Abstract
Chronic HIV infection is characterized by persistent inflammation despite antiretroviral therapy (ART). Cannabinoids may help reduce systemic inflammation in people with HIV (PWH). To assess the effects of oral cannabinoids during HIV, ten PWH on ART were randomized (n = 5/group) to increasing doses of oral Δ9-tetrahydrocannabinol (THC): cannabidiol (CBD) combination (2.5:2.5–15:15 mg/day) capsules or CBD-only (200–800 mg/day) capsules for 12 weeks. Blood specimens were collected prospectively 7–21 days prior to treatment initiation and at weeks 0 to 14. Plasma cytokine levels were determined via Luminex and ELISA. Immune cell subsets were characterized by flow cytometry. HIV DNA/RNA were measured in circulating CD4 T-cells and sperm by ultra-sensitive qPCR. Results from both arms were combined for statistical analysis. Plasma levels of IFN-, IL-1, sTNFRII, and REG-3α were significantly reduced at the end of treatment (p ˂ 0.05). A significant decrease in frequencies of PD1+ memory CD4 T-cells, CD73+ regulatory CD4 T-cells, and M-DC8+ intermediate monocytes was also observed (p ˂ 0.05), along with a transient decrease in CD28–CD57+ senescent CD4 and CD8 T-cells. Ki-67+ CD4 T-cells, CCR2+ non-classical monocytes, and myeloid dendritic cells increased over time (p ˂ 0.05). There were no significant changes in other inflammatory markers or HIV DNA/RNA levels. These findings can guide future large clinical trials investigating cannabinoid anti-inflammatory properties.
Keywords: cannabinoids, Δ9-tetrahydrocannabinol (THC), cannabidiol (CBD), systemic inflammation, immune response, HIV reservoir
1. Introduction
Despite effective antiretroviral therapy (ART), people with HIV (PWH) continue to suffer from chronic systemic inflammation and persistent immune activation [1,2,3]. This deleterious inflammatory state is thought to result from multifactorial and progressive events, including CD4 T-cell depletion in gut-associated lymphoid tissue (GALT) during acute infection, which induces persistent dysregulation of intestinal T-cell homeostasis and promotes long-lasting disruption of the gut epithelial mucosa [1,4]. Increased gut mucosal permeability leads to microbial antigen translocation from the gut lumen into the bloodstream [1,2,3,5,6]. These antigens, in turn, trigger immune cell activation and subsequent release of high amounts of pro-inflammatory soluble factors. Gut mucosal damage in PWH is chronic, hence it persists in PWH despite HIV infection being controlled with ART and regardless of the timing of ART initiation [7,8]. It leads to persistent T-cell activation and their subsequent exhaustion and immunosenescence [1,2,3,5,9]. Together, these factors contribute to the establishment of a vicious cycle that fuels a chronic inflammatory state, leading to early ageing and predisposing PWH to an increased risk of non-AIDS co-morbidities such as metabolic syndrome, cardiovascular diseases, cancers, and neurological disorders [1,2,3,5,6]. In addition, heightened levels of inflammation during ART are associated with the persistence of HIV reservoirs [10,11], the major obstacle to HIV eradication. Thus, targeting various players involved in this vicious cycle could help dampen chronic inflammation, reduce the occurrence rate of non-AIDS comorbidities in PWH, and facilitate the clearance of HIV reservoirs.
Primary phytocannabinoids 9-tetrahydrocannabinol (THC) and cannabidiol (CBD) displayed anti-inflammatory properties in experimental models in both in vitro [12,13,14,15,16] and in vivo studies involving mice [17], simian immunodeficiency virus (SIV) infection of non-human primates [18,19,20,21] and in humans [22]. Cannabinoids improved experimental inflammation by suppressing the release of pro-inflammatory cytokines and reactive oxygen species [23], reducing frequencies of inflammatory T helper (Th) 17 lymphocytes, and increasing frequencies of regulatory T-cells (Tregs) [24,25]. Cannabinoids also attenuated SIV-associated intestinal inflammation [18], as well as SIV-induced neuroinflammation, by reducing gut microbiome dysbiosis [26]. The latter occurred via increased gut bacterial diversity, decreased expression of pro-inflammatory genes, and increased production of anti-inflammatory regulatory micro-RNA in the gastrointestinal tract [20,26,27,28]. THC and CBD exert their anti-inflammatory properties largely through the activation of receptors of the endocannabinoid system, particularly the cannabinoid receptor type 1 (CB1R) and 2 (CB2R), which are mainly expressed, although at different levels, in the brain and the central nervous system (CNS) [29,30], the gastrointestinal tract [25,31,32] and on immune cells [33]. By alleviating gut epithelial damage and systemic inflammation, cannabinoid treatment could have the potential to reduce systemic inflammation in PWH [34,35]. Furthermore, oral administration of lipid-based formulations of cannabinoids could substantially facilitate the exposure of intestinal tissue-resident immune cells to these molecules and promote their anti-inflammatory effects [36].
Recreational cannabis was legalized in Canada in 2018, making it possible to possess cannabis without the need for a prescription. Cannabis use is common among PWH for both recreational and medicinal purposes. Many PWH use it to alleviate various symptomatologies such as anxiety, depression, and chronic pain [37,38,39,40]. Similarly to purified cannabinoids, consumption of the cannabis plant via inhalation has also been associated with anti-inflammatory outcomes in human observational studies [32,41,42,43,44,45]. However, because the cannabis plant contains more than 120 bioactive compounds, including phytocannabinoids [46], disentangling the effects of specific compounds, their doses, and their observed effects is challenging if not impossible. In addition, one of the main phytocannabinoids, THC, is well known for its psychotropic effects, which has been associated with adverse effects [46]. Cannabis consumption has also been associated with some negative outcomes, such as decreased adherence to ART in older PWH [47,48]. In adolescents, cannabis use has been associated with impaired cognitive abilities, an observation which has been attributable to CB1R activation in the CNS and the brain by THC [49].
Given the discrepancies across studies and the inherent risk of confounding in observational studies, it is important to clearly delineate whether cannabinoids may have a role in reducing systemic inflammation in PWH. In addition, it is necessary to determine whether cannabinoids, when administered at safe and tolerable doses to PWH, can have an impact on markers of systemic inflammation. We recently reported that cannabinoids administered in a randomized, open-label, interventional clinical trial (CTN PT028 study) were generally safe and well tolerated in PWH with well-controlled HIV on ART [50]. Herein, we report on the effects of oral cannabinoids on systemic inflammation, markers of gut mucosal damage, lymphoid and myeloid immune cell subsets, and markers of HIV persistence in PWH under suppressive ART. Given their synergistic effect, and the fact that CBD tends to improve the tolerability of THC when administered together [51,52,53,54], a THC:CBD combination was selected for one arm of this study. A formulation containing only CBD was also selected, in order to document effects related to the use of CBD in isolation.
2. Materials and Methods
2.1. Study Population and Design
This study was part of a randomized, open-label, interventional pilot clinical trial (CIHR Canadian HIV Trials Network (CTN) PT028, Trial registration number: NCT03550352) aiming to assess the safety and tolerability of oral THC:CBD combined, or CBD-only capsules consumed daily for 12 weeks with an initial target sample size of n = 26 (n = 13 per study arm) [50]. Participants were enrolled between September 2021 and February 2022. PWH (18 years and older) on ART for at least 3 years with suppressed viral loads (VL < 40 copies/mL) were recruited at the Chronic Viral Illness Service, Royal Victoria Hospital of the McGill University Health Centre in Montreal, Canada. Participants were randomized in a 1:1 ratio and received oral capsules of highly purified (>98%) cannabinoid oil (Tilray Brands, Inc., New York City, NY, USA), consisting of either TN-TC11M2 formulation, a THC:CBD combination in a 1:1 ratio (CBD: 2.5/THC: 2.5 mg), or TN-C200M2 formulation, containing CBD only (200 mg), for a maximum of 12 weeks. The following up-titration schedule was recommended to participants in THC:CBD study arm: 5 mg THC/5 mg CBD (1 capsule twice daily) during week 0 and week 1; 10 mg THC/10 mg CBD (2 capsules twice daily) week 2 and week 3; and 15 mg THC/15 mg CBD (2 capsules three times daily) week 4 to week 12 (end of treatment). For the CBD-only study arm, participants were advised to take 200 mg CBD (1 capsule once daily) from week 0 to week 1; 400 mg CBD (1 capsule twice daily) from week 2 to week 3; and 800 mg CBD (2 capsules twice daily) from week 4 to week 12. The dosages for the THC/CBD combination (2.5–15 mg/day) and the CBD-only formulation (200–800 mg/day) have been determined based on other clinical trials that have shown their safety, tolerability, and efficacy for the management of chronic pain, epilepsy, schizophrenia or multiple sclerosis [55,56,57,58]. The decision to use these specific formulations was also influenced in part by availability from the supplier. Of note, we chose a formulation of oral capsules for this study in order to have accurate information regarding the doses of cannabinoids participants were ingesting, to avoid the pulmonary toxicity associated with inhalation, and to determine whether oral administration can have an impact on markers associated with gut mucosal translocation. Participants were excluded if they used cannabinoid-containing products outside of this study or within 4 weeks before starting this study. Cannabinoids were permanently discontinued when severe adverse events occurred [50]. Further details on study design, participant recruitment, and inclusion/exclusion criteria can be found in the study protocol [59] and our previous publication on this trial which reported on safety and tolerability [50].
2.2. Blood and Semen Specimens Processing
Blood specimens were collected from each participant 7 to 21 days prior to treatment initiation, at week 0, and weeks 1, 2, 6, 8, 12, and 14 (end of study) for hematological/biochemical profile assessment [50], plasma cytokine and gut damage measurements, characterization of immune cell subsets, and quantification of HIV reservoir markers. Blood plasma was isolated via centrifugation, and stored at −80 °C to be analyzed in one batch at the end of this study. Peripheral blood mononuclear cells (PBMCs) were isolated using lymphocyte separation medium (WISENT Inc., Quebec City, QC, Canada) and cryopreserved in fetal bovine serum (FBS; WISENT Inc., Quebec City, QC, Canada) containing 10% dimethyl sulfoxide (DMSO) to be analyzed after study completion.
In parallel, semen samples were collected in sterile containers with 10 mL of Roswell Park Memorial Institute medium (RPMI; WISENT Inc., Quebec City, QC, Canada) with 100 U/mL penicillin and 100 mg/mL streptomycin (WISENT Inc., Quebec City, QC, Canada) were obtained from male participants (21 to 7 days prior to treatment initiation and at treatment completion at week 12). Upon arrival at the clinic, semen specimens were sent to the lab and centrifuged. The supernatant and cell pellets were separated and stored at −80 °C until HIV reservoir quantification.
2.3. Measurements of Soluble Markers and Cytokines in Plasma
Tumor necrosis factor alpha (TNF-), interferon-gamma (IFN-), interleukin 1-beta (IL-1), IL-6, IL-8, interferon gamma inducible protein-10 (IP-10) and IL-10 were measured in 25 μL of plasma using the MILLIPLEX® Human Cytokine/Chemokine/Growth Factor Panel A according to the manufacturer’s instructions (MilliporeSigma, Burlington, MA, USA). ELISA kits were used to quantify lipopolysaccharide (LPS) (Cusabio Technology LLC, Houston, TX, USA), soluble CD14 (sCD14) (Hycult Biotech, Uden, Netherlands), soluble CD27 (sCD27) (Thermo Fisher Scientific, Waltham, MA, USA), soluble receptor for tumor necrosis factor type II (sTNFRII) (R&D Systems, Inc., Minneapolis, MN, USA), intestinal fatty acid binding protein (I-FABP) (Hycult Biotech, Uden, Netherlands), and human regenerating islet derived protein 3 alpha (REG-3α) (R&D Systems, Inc., Minneapolis, MN, USA) according to the manufacturer’s protocols. All measurements were performed in duplicate.
2.4. Ex Vivo Immunophenotyping of T-Cells, Monocytes, and Dendritic Cells
Multiparametric flow cytometry was used for the immunophenotyping of T-cells, monocytes, and dendritic cells. 1 × 106 PBMCs were stained with extracellular antibodies in phosphate-buffered saline (PBS) + 2% FBS for 1 h at 4 °C protected from light. For intracellular staining, cells were fixed and permeabilized using the Transcription Factor Buffer Set according to the manufacturer’s instructions (BD Bioscience, ON, Canada), and incubated with the appropriate antibodies for 1 h at 4 °C protected from light. Antibody-labeled cells were acquired on a 3-laser BD Fortessa-X20. Antibodies used for phenotyping are listed in Table S1, and fluorochrome minus one control (FMO) were used for some markers such as CCR6, CCR4, CXCR3, CX3CR1, CD163, and CCR2. All flow cytometric data were analyzed using FlowJo V10.8.1 (FlowJo LLC, Ashland, OR, USA).
2.5. HIV DNA and Cell-Associated HIV RNA Quantification
Prior to quantification, genomic DNA and cellular RNA were extracted from PBMCs and semen cell pellets using the QIAamp DNA mini kit and QIAamp RNA mini kit (Qiagen, Hilden, Germany), respectively. Total HIV DNA and cell-associated HIV RNA targeting the LTR-gag region were measured in blood CD4 T-cells and sperm cells by ultra-sensitive nested real-time PCR, as previously described [60,61]. Cell-free viral RNA was also measured in semen supernatant by ultra-sensitive qPCR [60,61]. Detailed methodology is provided in Supplementary Material S1.
2.6. Statistical Analyses
Descriptive statistics of quantitative variables were presented as the means with standard deviations and medians with interquartile range (IQR). The Wilcoxon matched-pairs signed-rank test was used to compare paired repeated measurements between two visits. Due to the small number of participants who have completed the full treatment course (n = 8), and because each participant underwent a personalized cannabinoid titration schedule according to their tolerability, study results from both arms were pooled together for non-parametric paired statistical analysis. The primary analysis has been performed between the initiation of the cannabinoid treatment (week 0) versus the treatment interruption (week 12) as well as two weeks after treatment interruption (week 14) to assess the persistence of cannabinoid effects. Secondly, a week-to-week comparison is also provided as a supplementary analysis. GraphPad Prism Software (version 9.0.0, San Diego, CA, USA) was used for statistical analyses.
3. Results
3.1. Study Participants
Despite the initial sample size target of n = 26, this study was ended prematurely due to the rupture of cannabinoid capsules stock, the impossibility of renewing the stock of capsules with the same manufacturing criteria, and enrolment challenges as previously reported [50]. Thus, 10 PWH (median age: 57.5 years, IQR: 55–62), 8 males and 2 females, were included over a 6-month period, randomized in a 1:1 ratio to either TN-TC11M2 (arm 1) or TN-C200M2 (arm 2) [50]. Their baseline characteristics are summarized in Table 1. Eight study participants successfully completed the treatment and 2 were withdrawn for safety reasons as described previously [50]. CD4 T-cell count and CD4/CD8 ratio were stable and HIV viral load remained suppressed throughout the study [50]. All participants abstained from cannabis smoking and cannabis edibles for at least 4 weeks before study initiation and over time of the study duration. The majority (8/10) of participants reported at least one adverse event, and most of them were of mild to moderate severity. The frequently reported were somnolence (50%), diarrhea (20%), difficulty concentrating (20%), transaminitis (20%), and worsened diabetes type 2 (20%). A complete list of adverse event that have occurred during this study was previously reported [50]. Somnolence, difficulty concentrating, cognitive impairment, and increased appetite were considered definitively related to cannabinoids intake.
Table 1.
