Abstract
ETV6-ABL1 gene fusion is a rare genetic rearrangement in a variety of malignancies, including myeloproliferative neoplasms (MPN), acute lymphoblastic leukemia (ALL), and acute myeloid leukemia (AML). Here, we report the case of a 16-year-old male diagnosed with a MPN, 7 months post-completion of treatment for Burkitt leukaemia. RNA sequencing analysis confirmed the presence of an ETV6-ABL1 fusion transcript, with an intact, in-frame ABL tyrosine–kinase domain. Of note, secondary ETV6-ABL1-rearranged neoplastic diseases have not been reported to date. The patient was started on a tyrosine kinase inhibitor (TKI; imatinib) and, subsequently, underwent a 10/10 matched unrelated haematopoietic stem cell transplant. He is disease-free five years post-transplant. Definitive evidence of the prognostic influence of the ETV6-ABL1 fusion in haematological neoplasms is lacking; however, overall data suggest that it is a poor prognostic factor, particularly in patients with ALL and AML. The presence of this ETV6-ABL1 fusion should be more routinely investigated, especially in patients with a CML-like picture. More routine use of whole-genome and RNA sequencing analyses in clinical diagnostic care, in conjunction with conventional cytogenetics, will facilitate these investigations.
Keywords: pediatric malignancies, oncology, myeloproliferative syndrome
1. Introduction
ETV6-ABL1 gene fusion is a rare genetic rearrangement in a variety of malignancies, including myeloproliferative neoplasms (MPN), acute lymphoblastic leukemia (ALL), and acute myeloid leukemia (AML) [1]. This fusion results in enhanced tyrosine kinase activity of ABL1 and thus neoplastic transformation, producing molecular properties and mirroring the BCR-ABL1 rearrangement [2,3].
Here, we report the case of a 16-year-old male diagnosed with a MPN, 7 months post-completion of treatment for Burkitt leukaemia. Detailed genomic analysis, including whole genome and transcriptome sequencing, was carried out through the SickKids Cancer Sequencing (KiCS) Program to further clarify the diagnosis of the MPN and its relationship to the original Burkitt leukemia.
2. Case Report
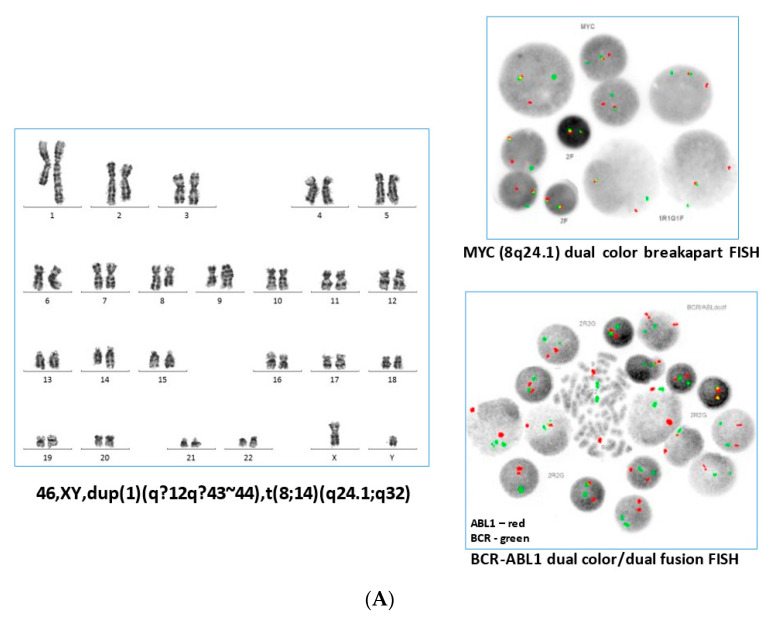
The patient presented with a diffuse petechial rash, bilateral subconjunctival haemorrhages, and haemoptysis, on a background of a history of several months of fatigue, bone pain, and night sweats. In their family history, a maternal grandmother had died of pancreatic cancer at age 67, and a maternal uncle had been diagnosed with childhood leukaemia. Complete blood count (CBC) showed a white blood cell (WBC) count of 101 × 109/L, haemoglobin at 106 g/L, and a platelet count of 12 × 109/L. A peripheral smear showed 75% leukemic blasts. Immunophenotyping results can be found in Supplementary Data S1. Fluorescence in situ hybridization (FISH) analysis with a MYC breakapart FISH probe (Abbott Molecular, Abbott Park, IL, USA) was consistent with the presence of an MYC gene rearrangement in 170/200 (85%) cells, and G-band analysis showed an abnormal male karyotype of 46,XY,dup(1)(q12q43~44),t(8;14)(q24.1;q32)[18]/46,XY[2] (Figure 1A, left and upper right panel). A diagnosis of Burkitt leukaemia was made, and treatment was started according to the institutional standard of care (Supplementary Data S2).
