Abstract
The entire sequence of the 3.5-kb fragment of genomic DNA from Rhodobacter capsulatus which contains the sqr gene and a second complete and two further partial open reading frames has been determined. A correction of the previously published sqr gene sequence (M. Schütz, Y. Shahak, E. Padan, and G. Hauska, J. Biol. Chem. 272:9890–9894, 1997) which in the deduced primary structure of the sulfide-quinone reductase changes four positive into four negative charges and the number of amino acids from 425 to 427 was necessary. The correction has no further bearing on the former sequence analysis. Deletion and interruption strains document that sulfide-quinone reductase is essential for photoautotrophic growth on sulfide. The sulfide-oxidizing enzyme is involved in energy conversion, not in detoxification. Studies with an alkaline phosphatase fusion protein reveal a periplasmic localization of the enzyme. Exonuclease treatment of the fusion construct demonstrated that the C-terminal 38 amino acids of sulfide-quinone reductase were required for translocation. An N-terminal signal peptide for translocation was not found in the primary structure of the enzyme. The possibility that the neighboring open reading frame, which contains a double arginine motif, may be involved in translocation has been excluded by gene deletion (rather, the product of this gene functions in an ATP-binding cassette transporter system, together with the product of one of the other open reading frames). The results lead to the conclusion that the sulfide-quinone reductase of R. capsulatus functions at the periplasmic surface of the cytoplasmic membrane and that this flavoprotein is translocated by a hitherto-unknown mechanism.
Inorganic reduced sulfur compounds serve as electron donors in many phototrophic and chemotrophic bacteria, mostly with sulfate as the major oxidation product (reviewed in references 7 and 13). The initial step in the metabolism of hydrogen sulfide, the most reduced sulfur compound, is the conversion to sulfur or polysulfide, the first observable intermediate. Mainly, two enzymatic systems are considered to be involved in this step: flavocytochrome c and sulfide-quinone reductase. Flavocytochromes c, located in the periplasm of several species, are soluble (7) or membrane-bound (40) enzymes showing sulfide: cytochrome c oxidoreductase activity in vitro. For that reason, it has been suggested that this enzyme plays an essential role in sulfide oxidation in vivo. However, flavocytochrome c does not occur in a variety of sulfide-oxidizing bacteria and seems to be confined to species additionally capable of thiosulfate oxidation (13).
During the last 20 years, evidence for a second, membrane-bound sulfide-oxidizing system in bacteria has accumulated (reviewed in references 13 and 34). It has been identified as sulfide-quinone oxidoreductase (SQR; EC 1.8.5.′) and was first detected in thylakoids of the filamentous cyanobacterium Oscillatoria limnetica (8). SQR activity has been attributed to an inducible, membrane-bound flavoprotein with an apparent molecular mass of approximately 57 kDa in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (2). Meanwhile, the presence of this enzyme or SQR activity in a variety of phototrophic and chemotrophic bacteria has been established (22, 30, 31, 33, 34).
The purple nonsulfur bacterium Rhodobacter capsulatus (DSMZ 155) appears to convert sulfide exclusively into extracellular elemental sulfur (15), and it has been suggested that an SQR, present in membranes of this phototrophic bacterium (33), is the enzyme responsible for this activity. The enzyme has been purified from R. capsulatus and has been characterized (32). Similar to the enzyme of O. limnetica, it is a membrane-bound protein with an apparent molecular mass of 55 kDa by SDS-PAGE showing fluorescence spectra characteristic of flavoproteins. In contrast to the cyanobacterial enzyme, SQR of R. capsulatus is more loosely bound to the membrane. The sqr gene, cloned on a 3.5-kb fragment of genomic DNA of R. capsulatus, was sequenced and functionally expressed in Escherichia coli. The published sequence comprises 1,275 bp encoding a protein of 425 amino acid residues. In the deduced amino acid sequence, three flavin adenine dinucleotide (FAD)-binding domains, which are present in flavocytochrome c and in pyridine nucleotide disulfide oxidoreductases like glutathione reductase, which reduces disulfide bonds, were found. Predictions of secondary structure did not indicate any membrane-spanning or anchoring helix in the SQR. Therefore, the question of which way the enzyme is attached to the membrane and gets into contact with the quinone in the lipid bilayer arose.
Since R. capsulatus deposits sulfur outside the cells, it seems reasonable that the SQR is attached to the periplasmic surface of the cytoplasmic membrane. However, the amino acid sequence of SQR lacks an N-terminal signal peptide for translocation. On the other hand, if SQR is bound to the cytoplasmic surface of the membrane, a transporter which translocates the produced sulfur out of the cytoplasm must exist. Another possibility might be that SQR is not the major sulfide-oxidizing enzyme and that a second sulfide dehydrogenase exists in the periplasmic space. However, if SQR is the only sulfide-oxidizing entity in Rhodobacter, then either a signal for translocation, different from N-terminal signal peptides, must be present in the amino acid sequence of SQR or the enzyme might be cotranslocated with a protein bearing such a signal, as was found for the catalytic subunits of several periplasmic redox proteins (reviewed in reference 4).