Demographic and biological characteristics of study participants at inclusion.
| Total Population (n = 10) |
|
|---|---|
| Age (Years), median (±IQR) | 57.5 (54.75–61.75) |
| Sex assigned at birth (n (%)) | |
| Male | 8 (80%) |
| Female | 2 (20%) |
| Ethnicity (n (%)) | |
| White-North American | 6 (60%) |
| Black-African | 1 (10%) |
| Asian | 1 (10%) |
| Mixed ethnicity | 2 (20%) |
| Antiretroviral regimens (n (%)) | |
| Biktarvy® (Bictegravir/Tenofovir alafenamide/Emtricitabine) | 5 (50%) |
| Triumeq® (Dolutegravir/Abacavir/Lamivudine) | 1 (10%) |
| Truvada®/Viramure® (Tenofovir/Emtricitabine/Nevirapine) | 1 (10%) |
| Raltegravir/Kivexa®/Biktarvy®
(Raltegravir/Abacavir/Lamivudine/Bictegravir/Tenofovir alafenamide/Emtricitabine) |
1 (10%) |
| Genvoya® (Elvitegravir/Cobicistat/Emtricitabine/Tenofovir alafenamide) | 1 (10%) |
| Delstrigo® (Doravirine/Lamivudine/Tenofovir disoproxil fumarate) | 1 (10%) |
| Oral cannabinoids regimens (n (%)) | |
| TN-TC11M2 formulation (CBD: 2.5/THC: 2.5 mg) | 5 (50%) |
| TN-C200M2 formulation (CBD: 200 mg) | 5 (50%) |
| Cannabis use in the past 6 months before study initiation (n (%)) | |
| No | 3 (30%) |
| Yes | 7 (70%) |
| Monthly | 5 (72.43%) |
| Weekly | 2 (28.57%) |
| Daily | 0 (0%) |
| Alcohol use in the past 6 months (n (%)) | |
| No | 5 (50%) |
| Yes | 5 (50%) |
| Drug use in the past 6 months (n (%)) | |
| No | 3 (30%) |
| Yes | 7 (70%) |
| History of infectious diseases (n (%)) | |
| Syphilis (treated) | 2 (20%) |
| Hepatitis B (Anti-HBc Antibodies) | 4 (40%) |
| Hepatitis C (Anti-HCV Antibodies) | 0 (0%) |
3.2. Effect of Oral Cannabinoids on Plasma Markers of Gut Epithelial Damage, Microbial Translocation, and Systemic Inflammation
Reduced Levels of Soluble Markers of Gut Epithelial Damage, Microbial Translocation, Immune Activation, and Pro-Inflammatory Cytokines
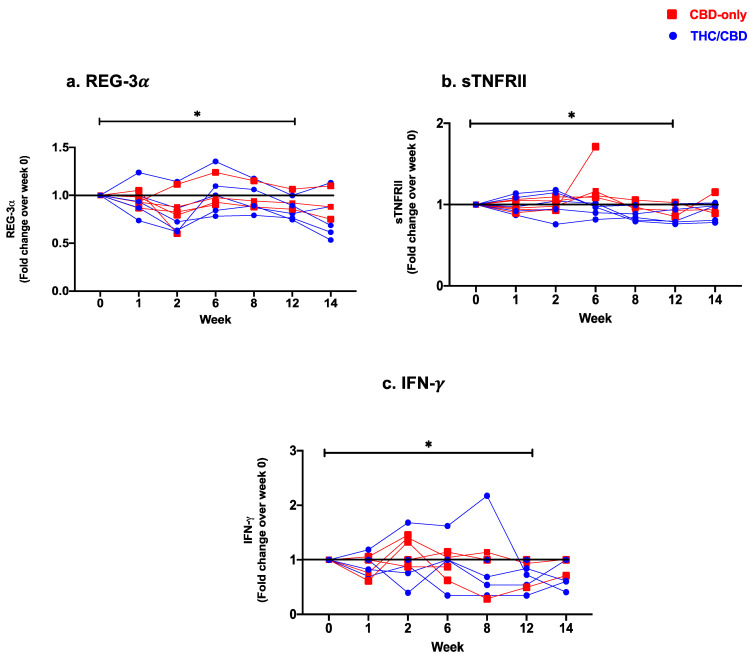
The effects of cannabinoid treatment on the integrity of the gut mucosal barrier were evaluated through changes in plasma levels of I-FABP and REG-3 as markers of gut epithelial damage in PWH [7,62]. The overall plasma levels of REG-3 were significantly lower after treatment completion (week 0 vs. week 12: p = 0.04, Figure 1a). Following treatment initiation, a significant decrease in levels of REG-3 was observed at week 2 (week 0 vs. week 2: p = 0.001), which increased transiently at week 6 (week 2 vs. week 6: p = 0.004), and then continued to drop until the end of treatment (week 6 vs. week 12: p = 0.008). No significant changes in levels of I-FABP were observed (Table 2).
Figure 1.
Fold change expression over 12 weeks versus weeks 0 of CBD-only (red square) and THC/CBD combination (blue circle), (a) Regenerating islet derived protein 3 alpha (REG-3α), (b) Soluble receptor for tumor necrosis factor type II (sTNFRII), and (c) Interferon-gamma (IFN-γ). * p < 0.05.
Table 2.
Dynamic of plasma soluble markers of gut mucosal damages (REG-3α, I-FABP), microbial translocation and immune activation (LPS, sCD14, and sCD27, sTNFRII), and pro-inflammatory (TNF-α, IFN-γ, IL-1β, IL-6, IL-8, IP-10) and anti-inflammatory (IL-10) cytokines during cannabinoids treatment, from treatment initiation to study termination.
| Plasma Markers | Study Time-Line | ||||||
|---|---|---|---|---|---|---|---|
|
Week 0 Treatment Initiation n = 10 |
Week 1 n = 10 |
Week 2 n = 10 |
$ Week 6 n = 9 |
$ Week 8 n = 8 |
$ Week 12 End of Treatment n = 8 |
$ Week 14 Study Termination n = 8 |
|
| Gut mucosal damage | |||||||
| REG-3(pg/mL) [Mean (SD)] |
5621 (3299) b,e | 5390 (3294) g | 4663 (2970) b,g,l,m | 5610 (3403) l,q,r | 5315 (3140) m,s,t | 4950 (3018) e,q,s | 4757 (3366) r,t |
| [Median (IQR)] | 5608 (2617–7039) b,e | 5157 (2533–6751) g | 4893 (2220–5740) b,g,l,m | 5620 (2582- 6762) l,q,r | 5784 (2451–6430) m,s,t | 5612 (2074–6013) e,q,s | 4834 (1705–6048) r,t |
| I-FABP (pg/mL) [Mean (SD)] |
895.6 (774.2) | 1129 (1026) | 1117 (1160) | 794.5 (455.6) | 887.4 (768.6) | 802.3 (581.7) | 709.3 (526.4) |
| [Median (IQR)] | 660.6 (436.6–1275) | 731.1 (422.2–2001) | 717.8 (549.3–1174) | 557.5 (354.6–1202) | 665.8 (316.6–1368) | 658.0 (288.6–1338) | 517.6 (388.1–916.1) |
| Microbial translocation and immune activation | |||||||
| LPS (pg/mL) [Mean (SD)] |
98.7 (47.76) d | 99.75 (54.67) i | 89.3 (45.11) m | 104.7 (43.97) p | 138.3 (63.09) d,i,m,p,s,t | 112.2 (45.7) s | 94.73 (37.82) t |
| [Median (IQR)] | 88.29 (60.6–144.0) d | 93.18 (55.7–139.7) i | 76.56 (56.2–142.3) m | 102.8 (64.8–146.4) p | 131.9 (98.8–197.7) d,i,m,p,s,t | 115.8 (80.7–136.2) s | 98.78 (72.6–125.4) t |
| sCD14 (ng/mL) [Mean (SD)] |
2583 (536.9) | 2446 (447.3) h | 2605 (472.9) | 2855 (676.3) h,p,q | 2407 (399.2) p | 2491 (449.7) q | 2650 (602.7) |
| [Median (IQR)] | 2472 (2166–2825) | 2344 (2110–2877) h | 2493 (2248–2995) | 2766 (2371–3257) h,p,q | 2327 (2034–2797) p | 2371 (2181–2800) q | 2523 (2144–2945) |
| sCD27 (U/mL) [Mean (SD)] |
110.9 (22.73) | 113.3 (31.55) | 111.4 (26.81) | 113.3 (28.88) | 112.8 (35.98) | 109.3 (35.76) | 108.6 (28.85) |
| [Median (IQR)] | 114.6 (87.3–127.2) | 110.5 (93.4–122.3) | 105.7 (88.2–135.2) | 103.4 (88.8–137.8) | 108.6 (79.5–136.2) | 91.20 (83.5–141.9) | 106.3 (80.6–132.8) |
| sTNFRII (pg/mL) [Mean (SD)] |
2297 (665.4) d,e | 2279 (713.7) i,j | 2329 (800.1) n | 2631 (1076) | 2096 (481.1) d,i | 2027 (410.0) e,j,n | 2215 (740.8) |
| [Median (IQR)] | 2202 (1708–2823) d,e | 2037 (1680–3061) i,j | 2115 (1639–3158) n | 2608 (1752–3321) | 1971 (1727–2626) d,i | 1938 (1674–2353) e,j,n | 1957 (1719–3002) |
| Pro-inflammatory cytokines | |||||||
| TNF- (pg/mL) [Mean (SD)] |
2.61 (1.64) f | 2.18 (1.19) | 2.57 (2.17) o | 2.06 (1.10) | 2.27 (0.77) | 2.03(0.71) | 2.13 (1.48) f,o |
| [Median (IQR)] | 2.38 (1.38–3.63) f | 2.16 (0.87–3.21) | 1.82 (0.89–3.58) o | 1.93 (1.27–2.69) | 2.09 (1.54–2.91) | 1.94 (1.39–2.67) | 1.56 (0.95–3.78) f,o |
| INF- (pg/mL) [Mean (SD)] |
10.98 (18.11) e | 8.24 (11.43) | 14.31 (24.62) | 9.9 (13.52) q | 9.26 (11.73) | 8.55 (11.63) e,q | 9.93 (15.35) |
| [Median (IQR)] | 3.38 (0.53–13.63) e | 3.2 (0.53–13.28) | 2.28 (0.48–21.01) | 1.63 (0.52–21.62) q | 2.79 (0.54–18.82) | 3.32 (0.54–20.81) e,q | 2.66 (0.67–21.74) |
| IL-1 (pg/mL) [Mean (SD)] |
0.52 (0.45) | 0.62 (0.64) h,j | 0.58 (0.58) | 0.43 (0.34) h | 0.40 (0.20) | 0.40 (0.24) j | 0.62 (0.72) |
| [Median (IQR)] | 0.44 (0.24–0.59) | 0.47 (0.18–0.68) h,j | 0.46 (0.26–0.59) | 0.38 (0.14–0.65) h | 0.46 (0.17–0.60) | 0.44 (0.15–0.52) j | 0.42 (0.19–0.66) |
| IL-6 (pg/mL) [Mean (SD)] |
1.44 (1.00) | 1.11 (0.75) | 1.36 (0.97) | 1.36 (0.91) | 1.40 (1.15) | 1.22 (0.97) | 1.29 (1.13) |
| [Median (IQR)] | 1.14 (0.51–2.30) | 0.91 (0.41–1.91) | 1.30 (0.55–1.86) | 0.92 (0.63–2.30) | 0.87(0.63–1.92) | 0.88 (0.47–1.69) | 1.07 (0.38–1.72) |
| IL-8 (pg/mL) [Mean (SD)] |
5.50 (3.5) | 5.79 (5.6) | 5.12 (3.18) | 5.53 (2.88) | 6.20 (2.41) t | 5.61 (2.99) | 4.21 (1.42) t |
| [Median (IQR)] | 4.97 (3.05–7.38) | 3.88 (2.08–8.10) | 4.89 (2.80–7.67) | 5.94 (2.74–7.92) | 5.84 (4.00–8.37) t | 5.76 (2.66–7.59) | 4.68 (2.86–5.32) t |
| IP-10 (pg/mL) [Mean (SD)] |
55.99 (36.72) | 55.5 (33.47) h | 51.26 (24.24) | 50.55 (26.04) h,p | 61.80 (35.97) p | 56.68 (25.52) | 52.59 (28.58) |
| [Median (IQR)] | 43.74 (30.11–77.91) | 43.18 (31.62–76.80) h | 42.97 (32.30–74.06) | 41.51 (35.77–69.95) h,p | 50.05 (36.14–95.64) p | 48.65 (42.49–81.62) | 41.80 (33.77–75.17) |
| Anti-inflammatory cytokine | |||||||
| IL-10 (pg/mL) [Mean (SD)] |
1.26 (1.37) | 0.92 (0.54) | 1.0 (0.68) | 0.93 (0.44) | 1.1 (0.85) | 1.1 (0.74) | 0.93 (0.49) |
| [Median (IQR)] | 0.77 (0.49–1.55) | 0.79 (0.51–1.28) | 0.85 (0.47–1.26) | 0.86 (0.53–1.29) | 0.82 (0.50–1.36) | 0.87 (0.60–1.52) | 0.90 (0.52–1.18) |
Results are shown as the mean and standard deviation (SD) and as the median and interquartile range (IQR). Significant differences (p < 0.05) following the Wilcoxon matched-pairs signed-rank test are mentioned as follows: a: week 0 vs. week 1; b: week 0 vs. week 2; c: week 0 vs. week 6; d: week 0 vs. week 8; e: week 0 vs. week 12; f: week 0 vs. week 14; g: week 1 vs. week 2; h: week 1 vs. week 6; i: week 1 vs. week 8; j: week 1 vs. week 12; k: week 1 vs. week 14; l: week 2 vs. week 6; m: week 2 vs. week 8; n: week 2 vs. week 12; o: week 2 vs. week 14; p: week 6 vs. week 8; q: week 6 vs. week 12; r: week 6 vs. week 14; s: week 8 vs. week 12; t: week 8 vs. week 14; u: week 12 vs. week 14. Significant values are presented in bold. $: Two participants from CBD-only were excluded from analyses at week 6 (one participant) and weeks 8, 12, and 14 (two participants) because they were withdrawn for safety concerns.
Plasma levels of bacterial LPS did not significantly change from baseline until treatment completion. However, plasma levels of LPS showed a transient increase at week 8 (week 0 vs. week 8: p = 0.008), followed by a significant drop up until treatment termination (week 8 vs. week 12: p = 0.02) (Table 2). Plasma markers of immune activation (sCD14, sCD27, sTNFRII) were also assessed. sCD14 is shed by activated monocytes and serves as a marker of monocyte activation and inflammation during HIV infection [7,62]. As with LPS, no significant changes in the plasma level of sCD14 were observed from baseline until treatment completion. sCD14 also followed a biphasic pattern with a slight increase after cannabinoid treatment initiation (week 1 vs. week 6 p = 0.02), followed by a significant drop in its plasma concentration until the end of treatment (week 6 vs. week 8: p = 0.02; week 6 vs. week 12: p = 0.02). sTNFRII is a soluble form of the TNF-α receptor and serves also as a marker of inflammation in HIV infection [63]. Levels of sTNFRII dropped significantly at week 8 (week 0 vs. week 8: p = 0.04) and remained low until the end of treatment (week 0 vs. week 12: p = 0.04, Figure 1b). Lastly, we have assessed plasma levels of sCD27, which is shed from the surface of activated lymphocytes, and serves as a marker of T-cell–mediated inflammation [64]. There were no significant changes in levels of sCD27 over the course of treatment (Table 2). Plasma levels of pro-inflammatory (TNF-α, IFN-γ, IL-1β, IL-6, IL-8, IP-10) and anti-inflammatory (IL-10) cytokines were measured by Luminex. A significant decrease in plasma levels of TNF- (week 0 vs. week 14; p = 0.02), IFN- (week 0 vs. week 12: p = 0.03, Figure 1c), IL-1 (week 1 vs. week 12; p = 0.02), and IL-8 (week 8 vs. week 14; p = 0.03) was observed (Table 2). Other soluble markers showed a biphasic pattern with an early increase followed by a reduction until the end of the treatment. Plasma levels of IP-10 showed a transient drop at week 6 (week 1 vs. week 6: p = 0.01) and a subsequent rebound at week 8 (week 6 vs. week 8: p = 0.04), with no overall significant change between initiation and end of treatment. No significant changes in levels of IL-6 or IL-10 were observed (Table 2).
Altogether, over the treatment period, the dynamic of most of the soluble plasma markers showed a biphasic pattern, with a transient increase during the first weeks of cannabinoids uptake, followed by a significant reduction until treatment completion.