Figure 1.
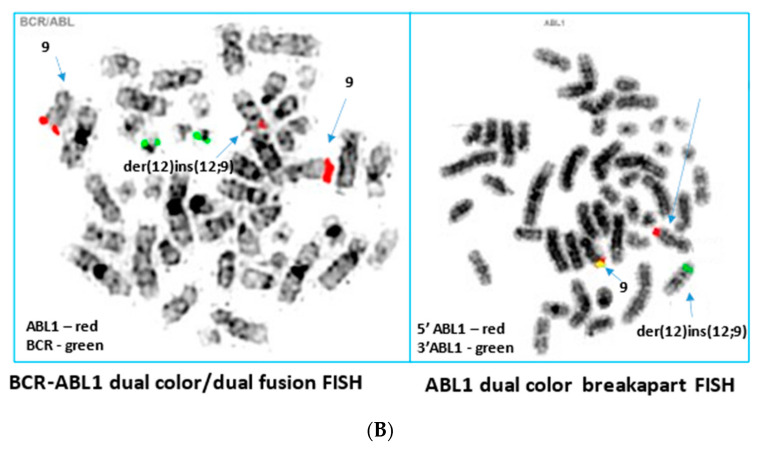
(A) Representative karyotype from the blood sample obtained at initial diagnosis with Burkitt Leukaemia (left panel), demonstrating a t(8;14) and duplication of 1q. Interphase FISH analysis with a dual colour breakapart probe for the MYC gene (8q24.1) showed MYC gene rearrangement in the majority of cells (upper right panel). After the patient subsequently presented with MPN, BCR-ABL1 FISH performed on an archived Burkitt Leukaemia sample yielded normal results (lower right panel). (B) By G-band analysis of the myeloproliferative neoplasm, the karyotype appeared normal. Sequential BCR-ABL1 metaphase FISH analysis on G-banded cells showed insertion of the ABL1 signal into 12p13, with no involvement of the BCR gene (left panel). Metaphase FISH analysis using an ABL1 breakapart FISH probe (right panel) showed the insertion of 3′ABL1 into 12p13. Results of the ETV6 breakapart as well as the subtelomeric 9q and 12p FISH testing were normal, consistent with an insertion mechanism rather than a translocation mechanism in the generation of the ETV6-ABL1 fusion identified by molecular analysis.
A CBC 7 months after the completion of therapy showed WBC 29.2 × 109/L, with a significant left shift, eosinophilia (2.92 × 109/L), monocytosis (4.38 × 109/L), thrombocytopenia (100 × 109/L), and no peripheral blasts. The patient was asymptomatic. Viral workup was negative, and EBV serology was consistent with past infection. There was no travel history. Bone marrow aspirate (BMA) revealed marked hypercellularity with significant eosinophilia and an absence of elevated blasts consistent with a myeloproliferative disorder. Immunophenotyping showed no abnormal cell population. FISH analyses were negative for MYC, PDGFRA (Cytocell OGT, Cambridge), and PDGFRB (Cytocell OGT, Cambridge) gene rearrangements, and the karyotype was normal. Molecular analysis was negative for the BCR-ABL1, FLT3-ITD, JAK2 V617F, and CALR variants.
The leucocytosis increased to 192 × 109/L over 3 months, accompanied by persistent thrombocytopenia (range 50–100 × 109/L). The patient became progressively symptomatic with fatigue, generalized bone pain, as well as significant spontaneous bruising. He was started empirically on Hydroxyurea 25 mg/kg/dose, which normalized his WBC count and improved his symptoms within one month.