In the present study, the sequence analysis of the adjacent regions of the sqr gene on the cloned 3.5-kb PstI fragment, including corrections of minor sequencing errors within the sqr sequence, is presented. A complete open reading frame (ORF2) upstream of the sqr gene and the 3′ end of a putative gene at each end of the cloned region were found (Fig. 1). Results from mutational analysis that indicate the essential role of SQR in sulfide oxidation are presented. Additionally, the functional relationship between the upstream ORF and sqr was investigated, and a possible involvement of a second protein in SQR function will be discussed. The localization of SQR was investigated by translational PhoA fusions.
FIG. 1.
Physical and genetic map of the sqr gene region of R. capsulatus wild-type strain and mutant strains 22/11, 22/17, F14, and A10 as determined by Southern blot analysis. (A) The 3.5-kb PstI fragment of genomic DNA in pUC19 (pUSQR). For complementation studies, the Smr Spcr cassette was inserted into the NotI site, resulting in pUSQRΩ. (B to D) sqr gene region of mutant strains 22/11 (B), 22/17 (C), and F14 and A10 (D). The inserted plasmids pRN4C and pIM101 are indicated by double-headed arrows. The sqr gene and sqr fragments are marked by vertically striped boxes; ORF2 is indicated by obliquely hatched boxes. The fragments sqr-nt and sqr-ct(−75) are described in Materials and Methods; sqr(−75), sqr lacking the 3′-end 75 bp; sqr-ct, 3′ part of sqr behind the EcoRI site; luxA and luxB, the genes encoding luciferase from Vibrio fischeri; oriT, origin of transfer; the npt gene encodes neomycin phosphotransferase, which confers resistance to neomycin and kanamycin. Abbreviations for restriction sites: Bs, BsiWI; E, EcoRI; H, HindIII; M, MscI; No, NotI; Nd, NdeI; P, PstI; S, StuI; S2, SacII; Sm, SmaI; X, XbaI.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. R. capsulatus (DSMZ 155) was cultured under photosynthetic conditions at 30°C in RCV (42), modified by the addition of FeSO4 to a final concentration of 40 μM. Under photoautotrophic conditions, malate was omitted. Sulfide was added as Na2S. For growth on plates, media were supplemented with 0.15% purified agar (Merck, Darmstadt, Germany). Growth under sulfidic atmosphere was performed in an Oxoid anaerobic jar (Unipath Limited, Basingstoke, Hampshire, United Kingdom) briefly flushed with CO2. Sulfidic atmosphere was produced as described by Irgens (17) with 0.1 g of thioacetamide resolved in 1 ml of 0.2 N HCl. Culturing of E. coli was done in Luria-Bertani medium (24) at 37°C.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Rhodobacter capsulatus | ||
| DSMZ 155 | Wild type | Deutsche Sammlung von Mikroorganismen und Zellkulturen |
| F14, A10 | Δ(orf2-sqr) Kmr SQR− | This study |
| 22/11 | sqr::pRN4C Kmr LuxAB+ SQR− | This study |
| 22/17 | sqr::pRN4C Kmr LuxAB+ SQR+ | This study |
| F14n | Kmr Tcr Smr Spcr SQR+; strain F14 transformed with pPSQR | This study |
| F14sn | Kmr Tcr Smr Spcr SQR+; strain F14 transformed with pPStuSQR | This study |
| F14(pPSAPo) | Kmr Tcr Smr Spcr SQR+ PhoA+; strain F14 transformed with pPSAPo | This study |
| Escherichia coli | ||
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm met (DE3) | 35 |
| DH10B | mcrA Δ(mrr-hsdRMS-mcrBC) φ80d lacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK rspL nupG[p3::Km(Am) Amp (Am) Tet] | 14 |
| HB101 | lacY1 galK2 supE44 ara-14 proA2 rpsL20 recA13 xyl-5 mtl-1 hsdS20 mcrB mrr | 5 |
| J-53 | Apr Kmr Tcr RP-4 | 9 |
| Plasmids | ||
| pGEM-T | AprlacZ′ | Promega |
| pGSAP | Apr 2,900-bp sqr::phoA fusion in pGEM-T | This study |
| pHP45Ω | Apr Smr Spcr | 28 |
| pIM101 | KmroriTRP4, ColE1oriluxAB pRL488 without pDU1 | This study |
| pPHU233 | TcrlacZ′ oriTRP4 | |
| pPSQR | Tcr Smr SpcroriT′RP4; 5,400-bp PstI fragment from pUSQRΩ bearing ORF2 and sqr in pPHU233 | This study |
| pPStuSQR | Tcr Smr SpcroriTRP4; 4,500-bp StuI-PstI fragment from pUSQRΩ bearing sqr in pPHU233 | This study |
| pPSAPo | Tcr Smr SpcroriTRP4; 5,350-bp StuI-PstI fragment bearing the sqr::phoA fusion in pPHU233 | This study |