3.3. Effect of Oral Cannabinoids on Blood T-Cell, Monocyte, and Dendritic Cell Subsets
3.3.1. Changes in Circulating CD4 T-Cell Subsets
While no significant changes in frequencies of naïve (CD45RA+CD28+CCR7+), central memory (CD45RA−CD28+CCR7+), and transitional memory (CD45RA−CD28+CCR7−) CD4 T-cells were observed, levels of effector memory (CD45RA−CD28−CCR7−) CD4 T-cells were significantly lower at weeks 6 and 8 following treatment initiation (week 0 vs. week 6; p = 0.04, week 1 vs. week 8; p = 0.02) (Table 3, Figure S1). Similarly, frequencies of terminally differentiated (CD45RA+CD28−CCR7−) CD4 T-cells were significantly lower at week 8 (week 1 vs. week 8: p = 0.02). However, in both cases, the overall change in these memory subsets between week 0 and week 12 was not significant.
Table 3.
Dynamic of CD4 and CD8 T-cells, monocytes and dendritic cells populations during cannabinoids treatment.
| Cellular Immune Markers | Study Time-Line | ||||||
|---|---|---|---|---|---|---|---|
|
Week 0 Treatment Initiation n = 10 |
Week 1 n = 10 |
Week 2 n = 10 |
$
Week 6 n = 9 |
$
Week 8 n = 8 |
$
Week 12 End of Treatment n = 8 |
$
Week 14 Study Termination n = 8 |
|
| CD4 T-Cells | |||||||
| Memory T-Cell Subsets | |||||||
| Naïve (CD45RA+CD28+CCR7+) % [Mean (SD)] |
78.2 (13.1) | 79.6 (11.0) | 79.4 (12.5) | 78.0 (14.3) | 76.7 (13.5) | 76.2 (13.6) | 73.9 (15.6) |
| [Median (IQR)] | 83.4 (66.3–87.4) | 83.8 (74.2–86.6) | 82.0 (74.4–87.4) | 86.2 (65.4–87.7) | 81.9 (63.1–85) | 80.7 (63.9–84.8) | 78.7 (59.1–85.4) |
| Central Memory (CD45RA−CD28+CCR7+) % [Mean (SD)] |
32.9 (11.7) | 33.6 (10.6) | 35.3 (12.7) | 34.4 (12.7) | 32.5 (11.5) | 32.7 (12.8) | 32.7 (12.4) |
| [Median (IQR)] | 36.6 (22.3–39.6) | 33.3 (28.3–40.1) | 34.5 (29.3–42.2) | 34.4 (25.8–44.1) | 34.7 (24.8–38.6) | 34.5 (21.1–40.9) | 33.8 (22.8–39.7) |
| Transitional Memory (CD45RA−CD28+CCR7−) % [Mean (SD)] |
64.1 (11.1) | 63.8 (10.6) | 62.4 (12.5) | 63.0 (12.6) | 64.9 (11.4) | 64.4 (12.7) | 63.6 (13.3) |
| [Median (IQR)] | 62 (57.2–71.7) | 63.4 (57.6–69.6) | 62.6 (52.2–68.3) | 60.9 (52.2–73.2) | 63.6 (56.6–73.0) | 61.5 (55.3–77.4) | 62.3 (50.7–75.7) |
| Effector Memory (CD45RA−CD28−CCR7−) % [Mean (SD)] |
2.6 (3.2) c | 2.3 (2.8) i | 2.1 (2.5) | 2.3 (2.7) c | 2.4 (2.7) i | 2.6 (2.3) | 3.2 (3.9) |
| [Median (IQR)] | 1.0 (0.3–5.9) c | 1.0 (0.4–4.6) i | 1.1 (0.3–3.7) | 0.8 (0.3–4.5) c | 1.2 (0.4–5.1) i | 1.4 (0.8–5.3) | 1.6 (0.5–5.6) |
| Terminally Differentiated (CD45RA+CD28−CCR7−) % [Mean (SD)] |
2.1 (4.4) b | 2.1 (4.6) g,i | 1.6 (3.9) b,g | 2.1 (4.4) | 1.8 (4.2) i | 1.8 (3.6) | 2.4 (5.6) |
| [Median (IQR)] | 0.2 (0.04–1.9) b | 0.2 (0.04–1.6) g,i | 0.1 (0.03–1.1) b,g | 0.2 (0.05–2.2) | 0.1 (0.09–1.3) i | 0.2 (0.07–1.8) | 0.1 (0.05–1.8) |
| T-cell functions | |||||||
| HLADR+CD38+ (%) [Mean (SD)] |
3.1 (1.6) | 4.0 (3.1) | 3.8 (4.1) | 2.8 (1.7) | 2.7 (1.7) | 3.6 (2.7) | 3.2 (3.6) |
| [Median (IQR)] | 2.9 (1.6–4.8) | 3.8 (1.6–4.9) | 2.3 (1.6–4.6) | 1.7 (1.3–4.4) | 2.2 (1.5–4.6) | 2.5 (1.3–6.5) | 1.8 (1.2–4.2) |
| CD45RA−Ki-67+ (%) [Mean (SD)] |
0.8 (0.3) e | 0.9 (0.4) | 0.9 (0.4) | 1.0 (0.5) | 1.0 (0.6) | 1.0 (0.4) e | 0.8 (0.3) |
| [Median (IQR)] | 0.7 (0.5–1.0) e | 0.8 (0.5–1.3) | 0.8 (0.5–1.2) | 1.0 (0.6–1.2) | 0.8 (0.6–1.2) | 0.9 (0.7–1.2) e | 0.7 (0.7–0.9) |
| CD45RA−PD-1+ (%) [Mean (SD)] |
32.3 (9.5) f | 32.4 (9.5) j,k | 31.4 (9.5) n,o | 32.2 (10.1) r | 32.6 (10.3) | 30.6 (11.0) j,n | 30.8 (9.2) f,k,o,r |
| [Median (IQR)] | 30.5 (26.3–39.0) f | 30.7 (24.9–38.3) j,k | 30.6 (23.8–37.5) n,o | 32.2 (23.0–37.8) r | 31.2 (24.5–41.7) | 31.3 (22.6–39.2) j,n | 30.5 (22.6–37.8) f,k,o,r |
| CD45RA−CTLA-4+ (%) [Mean (SD)] |
3.0 (1.4) | 3.2 (1.5) | 3.3 (1.7) | 3.6 (2.4) | 3.9 (2.2) | 3.6 (1.4) | 3.8 (1.6) |
| [Median (IQR)] | 3.1 (1.8–4.2) | 2.5 (2.0–4.9) | 2.5 (2.0–4.9) | 3.2 (1.7–5.0) | 3.2 (2.4–5.4) | 3.8 (2.3–5.1) | 3.9 (2.2–4.7) |
| Senescent (CD28−CD57+) (%) [Mean (SD)] |
1.8 (2.5) | 1.5 (2.1) i | 1.3 (1.7) | 1.5 (2.0) | 1.4 (1.6) i | 1.3 (1.2) | 2.1 (3.2) |
| [Median (IQR)] | 0.5 (0.07–4.0) | 0.7 (0.05–3.0) i | 0.7 (0.04–2.3) | 0.7 (0.1–2.6) | 0.7 (0.2–2.2) i | 0.8 (0.5–2.0) | 0.9 (0.3–2.2) |
| CD4+CD39+ (%) [Mean (SD)] |
6.1 (4.7) | 6.3 (5.1) g | 5.9 (4.8) g | 6.4 (5.1) r | 6.4 (5.3) t | 5.9 (5.7) | 5.5 (4.8) r,t |
| [Median (IQR)] | 5.1 (2.1–9.0) | 5.0 (1.9–9.0) g | 4.9 (1.8–8.1) g | 5.8 (1.5–10.3) r | 5.0 (1.8–11.4) t | 5.1 (1.3–7.9) | 4.8 (1.3–7.8) r,t |
| CD4+CD73+ (%) [Mean (SD)] |
6.9 (3.1) d,f | 6.4 (2.6) | 6.6 (2.8) o | 6.2 (3.0) | 6.1 (2.8) d | 6.3 (3.7) | 5.8 (2.9) f,o |
| [Median (IQR)] | 7.1 (4.6–9.2) d,f | 6.8 (4.5–7.9) | 6.5 (4.9–7.9) o | 6.2 (3.9–7.3) | 6.6 (3.6–7.0) d | 6.1 (3.4–6.8) | 5.6 (3.3–6.9) f,o |
| Chemokine receptors expression | |||||||
| CD45RA−CCR4+ (%)[Mean (SD)] | 29.7 (11.4) | 29.2 (10.2) | 30.1 (10.8) | 31.4 (9.5) | 30.5 (9.8) | 29.4 (10.1) | 29.1 (10.4) |
| [Median (IQR)] | 31 (20.5–36.6) | 31.6 (21.1–36.0) | 31.7 (20.8–38.0) | 34.4 (23.2–36.1) | 31.8 (20.3–36.6) | 30.1 (18.6–38.4) | 30.6 (18.3–38.1) |
| CD45RA−CCR6+ (%) [Mean (SD)] |
9.8 (6.1) | 12.6 (7.1) k | 11.7 (5.0) | 15.0 (10.8) | 12.0 (7.2) | 10.9 (4.3) | 9.7 (6.4) k |
| [Median (IQR)] | 8.8 (5.7–12.4) | 11.8 (7.1–19.0) k | 11.2 (8.3–16.2) | 10.6 (7.4–20.8) | 11.4 (8.3–13.3) | 10.3 (8.5–13.2) | 6.7 (5.7–15.5) k |
| CD45RA−CXCR3+ (%) [Mean (SD)] |
2.6 (2.9) | 3.8 (5.2) | 4.1 (3.8) | 5.7 (3.6) | 2.4 (3.1) | 3.5 (3.2) | 3.1 (4.7) |
| [Median (IQR)] | 1.7 (0.3–4.1) | 2.4 (0.4–4.1) | 3.0 (1.8–6.1) | 4.9 (3.0–9.1) | 0.8 (0.5–5.2) | 2.3 (1.7–5.3) | 1.2 (0.9–3.5) |
| Th subsets | |||||||
| Th17 (CD45RA−CCR4+CCR6+ CXCR3−) (%) [Mean (SD)] |
5.2 (3.0) | 6.8 (4.0) k | 6.4 (2.8) | 8.0 (5.9) | 6.3 (3.6) | 5.8 (2.5) | 5.2 (3.5) k |
| [Median (IQR)] | 5.2 (2.8–7.2) | 5.5 (4.4–10.3) k | 6.0 (4.8–9.2) | 6.9 (3.4–10.7) | 5.7 (4.8–7.1) | 5.4 (5.0–6.7) | 3.8 (2.4–8.2) k |
| Th1-Th17 (CD45RA−CCR4−CCR6+CXCR3+) (%) [Mean (SD)] |
0.36 (0.6) c | 0.62 (0.9) k | 0.6 (0.9) | 1.0 (1.1) c | 0.5 (0.9) | 0.3 (0.3) | 0.2 (0.3) k |
| [Median (IQR)] | 0.2 (0.02–0.4) c | 0.3 (0.05–0.8) k | 0.3 (0.1–0.7) | 0.4 (0.3–1.7) c | 0.06 (0.03–0.9) | 0.2 (0.1–0.4) | 0.1 (0.04–0.2) k |
| Th2 (CD45RA−CCR4+CCR6−CXCR3−) (%) [Mean (SD)] |
23.0 (10.5) c | 20.7 (7.6) | 21.9 (8.9) | 20.9 (6.5) c | 22.9 (10.4) | 22.1 (8.9) | 22.6 (8.5) |
| [Median (IQR)] | 21.9 (13.7–32.6) c | 20.0 (14.6–29.1) | 20.7 (14.7–30.7) | 20.9 (14.7–26.9) c | 21.7 (13.4–31.9) | 20.9 (14.0–30.0) | 23.7 (13.6–29.6) |
| Th1 (CD45RA−CCR4−CCR6−CXCR3+) (%) [Mean (SD)] |
1.7 (1.7) | 2.4 (3.6) | 2.7 (2.5) | 3.2 (1.8) p | 1.4 (1.6) p | 2.5 (2.3) | 2.4 (3.8) |
| [Median (IQR)] | 1.3 (0.3–3.0) | 1.3 (0.3–2.5) | 2.0 (1.0–4.5) | 3.6 (1.9–4.7) p | 0.6 (0.4–2.9) p | 1.8 (1.0–4.4) | 0.9 (0.7–2.5) |
| Regulatory T-cells | |||||||
| Treg (CD25hi CD127lo FoxP3+) (%) [Mean (SD)] |
2.9 (1.6) | 2.7 (1.4) | 2.9 (1.2) | 3.2 (1.6) | 3.4 (1.7) | 2.6 (1.1) | 2.9 (1.1) |
| [Median (IQR)] | 2.0 (1.5–4.5) | 2.6 (1.5–3.6) | 2.5 (1.9–3.9) | 2.7 (1.8–4.4) | 3.7 (1.6–4.8) | 2.0 (1.8–3.8) | 2.6 (2.2–4.1) |
| CD73+ Treg (%) [Mean (SD)] |
5.0 (2.4) f | 4.9 (2.6) j,k | 4.8 (2.5) | 4.2 (2.2) | 4.5 (2.1) | 4.2 (1.7) j | 4.2 (1.7) f,k |
| [Median (IQR)] | 4.9 (3.0–7.4) f | 4.1 (3.3–7.3) j,k | 4.4 (2.9–7.2) | 3.5 (3.0–5.8) | 4.3 (3.0–6.6) | 3.9 (3.5–5.7) j | 3.9 (3.4–5.9) f,k |
| CD39+ Treg (%) [Mean (SD)] |
45.1 (21.1) | 45.4 (23.0) | 44.0 (22.2) n | 44.1 (24.5) | 41.3 (23.3) | 41.3 (21.7) n | 41.0 (20.4) |
| [Median (IQR)] | 41.5 (28.5–62.5) | 46.3 (23.4–65.6) | 43.4 (25.2–65.4) n | 43.2 (24.1–69.2) | 36.5 (21.5–66.6) | 37.4 (23.3–59.8) n | 38.1 (25.1–63.3) |
| CD8 T-cells | |||||||
| Memory T-cell subsets | |||||||
| Naïve (CD45RA+CD28+CCR7+) % [Mean (SD)] |
50.4 (25.3) b,d | 52.3 (24.7) i | 54.6 (25.3) b | 51.8 (24.7) | 52.8 (26.8) d,i | 48.2 (25.9) | 52.8 (25.6) |
| [Median (IQR)] | 48.7 (24.4–77.7) b,d | 53.4 (26.5–77.4) i | 55.3 (29.8–80.1) b | 51.2 (27.4–74.2) | 57.5 (25.9–77.7) d,i | 47.0 (24.0–67.5) | 56.2 (28.9–77.8) |
| Central Memory (CD45RA−CD28+CCR7+) % [Mean (SD)] |
9.7 (6.1) | 10.0 (4.7) k | 10.8 (5.3) | 12.1 (6.3) | 11.8 (4.5) | 11.5 (4.5) | 12.1 (5.3) k |
| [Median (IQR)] | 7.6 (4.9–15.9) | 8.6 (5.7–14.6) k | 8.2 (6.5–17.4) | 9.5 (5.8–18.4) | 12.9 (6.8–16.0) | 10.8 (7.3–15.1) | 12.6 (6.4–17.6) k |
| Transitional Memory (CD45RA−CD28+CCR7−) % [Mean (SD)] |
66.7 (12.0) | 67.3 (11.1) | 67.5 (10.7) | 65.2 (9.7) | 67.4 (9.5) | 66.3 (8.1) | 64.7 (13.1) |
| [Median (IQR)] | 70.5 (58.7–75.2) | 72.0 (58.5–74.7) | 69.4 (58.0–74.7) | 69.2 (59.0–72.8) | 72.1 (61.3–73.6) | 70.7 (59.2–72.0) | 70.6 (57.0–72.5) |
| Effector Memory (CD45RA−CD28−CCR7−)% [Mean (SD)] |
22.8 (15.6) | 21.9 (13.9) | 21.0 (13.6) | 21.7 (14.2) | 19.8 (12.8) | 21.2 (10.6) | 21.8 (15.8) |
| [Median (IQR)] | 17.1 (9.8–35.7) | 15.6 (9.8–33.8) | 14.8 (9.3–34.5) | 19.2 (7.9–32.2) | 14.1 (9.9–30.9) | 16.9 (13.3–33.4) | 14.1 (10.7–35.3) |
| Terminally Differentiated (CD45RA+CD28−CCR7−) % [Mean (SD)] |
31.2 (22.2) | 30.0 (22.7) | 28.5 (22.0) | 31.4 (21.6) | 29.8 (23.4) | 33.0 (23.9) | 29.6 (22.5) |
| [Median (IQR)] | 35.5 (5.7–49.3) | 32.1 (4.4–54.1) | 29.5 (4.2–51.6) | 30.6 (9.7–53.1) | 26.5 (6.4–55.4) | 38.4 (6.5–55.4) | 28.6 (6.9–48.2) |
