the arp1 deletant. Interestingly, when the wild-type strain was grown on Aspergillus minimal medium containing 8 μg of tricyclazole per ml, it produced reddish pink conidia similar to those of the arp2 deletants (Fig. 3C). Tricyclazole is a fungicide which specifically inhibits the HN reductases involved in DHN-melanin biosynthesis in brown and black fungi (33, 40). Thus, disruption of arp2 revealed a phenotype similar to that obtained by growth in the presence of this specific reductase inhibitor. Collectively, the data indicated that arp2 encodes an HN reductase involved in conidial pigment biosynthesis.
Mapping the gene cluster associated with conidiation.
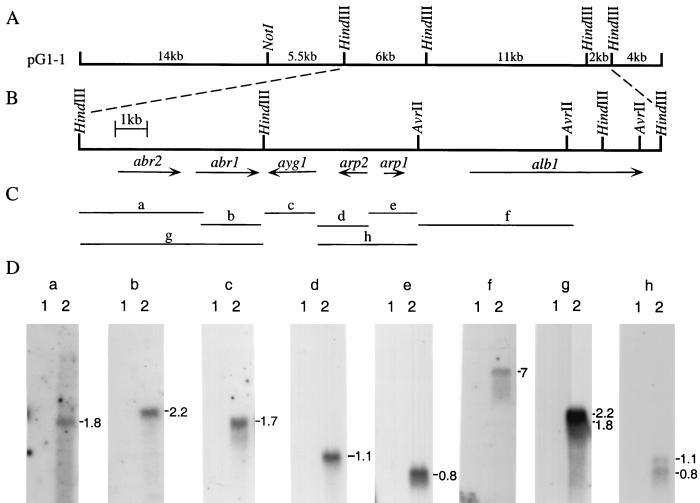
Because the genes alb1, arp1, and arp2 are adjacent to each other, the possibility exists that other genes involved in conidial pigment biosynthesis are located in close proximity. The structure and sequences of pG1-1 were further analyzed. No DNA rearrangement had occurred in pG1-1 during its cloning process, indicated by the same restriction enzyme digestion patterns of pG1-1 and the genomic DNA of B-5233 (Southern blot data not shown). Northern blot analysis was used to detect the transcripts that might be encoded by pG1-1. Eight DNA fragments, a, b, c, d, e, f, g, and h, spanning a 15.5-kb region of pG1-1, were used as DNA probes to analyze RNA from conidiating and nonconidiating cultures (Fig. 4). Six transcripts hybridizing to the DNA probes used were found to be expressed in 14-h conidiating cultures but not in nonconidiating cultures (Fig. 4C and D). Therefore, these six transcripts ranging from 0.8 to 7 kb in size are associated with conidiation. Similar Northern analysis was carried out with probes of the 4-kb HindIII DNA fragment located adjacent to alb1 and the 5.5-kb NotI-HindIII DNA fragment adjacent to fragment g (Fig. 4A to C). The 4-kb HindIII DNA probe detected two transcripts expressed in both conidiating and nonconidiating cultures, while the 5.5-kb NotI-HindIII DNA probe did not detect any transcript at either developmental stage (data not shown). The difference in gene expression patterns suggested that only the 19-kb region of pG1-1 (Fig. 4B) encodes transcripts which are associated with conidiation.
FIG. 4.
Gene cluster for conidial pigment biosynthesis in A. fumigatus. (A) Physical map of pG1-1. (B) Map of transcripts associated with conidiation. Arrows indicate the direction of transcripts. All of the six transcripts are found within a 19-kb DNA fragment. (C) Probes used for Northern analyses. The DNA fragments used as probes are labeled a, b, c, d, e, f, g, and h. (D) Northern blot analyses of gene expression at two different developmental stages. The sizes of the transcripts are labeled on the right of the blot in kilobases. Total RNA from B-5233 was isolated from mycelia 0 h (lane 1) or 14 h (lane 2) after induction of conidiation. A 12-μg portion of total RNA was fractionated on the 1% formaldehyde–agarose gel. The sizes of the hybridizing fragments are indicated at the side. rRNA stained with ethidium bromide was used as a control for the quantity and quality of the RNA preparation (data not shown).
Characterization of the conidiation-associated gene cluster.