| pPSAP389 | Tcr Smr SpcroriTRP4; 5,236-bp StuI-PstI fragment bearing the truncated sqr(398)::phoA fusion in pPHU233 | This study |
| pPSAP108 | Tcr Smr SpcroriTRP4; 4,379-bp StuI-PstI fragment bearing the sqr(108)::phoA fusion in pPHU233 | This study |
| pRL488 | KmroriTRP4 ColE1ori pDU1 luxAB | 10 |
| pRN4C | KmroriTRP4 ColE1ori luxAB; derivative of pRL488 | This study |
| pRU4C | Kmr Smr SpcroriTRP4 ColE1ori luxAB; derivative of pRL488 | This study |
| pRL528 | Cmr | 9 |
| pTSAP | Apr, in-frame fusion of sqr and phoA of E. coli in pT7-7 | This study |
| pTSAP398 | Apr, in-frame fusion of phoA of E. coli and sqr(398), which encodes the N-terminal 389 amino acid residues of SQR, in pT7-7 | This study |
| pTSAP108 | Apr, in-frame fusion of phoA of E. coli and sqr(108), which encodes the N-terminal 108 amino acid residues of SQR, in pT7-7 | This study |
| pTSQR | Apr; 2,047-bp NdeI-BamHI fragment of pUSQR bearing R. capsulatus sqr in the expression vector pT7-7 | 32 |
| pUSQR | Apr; 3,492-bp PstI fragment bearing R. capsulatus sqr in pUC19 | 32 |
| pUSQRΩ | Apr Smr Spcr pUSQR with the Ω cassette from pHP45Ω in the NotI site | This study |
When appropriate, media were supplemented with antibiotics either to maintain selection for plasmids or to select for various recombinant strains. For R. capsulatus, final concentrations were 25 μg of kanamycin, 25 μg of spectinomycin, and 1 μg of tetracycline per ml; for E. coli, final concentrations were 50 μg of ampicillin, 25 μg of chloramphenicol, 50 μg of kanamycin, 50 μg of spectinomycin, and 10 μg of tetracycline per ml. All antibiotics were of reagent grade and were purchased from Sigma Chemical Co. (St. Louis, Mo.).
DNA manipulations, amplification, sequencing, and conjugation.
DNA was purified from R. capsulatus as described by Klug and Drews (21). Other techniques used for manipulation and Southern analysis of DNA were standard (24). Oligonucleotides were obtained from MWG-Biotech (Ebersberg, Germany). DNA sequencing was performed with the T7Sequencing kit (Pharmacia, Uppsala, Sweden) or by automated sequencing done by SEQLAB (Sequence Laboratories, Göttingen, Germany). Sequence analysis was performed with University of Wisconsin GCG version 7.3. Plasmids were mobilized into R. capsulatus by triparental mating with E. coli J-53 bearing the conjugative plasmid RP4, E. coli HB101 bearing both the helper plasmid pRL528 and the mobilizable plasmid, and strains of R. capsulatus (Table 1). Presumptive exconjugants were freed from E. coli by streaking on selective medium.
Construction of mutant strains of R. capsulatus and construction of plasmids for complementation.
For insertional inactivation of sqr in R. capsulatus, plasmid pRN4C (Fig. 1) was constructed as follows. The cyanobacterial replicon (pDU1) was removed from plasmid pRL488 (10) by cleavage with EcoRV, resulting in pIM101. The 1,650-bp XbaI-MscI fragment from pUSQR bearing ORF2 together with the putative promoter and the 5′ region of sqr (32) was inserted in front of the luxAB genes into the XbaI-SmaI site of pIM101. The 1,360-bp XbaI-SacII fragment was removed, and the 706-bp EcoRI-SacII fragment [sqr-ct(−75) in Fig. 1] of pUSQR bearing the 3′ part of sqr was then inserted in front of the remaining fragment (sqr-nt in Fig. 1).
The plasmid pRU4C for deletion of the 3′ part of ORF2 and the 5′ part of sqr was constructed as follows. The 920-bp XbaI-StuI fragment of pUSQR (Fig. 1) bearing the 5′ part of ORF2 was inserted into the XbaI-SmaI site of pIM101. Then, a 2,730-bp ClaI-XbaI fragment consisting of the 1,900-bp Ω Smr Spcr cassette from pHP45Ω and the 830-bp HindIII-PstI fragment of pUSQR, which bears the 3′ end of sqr and ORF3, was inserted into the XbaI site. Double recombinants were selected for resistance to kanamycin and sensitivity to spectinomycin.
Construction of plasmids for complementation was as follows. The 1,900-bp Ω Smr Spcr cassette from pHP45Ω was inserted into the NotI site of pUSQR (Fig. 1A), resulting in pUSQRΩ. After the 5,400-bp PstI fragment and the 4,500-bp StuI-PstI fragments had been inserted into pPHU233, the resulting plasmids pPSQR and pPStuSQR were mobilized into R. capsulatus strains.
Construction of SQR-PhoA fusion proteins.