| T-cell functions | |||||||
| HLA-DR+CD38+ (%) [Mean (SD)] |
4.9 (4.1) | 5.3 (4.3) | 5.2 (5.0) | 5.1 (3.8) | 6.1 (6.7) | 6.1 (5.7) | 5.4 (5.0) |
| [Median (IQR)] | 3.6 (2.0–7.4) | 4.3 (2.6–6.9) | 3.5 (1.6–8.2) | 4.4 (2.2–7.8) | 3.5 (1.8–9.0) | 3.4 (2.7–11.1) | 3.8 (2.1–8.5) |
| CD45RA−Ki-67+ (%) [Mean (SD)] |
0.39 (0.19) a | 0.51 (0.25) a | 0.49 (0.27) | 0.55 (0.40) | 0.58 (0.63) | 0.52 (0.18) | 0.58 (0.46) |
| [Median (IQR)] | 0.41 (0.25–0.56) a | 0.47 (0.33–0.73) a | 0.47 (0.32–0.74) | 0.42 (0.35–0.54) | 0.36 (0.28–0.49) | 0.53 (0.34–0.68) | 0.36 (0.30–0.96) |
| CD45RA−PD-1+ (%) [Mean (SD)] |
37.9 (13.1) | 37.9 (11.9) k | 37.7 (14.9) | 35.2 (10.8) | 37.1 (11.6) t | 34.9 (12.8) | 34.7 (10.2) k,t |
| [Median (IQR)] | 37.0 (31.0–45.3) | 38.1 (29.5–46.5) k | 35.3 (27.5–47.8) | 35.9 (27.1–45.7) | 36.4 (29.4–49.2) t | 37.2 (25.1–47.1) | 36.7 (27.2–42.9) k,t |
| CD45RA−CTLA-4+ (%) [Mean (SD)] |
1.0 (0.7) | 1.3 (1.0) | 1.4 (0.9) | 2.1 (3.1) | 1.1 (1.0) | 1.0 (0.6) | 1.3 (0.9) |
| [Median (IQR)] | 0.9 (0.5–1.2) | 1.0 (0.6–1.7) | 1.2 (0.6–2.2) | 0.8 (0.5–2.7) | 0.9 (0.4–1.4) | 1.0 (0.6–1.1) | 1.0 (0.7–2.0) |
| Senescent (CD28−CD57+) (%) [Mean (SD)] |
18.5 (14.5) c,d | 17.0 (13.5) | 16.4 (12.6) m | 17.9 (13.5) c | 16.2 (12.5) d,m | 17.0 (11.1) | 18.1 (15.4) |
| [Median (IQR)] | 16.1 (4.9–31.8) c,d | 13.6 (4.5–31.4) | 12.5 (4.6–29.6) m | 16.8 (5.7–31.6) c | 10.6 (6.4–27.7) d,m | 14.9 (5.8–28.2) | 11.4 (7.2–29.9) |
| CD8+CD39+ (%) [Mean (SD)] |
2.3 (1.4) | 2.6 (1.9) | 2.5 (1.9) | 2.6 (2.2) r | 2.3 (1.9) | 2.2 (1.6) | 2.0 (1.6) r |
| [Median (IQR)] | 2.8 (0.5–3.4) | 2.6 (0.6–3.9) | 2.7 (0.4–3.8) | 2.8 (0.3–4.5) r | 2.1 (0.4–3.8) | 2.1 (0.6–3.4) | 2.2 (0.3–3.3) r |
| CD8+CD73+ (%) [Mean (SD)] |
36.6 (24.4) b | 36.9 (24.6) | 38.1 (24.7) b | 32.9 (19.8) | 32.9 (20.7) | 30.9 (21.4) | 32.1 (21.3) |
| [Median (IQR)] | 35.3 (10.4–54.4) b | 35.8 (10.5–54.3) | 39.5 (11.7–55.6) b | 36.6 (10.8–50.2) | 36.6 (10.8–51.8) | 30.6 (9.2–52.9) | 34.8 (9.3–50.1) |
| CD8+FoxP3+ (%) [Mean (SD)] |
0.29 (0.23) | 0.28 (0.19) | 0.40 (0.29) n | 0.39 (0.35) q | 0.42 (0.51) | 0.20 (0.19) n,q | 0.28 (0.16) |
| [Median (IQR)] | 0.19 (0.13–0.56) | 0.21 (0.17–0.35) | 0.37 (0.12–0.60) n | 0.29 (0.11–0.67) q | 0.14 (0.08–0.80) | 0.12 (0.09–0.29) n,q | 0.25 (0.13–0.44) |
| Chemokine receptors expression | |||||||
| CD45RA−CCR4+ (%) [Mean (SD)] |
15.8 (10.6) | 15.2 (7.5) | 15.6 (8.4) | 18.2 (10.3) | 17.6 (9.5) | 17.7 (11.4) | 18.2 (9.3) |
| [Median (IQR)] | 13.7 (8.1–21.9) | 13.2 (8.6–21.6) | 13.9 (9.0–20.9) | 15.5 (8.1–28.8) | 17.2 (10.2–26.8) | 14.1 (9.4–31.0) | 17.0 (9.6–26.0) |
| CD45RA−CCR6+ (%) [Mean (SD)] |
4.4 (4.2) | 4.9 (3.8) | 4.5 (3.0) | 5.3 (4.2) | 5.1 (4.4) t | 4.0 (2.4) | 4.0 (3.8) t |
| [Median (IQR)] | 3.8 (1.4–5.5) | 3.5 (1.6–6.7) | 3.0 (2.1–7.5) | 4.5 (2.4–6.8) | 3.9 (2.1–6.2) t | 3.2 (2.5–5.7) | 3.1 (1.9–3.8) t |
| CD45RA−CXCR3+ (%) [Mean (SD)] |
4.1 (3.1) b | 6.0 (4.7) | 7.1 (5.5) b | 9.3 (6.4) | 4.7 (4.1) | 6.4 (5.2) | 5.9 (4.2) |
| [Median (IQR)] | 4.0 (1.5–6.3) b | 5.1 (1.8–9.2) | 5.6 (3.2–11.8) b | 8.7 (3.7–14.7) | 2.7 (2.1–8.6) | 4.6 (2.8–10.3) | 3.9 (2.7–10.0) |
| Monocytes | |||||||
| Classical monocytes | |||||||
| Classical (CD14++CD16−) (%) [Mean (SD)] |
78.0 (13.0) | 78.2 (11.8) | 77.5 (10.8) | 75.1 (16.7) | 74.9 (14.5) | 74.7 (14.6) | 73.0 (16.0) |
| [Median (IQR)] | 78.6 (69.7–90.9) | 80.3 (65.4–86.2) | 80.4 (65.5–87.1) | 83.5 (57.6–89.2) | 77.1 (58.4–89.1) | 69.2 (62.6–91.8) | 80.4 (59.9–86.1) |
| Classical CD163+ (%) [Mean (SD)] |
46.4 (19.2) | 44.7 (23.3) | 54.1 (26.7) | 50.8 (19.3) | 56.0 (20.4) t | 43.5 (19.0) | 46.1 (21.0) t |
| [Median (IQR)] | 40.0 (32.0–62.2) | 39.3 (31.6–64.8) | 56.5 (30.3–79.1) | 48.2 (33.8–62.1) | 57.2 (41.0–73.5) t | 49.2 (24.7–53.3) | 41.1 (32.0–64.0) t |
| Classical CX3CR1+ (%) [Mean (SD)] |
51.2 (14.3) b | 52.7 (14.4) | 60.3 (15.8) b | 51.5 (13.4) | 52.3 (11.9) | 52.1 (11.9) | 55.7 (12.9) |
| [Median (IQR)] | 53.8 (38.3–61.3) b | 56.6 (40.8–62.3) | 59.7 (54.3–69.0) b | 56.5 (38.1–63.1) | 51.9 (44.8–64.4) | 56.4 (41.6–61.8) | 57.4 (47.1–66.7) |
| Classical M-DC8+ (%) [Mean (SD)] |
2.56 (3.72) | 3.24 (6.73) k | 3.53 (6.94) o | 1.61 (2.00) | 3.18 (6.28) t | 2.11 (2.93) | 2.64 (5.78) k,o,t |
| [Median (IQR)] | 0.61 (0.42–4.26) | 0.75 (0.48–2.62) k | 0.64 (0.56–3.90) o | 0.72 (0.41–2.31) | 0.74 (0.46–2.42) t | 0.61 (0.38–4.86) | 0.49 (0.33–1.43) k,o,t |
| Classical CCR2+ (%) [Mean (SD)] |
91.6 (3.1) | 92.6 (2.6) | 92.9 (3.2) | 92.1 (3.6) | 93.5 (2.9) | 92.5 (4.6) | 92.2 (5.7) |
| [Median (IQR)] | 92.3 (89.9–93.3) | 92.3 (90.8–94.8) | 93.4 (91.1–95.7) | 91.1 (89.7–95.0) | 94.0 (91.6–95.1) | 93.4 (91.0–95.2) | 94.4 (89.0–95.3) |
| Intermediate monocytes | |||||||
| Intermediate (CD14+CD16+) (%) [Mean (SD)] |
11.0 (4.9) | 10.4 (4.6) | 10.5 (4.4) | 12.0 (7.2) | 12.0 (5.0) | 12.9 (6.7) | 12.7 (5.7) |
| [Median (IQR)] | 11.0 (6.0–15.3) | 10.5 (6.4–14.9) | 12.0 (6.2–13.7) | 9.1 (6.5–21.0) | 14.3 (6.4–15.2) | 14.4 (5.5–18.9) | 11.5 (8.1–18.0) |
| Intermediate CD163+ (%) [Mean (SD)] |
66.2 (11.7) | 66.7 (15.5) | 74.0 (17.6) | 70.4 (12.5) | 73.5 (12.4) t | 64.5 (14.2) | 65.9 (15.0) t |
| [Median (IQR)] | 65.2 (59.4–72.2) | 69.7 (58.1–76.2) | 74.0 (60.4–88.1) | 69.4 (60.3–77.5) | 71.4 (64.9–84.2) t | 59.6 (54.2–78.9) | 64.7 (54.6–78.6) t |
| Intermediate CX3CR1+ (%) [Mean (SD)] |
86.1 (8.2) | 86.8 (8.4) | 88.3 (8.6) | 88.3 (7.3) p | 86.0 (10.5) p | 85.8 (9.8) | 88.8 (7.2) |
| [Median (IQR)] | 87.0 (84.7–91.2) | 87.2 (83.6–94.1) | 89.3 (83.6–94.9) | 91.3 (85.7–92.8) p | 89.3 (84.5–92.2) p | 89.3 (78.6–91.6) | 90.6 (85.9–93.4) |
| Intermediate M-DC8+ (%) [Mean (SD)] |
15.5 (7.7) | 17.9 (10.6) j | 18.8 (13.0) | 15.2 (6.5) | 17.2 (11.2) | 13.4 (7.7) j | 14.6 (6.6) |
| [Median (IQR)] | 11.9 (9.1–23.6) | 15.7 (7.8–26.6) j | 16.1 (9.3–26.1) | 14.1 (11.1–21.1) | 16.0 (7.1–25.5) | 13.4 (6.7–20.0) j | 14.7 (8.3–19.1) |
| Intermediate CCR2+ (%) [Mean (SD)] |
89.3 (2.9) | 88.6 (2.1) | 89.0 (5.5) | 89.6 (1.6) | 90.1 (3.4) | 89.6 (4.5) | 89.6 (4.3) |
| [Median (IQR)] | 88.8 (87.8–91.2) | 88.8 (87.2–90.6) | 91.3 (86.5–92.4) | 90.0 (88.3–91.1) | 90.6 (87.7–92.9) | 91.0 (87.9–92.2) | 89.4 (88.9–93.2) |
| Non classical monocytes | |||||||
| Non classical (CD16++CD14−) (%) [Mean (SD)] |
11.0 (8.8) | 11.4 (7.5) | 12.1 (7.6) | 12.9 (10.2) | 13.0 (10.9) | 12.3 (9.5) | 14.3 (12.2) |
| [Median (IQR)] | 9.3 (3.7–16.5) | 10.7 (5.2–19.9) | 10.2 (5.0–19.4) | 7.1 (5.4–23.1) | 8.3 (4.7–25.7) | 12.2 (3.3–18.7) | 8.6 (6.7–19.1) |
| Non classical CD163+ (%) [Mean (SD)] |
31.2 (7.2) d | 33.7 (9.1) | 38.3 (15.9) | 35.3 (8.2) r | 35.7 (9.5) d | 32.6 (6.3) | 30.9 (6.8) r |
| [Median (IQR)] | 29.7 (26.3–38.0) d | 35.0 (25.8–41.4) | 31.6 (29.3–42.8) | 35.7 (30.0–40.0) r | 31.7 (30.2–46.4) d | 31.8 (27.6–38.5) | 30.2 (25.6–35.7) r |
| Non classical CX3CR1+ (%) [Mean (SD)] |
75.6 (11.5) | 74.8 (14.0) | 76.3 (14.8) | 76.9 (12.2) | 74.6 (15.5) | 75.5 (12.4) | 77.6 (14.9) |
| [Median (IQR)] | 74.7 (71.6–84.0) | 73.7 (67.2–85.9) | 81.9 (61.1–87.6) | 78.9 (65.5–86.6) | 81.0 (57.4–83.8) | 75.5 (63.6–87.9) | 84.5 (60.3–88.7) |
| Non classical M-DC8+ (%) [Mean (SD)] |
28.4 (10.3) | 30.8 (15.3) | 31.3 (18.6) | 25.6 (7.5) | 28.8 (10.8) | 23.2 (7.0) | 26.8 (10.5) |
| [Median (IQR)] | 24.7 (21.5–39.8) | 28.5 (17.8–37.7) | 25.1 (20.5–34.4) | 27.2 (19.1–32.5) | 31.0 (17.9–37.6) | 23.4 (17.6–26.8) | 24.7 (18.7–37.5) |
| Non classical CCR2+ (%) [Mean (SD)] |
7.2 (3.3) e | 8.1 (2.4) | 8.1 (4.7) | 7.2 (5.9) | 7.9 (3.3) | 12.1 (8.2) e | 7.9 (3.8) |
| [Median (IQR)] | 6.3 (4.5–10.5) e | 7.8 (6.8–9.6) | 8.3 (5.4–9.3) | 5.0 (3.9–8.8) | 6.8 (5.7–11.3) | 9.3 (6.7–18.1) e | 7.9 (5.1–11.2) |
| Dendritic cells (DC) | |||||||
| Plasmacytoid DC (CD123+CD11c−) (%) [Mean (SD)] |
4.4 (2.1) | 3.6 (1.1) | 3.8 (1.3) | 3.6 (1.5) | 4.2 (1.1) | 3.8 (1.7) | 4.1 (1.1) |
| [Median (IQR)] | 3.7 (2.7–5.5) | 3.8 (2.5–4.4) | 3.7 (3.1–5.1) | 3.7 (2.4–4.4) | 4.1 (3.2–5.0) | 3.8 (2.5–4.7) | 4.2 (3.3–4.9) |
| Myeloid DC (CD123−CD11c+) (%) [Mean (SD)] |
8.8 (3.2) b,e | 11.5 (2.7) | 12.3 (4.0) b | 10.3 (7.9) | 8.8 (3.9) s | 12.5 (4.8) e,s | 12.6 (6.6) |
| [Median (IQR)] | 8.1 (7.0–10.4) b,e | 11.2 (9.4–14.1) | 11.8 (9.9–13.7) b | 7.8 (5.4–12.0) | 8.1 (5.7–10.5) s | 11.5 (9.4–15.6) e,s | 13.0 (6.4–17.7) |
Results are shown as the mean and standard deviation (SD) and as the median and interquartile range (IQR). Significant differences (p < 0.05) following the Wilcoxon matched-pairs signed-rank test are mentioned as follows: a: week 0 vs. week 1; b: week 0 vs. week 2; c: week 0 vs. week 6; d: week 0 vs. week 8; e: week 0 vs. week 12; f: week 0 vs. week 14; g: week 1 vs. week 2; h: week 1 vs. week 6; i: week 1 vs. week 8; j: week 1 vs. week 12; k: week 1 vs. week 14; l: week 2 vs. week 6; m: week 2 vs. week 8; n: week 2 vs. week 12; o: week 2 vs. week 14; p: week 6 vs. week 8; q: week 6 vs. week 12; r: week 6 vs. week 14; s: week 8 vs. week 12; t: week 8 vs. week 14; u: week 12 vs. week 14. Significant values are presented in bold. $: Two participants from CBD-only were excluded from analyses at week 6 (one participant) and weeks 8, 12, and 14 (two participants) because they were withdrawn at week 6 for safety concerns.