The cDNAs corresponding to these conidiation-associated transcripts were isolated by using DNA fragments g (6-kb HindIII fragment) and c (1.6-kb HindIII-BamHI fragment) as probes, respectively. The sequences of these cDNA clones and the 19-kb region of pG1-1 were determined. The corresponding ORFs were designated abr1, abr2, and ayg1 (Fig. 4B). Therefore, including alb1, arp1, and arp2, a total of six ORFs were clustered in the 19-kb genomic fragment. Comparison of the cDNA sequences with the 19-kb genomic sequences revealed the genomic arrangement of these ORFs in the cluster (Fig. 4B). The alb1, arp1, abr1, and abr2 genes were transcribed in the opposite direction to arp2 and ayg1 (Fig. 4B). Within this arrangement, abr1 and ayg1 were convergently transcribed whereas arp1 and arp2 are divergently transcribed. These ORFs are closely spaced: for instance, abr1 was separated from ayg1 by 427 nucleotides. The largest intergenic distance, 1,101 nucleotides, was found between arp1 and alb1 (Fig. 4B).
Further sequence analysis revealed that abr1 encodes a putative protein (Abr1p) of 664 amino acids. A BLAST search with Abr1p amino acid sequences revealed that it possesses two multicopper oxidase signatures and has 34% identity and 43% similarity to FET3, an iron multicopper oxidase from Candida albicans (GenBank accession no. Y09329) (Table 1). Two introns, of 47 and 52 nucleotides, were present in the abr1 gene. A putative protein of 587 amino acids (Abr2p) was encoded by abr2. Abr2p had 41% identity and 65% similarity to a laccase encoded by yA of A. nidulans (2). Six introns were observed in the abr2 gene, and the sizes of these introns ranged from 43 to 59 nucleotides. All of these introns were located close to the 5′ end of abr2. The ayg1 gene encoded a putative protein (Ayg1p) of 406 amino acids. Four introns were present in the ayg1 gene, and the sizes of these introns ranged from 45 to 56 nucleotides. A BLAST search with either nucleotide or amino acid sequences of ayg1 did not identify any homolog.
TABLE 1.
Analysis of genes involved in conidial pigmentation in A. fumigatus
| Gene | Putative activity | Size of transcripts (kb) | Size of deduced protein (amino acids) | % Identitya | Reference protein |
|---|---|---|---|---|---|
| alb1 | Polyketide synthase | 7.0 | 2,146 | 66 | wA (24) |
| arp1 | Scytalone dehydratase | 0.8 | 168 | 55 | SCD1 (17) |
| arp2 | HN reductase | 1.1 | 273 | 49 | THR1 (26) |
| abr1 | Multicopper oxidase | 2.2 | 664 | 34 | FET3b |
| ayg1 | Unknown | 1.7 | 406 | NAc | NA |
| abr2 | Laccase | 1.8 | 587 | 41 | yA (2) |
Percent identity of each putative protein to the reference protein is based on analysis with the Genetics Computer Group Gap program and the default parameter.
GenBank accession no. Y09329.
NA, not applicable.
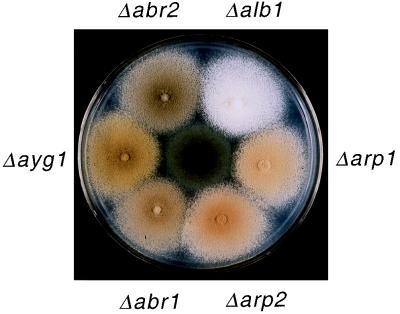
Disruption of each abr1, abr2, and ayg1 in A. fumigatus resulted in an alteration of the conidial color phenotype. Both abr1 and abr2 single-gene deletants produced brown conidia, while the ayg1 deletant produced yellowish green conidia (Fig. 5). It is of interest that the ayg1 deletant produced yellow conidia at an early growth stage. However, as the cultures aged, the yellow conidial color gradually converted to green, which was distinct from the bluish green color of wild-type conidia.
FIG. 5.
Sporulating cultures of the A. fumigatus wild-type strain and six single-gene deletants. The wild-type strain, producing bluish green conidia, is at the center of the plate. Each gene deletant is designated by the name of the deleted gene.
The presence of these three genes in addition to alb1, arp1, and arp2 suggested that conidial pigment biosynthesis in A. fumigatus is more complex than the known DHN-melanin pathway in brown and black fungi.