With plasmid pT7-5/lacY-PhoA (United States Biochemicals, Cleveland, Ohio) as template, phoA of E. coli lacking the 5′ portion encoding the N-terminal translocation signal sequence was amplified by PCR with oligonucleotides apn2 (5′-GGATCCCCGGGTACCGCTAGCGACTCTTATACACAA-3′) and apc (5′-AGATCTTCATGTTTTAACCATG-3′). By using oligonucleotides sqrn (5′-TTCCCATATATGCATCTG-3′) and sqrc (5′-GCGGATCCCTTCTTCACGGCCTT-3′), sqr was amplified. In-frame fusion of the two fragments via the BamHI sites in sqrc and apn2 and subsequent insertion of the fusion into pGEM-T (Promega, Madison, Wis.) resulted in pGSAP. For the construction of fusions with truncated sqr, the 1,300-bp BsiWI-SacI fragment of pTSQR (32) was replaced by the 2,840-bp BsiWI-SacI fragment of pGSAP, resulting in pTSAP. Exonuclease treatment was done with the Erase-a-Base kit (Promega) according to the manufacturer’s instructions by using the SmaI site and the KpnI site, which had been inserted behind the BamHI site by the oligonucleotide apn2. Two of the obtained truncated fusions were utilized in this study: pTSAP389, bearing the 1,167-bp fragment, which encodes the N-terminal 389 amino acid residues of SQR, fused to phoA, and pTSAP108, bearing the 324-bp fragment, which encodes the N-terminal 108 amino acid residues of SQR, fused to phoA. The BsiWI-SacI fragment of pUSQR was exchanged for the BsiWI-SacI fragments from pGSAP, pTSAP389, and pTSAP108, and the Ω cassette (SmaI) from pHP45Ω was inserted into the SpeI site in each case. Subsequently, the StuI-PstI fragments had been cloned into the ScaI-PstI site of the mobilizable plasmid pPHU233, and the obtained plasmids pPSAPo, pPSAP389, and pPSAP108 were transformed into Rhodobacter wild type and strain F14.
Isolation of membranes and enzymatic assay of SQR.
Chromatophores were isolated as previously described (33); spheroplasts were isolated according to the work of Kabak (18). The bacteriochlorophyll a content of membranes was measured as described elsewhere (3). The activity of SQR was measured as previously described (33). For inactivation by proteinase K treatment or inhibition by anti-SQR antiserum, membranes equivalent to 100 μg of bacteriochlorophyll a were suspended in 500 μl of 50 mM glycylglycine, pH 7.0. Proteinase K was added to a concentration of 1 mg ml−1. Samples and the control without protease were incubated at room temperature in the dark for 30 min before SQR activity was measured. Anti-SQR antiserum was added in a dilution of 1:100. Control samples were supplemented with preimmune serum. Both were incubated on ice for 2 h before SQR activity was measured.
Enzymatic assay of alkaline phosphatase.
For the enzymatic assay of alkaline phosphatase (PhoA), cells were washed with 1 M Tris HCl, pH 8.0, supplemented with 35 μg of chloramphenicol ml−1 and were resuspended in the same buffer. Cells equivalent to an optical density at 770 nm (OD770) of 0.05 were used for the enzymatic assay performed according to the procedure in reference 6. The reaction was terminated by addition of 0.2 ml of 1 M K2HPO4 after incubation for 1 h at 37°C. In control samples, the reaction was stopped by the addition of 0.2 ml of 1 M K2HPO4, immediately after p-nitrophenol phosphate had been added. The activity of the phosphatase is given as the difference between the OD420 of the samples and that of the control.
SDS-PAGE and Western blotting.
SDS-PAGE was carried out according to the method of Laemmli (23). Western blotting was carried out as described in the work of Towbin et al. (37). Rabbit antiserum against purified SQR was obtained from Eurogentec (Seraing, Belgium) and was used diluted 1:3,000; anti-E. coli PhoA antiserum was obtained from CP Laboratories and was used diluted 1:5,000. For detection, the BM chromogenic Western blotting kit (Boehringer Mannheim, Mannheim, Germany) was used.
Other assays and chemicals.
The concentration of sulfide was determined as described by Trüper and Schlegel (38). Protein was determined with the bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.); p-nitrophenol phosphate was purchased from Fluka Chemie (Buchs, Switzerland). All other chemicals were of reagent grade and were purchased from commercial sources.
Nucleotide sequence accession number.
The sqr sequence has been updated in the EMBL nucleotide sequence database (accession no. X97478).
RESULTS
Sequence analysis of the sqr region.
The cloned PstI fragment (EMBL nucleotide sequence database accession no. X97478) comprises 3,492 bp with a G+C content of 66% that is within the range of 65.5 to 66.5% given for R. capsulatus (16). Besides the sqr gene, one complete ORF (ORF2) and parts of two putative genes were found (Fig. 1A).
Some minor errors in the published sequence of the sqr gene (32) were corrected. The coding region of 1,281 bp extends in the middle of the cloned fragment from position 1442 to position 2722. The deduced amino acid sequence consists of 427 residues with a molecular weight of 46,929 and a net charge of +1. The former published amino acid sequence has altered as follows: (i) the peptide from amino acid position 106 to 117 (ARNWPSTRSRLR) was replaced by 106-GPELAFDEIEGF-118, (ii) the change of four positive to four negative charges changed the net charge from +9 to +1, and (iii) an alanine was inserted at position 153. None of the other features of SQR published in reference 32 were altered after correction.