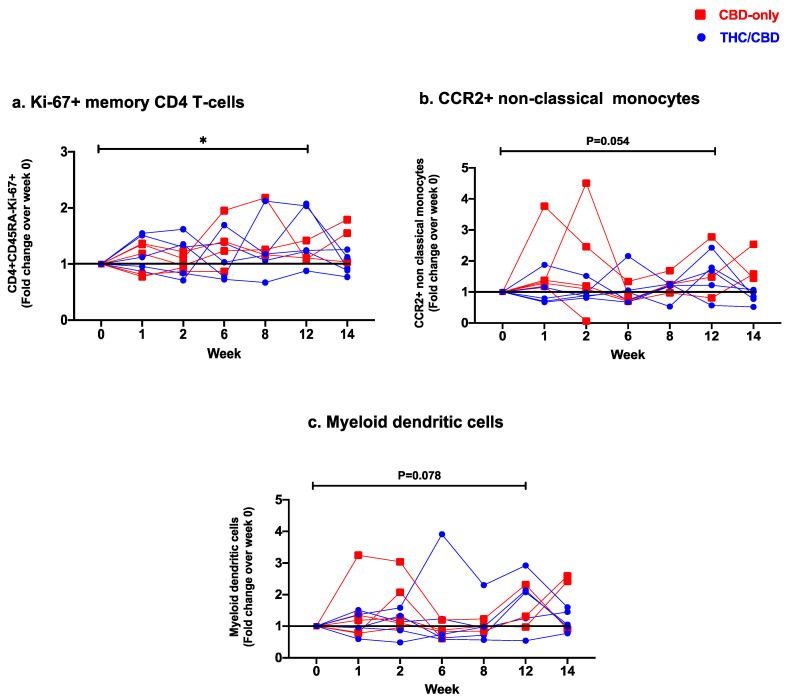
Frequencies of senescent (CD28−CD57+) CD4 T-cells dropped significantly between weeks 1 and 8 (p = 0.04). Levels of PD1+ CD45RA−CD4 T-cells also decreased significantly over the treatment course (week 0 vs. week 14: p = 0.02, week 1 vs. week 12: p = 0.02, week 1 vs. week 14: p = 0.01, week 2 vs. week 12: p = 0.03, week 2 vs. week 14: p = 0.01, week 6 vs. week 14: p = 0.02; Table 3). We observed a significant increase in frequencies of proliferating Ki67+ CD45RA−CD4 T-cells at the end of treatment (week 0 vs. week 12: p = 0.047, Figure 2a). Frequencies of CCR6+ CD45RA−CD4 T-cells, as well as expression of both ectonucleotidases CD39/CD73 by CD4 T-cells, were significantly lower after treatment termination (CCR6: week 1 vs. week14 p = 0.02; CD39: week 6 vs. week 14 p = 0.04; CD73 week 0 vs. week 14 p = 0.02). No significant differences in expression levels of HLA-DR/CD38, CTLA-4, and chemokine receptors CCR4/CXCR3 were observed (Table 3, Figure S1).
Figure 2.
Fold change expression over 12 weeks versus weeks 0 of CBD-only (red square) and THC/CBD combination (blue circle), (a) Ki-67+ memory CD4 T-cells, (b) CCR2+ non-classical monocytes, and (c) Myeloid dendritic cells. * p < 0.05.
Levels of Th and Treg subsets showed significant alterations after treatment termination. We observed a significant decrease in levels of pro-inflammatory Th17 cells (week 1 vs. week 14: p = 0.02) and Th1Th17 cells (week 1 vs. week 14: p = 0.04). Frequencies of Th1 showed a significant decrease between weeks 6 and 8 (p = 0.04) but with no significant change between the start and end of treatment. Frequencies of Th2 cells decreased significantly at week 6 (week 0 vs. week 6: p = 0.04). Lastly, we observed no significant changes in Treg frequency, but their expression levels of CD39+ (week 2 vs. week 12; p = 0.008) and CD73+ (week 0 vs. week 14; p = 0.008, week 1 vs. week 12, p = 0.05) decreased throughout the treatment (Table 3, Figure S2).
3.3.2. Changes in Circulating CD8 T-Cell Subsets
Significant increases in frequencies of naïve and central memory CD8 T-cells were observed after treatment initiation (naïve: week 0 vs. week 8 p = 0.04; central memory: week 1 vs. week 14: p = 0.008) with no significant changes in other memory CD8 T-cell subsets over the course of treatment (Table 3). Similar to CD4 T-cells, frequencies of senescent CD8 T-cells showed a significant decrease over the treatment period, reaching statistical significance at week 8 (week 0 vs. week 8: p = 0.04). Frequencies of PD-1+CD45RA−CD8 T-cells were also significantly reduced after treatment termination (week 1 vs. week 14: p = 0.02). In line with the decrease in CD8 T-cell senescence and exhaustion, we observed an increase in Ki67 expression during the first week of treatment (week 0 vs. week 1: p = 0.047). There were no significant changes in frequencies of HLA-DR+CD38+ and CTLA4+CD45RA−CD8 T-cells between the start and the end of treatment (Table 3, Figure S3a,b).
Within CD45RA−CD8 T-cells, expression levels of CCR6 decreased between weeks 8 and 14 (p = 0.02), while levels of CXCR3 increased at week 2 (week 0 vs. week 2: p = 0.049) with no overall significant change between the start and the end of treatment. There were no significant alterations in CCR4 over the treatment period. Frequencies of FoxP3+ CD8 T-cells decreased significantly between weeks 2 and 12 (p = 0.02). Levels of CD73+ CD8 T-cells increased significantly at week 2 (week 0 vs. week 2: p = 0.03). No significant changes in CD39 expression were observed (Table 3, Figure S3c).
3.3.3. Changes in Monocyte Subsets and Dendritic Cell Frequencies
Levels of classical (CD14++CD16−), intermediate (CD14+CD16+), and non-classical (CD14-CD16++) monocytic subsets remained stable over time (Table 3, Figure S4). Within these subsets, we observed differences in the expression of CD163, CX3CR1, CCR2, and M-DC8 over the treatment duration (Table 3, Figure S5). Expression levels of CD163 in non-classical monocytes showed a significant increase at week 8 (week 0 vs. week 8: p = 0.03), followed by a return to baseline after treatment termination (week 6 vs. week 14: p = 0.008). Frequencies of CX3CR1+ classical monocytes increased significantly during the first two weeks of treatment (week 0 vs. week 2: p = 0.04) but with no overall significant changes before and after treatment course completion. Levels of intermediate and non-classical monocytes expressing CX3CR1 remained stable over time. Expression levels of CCR2 in classical and intermediate monocytes also remained unchanged. In contrast, a significant increase in CCR2+ non-classical monocytes was observed at the end of the treatment period (week 0 vs. week 12: p = 0.04, Table 3) with a trend in their fold change increase (Figure 2b). Levels of M-DC8+ cells remained unchanged in non-classical monocytes, whereas in both classical and intermediate monocytes this subpopulation was significantly reduced at the end of treatment (classical: week 1 vs. week 14: p = 0.04; intermediate: week 1 vs. week 12: p = 0.02, Table 3).
A significant increase in frequencies of myeloid dendritic cells (mDC: CD123−CD11c+), was observed after 12 weeks of oral cannabinoid treatment (week 0 vs. week 12: p = 0.04, Table 3) with trend in their fold-change increase (Figure 2c), while no significant changes were observed in levels of plasmacytoid dendritic cells (pDC: CD123+CD11c−) (Table 3, Figure S4).
3.4. Effect of Oral Cannabinoids on Total HIV DNA and Cell-Associated HIV RNA in CD4 T-Cells from Blood and Semen
The impact of oral cannabinoids on HIV reservoir markers was assessed by quantifying total HIV DNA and cell-associated HIV RNA in PBMCs over the treatment period as presented in Table 4. Levels of HIV DNA and cell-associated HIV RNA remained overall stable in circulating CD4 T-cells, although some modest variations were observed between weeks 8 and 12 (Table 4). In only two individuals, HIV DNA was detectable in sperm cells and remained unchanged after study completion. HIV RNA was not detected in the seminal supernatant (not shown).
Table 4.
Dynamic of HIV DNA and cell-associated RNA levels in CD4 T-cells isolated from blood and semen during cannabinoids treatment.
| HIV DNA and RNA | Study Timeline | ||||||
|---|---|---|---|---|---|---|---|
| Week 0 Treatment Initiation n = 10 |
Week 1 n = 10 |
Week 2 n = 10 |
$ Week 6 n = 9 |
$ Week 8 n = 8 |
$ Week 12 End of Treatment n = 8 |
$ Week 14 Study Termination n = 8 |
|
| Total HIV DNA (copies/106 CD4) [Mean (SD)] |
1016 (1081) | 908.1 (817.3) | 920.4 (816.5) | 1053 (1121) | 1292 (1310) | 979.1 (1015) | 1143 (1318) |
| [Median (IQR)] | 708.1 (125.1–1679) | 765.3 (239.3–1449) | 819.8 (176.2–1529) | 911.9 (141- 1886) | 790.3 (165.6–2843) | 629.9 (155.5–2156) | 658.5 (152.3–2317) |
| LTR-gag cell-associated RNA (copies/106 CD4) [Mean (SD)] |
493.5 (557.3) | 593.9 (817.3) i | 529.2 (630.3) | 476.3 (479.6) | 1370 (1687) i,s | 572.9 (752.8) s | 751.9 (858.3) |
| [Median (IQR)] | 308.8 (4.9–915.8) | 224.3 (3.5–1088) i | 271.2 (13.4–1081) | 281.1 (21.9–938.8) | 142 (10.1–3151) i,s | 283.9 (2.1–1290) s | 386.9 (26.0–1770) |
| RNA/DNA ratio [Mean (SD)] |
0.47 (0.45) | 0.56 (0.55) | 0.43 (0.41) | 0.43 (0.37) | 0.62 (0.62) s | 0.44 (0.53) s | 1.85 (3.53) |
| [Median (IQR)] | 0.3 (0.2–1.0) | 0.46 (0.08–0.87) | 0.37 (0.08–0.76) | 0.33 (0.12–0.84) | 0.44 (0.05–1.26) s | 0.17 (0.01–1.07) s | 0.50 (0.28–1.54) |
Results are shown as the mean and standard deviation (SD) and as the median and interquartile range (IQR). Significant differences (p < 0.05) following the Wilcoxon matched-pairs signed-rank test are mentioned as follows: a: week 0 vs. week 1; b: week 0 vs. week 2; c: week 0 vs. week 6; d: week 0 vs. week 8; e: week 0 vs. week 12; f: week 0 vs. week 14; g: week 1 vs. week 2; h: week 1 vs. week 6; i: week 1 vs. week 8; j: week 1 vs. week 12; k: week 1 vs. week 14; l: week 2 vs. week 6; m: week 2 vs. week 8; n: week 2 vs. week 12; o: week 2 vs. week 14; p: week 6 vs. week 8; q: week 6 vs. week 12; r: week 6 vs. week 14; s: week 8 vs. week 12; t: week 8 vs. week 14; u: week 12 vs. week 14. Significant values are presented in bold. $: Two participants from CBD-only were excluded from analyses at week 6 (one participant) and weeks 8, 12, and 14 (two participants) because they were withdrawn at week 6 for safety concerns.
4. Discussion
To our knowledge, we report for the first time on in vivo anti-inflammatory effects of orally administrated cannabinoid-based treatment in humans in an interventional pilot clinical trial in PWH on ART. Pooled results from eight participants who successfully completed the oral cannabinoid treatment course showed a significant reduction in surrogate markers of gut mucosal damage, systemic inflammation, as well as cellular immune activation, exhaustion, and senescence. These preliminary findings support the use of further evaluation of orally administrated cannabinoid capsules in larger clinical trials as a potential strategy to help alleviate chronic inflammation experienced by PWH despite ART.
In this pilot clinical trial, our primary objective was the assessment of safety and tolerability, as previously reported [50]. In the current set of analyses, we assessed the effects of oral cannabinoids on the integrity of the gut mucosal barrier by measuring the dynamics of REG-3α and I-FABP over the treatment period. I-FABP is an intracellular protein expressed by enterocytes that is released upon cell death, while REG-3α is an antimicrobial peptide secreted by Paneth cells in the gut lumen that is crucial for the regulation of interplay between the microbiota and the host [65,66]. Notably, our team has previously reported that plasma levels of both markers are known to be elevated in PWH despite ART [7,62]. In the present study, REG-3 plasma levels decreased after 12 weeks of oral cannabinoid treatment. These results are promising since increased gut mucosal permeability is one of the major contributors to chronic inflammation in PWH [5,6]. These findings are also in line with previous observations on the effects of CBD on aspirin-induced gut mucosal permeability where participants treated with CBD and palmitoylethanolamide (an endocannabinoid-like fatty acid) showed significant improvement in gut epithelial integrity [22]. The authors have attributed this effect to the activation of the endocannabinoid system mediated by CBD and the CB1R [22]. Other evidence put forth by in vitro and animal studies also supports the CB1R-mediated reduction in gut epithelial permeability induced by CBD [23,67,68,69,70,71]. It is worth noting that the CB1R signaling cascade is highly complex, where THC acts as a partial CB1R agonist and CBD as a CB1R antagonist [72]. Furthermore, THC and CBD can also bind to other endocannabinoid receptors to exert their anti-inflammatory properties [73]. While we do observe measurable anti-inflammatory effects of oral cannabinoids in our study, the exact mechanism of action remains to be elucidated.
In our study, plasma levels of LPS and sCD14 showed a transient increase during the first weeks of treatment, followed by a significant reduction later during the treatment course. This observation was unexpected since other groups have reported that CB1R antagonists, such as CBD, reduce plasma LPS levels in diet-induced-obesity models [74]. Cannabidiol-rich cannabis extracts can alter gut microbial composition [75]. Furthermore, diarrhea and abdominal pain are common oral cannabinoid-related adverse effects, indicative of gastrointestinal imbalances that may occur when intake of cannabinoids is first initiated, as it also occurred in our study participants [50]. Furthermore, CBD has been shown to reduce peristalsis via modulation of motor and sensory pathways of the peristaltic reflex [76]. It is plausible that transient gut dysbiosis along with increased retention time of colonic contents may contribute to a temporary increase in LPS absorption that we observe. However, an in-depth analysis of gut microbiome changes over the course of treatment is required to validate this hypothesis.
Over the treatment course, we also observed a gradual, significant decrease in the sTNFRII inflammatory marker. Interestingly, similar observations on sTNFRII have been reported in PWH after recent cannabis use, although the authors report no change in microbial translocation makers [41]. We also observed a significant reduction in plasma levels of pro-inflammatory cytokines IFN- and IL-1 following 12 weeks of treatment which is in line with previous studies that reported a direct inhibitory effect of cannabinoids on pro-inflammatory cytokine production by immune cells in vitro [15] and in animal models [77]. This effect is likely driven by CB2R activation, which is highly expressed in immune cells, including monocytes [16].