Biochemical evidence for a melanin pathway in A. fumigatus.
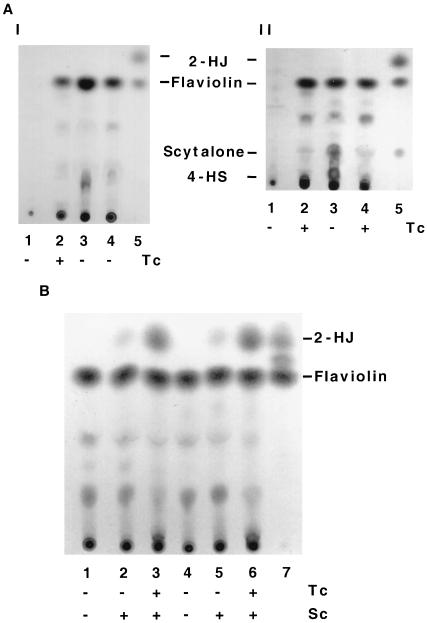
To confirm that the arp1 and arp2 putative proteins (Arp1p and Arp2p) are homologs of scytalone dehydratase and HN reductase respectively, extracts obtained from cultures grown on ASA medium were analyzed by TLC for pigment biosynthetic intermediates. Based on the melanin studies that have been carried out with brown and black fungi, blockage of the 1,3,6,8-THN reduction step results in the accumulation of flaviolin (Fig. 1). Blockage of the scytalone dehydration step results in accumulation of stable intermediates or branch products, i.e., scytalone, flaviolin, and 4-HS (Fig. 1). TLC analysis showed that B-5233 did not produce any detectable flaviolin (Fig. 6A, panel I, lane 1). In contrast, the arp2 deletants, B-5233/pRGD3-1 and B-5233/RGD10-1, produced flaviolin abundantly (lanes 3 and 4). When B-5233 was grown on ASA medium containing 30 μg of tricyclazole per ml, it also produced flaviolin abundantly (lane 2).
FIG. 6.
Biochemical analysis of accumulated intermediates and branch products of the DHN-melanin biosynthetic pathway of A. fumigatus. (A) (I) TLC analysis. Lanes: 1 and 2, wild-type strain B-5233; 3 and 4, arp2 deletants B-5233/RGD3-1 and B-5233/RGD10-1; 5, standards. (II) Lanes: 1 and 2, B-5233; 3 and 4, arp1 deletant B-5233/RGD4-2; 5, standards. (B) Scytalone feeding experiment. Lanes: 1 to 3, B-5233/RGD3-1; 4 to 6, B-5233/RGD10-1; 7, standards. The strains were grown on ASA medium for 6 days. Sc, 10−3 M scytalone in ASA medium; Tc, 30 μg of tricyclazole per ml in ASA medium.
Tricyclazole did not have an apparent effect on the arp2 deletants, and culture extracts of these strains appeared identical to those of tricyclazole-treated B-5233. Scytalone, 4-HS, and 2-HJ, which are stable intermediates and branch products of the DHN-melanin pathway, were not detected in the culture extracts. These data suggested that Arp2p is a tricyclazole-sensitive HN reductase which converts 1,3,6,8-THN to scytalone. Culture extracts of the arp1 deletant (B-5233/RGD4-2) (34) were also analyzed by TLC. High levels of scytalone and flaviolin and a detectable amount of 4-HS were present in the arp1 deletant but not in B-5233 (Fig. 6A, panel II, lanes 1 and 3). The identities of scytalone, flaviolin, and 4-HS were confirmed by mass spectrometry (data not shown). Accumulation of these compounds indicated that the arp1 deletant was incapable of converting scytalone to 1,3,8-THN. This agrees with the notion that Arp1p is a scytalone dehydratase. Tricyclazole treatment prevented accumulation of scytalone and 4-HS by the arp1 deletant (lane 4). This indicates that Arp2p is upstream of Arp1p in the pathway.
The accumulation of scytalone and flaviolin by the arp1 deletant and flaviolin by the arp2 deletants clearly confirmed the presence of both a scytalone dehydratase and an HN reductase in the conidial pigment biosynthetic pathway of A. fumigatus. This strongly supported the notion that A. fumigatus utilizes a pathway similar to the DHN-melanin pathway for conidial pigment biosynthesis.