On the cloned fragment, a second gene (ORF2) extends from position 196 to position 1278 in the same orientation as sqr. The nucleotide sequence from position −6 to −13 upstream of ORF2 matches well-known ribosome binding sites of R. capsulatus (1). ORF2 encodes a protein of 360 amino acid residues with a molecular mass of 38.5 kDa. Comparison to protein databases revealed a similarity of approximately 63% (identity, 47%) to the corresponding part of a protein of unknown function from Archaeoglobus fulgidus (coding region AF0890), 397 amino acid residues in length (20). In contrast to the protein from A. fulgidus, the N-terminal 26 amino acid residues of the protein from R. capsulatus (N-MDRRSFLKTTAATATLAAVGLPVAAA-) show the characteristic twin arginine motif for translocation by the Sec-independent targeting and translocation system (4). Interestingly, the next upstream gene (ORF1) is also related to A. fulgidus. The amino acid sequence deduced from the first 153 nucleotides of the cloned PstI fragment resembles the C-terminal part of RbsC2 (coding region AF0889) of A. fulgidus (20) with significant homology (similarity, 60%; identity, 45%). RbsC2 functions as the permease of a ribose transporter of the ATP-binding cassette traffic ATPase family. The rbsC2 gene from this archaeon precedes the gene encoding the protein homologous to ORF2, showing the same organization of those genes on the genomes of R. capsulatus and A. fulgidus. In opposite orientation to the first three genes, from the end of the PstI fragment to position 3097 the 3′ end of a further coding sequence (ORF3) could be found. Protein database analysis with the deduced amino acid sequence revealed more than 60% homology (>45% identity) to the C-terminal EAL motif pattern of many regulatory proteins (19, 36, 41).
Mutational analysis of the sqr region in R. capsulatus.
Different mutant strains of R. capsulatus were constructed. Figure 1 gives a view of the sqr region of the mutant strains as ascertained by Southern blot analysis (data not shown). By insertion of plasmid pRN4C into the genome, two types of mutants were obtained. In one type, represented by strain 22/11, no intact copy of sqr is present in the genome (Fig. 1B). In the other type, represented by strain 22/17, a complete sqr gene including the putative promoter sequence (32) is located behind the inserted plasmid (Fig. 1C). Insertion of the plasmid pRU4C into the genome by double recombination resulted in the identical mutant strains F14 and A10 (Fig. 1D). In these strains, part of ORF2 and almost the total sqr gene had been deleted.
Phenotypes of the sqr mutants.
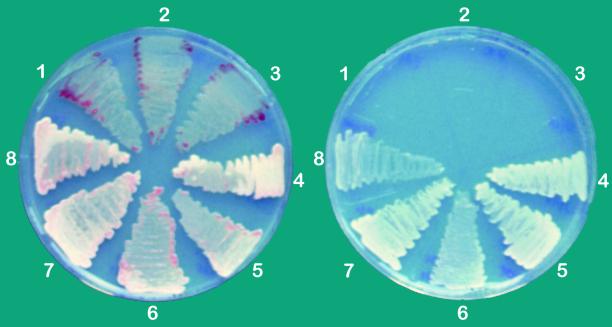
The different strains were streaked out on solidified medium supplemented with malate for heterotrophic growth and without any organic carbon source for photoautotrophic sulfide-dependent growth (Fig. 2). All strains grew well under heterotrophic conditions in the presence of sulfide, similar to the wild-type strain. The color of colonies of the sqr mutant 22/11 and the Δ(orf2-sqr) strains F14 and A10 was purple, in contrast to the pale yellow color of colonies of the other strains. The pale yellow coating is due to the deposition of sulfur outside the cells. The purple color of mutants 22/11, F14, and A10 indicates that they do not possess sulfide oxidation activity. Under sulfide-dependent photoautotrophic conditions, no growth of strains 22/11, F14, and A10 was observed, in contrast to the sqr+ strains 22/17, F14sn, and F14n, which grew as well as the wild-type strain under this condition (Fig. 2). Strain F14sn was obtained by transformation of strain F14 with the autonomously replicating plasmid pPStuSQR, which complements only the deletion of sqr. In strain F14n, the two partially deleted genes ORF2 and sqr were complemented by transformation of the plasmid pPSQR into strain F14 (Table 1).
FIG. 2.
Growth of mutant strains of R. capsulatus under sulfidic atmosphere. (left) Growth under photoheterotrophic conditions; (right) growth under photoautotrophic sulfide-dependent conditions. Plate segments: 1 and 2, Δ(orf2-sqr) strains F14 and A10; 3, sqr mutant strain 22/11; 4, sqr+ strain 22/17; 5, strain F14n; 6, strain F14sn; 7, strain F14(pPSAPo); 8, strain F14(pPSAP). Strains grew for 1 week as described in Materials and Methods. The genotypes are described in Table 1 and Fig. 1. In the case of F14n, the deletion of F14 was complemented by an intact copy of ORF2 and sqr on plasmid pPSQR; in the case of F14sn, only the deficiency of sqr was complemented by plasmid pPStuSQR (Table 1). F14(pPSAPo) bears a plasmid that encodes the SQR::PhoA fusion protein; F14(pPSAP) bears a plasmid that encodes the SQR::PhoA fusion protein and ORF2.