We further report a significant decrease in frequencies of senescent and PD1+ CD memory T-cells. This was further accompanied by an increase in proliferative memory Ki-67+ CD4 T-cells and lower levels of terminally differentiated CD4 T-cells. Our findings support previous observations made in SIV-infected monkeys, where early THC treatment was associated with decreased levels of PD-1-expressing T-cells [20]. This is of particular importance since T-cell exhaustion and senescence during chronic inflammation, characterized by increased PD1 and CD57 expression, reduced proliferative capacity, and antiviral functions, have been extensively characterized in PWH [5,9,78,79,80]. Tregs are a highly immunosuppressive cell subset that is known to be increased during HIV infection, which has also been linked to disease progression [4,81,82,83,84,85,86]. One way that these cells exert their anti-inflammatory properties is through CD39 and CD73 ectonucleotidases, which convert pro-inflammatory extracellular adenosine triphosphate into immunosuppressive adenosine [87]. Importantly, we previously showed that Tregs-expressing ectonucleotidases are involved in HIV pathogenesis and disease progression, and even early ART initiation could not reverse their increased frequencies and their migration into the gut lymphoid tissues [8,88,89,90]. In our study, we did not observe any significant changes in CD4+ Treg frequencies over the treatment course. However, we do report a significant decrease in CD73+ Tregs, suggesting their decreased immunosuppressive activity.
Activated monocytes are among the first sources of pro-inflammatory cytokines during HIV infection and are one of the main contributors to chronic inflammatory complications in PWH [6,91]. In our study, we did not observe any changes in intermediate, classical, or non-classical monocytic subsets. Previous studies have reported that cannabis use in HIV-infected individuals is associated with lower levels of inflammatory CD16+ (i.e., intermediate and non-classical) monocytes and lower expression of CD163, indicative of their decreased migration potential toward the brain [42,43]. Notably, those studies looked at cannabis smokers, where THC and CBD were inhaled, whereas in our study these compounds were ingested. It is also known that the bioavailability of inhaled THC and CBD is several fold higher than if they are administered orally [92]. Thus, it is likely that THC/CBD plasma concentrations were insufficient to have a measurable effect on CD16+ monocytes in this clinical trial. However, we did observe a decrease in intermediate M-DC8+ monocytes which are known to be a major source of TNF- during their response to microbial translocation [91]. Furthermore, oral cannabinoids also significantly increased CCR2-expressing non-classical monocytes, a loss of this subpopulation had been associated with cognitive impairment in PWH not taking ART [93]. The increase in these non-classical monocyte subpopulations during the course of the treatment would be indicative of a potential neuroprotective effect of oral cannabinoids in PWH. Furthermore, we also noticed a significant increase in the frequencies of mDCs during cannabinoid treatment, while the frequency of plasmacytoid dendritic cells remained stable. Although mDCs are well known for promoting the activation and expansion of effector T-cells during inflammatory conditions [94,95], these cells also play an immuno-regulatory role in the maintenance of tissue homeostasis and immune tolerance [94,95].
Lastly, given that cannabinoids were reported to be associated with decreased SIV and HIV viral replication [19,48,96,97,98,99], and since an observational study previously reported that cannabis use was associated with intermittent HIV shedding in the semen of men who have sex with men on ART [100], we explored if oral cannabinoids impact HIV reservoir size. While we found no significant changes in HIV DNA/RNA levels neither in blood or semen, larger clinical trials are required to decipher the impact of oral cannabinoids on HIV reservoir dynamics along with the changes in immune cells.
It is evident that there are several limitations in this pilot clinical trial. First, our initial target sample size included 26 participants randomized in a 1:1 ratio in two arms. Because the production of oral cannabinoid capsules used in this study was permanently discontinued, we were forced to stop the recruitment process prematurely [50]. Furthermore, 2 of the 10 participants were withdrawn from the trial due to adverse events [50]. Thus, unfortunately we lacked the statistical power to compare the different cannabinoid formulations (CBD:THC versus CBD-only), and therefore we had to pool the results of both study arms. However, since each study participant underwent a personalized tolerability titration, and the fact that each study participant was in her/his own control to assess the overtime changes, we believe a nonparametric paired statistical comparison between each two time-points was the only way to analyze our overtime data based on the limited sample size. Potential confounding effects related to the open-label design of this study may also have occurred. Some reports show that an open-label study design has been associated with increased chance of bias and potential overestimation of treatment-related adverse event occurrence rate compared to blinded studies [101,102]. Indeed, since the participants were aware of the formulation they were taking and the possible associated side effects, it might have affected their ability to reach and/or maintain the highest target dose until the end of the treatment period, leading to individual-dependent dosage variations in cannabinoid intake in this study. Moreover, oral cannabinoids have low bioavailability and there is high variability across studies depending on the vehicle of administration [103]. Cannabinoid bioavailability is also heavily impacted by food intake, and consumption of CBD with a high-fat meal can increase its bioavailability [104,105]. Another limitation is the fact that our observations are made on circulating immune cells from peripheral blood, which could underestimate the real effects of cannabinoids on tissue-resident immune cells in the GALT, one of the main sites permanently affected by HIV. Future studies using distal colon pinch biopsies could address the effect of cannabinoids on cytokine and immune cell dynamics in the gastrointestinal tract [106,107]. Finally, we were not able to provide a measure of the replication-competent HIV reservoir. The new intact proviral DNA assay (IPDA) would be a good strategy to measure and discriminate between intact replication-competent, defective, and total HIV genomes. However, we previously showed that total and integrated HIV DNA measures and IPDA measures correlate well with each other [108].
5. Conclusions
In summary, our findings further support the anti-inflammatory effects of oral cannabinoid capsules in PWH on ART. Oral cannabinoids show promise in improving the gut mucosal barrier, alleviating immune activation, and decreasing T-cell exhaustion and immunosenescence. These findings constitute supplemental evidence of cannabinoids’ therapeutic potential in combination with ART for PWH that could help reduce the rates of non-AIDS-related morbidity and mortality in this population. These findings suggest that larger clinical trials are warranted to further evaluate the role of oral cannabinoids as a strategy to reduce systemic inflammation and gut microbial translocation in PWH on ART.
Acknowledgments
First, we thank the participants for their participation in this study. We also thank the staff at the CTN, database management and study monitoring, and all CVIS/MUHC research and clinical staff. We are grateful to the Tilray staff, and the Research Centre for Cannabis at McGill University. This work has been supported by a team grant from the Canadian Institutes of Health Research (CIHR) Canadian HIV Trials Network (CTN PT028), and, the Lotte & John Hecht Memorial Foundation. Tilray provided study capsules as an in-kind service but did not provide study funding.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells12141811/s1, Table S1. List of antibodies used for ex vivo phenotyping of T-cell monocytes and dendritic cell subsets. Material S1. Detailed methodology for total HIV DNA and cell-associated and cell-free RNA quantification. Figure S1: Examples of gating on CD4 T-cell memory subsets (naïve: CD45RA+CD28+CCR7+, central memory: CD45RA−CD28+CCR7+, transitional memory: CD45RA−CD28+CCR7−, effector memory: CD45RA−CD28−CCR7−, and terminally differentiated: CD45RA+CD28−CCR7−) and markers involved in CD4 T-cell function. Figure S2: Examples of gating on (a) CD4 helper T-cell subsets (Th17: CD45RA−CCR4+CCR6+ CXCR3−, Th1-Th17: CD45RA−CCR4−CCR6+CXCR3+, Th2: CD45RA−CCR4+CCR6−CXCR3−, Th1: CD45RA−CCR4−CCR6−CXCR3+) and (b) regulatory T-cells (CD25hi CD127lo FoxP3+). Figure S3: Examples of gating on markers involved in CD8 T-cell function. (c) Examples of gating on CD8 T-cell memory subsets (naïve: CD45RA+CD28+CCR7+, central memory: CD45RA−CD28+CCR7+, transitional memory: CD45RA−CD28+CCR7−, effector memory: CD45RA−CD28−CCR7−, and terminally differentiated: CD45RA+CD28−CCR7−) and regulatory CD8 T-cells (FoxP3+). Figure S4: (Examples of gating on classical (CD14++CD16−), intermediates (CD14+CD16+) and non-classical (CD14−CD16++) monocytes and myeloid (HLA-DR+CD123−CD11c+) and plasmacytoid (HLA-DR+CD123+CD11c−) dendritic cells. Figure S5: Examples of gating on markers of cell migration and function in (a) classical, (b) intermediate, and (c) non-classical monocytes.
Author Contributions
Conceptualization: M.-A.J. and C.T.C.; participant recruitment and data acquisition: J.N., C.V., F.B., C.T.C., G.S., M.B.K., B.L., J.-P.R., S.M. and E.M.; methodology: C.T.C., M.-A.J., J.N., J.S., T.L., S.M. and E.M.; software: J.N., J.S. and T.L.; validation: J.S. and T.L.; formal analysis: R.-S.M.B., E.C., A.P., Y.A., A.Y., J.S., T.L., N.C., C.T.C. and M.-A.J.; investigation: R.-S.M.B., E.C., C.V., F.B., B.L., S.S., C.T.C., G.S., M.B.K. and J.-P.R.; resources: C.V., F.B., B.L., S.S., J.-P.R., G.S., M.B.K., E.M., S.M., C.T.C., M.-A.J. and J.N.; data curation: R.-S.M.B., S.S., E.C., A.P., N.C., Y.A., A.Y., C.T.C. and M.-A.J.; writing—original draft preparation: R.-S.M.B., Y.A., C.T.C. and M.-A.J.; writing—review and editing: all authors; supervision: C.T.C., M.-A.J., N.C., J.S. and J.N.; project administration: C.T.C., F.B., C.V., J.N. and M.-A.J.; funding acquisition: M.-A.J. and C.T.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of McGill University Health Centre (protocol code #2018-4336).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
Anonymized data may be available upon reasonable request of the author and the CTN.
Conflicts of Interest
G.S. has acted as speaker for Merck, Gilead, Abbvie, Pfizer, Novonordisk, served as an advisory board member for Merck, Novartis, Pfizer, Gilead and Intercept and has received unrestricted research funding from Theratecnologies. C.T.C. has served on advisory boards for Viiv Healthcare and Gilead, and received grant support from Merck, Gilead, Viiv and Tilray Inc. She has also received travel support to attend conferences from Gilead and Viiv Healthcare. B.L. has served on advisory boards for Viiv Healthcare, Gilead and Merck, and received grant support from Merck, Gilead, Viiv. He has also received travel support to attend conferences from Gilead.
Funding Statement
This research was funded by Canadian Institutes of Health Research (CIHR) Canadian HIV Trials Network, grant number (CTN PT028) and the Lotte & John Hecht Memorial Foundation R.-S.M.B. is supported by FRQ-S and CIHR postdoctoral fellowships. È.C. is supported by a CIHR MSc scholarship. A.Y. was supported by an FRQ-S doctoral scholarship. G.S. holds a FRSQ-S Chercheur Boursier Clinicien Senior career award. JPR holds the McGill University Louis Lowenstein Chair in Hematology and Oncology. MK is the holder of a Tier 1 Canada Research Chair in Clinical and Epidemiologic Studies of Chronic Viral Infections in Vulnerable Populations. BL is supported by 2 career awards: a Senior Salary Award from Fonds de recherche du Québec–Santé (FRQS) (#311200) and the LE 250 (from the Québec Ministry of Health for researchers in Family Medicine), and is the holder of the Patient-Oriented Research (CIHR SPOR) Mentorship Chair in Innovative Clinical Trials. NC is a recipient of an FRQ-S senior Chercheur Boursier career award. C.T.C. is a recipient of an FRQ-S Junior 2 Chercheur Boursier Clinicien career award. M.-A.J. holds the tier 2 CIHR Canada Research Chair in Immuno-Virology. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Deeks S.G. Immune dysfunction, inflammation, and accelerated aging in patients on antiretroviral therapy. Top. HIV Med. 2009;17:118–123. [PubMed] [Google Scholar]
- 2.Deeks S.G. HIV infection, inflammation, immunosenescence, and aging. Annu. Rev. Med. 2011;62:141–155. doi: 10.1146/annurev-med-042909-093756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zicari S., Sessa L., Cotugno N., Ruggiero A., Morrocchi E., Concato C., Rocca S., Zangari P., Manno E.C., Palma P. Immune Activation, Inflammation, and Non-AIDS Co-Morbidities in HIV-Infected Patients under Long-Term ART. Viruses. 2019;11:200. doi: 10.3390/v11030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yero A., Bouassa R.M., Ancuta P., Estaquier J., Jenabian M.A. Immuno-metabolic control of the balance between Th17-polarized and regulatory T-cells during HIV infection. Cytokine Growth Factor. Rev. 2023;69:1–13. doi: 10.1016/j.cytogfr.2023.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Deeks S.G., Tracy R., Douek D.C. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39:633–645. doi: 10.1016/j.immuni.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt P.W., Sinclair E., Rodriguez B., Shive C., Clagett B., Funderburg N., Robinson J., Huang Y., Epling L., Martin J.N., et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J. Infect. Dis. 2014;210:1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenabian M.A., El-Far M., Vyboh K., Kema I., Costiniuk C.T., Thomas R., Baril J.G., LeBlanc R., Kanagaratham C., Radzioch D., et al. Immunosuppressive Tryptophan Catabolism and Gut Mucosal Dysfunction Following Early HIV Infection. J. Infect. Dis. 2015;212:355–366. doi: 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- 8.Yero A., Farnos O., Rabezanahary H., Racine G., Estaquier J., Jenabian M.A. Differential Dynamics of Regulatory T-Cell and Th17 Cell Balance in Mesenteric Lymph Nodes and Blood following Early Antiretroviral Initiation during Acute Simian Immunodeficiency Virus Infection. J. Virol. 2019;93:e00371-19. doi: 10.1128/JVI.00371-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sokoya T., Steel H.C., Nieuwoudt M., Rossouw T.M. HIV as a Cause of Immune Activation and Immunosenescence. Mediators Inflamm. 2017;2017:6825493. doi: 10.1155/2017/6825493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin G.E., Pace M., Shearer F.M., Zilber E., Hurst J., Meyerowitz J., Thornhill J.P., Lwanga J., Brown H., Robinson N., et al. Levels of Human Immunodeficiency Virus DNA Are Determined Before ART Initiation and Linked to CD8 T-Cell Activation and Memory Expansion. J. Infect. Dis. 2020;221:1135–1145. doi: 10.1093/infdis/jiz563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoury G., Fromentin R., Solomon A., Hartogensis W., Killian M., Hoh R., Somsouk M., Hunt P.W., Girling V., Sinclair E., et al. Human Immunodeficiency Virus Persistence and T-Cell Activation in Blood, Rectal, and Lymph Node Tissue in Human Immunodeficiency Virus-Infected Individuals Receiving Suppressive Antiretroviral Therapy. J. Infect. Dis. 2017;215:911–919. doi: 10.1093/infdis/jix039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anil S.M., Shalev N., Vinayaka A.C., Nadarajan S., Namdar D., Belausov E., Shoval I., Mani K.A., Mechrez G., Koltai H. Cannabis compounds exhibit anti-inflammatory activity in vitro in COVID-19-related inflammation in lung epithelial cells and pro-inflammatory activity in macrophages. Sci. Rep. 2021;11:1462. doi: 10.1038/s41598-021-81049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeMarino C., Cowen M., Khatkar P., Cotto B., Branscome H., Kim Y., Sharif S.A., Agbottah E.T., Zhou W., Costiniuk C.T., et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells. 2022;11:723. doi: 10.3390/cells11040723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriquez J.E., Rizzo M.D., Schulz M.A., Crawford R.B., Gulick P., Kaminski N.E. Δ9-Tetrahydrocannabinol Suppresses Secretion of IFNα by Plasmacytoid Dendritic Cells From Healthy and HIV-Infected Individuals. J. Acquir. Immune Defic. Syndr. 2017;75:588–596. doi: 10.1097/qai.0000000000001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suryavanshi S.V., Zaiachuk M., Pryimak N., Kovalchuk I., Kovalchuk O. Cannabinoids Alleviate the LPS-Induced Cytokine Storm via Attenuating NLRP3 Inflammasome Signaling and TYK2-Mediated STAT3 Signaling Pathways In Vitro. Cells. 2022;11:1391. doi: 10.3390/cells11091391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzo M.D., Crawford R.B., Bach A., Sermet S., Amalfitano A., Kaminski N.E. Δ(9)-Tetrahydrocannabinol Suppresses Monocyte-Mediated Astrocyte Production of Monocyte Chemoattractant Protein 1 and Interleukin-6 in a Toll-Like Receptor 7-Stimulated Human Coculture. J. Pharmacol. Exp. Ther. 2019;371:191–201. doi: 10.1124/jpet.119.260661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmona-Hidalgo B., González-Mariscal I., García-Martín A., Prados M.E., Ruiz-Pino F., Appendino G., Tena-Sempere M., Muñoz E. Δ9-Tetrahydrocannabinolic Acid markedly alleviates liver fibrosis and inflammation in mice. Phytomedicine. 2021;81:153426. doi: 10.1016/j.phymed.2020.153426. [DOI] [PubMed] [Google Scholar]