Two reductases involved in the melanin biosynthesis in A. fumigatus.
There is no general assertion that the same HN reductase catalyzes the two reduction steps in the DHN-melanin pathway in all DHN-melanin producing fungi, i.e., 1,3,6,8-THN to scytalone and 1,3,8-THN to vermelone (7, 26, 36). To delineate whether this is the case in A. fumigatus, we analyzed the accumulated intermediates and branch products in the arp2 deletants which were grown on ASA medium and ASA medium supplemented with scytalone or with scytalone and tricyclazole. Detection of scytalone and 4-HS in the arp1 deletant but not in the arp2 deletants indicated that Arp2p is required for production of both compounds (Fig. 1 and 6A). The branch product 2-HJ, derived from 1,3,8-THN due to blockage of the second reduction step in the pathway, was not detected in the arp2 deletants grown on medium without scytalone (Fig. 6B, lanes 1 and 4). A very small amount of 2-HJ was accumulated by the arp2 deletants grown on ASA medium containing only scytalone (lane 2 and 5). In contrast, 2-HJ was accumulated abundantly in the arp2 deletants grown on ASA medium containing both scytalone and tricyclazole (lanes 3 and 6). This indicated the presence of another tricyclazole-sensitive HN reductase for metabolizing 1,3,8-THN, which is derived from the exogenous scytalone in A. fumigatus.
DISCUSSION
A developmentally regulated six-gene cluster involved in conidial pigment biosynthesis in A. fumigatus was identified and characterized. DNA sequence, Northern blot, and biochemical analyses indicated that A. fumigatus synthesizes its conidial pigment through a pathway similar to the DHN-melanin pathway found in many brown and black fungi. To date, this is the largest gene cluster ever reported to be involved in fungal pigment biosynthesis. The six genes have been designated alb1, arp2, arp1, abr1, abr2, and ayg1. According to the deduced amino acid sequence data, the alb1, arp1, and arp2 gene products are homologs of polyketide synthase, scytalone dehydratase, and HN reductase, respectively, which are found in the brown and black fungi. Identification of these three genes suggests the presence of a similar DHN-melanin pathway in A. fumigatus. The notion of the presence of this melanin biosynthetic pathway in A. fumigatus was strengthened by detection of the accumulated intermediates characteristic of the pathway in the wild-type strain treated with tricyclazole and in the strains in which these three genes were individually disrupted. Furthermore, the altered conidial color in each single-gene-disrupted strain indicates that the gene cluster is indeed involved in conidial pigment biosynthesis in A. fumigatus.
Clustering of functionally related genes, a common feature in prokaryotes, was not recognized in filamentous fungi until 10 years ago (11). Gene clusters discovered in fungi have been mainly divided into two groups, those responsible for low-molecular-weight nutrient utilization and those responsible for secondary-metabolite production (14). Gene clusters of nutrient utilization pathways observed in filamentous fungi include nitrate assimilation and proline utilization in A. nidulans (1, 8, 12). Clusters of genes involved in secondary-metabolite biosynthesis include those producing mycotoxins and melanin. Mycotoxin-producing gene clusters include those of the aflatoxin pathway in A. flavus (5) and the sterigmatocystin pathway in A. nidulans (6). For melanin biosynthesis, the three known genes involved in conidial pigment biosynthesis are clustered in Alternaria alternata (15). However, the genes involved in DHN-melanin biosynthesis are not always clustered. In Colletotrichum lagenarium, for example, the three genes are dispersed in the genome (14). Furthermore, the wA and yA genes involved in conidial pigment biosynthesis in a green-spored fungus, A. nidulans, also are not linked.
Gene clustering is thought to be beneficial for gene regulation (14). Functionally associated genes work coordinately as a team, and so coregulation of these genes is an advantage for living organisms. This is especially true with developmentally regulated genes. Clustering of genes allows regulatory elements to be shared between genes, i.e., divergent promoters such as GAL1-GAL10 in yeast and niiA-niaD in Aspergillus spp. (1, 12, 39). In the pigment biosynthesis gene cluster in A. fumigatus, arp1 and arp2 are divergently transcribed genes. Their intergenic regions are very small, only 464 nucleotides. It is likely that these two genes share cis-acting elements for regulation of their expression. Indeed, Arp1p acts on the products produced by Arp2p. Regulator genes were observed in the gene clusters for nutrient and mycotoxin biosynthesis. However, we have yet to identify the regulator genes for the pigment biosynthetic gene cluster in A. fumigatus. Alternatively, gene clustering places genes in the same chromosomal region and may allow gene regulation through modulation of chromatin structure.