Consumption of sulfide, activity of SQR in membranes, and doubling times of the strains F14, 22/11, and F14sn and wild type were measured (Table 2). The amount of the consumption of sulfide by cultures of the strains 22/11 and F14 was significantly lower than the amount consumed by the wild type and was in the same range as was found for chemical oxidation of sulfide by traces of oxygen under microoxic conditions (data not shown). Strain F14sn oxidized four to six times the amount of sulfide oxidized by the wild-type strain due to multiple copies of the sqr-bearing self-replicating plasmid. Membranes from the four strains have been isolated, and their SQR activity has been measured. No SQR activity was found in membranes from mutants F14 and 22/11. Membranes of strain F14sn showed six to seven times the activity of the membranes from the wild-type strain. The doubling times of the wild-type strain and sqr mutants F14 and 22/11 were in the same range up to concentrations of 0.8 mM sulfide. In the presence of 1.2 mM sulfide, doubling times of the wild-type strain and the sqr mutants increased approximately 1.2- to 1.5-fold. The doubling time of strain F14sn was similar to that of the wild-type strain in the absence of sulfide and with 1.2 mM sulfide. At 0.4 and 0.8 mM sulfide, strain F14sn grew somewhat more slowly than the wild-type strain. The behavior of strain 22/17 (sqr+) was identical to that of the wild type (data not shown).
TABLE 2.
Consumption of sulfide, membranous SQR activities, and doubling times of R. capsulatus wild type, F14 [Δ(orf2-sqr)], 22/11 (sqr mutant), and complemented strain F14sn (sqr+)a
| Strain | Sulfide (μmol) consumed within 24 h after addition of mM sulfide:
|
Sp act of SQR (μmol of decyl-ubiquinone reduced mg of protein−1 min−1) | Doubling time (h) with mM sulfide:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0.4 | 0.8 | 1.2 | 0 | 0.4 | 0.8 | 1.2 | ||
| Wild type | 60 (20) | 130 (24) | 251 (172) | 0.68 | 65 (8) | 61 (12) | 62 (11) | 80 (13) |
| F14 | 9 (9) | 26 (45) | 24 (12) | 0.00 | 63 (11) | 58 (8) | 65 (14) | 97 (4) |
| 22/11 | 11 (19) | 19 (23) | 37 (32) | 0.00 | 59 (8) | 57 (8) | 61 (4) | 87 (13) |
| F14sn | 390 (130) | 620 (200) | 850 (290) | 4.48 | 59 (13) | 80 (20) | 82 (19) | 86 (13) |
Strains were cultured in microoxic liquid RCV medium. Sulfide was added in concentrations as indicated. Values for consumption of sulfide are averages of three experiments; those for doubling times are averages of six experiments. Membranes were prepared from cultures 3 days after induction by sulfide at a 0.8 mM concentration. SQR activity was measured as previously described (33). Rates were corrected for the rates with membranes inactivated by heat treatment. Values in parentheses are standard deviations.
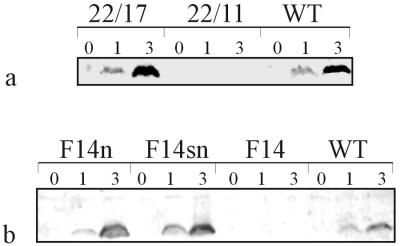
Western blot analysis was performed with cytoplasmic membranes isolated from all strains to test for expression of SQR (Fig. 3). A specific reaction with the anti-SQR antiserum could be obtained with membrane preparations from cultures of the sqr+ strains 22/17, F14n, and F14sn and the wild type, after incubation with sulfide for 3 h; without sulfide, no reaction occurred. In strains F14n and F14sn, the level of expression of SQR was higher than that in the wild-type strain, consistent with the higher sulfide consumption rates and the higher SQR activity of these strains (see above). No signals were observed with membranes from induced cultures of the sqr mutants F14 and 22/11.
FIG. 3.
Western blot analysis with membranes isolated from R. capsulatus wild-type and mutant strains. (a) Western blot analysis with membranes from strains 22/17 and 22/11 and the wild type (WT); (b) analysis with membranes from strains F14n, F14sn, F14, and wild type (WT). Genotypes of the strains are described in Fig. 1 and the legend to Fig. 2. Membranes were isolated from cultures prior to (0) addition of sulfide and 1 h (1) and 3 h (3) after addition of sulfide to a concentration of 0.5 mM. SQR was detected with rabbit antiserum against purified SQR. Every lane represents membranes equivalent to 40 μg of protein.
Subcellular localization of SQR.
Ambiguous results for SQR activity, which are not elaborated here, have been obtained by treatment of inside-out-oriented chromatophores and right-side-out-oriented spheroplasts with proteinase K and an SQR antiserum. In brief, after incubation with proteinase K, SQR activity in chromatophores and spheroplasts decreased to 75 to 80 and to 20 to 40% of the control, respectively, suggesting a localization of SQR at the periplasmic surface of the cytoplasmic membrane. However, after incubation with anti-SQR antiserum, differences in SQR activity were less significant. Values of 50 to 80 and 40 to 60% of the activity of the control were determined with chromatophores and spheroplasts, respectively. Therefore, we switched to the PhoA fusion technique. Since alkaline phosphatase of E. coli is active only when translocated to the periplasmic space (25), the plasmid pPSAPo, which bears the translational fusion sqr::phoA, was used (Fig. 4 and Table 3). Additionally, phoA was fused with truncated forms of sqr: from plasmid pPSAP389, a protein consisting of the N-terminal 389 amino acid residues of SQR fused to PhoA and, from plasmid pPSAP108, a protein consisting of the N-terminal 108 amino acid residues of SQR fused to PhoA can be expressed.