- 18.Chandra L.C., Kumar V., Torben W., Vande Stouwe C., Winsauer P., Amedee A., Molina P.E., Mohan M. Chronic administration of Δ9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J. Virol. 2015;89:1168–1181. doi: 10.1128/jvi.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar V., Torben W., Kenway C.S., Schiro F.R., Mohan M. Longitudinal Examination of the Intestinal Lamina Propria Cellular Compartment of Simian Immunodeficiency Virus-Infected Rhesus Macaques Provides Broader and Deeper Insights into the Link between Aberrant MicroRNA Expression and Persistent Immune Activation. J. Virol. 2016;90:5003–5019. doi: 10.1128/jvi.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar V., Torben W., Mansfield J., Alvarez X., Vande Stouwe C., Li J., Byrareddy S.N., Didier P.J., Pahar B., Molina P.E., et al. Cannabinoid Attenuation of Intestinal Inflammation in Chronic SIV-Infected Rhesus Macaques Involves T Cell Modulation and Differential Expression of Micro-RNAs and Pro-inflammatory Genes. Front. Immunol. 2019;10:914. doi: 10.3389/fimmu.2019.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDew-White M., Lee E., Alvarez X., Sestak K., Ling B.J., Byrareddy S.N., Okeoma C.M., Mohan M. Cannabinoid control of gingival immune activation in chronically SIV-infected rhesus macaques involves modulation of the indoleamine-2,3-dioxygenase-1 pathway and salivary microbiome. EBioMedicine. 2022;75:103769. doi: 10.1016/j.ebiom.2021.103769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couch D.G., Cook H., Ortori C., Barrett D., Lund J.N., O’Sullivan S.E. Palmitoylethanolamide and Cannabidiol Prevent Inflammation-induced Hyperpermeability of the Human Gut In Vitro and In Vivo-A Randomized, Placebo-controlled, Double-blind Controlled Trial. Inflamm. Bowel Dis. 2019;25:1006–1018. doi: 10.1093/ibd/izz017. [DOI] [PubMed] [Google Scholar]
- 23.Cocetta V., Governa P., Borgonetti V., Tinazzi M., Peron G., Catanzaro D., Berretta M., Biagi M., Manetti F., Dall’Acqua S., et al. Cannabidiol Isolated From Cannabis sativa L. Protects Intestinal Barrier From In Vitro Inflammation and Oxidative Stress. Front. Pharmacol. 2021;12:641210. doi: 10.3389/fphar.2021.641210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozela E., Juknat A., Kaushansky N., Rimmerman N., Ben-Nun A., Vogel Z. Cannabinoids decrease the th17 inflammatory autoimmune phenotype. J. Neuroimmune Pharmacol. 2013;8:1265–1276. doi: 10.1007/s11481-013-9493-1. [DOI] [PubMed] [Google Scholar]
- 25.Berg B.B., Soares J.S., Paiva I.R., Rezende B.M., Rachid M.A., Cau S.B.A., Romero T.R.L., Pinho V., Teixeira M.M., Castor M. Cannabidiol Enhances Intestinal Cannabinoid Receptor Type 2 Receptor Expression and Activation Increasing Regulatory T Cells and Reduces Murine Acute Graft-versus-Host Disease without Interfering with the Graft-versus-Leukemia Response. J. Pharmacol. Exp. Ther. 2021;377:273–283. doi: 10.1124/jpet.120.000479. [DOI] [PubMed] [Google Scholar]
- 26.McDew-White M., Lee E., Premadasa L.S., Alvarez X., Okeoma C.M., Mohan M. Cannabinoids modulate the microbiota-gut-brain axis in HIV/SIV infection by reducing neuroinflammation and dysbiosis while concurrently elevating endocannabinoid and indole-3-propionate levels. J. Neuroinflammation. 2023;20:62. doi: 10.1186/s12974-023-02729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaddour H., McDew-White M., Madeira M.M., Tranquille M.A., Tsirka S.E., Mohan M., Okeoma C.M. Chronic delta-9-tetrahydrocannabinol (THC) treatment counteracts SIV-induced modulation of proinflammatory microRNA cargo in basal ganglia-derived extracellular vesicles. J. Neuroinflammation. 2022;19:225. doi: 10.1186/s12974-022-02586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopcho S., McDew-White M., Naushad W., Mohan M., Okeoma C.M. Alterations in Abundance and Compartmentalization of miRNAs in Blood Plasma Extracellular Vesicles and Extracellular Condensates during HIV/SIV Infection and Its Modulation by Antiretroviral Therapy (ART) and Delta-9-Tetrahydrocannabinol (Delta(9)-THC) Viruses. 2023;15:623. doi: 10.3390/v15030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pak K., Kantonen T., Pekkarinen L., Nuutila P., Nummenmaa L. Association of CNR1 gene and cannabinoid 1 receptor protein in the human brain. J. Neurosci. Res. 2023;101:327–337. doi: 10.1002/jnr.25149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabon W., Rheims S., Smith J., Bodennec J., Belmeguenai A., Bezin L. CB2 receptor in the CNS: From immune and neuronal modulation to behavior. Neurosci. Biobehav. Rev. 2023;150:105226. doi: 10.1016/j.neubiorev.2023.105226. [DOI] [PubMed] [Google Scholar]
- 31.Stanzani A., Galiazzo G., Giancola F., Tagliavia C., De Silva M., Pietra M., Fracassi F., Chiocchetti R. Localization of cannabinoid and cannabinoid related receptors in the cat gastrointestinal tract. Histochem. Cell Biol. 2020;153:339–356. doi: 10.1007/s00418-020-01854-0. [DOI] [PubMed] [Google Scholar]
- 32.Mboumba Bouassa R.S., Sebastiani G., Di Marzo V., Jenabian M.A., Costiniuk C.T. Cannabinoids and Chronic Liver Diseases. Int. J. Mol. Sci. 2022;23:9423. doi: 10.3390/ijms23169423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzo M.D., Henriquez J.E., Blevins L.K., Bach A., Crawford R.B., Kaminski N.E. Targeting Cannabinoid Receptor 2 on Peripheral Leukocytes to Attenuate Inflammatory Mechanisms Implicated in HIV-Associated Neurocognitive Disorder. J. Neuroimmune Pharmacol. 2020;15:780–793. doi: 10.1007/s11481-020-09918-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costiniuk C.T., Jenabian M.A. Cannabinoids and inflammation: Implications for people living with HIV. Aids. 2019;33:2273–2288. doi: 10.1097/qad.0000000000002345. [DOI] [PubMed] [Google Scholar]
- 35.Hsu D.C., Sereti I. Serious Non-AIDS Events: Therapeutic Targets of Immune Activation and Chronic Inflammation in HIV Infection. Drugs. 2016;76:533–549. doi: 10.1007/s40265-016-0546-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zgair A., Lee J.B., Wong J.C.M., Taha D.A., Aram J., Di Virgilio D., McArthur J.W., Cheng Y.K., Hennig I.M., Barrett D.A., et al. Oral administration of cannabis with lipids leads to high levels of cannabinoids in the intestinal lymphatic system and prominent immunomodulation. Sci. Rep. 2017;7:14542. doi: 10.1038/s41598-017-15026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Canada H. Understanding the New Access to Cannabis for Medical Purposes Regulations. 2016. [(accessed on 7 July 2023)]. Available online: https://www.canada.ca/en/health-canada/services/publications/drugs-health-products/understanding-new-access-to-cannabis-for-medical-purposes-regulations.html.
- 38.Furler M.D., Einarson T.R., Millson M., Walmsley S., Bendayan R. Medicinal and recreational marijuana use by patients infected with HIV. AIDS Patient Care STDS. 2004;18:215–228. doi: 10.1089/108729104323038892. [DOI] [PubMed] [Google Scholar]
- 39.Harris G.E., Dupuis L., Mugford G.J., Johnston L., Haase D., Page G., Haldane H., Harris N., Midodzi W.K., Dow G. Patterns and correlates of cannabis use among individuals with HIV/AIDS in Maritime Canada. Can. J. Infect. Dis. Med. Microbiol. 2014;25:e1–e7. doi: 10.1155/2014/301713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawke L.D., Henderson J. Legalization of cannabis use in Canada: Impacts on the cannabis use profiles of youth seeking services for substance use. J. Subst. Abuse Treat. 2021;126:108340. doi: 10.1016/j.jsat.2021.108340. [DOI] [PubMed] [Google Scholar]
- 41.Ellis R.J., Peterson S.N., Li Y., Schrier R., Iudicello J., Letendre S., Morgan E., Tang B., Grant I., Cherner M. Recent cannabis use in HIV is associated with reduced inflammatory markers in CSF and blood. Neurol. Neuroimmunol. Neuroinflamm. 2020;7 doi: 10.1212/nxi.0000000000000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manuzak J.A., Gott T.M., Kirkwood J.S., Coronado E., Hensley-McBain T., Miller C., Cheu R.K., Collier A.C., Funderburg N.T., Martin J.N., et al. Heavy Cannabis Use Associated With Reduction in Activated and Inflammatory Immune Cell Frequencies in Antiretroviral Therapy-Treated Human Immunodeficiency Virus-Infected Individuals. Clin. Infect. Dis. 2018;66:1872–1882. doi: 10.1093/cid/cix1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizzo M.D., Crawford R.B., Henriquez J.E., Aldhamen Y.A., Gulick P., Amalfitano A., Kaminski N.E. HIV-infected cannabis users have lower circulating CD16+ monocytes and IFN-γ-inducible protein 10 levels compared with nonusing HIV patients. Aids. 2018;32:419–429. doi: 10.1097/qad.0000000000001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson C.W., Campbell L.M., Sun-Suslow N., Hong S., Umlauf A., Ellis R.J., Iudicello J.E., Letendre S., Marcotte T.D., Heaton R.K., et al. Daily Cannabis Use is Associated With Lower CNS Inflammation in People With HIV. J. Int. Neuropsychol. Soc. 2021;27:661–672. doi: 10.1017/s1355617720001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin L., Dinasarapu A.R., Borkar S.A., Chang K.F., De Paris K., Kim-Chang J.J., Sleasman J.W., Goodenow M.M. Anti-inflammatory effects of recreational marijuana in virally suppressed youth with HIV-1 are reversed by use of tobacco products in combination with marijuana. Retrovirology. 2022;19:10. doi: 10.1186/s12977-022-00594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner S.E., Williams C.M., Iversen L., Whalley B.J. Molecular Pharmacology of Phytocannabinoids. Prog. Chem. Org. Nat. Prod. 2017;103:61–101. doi: 10.1007/978-3-319-45541-9_3. [DOI] [PubMed] [Google Scholar]
- 47.Manuzak J.A., Granche J., Tassiopoulos K., Rower J.E., Knox J.R., Williams D.W., Ellis R.J., Goodkin K., Sharma A., Erlandson K.M. Cannabis Use Is Associated With Decreased Antiretroviral Therapy Adherence Among Older Adults With HIV. Open Forum Infect. Dis. 2023;10:ofac699. doi: 10.1093/ofid/ofac699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahji A., Li Y., Vickers-Smith R., Crystal S., Kerns R.D., Gordon K.S., Macmadu A., Skanderson M., So-Armah K., Sung M.L., et al. Self-Reported Cannabis Use and HIV Viral Control among Patients with HIV Engaged in Care: Results from a National Cohort Study. Int. J. Environ. Res. Public Health. 2022;19:5649. doi: 10.3390/ijerph19095649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chadwick B., Miller M.L., Hurd Y.L. Cannabis Use during Adolescent Development: Susceptibility to Psychiatric Illness. Front. Psychiatry. 2013;4:129. doi: 10.3389/fpsyt.2013.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mboumba Bouassa R.S., Needham J., Nohynek D., Singer J., Lee T., Bobeuf F., Samarani S., Del Balso L., Paisible N., Vertzagias C., et al. Safety and Tolerability of Oral Cannabinoids in People Living with HIV on Long-Term ART: A Randomized, Open-Label, Interventional Pilot Clinical Trial (CTNPT 028) Biomedicines. 2022;10:3168. doi: 10.3390/biomedicines10123168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergamaschi M.M., Queiroz R.H., Zuardi A.W., Crippa J.A. Safety and side effects of cannabidiol, a Cannabis sativa constituent. Curr. Drug Saf. 2011;6:237–249. doi: 10.2174/157488611798280924. [DOI] [PubMed] [Google Scholar]
- 52.Burstein S. Cannabidiol (CBD) and its analogs: A review of their effects on inflammation. Bioorganic Med. Chem. 2015;23:1377–1385. doi: 10.1016/j.bmc.2015.01.059. [DOI] [PubMed] [Google Scholar]
- 53.Burstein S.H., Zurier R.B. Cannabinoids, endocannabinoids, and related analogs in inflammation. AAPS J. 2009;11:109–119. doi: 10.1208/s12248-009-9084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iuvone T., Esposito G., De Filippis D., Scuderi C., Steardo L. Cannabidiol: A promising drug for neurodegenerative disorders? CNS Neurosci. Ther. 2009;15:65–75. doi: 10.1111/j.1755-5949.2008.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leweke F.M., Rohleder C., Gerth C.W., Hellmich M., Pukrop R., Koethe D. Cannabidiol and Amisulpride Improve Cognition in Acute Schizophrenia in an Explorative, Double-Blind, Active-Controlled, Randomized Clinical Trial. Front. Pharmacol. 2021;12:614811. doi: 10.3389/fphar.2021.614811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leweke F.M., Piomelli D., Pahlisch F., Muhl D., Gerth C.W., Hoyer C., Klosterkotter J., Hellmich M., Koethe D. Cannabidiol enhances anandamide signaling and alleviates psychotic symptoms of schizophrenia. Transl. Psychiatry. 2012;2:e94. doi: 10.1038/tp.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thiele E.A., Marsh E.D., French J.A., Mazurkiewicz-Beldzinska M., Benbadis S.R., Joshi C., Lyons P.D., Taylor A., Roberts C., Sommerville K. Cannabidiol in patients with seizures associated with Lennox-Gastaut syndrome (GWPCARE4): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2018;391:1085–1096. doi: 10.1016/S0140-6736(18)30136-3. [DOI] [PubMed] [Google Scholar]
- 58.Wade D.T., Makela P., Robson P., House H., Bateman C. Do cannabis-based medicinal extracts have general or specific effects on symptoms in multiple sclerosis? A double-blind, randomized, placebo-controlled study on 160 patients. Mult. Scler. 2004;10:434–441. doi: 10.1191/1352458504ms1082oa. [DOI] [PubMed] [Google Scholar]