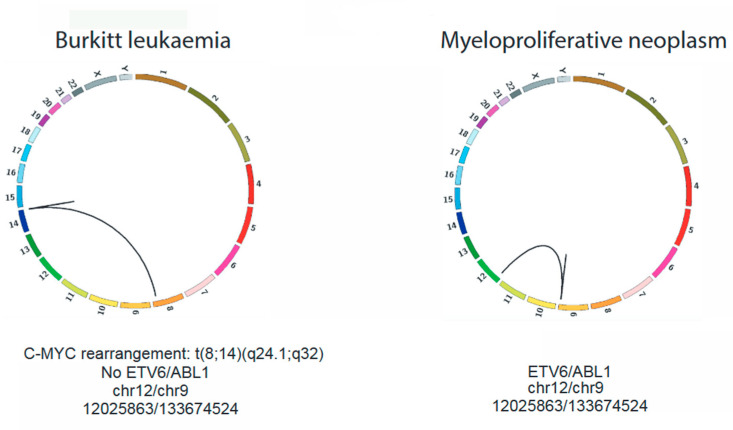
Whole-genome sequencing of his MPN BMA sample was performed on an Illumina HiSeqX platform with a depth of 22X, and RNAseq was performed on Illumina HiSeq 2500, with a depth of 294X. Structural variant analysis from whole-genome sequencing showed a dispersed duplication event of approximately 600 kb from ABL1 to PRRC2B (9q34), inserted into the ETV6 locus (12p13) (Figure 2, right panel). RNAseq analysis confirmed the presence of a corresponding ETV6-ABL1 fusion transcript, with an intact, in-frame ABL tyrosine–kinase domain.
Figure 2.
Circos plots generated from WGS of the Burkitt Leukaemia (left) and MPN (right) samples. Both samples represent diagnostic specimens (peripheral blood for the Burkitt leukaemia and bone marrow aspirate for the MPN). ETV6-ABL1 rearrangement (chr12:12025863-chr9:133674524) was confirmed only in the MPN sample.
Cytogenetic re-evaluation of the MPN BMA sample using sequential FISH analysis with a BCR-ABL1 probe (Cytocell OGT, Cambridge, UK) was suggestive of a cryptic insertion of a portion of the ABL1 locus into one chromosome 12 at 12p13 (Figure 1B, left panel). ABL1 gene rearrangement was present in 193/200 (96.5%) interphase cells. Interphase FISH analysis with a dual colour breakapart probe for the ETV6 locus (Abbott Molecular, Abbott Park, IL, USA) did not show ETV6 gene rearrangement in this patient, and metaphase FISH with subtelomeric probes for 9q and 12p yielded normal results, suggestive of the insertion of ABL1 into ETV6 rather than rearrangement by translocation. FISH analysis with a dual colour breakapart probe for ABL1 (Cytocell OGT, Cambridge, UK) demonstrated that the 3′ portion of ABL1 was inserted into 12p13 on the derivative chromosome 12, with the 5′ portion of ABL1 remaining on the derivative chromosome 9 (Figure 1B, right panel). The karyotype was updated to 46,XY[20]. ish ins(12;9)(p13;q34q34)(12ptel+,3′ABL1+;5′ABL1+,9qtel+)[5].
We were interested in further exploring the relationship between the patient’s two neoplastic processes. The WGS performed on the diagnostic Burkitt leukemia blood sample did not show evidence of the ETV6-ABL1 fusion (Figure 2, left panel). The RNA was not available for analysis. The results of the BCR-ABL1 FISH analysis of 200 stored fixed cells from the Burkitt leukemia sample were also negative for the ABL1 gene rearrangement (Figure 1A, lower right panel). The germline analysis by the WGS of the skin’s biopsy-derived fibroblasts did not reveal any pathogenic variants.
The patient was started on a tyrosine kinase inhibitor (TKI; imatinib) and, subsequently, underwent a 10/10 matched unrelated haematopoietic stem cell transplant. He is disease-free five years post-transplant.
3. Discussion
ETV6-ABL1-rearranged hematologic malignancies are rare entities [1]. To date, 51 cases have been reported [1,4,5]. The fusion was first described in a patient with ALL [6] and, subsequently, in patients with AML [7] and “CML-like” disease [8]. ETV6-ABL1 is found in <1% of all children and adults diagnosed with ALL and is less frequently in AML. The exact incidence of CML-like ETV6-ABL1 neoplasms is unknown, due to the lack of systematic screening [1].