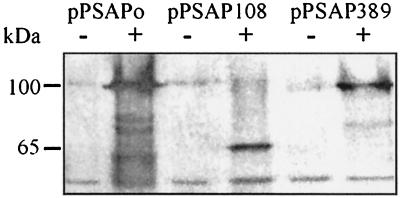
FIG. 4.
Western blot analysis with cell extracts of the double mutant F14 transformed with pPSAPo, pPSAP108, and pPSAP389. Plasmid pPSAPo encodes the fusion of total SQR (427 amino acid residues) with PhoA; plasmids pPSAP108 and pPSAP389 encode the fusions of the N-terminal 108 and 389 amino acid residues of SQR with PhoA, respectively. The markers indicate the sizes of the fusion proteins encoded by pPSAPo, pPSAP389 (approximately 100 kDa), and pPSAP108 (approximately 65 kDa). Samples were taken prior to (lanes 1, 3, and 5) and 3 h after (lanes 2, 4, and 6) addition of sulfide to a concentration of 0.5 mM. The fusion proteins were detected with anti-E. coli PhoA antiserum. Every lane represents 0.5 ml of a culture at an OD770 of 0.4.
TABLE 3.
Phosphatase activities of R. capsulatus wild type and F14 [Δ(orf2-sqr)] expressing protein fusions of SQR with PhoAa
| Strain with plasmid and protein fusion | PhoA activity (OD420)
|
|
|---|---|---|
| Without sulfide | With sulfide | |
| Wild type | ||
| None | 0.04 | 0.03 |
| pPSAPo and SQR::PhoA | 0.04 | 0.72 |
| pPSAP108 and SQR108::PhoA | 0.05 | 0.05 |
| pPSAP389 and SQR389::PhoA | 0.04 | 0.05 |
| F14 | ||
| None | 0.04 | 0.07 |
| pPSAPo and SQR::PhoA | 0.05 | 0.70 |
| pPSAP108 and SQR108::PhoA | 0.03 | 0.06 |
| pPSAP389 and SQR389::PhoA | 0.03 | 0.04 |
The fusion SQR::PhoA consists of the full-length SQR (427 amino acid residues) fused to PhoA. In SQR108::PhoA and in SQR389::PhoA, the N-terminal 108 and 389 amino acid residues, respectively, are fused with PhoA. Samples were taken from heterotrophically grown cultures without sulfide and cultures after incubation with 0.3 mM sulfide for 10 h. PhoA activity is given as the difference between the OD420 of the samples and that of the control as described in Materials and Methods.
The different fusion proteins were expressed in mutant F14 (Fig. 4) and the wild-type strain (data not shown) after incubation with sulfide. With cell extracts from the induced strains F14(pPSAPo), F14(pPSAP108), and F14(pPSAP389), the antibody reacted with proteins of approximately 100, 65, and 95 kDa in size, respectively, as expected for the different fusion proteins. The amount of fusion protein was somewhat lower in the case of both truncated proteins than with the full-length version. The same results were obtained with cultures of the wild type transformed with the three plasmids (data not shown). The alkaline phosphatase activity of noninduced cultures was compared to the PhoA activity of cells incubated with 300 μM sulfide for 10 h (Table 3). PhoA activity was observed only in strains that had been transformed with pPSAPo, which encodes the full-length fusion protein, and induction by sulfide was necessary. In these strains, phosphatase activity was more than 10-fold higher than the background activity. The noninduced cultures and the cultures transformed with pPSAP389 and pPSAP108 showed PhoA activity as low as that of nontransformed cultures. Besides phosphatase activity, strain F14 bearing pPSAPo was able to grow photoautotrophically on sulfide (Fig. 2), in contrast to mutant F14 transformed with pPSAP389 and pPSAP108 (data not shown). As was found for native SQR (32), approximately 90% of the total SQR activity of cell extracts of strain F14 expressing the protein fusion SQR::PhoA was associated with the membrane fraction, even after treatment with 100 mM sodium chloride (specific SQR activity: 0.25 μmol of decyl-ubiqinone reduced mg of protein−1 min−1). No SQR activity was found in cell extracts of strain F14 expressing either the fusion protein SQR108::PhoA or the fusion protein SQR389::SQR.
DISCUSSION
In the present study, mutants of R. capsulatus were constructed to examine whether SQR is the only sulfide-oxidizing enzyme in this bacterium. The insertion of plasmid pRN4C into the genome resulted in an SQR+ strain (22/17) and an SQR− strain (22/11). In strain F14, part of ORF2 and most of sqr had been deleted. Strain 22/11 and strain F14 were unable to grow photoautotrophically on sulfide, did not deposit any sulfur outside the cells under heterotrophic conditions, and did not consume any sulfide in liquid medium. Additionally, neither sulfide quinone reductase activity nor SQR was detected in membranes of these strains. These results clearly indicate that SQR is the only sulfide-oxidizing enzyme in R. capsulatus and that it is absolutely required for the sulfide-dependent growth of this bacterium.