- 59.Costiniuk C.T., Saneei Z., Routy J.P., Margolese S., Mandarino E., Singer J., Lebouché B., Cox J., Szabo J., Brouillette M.J., et al. Oral cannabinoids in people living with HIV on effective antiretroviral therapy: CTN PT028-study protocol for a pilot randomised trial to assess safety, tolerability and effect on immune activation. BMJ Open. 2019;9:e024793. doi: 10.1136/bmjopen-2018-024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Costiniuk C.T., Salahuddin S., Farnos O., Olivenstein R., Pagliuzza A., Orlova M., Schurr E., De Castro C., Bourbeau J., Routy J.P., et al. HIV persistence in mucosal CD4+ T cells within the lungs of adults receiving long-term suppressive antiretroviral therapy. Aids. 2018;32:2279–2289. doi: 10.1097/qad.0000000000001962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vandergeeten C., Fromentin R., Merlini E., Lawani M.B., DaFonseca S., Bakeman W., McNulty A., Ramgopal M., Michael N., Kim J.H., et al. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 2014;88:12385–12396. doi: 10.1128/jvi.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isnard S., Ramendra R., Dupuy F.P., Lin J., Fombuena B., Kokinov N., Kema I., Jenabian M.A., Lebouché B., Costiniuk C.T., et al. Plasma Levels of C-Type Lectin REG3α and Gut Damage in People With Human Immunodeficiency Virus. J. Infect. Dis. 2020;221:110–121. doi: 10.1093/infdis/jiz423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novelli S., Lecuroux C., Goujard C., Reynes J., Villemant A., Blum L., Essat A., Avettand-Fenoel V., Launay O., Molina J.M., et al. Persistence of monocyte activation under treatment in people followed since acute HIV-1 infection relative to participants at high or low risk of HIV infection. EBioMedicine. 2020;62:103129. doi: 10.1016/j.ebiom.2020.103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J., Jochems C., Anderson A.M., Talaie T., Jales A., Madan R.A., Hodge J.W., Tsang K.Y., Liewehr D.J., Steinberg S.M., et al. Soluble CD27-pool in humans may contribute to T cell activation and tumor immunity. J. Immunol. 2013;190:6250–6258. doi: 10.4049/jimmunol.1300022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marafini I., Di Sabatino A., Zorzi F., Monteleone I., Sedda S., Cupi M.L., Antenucci C., Biancheri P., Giuffrida P., Di Stefano M., et al. Serum regenerating islet-derived 3-alpha is a biomarker of mucosal enteropathies. Aliment. Pharmacol. Ther. 2014;40:974–981. doi: 10.1111/apt.12920. [DOI] [PubMed] [Google Scholar]
- 66.Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., Hooper L.V. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cuddihey H., MacNaughton W.K., Sharkey K.A. Role of the Endocannabinoid System in the Regulation of Intestinal Homeostasis. Cell Mol. Gastroenterol. Hepatol. 2022;14:947–963. doi: 10.1016/j.jcmgh.2022.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alhamoruni A., Lee A.C., Wright K.L., Larvin M., O’Sullivan S.E. Pharmacological effects of cannabinoids on the Caco-2 cell culture model of intestinal permeability. J. Pharmacol. Exp. Ther. 2010;335:92–102. doi: 10.1124/jpet.110.168237. [DOI] [PubMed] [Google Scholar]
- 69.Alhamoruni A., Wright K.L., Larvin M., O’Sullivan S.E. Cannabinoids mediate opposing effects on inflammation-induced intestinal permeability. Br. J. Pharmacol. 2012;165:2598–2610. doi: 10.1111/j.1476-5381.2011.01589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taschler U., Hasenoehrl C., Storr M., Schicho R. Cannabinoid Receptors in Regulating the GI Tract: Experimental Evidence and Therapeutic Relevance. Handb. Exp. Pharmacol. 2017;239:343–362. doi: 10.1007/164_2016_105. [DOI] [PubMed] [Google Scholar]
- 71.Wiley M.B., DiPatrizio N.V. Diet-Induced Gut Barrier Dysfunction Is Exacerbated in Mice Lacking Cannabinoid 1 Receptors in the Intestinal Epithelium. Int. J. Mol. Sci. 2022;23:10549. doi: 10.3390/ijms231810549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oláh A., Szekanecz Z., Bíró T. Targeting Cannabinoid Signaling in the Immune System: “High”-ly Exciting Questions, Possibilities, and Challenges. Front. Immunol. 2017;8:1487. doi: 10.3389/fimmu.2017.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pellati F., Borgonetti V., Brighenti V., Biagi M., Benvenuti S., Corsi L. Cannabis sativa L. and Nonpsychoactive Cannabinoids: Their Chemistry and Role against Oxidative Stress, Inflammation, and Cancer. Biomed. Res. Int. 2018;2018:1691428. doi: 10.1155/2018/1691428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mehrpouya-Bahrami P., Chitrala K.N., Ganewatta M.S., Tang C., Murphy E.A., Enos R.T., Velazquez K.T., McCellan J., Nagarkatti M., Nagarkatti P. Blockade of CB1 cannabinoid receptor alters gut microbiota and attenuates inflammation and diet-induced obesity. Sci. Rep. 2017;7:15645. doi: 10.1038/s41598-017-15154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skinner C.M., Nookaew I., Ewing L.E., Wongsurawat T., Jenjaroenpun P., Quick C.M., Yee E.U., Piccolo B.D., ElSohly M., Walker L.A., et al. Potential Probiotic or Trigger of Gut Inflammation—The Janus-Faced Nature of Cannabidiol-Rich Cannabis Extract. J. Diet. Suppl. 2020;17:543–560. doi: 10.1080/19390211.2020.1761506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grider J.R., Mahavadi S., Li Y., Qiao L.Y., Kuemmerle J.F., Murthy K.S., Martin B.R. Modulation of motor and sensory pathways of the peristaltic reflex by cannabinoids. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G539–G549. doi: 10.1152/ajpgi.00064.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henshaw F.R., Dewsbury L.S., Lim C.K., Steiner G.Z. The Effects of Cannabinoids on Pro- and Anti-Inflammatory Cytokines: A Systematic Review of In Vivo Studies. Cannabis Cannabinoid Res. 2021;6:177–195. doi: 10.1089/can.2020.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christensen-Quick A., Massanella M., Frick A., Rawlings S.A., Spina C., Vargas-Meneses M., Schrier R., Nakazawa M., Anderson C., Gianella S. Subclinical Cytomegalovirus DNA Is Associated with CD4 T Cell Activation and Impaired CD8 T Cell CD107a Expression in People Living with HIV despite Early Antiretroviral Therapy. J. Virol. 2019;93:10-1128. doi: 10.1128/jvi.00179-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maidji E., Somsouk M., Rivera J.M., Hunt P.W., Stoddart C.A. Replication of CMV in the gut of HIV-infected individuals and epithelial barrier dysfunction. PLoS Pathog. 2017;13:e1006202. doi: 10.1371/journal.ppat.1006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fenwick C., Joo V., Jacquier P., Noto A., Banga R., Perreau M., Pantaleo G. T-cell exhaustion in HIV infection. Immunol. Rev. 2019;292:149–163. doi: 10.1111/imr.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim A., French M.A., Price P. CD4+ and CD8+ T cells expressing FoxP3 in HIV-infected patients are phenotypically distinct and influenced by disease severity and antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 2009;51:248–257. doi: 10.1097/QAI.0b013e3181a74fad. [DOI] [PubMed] [Google Scholar]
- 82.Lim A., Tan D., Price P., Kamarulzaman A., Tan H.Y., James I., French M.A. Proportions of circulating T cells with a regulatory cell phenotype increase with HIV-associated immune activation and remain high on antiretroviral therapy. Aids. 2007;21:1525–1534. doi: 10.1097/QAD.0b013e32825eab8b. [DOI] [PubMed] [Google Scholar]
- 83.Nigam P., Velu V., Kannanganat S., Chennareddi L., Kwa S., Siddiqui M., Amara R.R. Expansion of FOXP3+ CD8 T cells with suppressive potential in colorectal mucosa following a pathogenic simian immunodeficiency virus infection correlates with diminished antiviral T cell response and viral control. J. Immunol. 2010;184:1690–1701. doi: 10.4049/jimmunol.0902955. [DOI] [PubMed] [Google Scholar]
- 84.Yero A., Shi T., Routy J.P., Tremblay C., Durand M., Costiniuk C.T., Jenabian M.A. FoxP3(+) CD8 T-cells in acute HIV infection and following early antiretroviral therapy initiation. Front. Immunol. 2022;13:962912. doi: 10.3389/fimmu.2022.962912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulze Zur Wiesch J., Thomssen A., Hartjen P., Tóth I., Lehmann C., Meyer-Olson D., Colberg K., Frerk S., Babikir D., Schmiedel S., et al. Comprehensive analysis of frequency and phenotype of T regulatory cells in HIV infection: CD39 expression of FoxP3+ T regulatory cells correlates with progressive disease. J. Virol. 2011;85:1287–1297. doi: 10.1128/jvi.01758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song J.W., Huang H.H., Zhang C., Yang H.G., Zhang J.Y., Xu R.N., Jin L., Shi M., Wang F.S., Jiao Y.M. Expression of CD39 Is Correlated With HIV DNA Levels in Naïve Tregs in Chronically Infected ART Naïve Patients. Front. Immunol. 2019;10:2465. doi: 10.3389/fimmu.2019.02465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Antonioli L., Pacher P., Vizi E.S., Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol. Med. 2013;19:355–367. doi: 10.1016/j.molmed.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jenabian M.A., Seddiki N., Yatim A., Carriere M., Hulin A., Younas M., Ghadimi E., Kok A., Routy J.P., Tremblay A., et al. Regulatory T cells negatively affect IL-2 production of effector T cells through CD39/adenosine pathway in HIV infection. PLoS Pathog. 2013;9:e1003319. doi: 10.1371/journal.ppat.1003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nikolova M., Carriere M., Jenabian M.A., Limou S., Younas M., Kok A., Hue S., Seddiki N., Hulin A., Delaneau O., et al. CD39/adenosine pathway is involved in AIDS progression. PLoS Pathog. 2011;7:e1002110. doi: 10.1371/journal.ppat.1002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yero A., Shi T., Farnos O., Routy J.P., Tremblay C., Durand M., Tsoukas C., Costiniuk C.T., Jenabian M.A. Dynamics and epigenetic signature of regulatory T-cells following antiretroviral therapy initiation in acute HIV infection. EBioMedicine. 2021;71:103570. doi: 10.1016/j.ebiom.2021.103570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dutertre C.A., Amraoui S., DeRosa A., Jourdain J.P., Vimeux L., Goguet M., Degrelle S., Feuillet V., Liovat A.S., Müller-Trutwin M., et al. Pivotal role of M-DC8⁺ monocytes from viremic HIV-infected patients in TNFα overproduction in response to microbial products. Blood. 2012;120:2259–2268. doi: 10.1182/blood-2012-03-418681. [DOI] [PubMed] [Google Scholar]
- 92.Chayasirisobhon S. Mechanisms of Action and Pharmacokinetics of Cannabis. Perm. J. 2020;25:1–3. doi: 10.7812/tpp/19.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ndhlovu L.C., D’Antoni M.L., Ananworanich J., Byron M.M., Chalermchai T., Sithinamsuwan P., Tipsuk S., Ho E., Slike B.M., Schuetz A., et al. Loss of CCR2 expressing non-classical monocytes are associated with cognitive impairment in antiretroviral therapy-naïve HIV-infected Thais. J. Neuroimmunol. 2015;288:25–33. doi: 10.1016/j.jneuroim.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amodio G., Gregori S. Dendritic cells a double-edge sword in autoimmune responses. Front. Immunol. 2012;3:233. doi: 10.3389/fimmu.2012.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amodio G., Cichy J., Conde P., Matteoli G., Moreau A., Ochando J., Oral B.H., Pekarova M., Ryan E.J., Roth J., et al. Role of myeloid regulatory cells (MRCs) in maintaining tissue homeostasis and promoting tolerance in autoimmunity, inflammatory disease and transplantation. Cancer Immunol. Immunother. 2019;68:661–672. doi: 10.1007/s00262-018-2264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Molina P.E., Winsauer P., Zhang P., Walker E., Birke L., Amedee A., Stouwe C.V., Troxclair D., McGoey R., Varner K., et al. Cannabinoid administration attenuates the progression of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses. 2011;27:585–592. doi: 10.1089/aid.2010.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milloy M.J., Marshall B., Kerr T., Richardson L., Hogg R., Guillemi S., Montaner J.S., Wood E. High-intensity cannabis use associated with lower plasma human immunodeficiency virus-1 RNA viral load among recently infected people who use injection drugs. Drug Alcohol. Rev. 2015;34:135–140. doi: 10.1111/dar.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okafor C.N., Zhou Z., Burrell L.E., 2nd, Kelso N.E., Whitehead N.E., Harman J.S., Cook C.L., Cook R.L. Marijuana use and viral suppression in persons receiving medical care for HIV-infection. Am. J. Drug Alcohol. Abuse. 2017;43:103–110. doi: 10.1080/00952990.2016.1191505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Slawek D.E., Arnsten J., Sohler N., Zhang C., Grossberg R., Stein M., Cunningham C.O. Daily and near-daily cannabis use is associated with HIV viral load suppression in people living with HIV who use cocaine. AIDS Care. 2021;33:1350–1357. doi: 10.1080/09540121.2020.1799922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghosn J., Leruez-Ville M., Blanche J., Delobelle A., Beaudoux C., Mascard L., Lecuyer H., Canestri A., Landman R., Zucman D., et al. HIV-1 DNA levels in peripheral blood mononuclear cells and cannabis use are associated with intermittent HIV shedding in semen of men who have sex with men on successful antiretroviral regimens. Clin. Infect. Dis. 2014;58:1763–1770. doi: 10.1093/cid/ciu187. [DOI] [PubMed] [Google Scholar]
- 101.Trone J.C., Ollier E., Chapelle C., Bertoletti L., Cucherat M., Mismetti P., Magne N., Laporte S. Statistical controversies in clinical research: Limitations of open-label studies assessing antiangiogenic therapies with regard to evaluation of vascular adverse drug events-a meta-analysis. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018;29:803–811. doi: 10.1093/annonc/mdy035. [DOI] [PubMed] [Google Scholar]
- 102.Chan A.W., Tetzlaff J.M., Gotzsche P.C., Altman D.G., Mann H., Berlin J.A., Dickersin K., Hrobjartsson A., Schulz K.F., Parulekar W.R., et al. SPIRIT 2013 explanation and elaboration: Guidance for protocols of clinical trials. BMJ. 2013;346:e7586. doi: 10.1136/bmj.e7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grotenhermen F. Pharmacokinetics and pharmacodynamics of cannabinoids. Clin. Pharmacokinet. 2003;42:327–360. doi: 10.2165/00003088-200342040-00003. [DOI] [PubMed] [Google Scholar]
- 104.Birnbaum A.K., Karanam A., Marino S.E., Barkley C.M., Remmel R.P., Roslawski M., Gramling-Aden M., Leppik I.E. Food effect on pharmacokinetics of cannabidiol oral capsules in adult patients with refractory epilepsy. Epilepsia. 2019;60:1586–1592. doi: 10.1111/epi.16093. [DOI] [PubMed] [Google Scholar]
- 105.Franco V., Gershkovich P., Perucca E., Bialer M. The Interplay Between Liver First-Pass Effect and Lymphatic Absorption of Cannabidiol and Its Implications for Cannabidiol Oral Formulations. Clin. Pharmacokinet. 2020;59:1493–1500. doi: 10.1007/s40262-020-00931-w. [DOI] [PubMed] [Google Scholar]
- 106.Tyler C.J., Guzman M., Lundborg L.R., Yeasmin S., Perez-Jeldres T., Yarur A., Behm B., Dulai P.S., Patel D., Bamias G., et al. Inherent Immune Cell Variation Within Colonic Segments Presents Challenges for Clinical Trial Design. J. Crohn’s Colitis. 2020;14:1364–1377. doi: 10.1093/ecco-jcc/jjaa067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bowcutt R., Malter L.B., Chen L.A., Wolff M.J., Robertson I., Rifkin D.B., Poles M., Cho I., Loke P. Isolation and cytokine analysis of lamina propria lymphocytes from mucosal biopsies of the human colon. J. Immunol. Methods. 2015;421:27–35. doi: 10.1016/j.jim.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Papasavvas E., Azzoni L., Ross B.N., Fair M., Yuan Z., Gyampoh K., Mackiewicz A., Sciorillo A.C., Pagliuzza A., Lada S.M., et al. Intact Human Immunodeficiency Virus (HIV) Reservoir Estimated by the Intact Proviral DNA Assay Correlates With Levels of Total and Integrated DNA in the Blood During Suppressive Antiretroviral Therapy. Clin. Infect. Dis. 2021;72:495–498. doi: 10.1093/cid/ciaa809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized data may be available upon reasonable request of the author and the CTN.