Despite minor genetic differences, the DHN-melanin pathways in many brown and black fungi require only polyketide synthase, HN reductase, and scytalone dehydratase to synthesize DHN, which forms DHN-melanin through oxidative polymerization (40). It has been suggested that the HN reductase catalyzes the conversion of 1,3,6,8-THN to scytalone as well as the conversion of 1,3,8-THN to vermelone in M. grisea (36). Also, scytalone dehydratase was suggested to catalyze two different dehydration steps in the DHN-melanin biosynthetic pathway in M. grisea: the conversion of scytalone to 1,3,8-THN and the conversion of vermelone to 1,8-DHN (23). These three enzymes are conserved among the known DHN-melanin pathways in fungi (15–18, 26, 31, 36, 40). The presence of homologs of the three genes in A. fumigatus suggests a similarity of melanin biosynthesis in A. fumigatus to that of brown and black fungi. However, the final pigments produced by A. fumigatus are bluish green. Although A. fumigatus and brown-to-black fungi have similarities in their melanin biosynthetic pathways, their melanin pathways must have diverged at steps prior to the final melanin products. Discovery of three additional genes, abr1, abr2, and ayg1, associated with conidial pigmentation supports this hypothesis. Since abr1 and abr2 have signature characteristics of oxidases, their existence indicates the presence of oxidation steps in the biosynthetic pathway. A laccase was suggested to polymerize and oxidize DHN into melanin based on previous biochemical studies (40). However, the gene encoding the putative oxidase has not yet been identified in brown and black fungi. Since Abr2p has sequence similarity to laccases, it is possible that the Abr2p is involved in oxidation steps. A BLAST search with ayg1 sequences did not identify any homolog; thus, the function of ayg1 remains unknown. Further genetic and biochemical characterization of the mutant strains will help us to uncover the role of ayg1 in conidial pigment biosynthesis.
It has been suggested that one reductase is responsible for both reduction steps in the DHN-melanin pathway in Magnaporthe grisea (36). However, a second reductase may also be present in the DHN-melanin pathway of other brown and black fungi, as suggested for Colletotrichum lagenarium (26). Our TLC analysis of arp2 deletants, however, detected the accumulation of flaviolin, a well-known branch product of the DHN-melanin pathway, but not of 2-HJ. Supplementing the arp2 deletant with scytalone and tricyclazole resulted in abundant accumulation of 2-HJ. This suggests the presence of a second reductase involved in conidial pigment biosynthesis of A. fumigatus. However, this putative reductase in A. fumigatus was not found in the six-gene cluster. These identified six genes can be used for further investigation of pigment biosynthetic pathway in other fungi.
Disruption of alb1 significantly reduces the virulence of A. fumigatus in mice (22, 35). Furthermore, disruption of either alb1 or arp1 greatly increased the ability of complement component C3 to bind to A. fumigatus conidia (34, 35), a process important for phagocytosis of the inhaled conidia. These effects may be due to alteration of the conidial surface structure or loss of certain intermediates or branch products of the melanin pathway. However, preliminary studies with the murine model showed that the arp1 deletant but not the arp2 deletant had significantly reduced virulence relative to the wild-type strain (unpublished data). This suggests that accumulated metabolites may be more important than the conidial pigment itself in influencing the virulence of A. fumigatus. Identification of this six-gene cluster will facilitate further investigation of the mechanisms involved in the pathogenesis of A. fumigatus. Availability of these color mutants with disruptions in these genes will allow us to identify accumulated intermediates or products. In addition, it will help us to further delineate the melanin biochemical pathway.
ACKNOWLEDGMENTS
We thank Herman Edskes and Ashok Varma for their critical reviews and helpful suggestions, R. D. Stipanovic for his assistance in mass-spectral analysis, Lisa Penoyer for her assistance in DNA sequencing, and P. Silar for providing plasmid pBC-hygro.