Conventional clinical diagnostic tests often fail to detect this fusion, given that there is no commercial ETV6-ABL1 FISH probe available, and the commonly used BCR-ABL1 or ETV6-RUNX1 probes can miss ETV6-ABL1 [9]. An ABL1 breakapart FISH probe would better enable the detection of all the ABL1 gene rearrangements, including ETV6-ABL1. RT-PCR analysis can also produce false negative results unless the exact breakpoints are known. RNAseq has been increasingly recognized as the most useful diagnostic tool for these cases [1].
The unique presentation of our patient and the fact that ETV6-ABL1 aberration can present in the form of immature lymphoid or myeloid neoplasms raise a number of possible pathogenetic mechanisms. First, it is possible that this patient had independent but simultaneous bi-lineage disease with two clones: the MYC-positive Burkitt leukemia and the ETV6-ABL1 MPN, the former being controlled by the initial Burkitt-targeted chemotherapy, allowing the other partially treated clone with ETV6-ABL1 to expand following therapy completion. Of note, our patient had marked eosinophilia at the presentation of both the Burkitt leukemia (1.01 × 109/L) and the ETV6-ABL1 MPN (4.67 × 109/L)—a finding that is commonly associated with MPNs and not with Burkitt disease [10]. Furthermore, FISH and WGS may have missed a subclone of ETV6-ABL1 MPN in the Burkitt leukemia sample due to low sensitivity; the lack of RNA for in-depth RNAseq did not allow us to confirm or refute this hypothesis. Of note, no pathogenic variants were identified on germline analysis to suggest an underlying cancer predisposition syndrome, which must be considered in the setting of two independent primary tumors.
The second possible molecular mechanism is that the ETV6-ABL1 was an early clonal event that occurred in a common hematopoietic progenitor cell, followed by a second hit in the MYC, resulting in the initial Burkitt leukemia. However, we would have expected the clonal presence of the ETV6-ABL1 fusion in the diagnostic Burkitt leukemia specimen in this circumstance.
Alternatively, the MPN may have been a therapy-induced secondary event associated with the alkylator chemotherapy included in the treatment of Burkitt leukemia. However, secondary ETV6-ABL1-rearranged neoplastic diseases have not been reported to date. Furthermore, such an early secondary malignancy in a paediatric patient after exposure to alkylators would be very unlikely, as the latency is typically several years post-completion of therapy [11].
Definitive evidence of the prognostic influence of the ETV6-ABL1 fusion in haematological neoplasms is lacking; however, overall data suggests that it is a poor prognostic factor, particularly in patients with ALL and AML [1,12]. There also are no definitive data to standardize the treatment for ETV6-ABL1 MPNs, although multiple reports advocate for the early introduction of first and/or second generation TKIs [13,14,15]. Recently, Zhang et al. published a retrospective series of 42 adult patients with a confirmed myeloproliferative neoplasm associated with eosinophilia and a tyrosine kinase gene fusion. The study showed an excellent response to the upfront tyrosine kinase inhibition, regardless of the type of genetic rearrangement; however, none of the patients in this series had an ETV6-ABL1 gene fusion [16]. It should also be noted that monotherapy with TKIs may result in transient responses: Brien et al. reported the case of a 38-year-old man with an ETV6-ABL1-driven chronic myeloid leukemia who initially responded to imatinib but who rapidly deteriorated over the following 2 weeks and required conventional chemotherapy [17]. The mechanisms responsible for TKI resistance remain unclear; Zimmermannova et al. have suggested that an activating mutation in the GNB1 gene could result in a resistance to TKI in the context of ETV6-ABL1-associated neoplasms, via the restoration of signalling through the phosphoinositide-3-kinase (PI3K)/Akt/mTOR and mitogen-activated protein kinase (MAPK) pathways [18]. With regard to other ABL-class fusions, such as BCR-ABL1, it is well-established that point mutations in the kinase domain of BCR-ABL1 are the prevalent mechanisms of resistance to ABL kinase inhibition (60% of cases; termed “BCR-ABL1 dependent resistance”) [19]. In other cases, other factors have been postulated, such as the activation of alternative signalling pathways, including the RAS/MAPK and JAK/STAT pathways, or changes in epigenetic regulation (termed BCR-ABL1 independent resistance) [20,21].