REFERENCES
- 1.Amaar Y G, Moore M M. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Genet. 1998;33:206–215. doi: 10.1007/s002940050328. [DOI] [PubMed] [Google Scholar]
- 2.Aramayo R, Timberlake W E. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 1990;18:3415. doi: 10.1093/nar/18.11.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell A A, Stipanovic R D, Puhalla J E. Pentaketide metabolites of Verticillium dahliae: identification of (+)-scytalone as a natural precursor to melanin. Tetrahedron. 1976;32:1353–1356. [Google Scholar]
- 4.Bell A A, Wheeler M H. Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol. 1986;24:411–451. [Google Scholar]
- 5.Bennett J W, Papa K E. The aflatoxigenic Aspergillus species. Adv Plant Pathol. 1988;6:263–280. [Google Scholar]
- 6.Brown D W, Yu J H, Kelkar H S, Fernandes M, Nesbitt T C, Keller N P, Adams T H, Leonard T J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler M J, Day A W. Fungal melanins: a review. Can J Microbiol. 1998;44:1115–1136. [Google Scholar]
- 8.Chang P K, Ehrlich K C, Linz J E, Bhatnagar D, Cleveland T E, Bennett J W. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr Genet. 1996;30:68–75. doi: 10.1007/s002940050102. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon D M, Polak A, Szaniszlo P J. Pathogenicity and virulence of wild-type and melanin-deficient Wangiella dermatitidis. J Med Vet Mycol. 1987;25:97–106. doi: 10.1080/02681218780000141. [DOI] [PubMed] [Google Scholar]
- 11.Hull E P, Green P M, Arst H N, Jr, Scazzocchio C. Cloning and physical characterization of the l-proline catabolism gene cluster of Aspergillus nidulans. Mol Microbiol. 1989;3:553–559. doi: 10.1111/j.1365-2958.1989.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 12.Johnstone I L, McCabe P C, Greaves P, Gurr S J, Cole G E, Brow M A, Unkles S E, Clutterbuck A J, Kinghorn J R, Innis M A. Isolation and characterisation of the crnA-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene. 1990;90:181–192. doi: 10.1016/0378-1119(90)90178-t. [DOI] [PubMed] [Google Scholar]
- 13.Käfer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 14.Keller N P, Hohn T M. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 15.Kimura N, Tsuge T. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J Bacteriol. 1993;175:4427–4435. doi: 10.1128/jb.175.14.4427-4435.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubo Y, Nakamura H, Kobayashi K, Okuno T, Furusawa I. Cloning of a melanin biosynthetic gene essential for appressorial penetration of Colletotrichum lagenarium. Mol Plant-Microbe Interact. 1991;3:135–143. doi: 10.1094/mpmi-9-0323. [DOI] [PubMed] [Google Scholar]
- 17.Kubo Y, Takano Y, Endo N, Yasuda N, Tajima S, Furusawa I. Cloning and structural analysis of the melanin biosynthesis gene SCD1 encoding scytalone dehydratase in Colletotrichum lagenarium. Appl Environ Microbiol. 1996;62:4340–4344. doi: 10.1128/aem.62.12.4340-4344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo Y, Takano Y, Furusawa I. Molecular genetic analysis of melanin biosynthetic genes essential for appressorium function in Colletotrichum lagenarium. In: Mills D, Kunoh H, Keen N, Mayama S, editors. Molecular aspects of pathogenicity: requirements for signal transduction. St. Paul, Minn: APS Press; 1996. pp. 73–82. [Google Scholar]
- 19.Kwon-Chung K J, Bennett J E. Medical mycology. 1992. pp. 201–247. and 620–677. Lea & Febiger, Philadelphia, Pa. [Google Scholar]
- 20.Kwon-Chung K J, Polacheck I, Popkin T J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage A A. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol (Berlin) 1998;187:79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- 23.Lundqvist T, Rice J, Hodge C N, Basarab G S, Pierce J, Lundqvist Y. Crystal structure of scytalone dehydratase—a disease determinant of rice pathogen, Magnaporthe grisea. Structure. 1994;15:937–944. doi: 10.1016/s0969-2126(94)00095-6. [DOI] [PubMed] [Google Scholar]
- 24.Mayorga M E, Timberlake W E. The developmentally regulated Aspergillus nidulans wA gene encodes a polypeptide homologous to polyketide and fatty acid synthases. Mol Gen Genet. 1992;235:205–212. doi: 10.1007/BF00279362. [DOI] [PubMed] [Google Scholar]
- 25.Parta M, Chang Y, Rulong S, Pinto-DaSilva P, Kwon-Chung K J. HYP1, a hydrophobin gene from Aspergillus fumigatus, complements the rodletless phenotype in Aspergillus nidulans. Infect Immun. 1994;62:4389–4395. doi: 10.1128/iai.62.10.4389-4395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perpetua N S, Kubo Y, Yasuda N, Takano Y, Furusawa I. Cloning and characterization of a melanin biosynthetic THR1 reductase gene essential for appressorial penetration of Colletotrichum lagenarium. Mol Plant-Microbe Interact. 1996;9:323–329. doi: 10.1094/mpmi-9-0323. [DOI] [PubMed] [Google Scholar]
- 27.Polacheck I, Hearing V J, Kwon-Chung K J. Biochemical studies of phenoloxidase and utilization of catecholamines in Cryptococcus neoformans. J Bacteriol. 1982;150:1212–1220. doi: 10.1128/jb.150.3.1212-1220.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Punt P J, Oliver R P, Dingemase M A, Pouwel P H, Van den Hondel C A. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 29.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Takano Y, Kubo Y, Shimizu K, Mise K, Okuno T, Furusawa I. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol Gen Genet. 1995;249:162–167. doi: 10.1007/BF00290362. [DOI] [PubMed] [Google Scholar]
- 32.Timberlake W E. Molecular genetics of Aspergillus development. Annu Rev Genet. 1986;24:5–36. doi: 10.1146/annurev.ge.24.120190.000253. [DOI] [PubMed] [Google Scholar]
- 33.Tokousbalides M C, Sisler H D. Site of inhibition by tricyclazole in the melanin biosynthetic pathway of Verticillium dahliae. Pestic Biochem Physiol. 1979;11:64–73. [Google Scholar]
- 34.Tsai H-F, Washburn R G, Chang Y C, Kwon-Chung K J. Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol Microbiol. 1997;26:175–183. doi: 10.1046/j.1365-2958.1997.5681921.x. [DOI] [PubMed] [Google Scholar]
- 35.Tsai H F, Chang Y C, Washburn R G, Wheeler M H, Kwon-Chung K J. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol. 1998;180:3031–3038. doi: 10.1128/jb.180.12.3031-3038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal-Cros A, Viviani F, Labesse G, Boccara M, Gaudry M. Polyhydroxynaphthalene reductase involved in melanin biosynthesis in Magnaporthe grisea. Eur J Biochem. 1994;219:985–992. doi: 10.1111/j.1432-1033.1994.tb18581.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washburn R G. Opportunistic mold infections. In: Esser L, Lemke P A, editors. The Mycota VI. Heidelberg, Germany: Springer-Verlag KG; 1996. pp. 147–158. [Google Scholar]
- 39.West R W, Jr, Chen S M, Putz H, Butler G, Banerjee M. GAL1-GAL10 divergent promoter region of Saccharomyces cerevisiae contains negative control elements in addition to functionally separate and possibly overlapping upstream activating sequences. Genes Dev. 1987;1:1118–1131. doi: 10.1101/gad.1.10.1118. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler M H, Bell A A. Melanins and their importance in pathogenic fungi. In: McGinnis M R, editor. Current topics in medical mycology. Vol. 2. New York, N.Y: Springer-Verlag; 1988. pp. 338–387. [DOI] [PubMed] [Google Scholar]
- 41.Wheeler M H, Klich M A. The effects of tricyclazole, pyroquilon, phthalide, and related fungicides on the production of conidial wall pigments by Penicillium and Aspergillus species. Pestic Biochem Physiol. 1995;52:125–136. [Google Scholar]
- 42.Wheeler M H, Stipanovic R D. Melanin biosynthesis and the metabolism of flaviolin and 2-hydroxyjuglone in Wangiella dermatitidis. Arch Microbiol. 1985;142:234–241. doi: 10.1007/BF00693396. [DOI] [PubMed] [Google Scholar]
- 43.Wheeler M H, Stipanovic R D. Melanin biosynthesis in Thielaviopsis basicola. Exp Mycol. 1979;3:340–350. [Google Scholar]
- 44.Yelton M M, Hamer J E, Timberlake W E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]