Further detailed molecular studies of other patients with the same genomic abnormalities will provide insight into the pathogenesis and clinical course of ETV6-ABL1-associated neoplasms. The presence of this fusion should be more routinely investigated, especially in patients with a CML-like picture, particularly as it can prompt therapy with a tyrosine kinase inhibitor, as in the patient presented here. More routine use of WGS and RNAseq analysis in clinical diagnostic care, in conjunction with conventional cytogenetics, will facilitate these investigations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/curroncol30070444/s1, Data S1: Immunophenotype of Burkitt’s Leukaemia; Data S2: Chemotherapy received for treatment of Burkitt’s Leukemia, based on institutional standard of care for B-cell Non-Hodgkin Lymphoma, group C.
Author Contributions
Conceptualization: S.R., F.A. and A.V.; Methodology, S.R., F.A., S.D., A.S. (Adam Shlien), D.M., M.S. and A.V.; writing—original draft preparation: S.R., F.A., S.D., M.S. and A.V.; writing—review and editing: S.R., F.A., S.D., K.P.S.L., F.F., S.A., N.A., L.B., J.B., M.A., A.N., K.B., A.S. (Andre Schuh), A.T., D.M., A.S. (Adam Shlien), M.S. and A.V.; funding acquisition: A.S. (Adam Shlien), D.M. and A.V. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical review and approval were waived for this study as per our Hospital’s policy.
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
This being a case report, data are confidential.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The KiCS program is supported by the Garron Family Cancer Centre at the Hospital for Sick Children through funding from the SickKids Foundation.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zaliova M., Moorman A.V., Cazzaniga G., Stanulla M., Harvey R.C., Roberts K.G., Heatley S.L., Loh M.L., Konopleva M., Chen I.M., et al. Characterization of leukemias with ETV6-ABL1 fusion. Haematologica. 2016;101:1082–1093. doi: 10.3324/haematol.2016.144345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song J.S., Shin S.Y., Lee S.T., Kim H.J., Kim S.H. A cryptic ETV6/ABL1 rearrangement represents a unique fluorescence in situ hybridization signal pattern in a patient with B acute lymphoblastic leukemia. Ann. Lab. Med. 2014;34:475–477. doi: 10.3343/alm.2014.34.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malinge S., Monni R., Bernard O., Penard-Lacronique V. Activation of the NF-kappaB pathway by the leukemogenic TEL-Jak2 and TEL-Abl fusion proteins leads to the accumulation of antiapoptotic IAP proteins and involves IKKalpha. Oncogene. 2006;25:3589–3597. doi: 10.1038/sj.onc.1209390. [DOI] [PubMed] [Google Scholar]
- 4.Choi S.I., Jang M.A., Jeong W.J., Jeon B.R., Lee Y.W., Shin H.B., Hong D.S., Lee Y.K. A Case of Chronic Myeloid Leukemia With Rare Variant ETV6/ABL1 Rearrangement. Ann. Lab. Med. 2017;37:77–80. doi: 10.3343/alm.2017.37.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kakadia P.M., Schmidmaier R., Völkl A., Schneider I., Huk N., Schneider S., Panzner G., Neidel U., Fritz B., Spiekermann K., et al. An ETV6-ABL1 fusion in a patient with chronic myeloproliferative neoplasm: Initial response to Imatinib followed by rapid transformation into ALL. Leuk Res. Rep. 2016;6:50–54. doi: 10.1016/j.lrr.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papadopoulos P., Ridge S.A., Boucher C.A., Stocking C., Wiedemann L.M. The novel activation of ABL by fusion to an ets-related gene, TEL. Cancer Res. 1995;55:34–38. [PubMed] [Google Scholar]
- 7.Golub T.R., Goga A., Barker G.F., Afar D.E., McLaughlin J., Bohlander S.K., Rowley J.D., Witte O.N., Gilliland D.G. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol. Cell. Biol. 1996;16:4107–4116. doi: 10.1128/MCB.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreasson P., Johansson B., Carlsson M., Jarlsfelt I., Fioretos T., Mitelman F., Hoglund M. BCR/ABL-negative chronic myeloid leukemia with ETV6/ABL fusion. Genes Chromosom. Cancer. 1997;20:299–304. doi: 10.1002/(SICI)1098-2264(199711)20:3<299::AID-GCC11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 9.Curtis C.E., Grand F.H., Waghorn K., Sahoo T.P., George J., Cross N.C. A novel ETV6-PDGFRB fusion transcript missed by standard screening in a patient with an imatinib responsive chronic myeloproliferative disease. Leukemia. 2007;21:1839–1841. doi: 10.1038/sj.leu.2404728. [DOI] [PubMed] [Google Scholar]
- 10.La Starza R., Trubia M., Testoni N., Ottaviani E., Belloni E., Crescenzi B., Martelli M., Flandrin G., Pelicci P.G., Mecucci C. Clonal eosinophils are a morphologic hallmark of ETV6/ABL1 positive acute myeloid leukemia. Haematologica. 2002;87:789–794. [PubMed] [Google Scholar]
- 11.Turcotte L.M., Neglia J.P., Reulen R.C., Ronckers C.M., van Leeuwen F.E., Morton L.M., Hodgson D.C., Yasui Y., Oeffinger K.C., Henderson T.O. Risk, Risk Factors, and Surveillance of Subsequent Malignant Neoplasms in Survivors of Childhood Cancer: A Review. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018;36:2145–2152. doi: 10.1200/JCO.2017.76.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuna J., Zaliova M., Muzikova K., Meyer C., Lizcova L., Zemanova Z., Brezinova J., Votava F., Marschalek R., Stary J., et al. Acute leukemias with ETV6/ABL1 (TEL/ABL) fusion: Poor prognosis and prenatal origin. Genes Chromosom. Cancer. 2010;49:873–884. doi: 10.1002/gcc.20796. [DOI] [PubMed] [Google Scholar]
- 13.Kawamata N., Dashti A., Lu D., Miller B., Koeffler H.P., Schreck R., Moore S., Ogawa S. Chronic phase of ETV6-ABL1 positive CML responds to imatinib. Genes Chromosom. Cancer. 2008;47:919–921. doi: 10.1002/gcc.20593. [DOI] [PubMed] [Google Scholar]
- 14.Nand R., Bryke C., Kroft S.H., Divgi A., Bredeson C., Atallah E. Myeloproliferative disorder with eosinophilia and ETV6-ABL gene rearrangement: Efficacy of second-generation tyrosine kinase inhibitors. Leuk. Res. 2009;33:1144–1146. doi: 10.1016/j.leukres.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Tiribelli M., Barraco D., Medeot M., Marin L., Ottaviani E., De Marchi F., Damiani D., Fanin R. Long-term efficacy and safety of nilotinib therapy after imatinib failure in eosinophilic myeloproliferative neoplasm and ETV6-ABL rearrangement. Ann. Hematol. 2015;94:1423–1424. doi: 10.1007/s00277-015-2381-4. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Nguyen L., Lu C.M., Wang E., Lauw M.I.S., Ball S., Dong N., Moscinski L., Chan O., Yun S., et al. Clinical response to Upfront Targeted Tyrosine Kinase Inhibitors among Patients with Myeloid/Lymphoid Neoplasms with Eosinophilia and Tyrosine Kinase Gene Fusions. Clin. Lymphoma Myeloma Leuk. 2023;23:e150–e163. doi: 10.1016/j.clml.2022.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Brien S.G., Vieira S., Connors S., Bown N., Chang J., Capdeville R., Melo J.V. Transient response to imatinib mesylate (STI571) in a patient with the ETV6-ABL t(9;12) translocation. Blood. 2022;99:3465–3467. doi: 10.1182/blood.V99.9.3465. [DOI] [PubMed] [Google Scholar]
- 18.Zimmermannova O., Doktorava E., Stuchly J., Kanderova V., Kuzilkova D., Strnad H., Starkova J., Alberich-Jorda M., Falkenburg J.H.F., Trka J., et al. An activating mutation of GNB1 is associated with resistance to tyrosine kinase inhibitors in ETV6-ABL1-positive leukemia. Oncogene. 2017;36:5985–5994. doi: 10.1038/onc.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel A., O’Hare T., Deninger M.W. Mechanisms of resistance to ABL kinase inhibition in CML and the development of next generation ABL kinase inhibitors. Hematol. Oncol. Clin. N. Am. 2017;31:589–612. doi: 10.1016/j.hoc.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves R., Goncalves A.C., Rutella S., Almeida A.M., De Las Rivas J., Trougakos I.P., Sarmento Ribeiro A.B. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Revelance. Cancers. 2021;13:4820. doi: 10.3390/cancers13194820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loscocco F., Visani G., Galimberti S., Curti A., Isidori A. BCR-ABL Indipendent Mechanisms of Resistance in Chronic Myeloid Leukemia. Front. Oncol. 2019;9:939. doi: 10.3389/fonc.2019.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This being a case report, data are confidential.