Sulfide is toxic even for organisms that require it for growth (26). The sulfide-dependent specific growth rate of R. capsulatus at 1 mM sulfide under autotrophic conditions was found to be half the maximal growth rate (39). In our experiments, we observed no inhibition of heterotrophic growth up to 0.8 mM sulfide, neither for the wild-type strain nor for the SQR− strains. This suggests that the sulfide tolerance of R. capsulatus is not due to oxidation of sulfide and that SQR does not function in sulfide detoxification up to this concentration. The lower inhibition of growth under the conditions that we used than of autotrophically grown cultures (39) is consistent with the increase of sulfide tolerance if yeast extract is added to the cultures, as found by Hansen and van Gemerden (15). The mechanism of this effect is unknown. Surprisingly, even 0.4 mM sulfide increased the doubling time of strain F14sn to the same as was found at 1.2 mM sulfide. Possibly, the high sulfide oxidation rate, due to the high level of SQR, might cause an overreduction of the quinone pool. This might decrease the cyclic electron transport across the photosystem and, therefore, decrease ATP production. However, more detailed studies are necessary to understand the mechanism of increased sulfide tolerance during heterotrophic growth and the decelerated growth of strain F14sn in the presence of sulfide.
Many data suggest that the conversion of hydrogen sulfide to sulfur or polysulfide takes place in the periplasmic space. Members of the families Rhodospirillaceae, Ectothiorhodospiraceae, and Chlorobiaceae deposit the sulfur from oxidation of sulfide outside the cells (7). Recently, it was found that members of the family Chromatiaceae, which store sulfur inside the cells, deposit the sulfur globules in vesicles equivalent to the periplasmic space (27). Additionally, in the chemotrophic bacterium Thiobacillus ferrooxidans, sulfur from the oxidation of hydrogen sulfide accumulates in the periplasmic space (29), and finally, all sulfide-oxidizing flavocytochromes c characterized so far are located in the periplasm (7, 30). In previous studies, it had been established that the SQR of R. capsulatus is peripherally bound to the cytoplasmic membrane (32, 33). In this study, we show that, despite the lack of an N-terminal signal peptide in SQR, strains of R. capsulatus which expressed the full-length fusion protein SQR::PhoA were PhoA active. In addition, strain F14(pPSAPo) grew photoautotrophically on sulfide (Fig. 2), and almost all SQR activity was associated with the membranes isolated from this strain. These confirm a localization of SQR at the periplasmic surface of the cytoplasmic membrane, where it gets into contact with the quinone within the membrane, as was suggested from our results obtained with proteinase K-treated vesicles. Localization of SQR at the periplasmic surface of the cytoplasmic membrane is consistent with the fact that R. capsulatus deposits sulfur, the product of oxidation of hydrogen sulfide, outside the cells (15). Therefore, a system for the export of sulfur across the cytoplasmic membrane is not necessary in this bacterium. The presumption that oxidation of sulfide in the periplasm might be a general feature in sulfide-oxidizing bacteria is supported by our data.
The protein encoded by ORF2, upstream of sqr, shows the N-terminal twin arginine motif, characteristic of proteins that are translocated by the Sec-independent pathway (4). Most of these proteins bear complex redox factors or mediate the translocation of redox proteins that lack a signal peptide. Because SQR lacks an N-terminal signal peptide, a possible involvement of ORF2 in the translocation of SQR was investigated. However, the deletion of the sqr gene and part of ORF2 in strain F14 could be complemented by an intact copy of the sqr gene alone on plasmid pPStuSQR (strain F14sn), restoring the phenotype of the wild-type strain. In the complemented strain F14sn, neither enzyme activity, attachment to the membrane, nor sulfide-dependent expression of SQR was affected by the lack of ORF2. In addition, strain F14 expressing the full-length protein fusion SQR::PhoA encoded by the plasmid pPSAPo showed phosphatase activity after induction by sulfide. Therefore, involvement of ORF2 in translocation and activity of SQR can be excluded. A distinct function of ORF2 is also suggested by the existence of a homologous ORF in A. fulgidus (20), an archaeon not capable of oxidation of sulfide. The function of the protein encoded by ORF2 remains unknown. It might participate in transport of ribose, because both ORF2 in the eubacterium R. capsulatus and the homologous ORF AF0890 in the archaeon A. fulgidus are connected to rbsC2, encoding a subunit of a ribose ATP-binding cassette transporter. The absence of a putative transcription initiation site between rbsC2 and ORF2 in R. capsulatus suggests cotranscription of both genes.
The protein fusion SQR::PhoA was translocated to the periplasmic side of the membrane. In contrast, the two truncated fusion proteins SQR389::PhoA and SQR108::PhoA were not translocated, although they had been expressed. From this, it can be concluded that the C terminus is essential for the translocation of SQR. So far, it is unknown whether the 38 amino acid residues at the C terminus bear a signal for translocation, or whether incorrect folding of SQR prevents interaction with a second protein participating in translocation. Like SQR of R. capsulatus (32), the dihydrolipoamide dehydrogenase of Synechocystis sp. is a flavoprotein (11), which binds FAD in a way similar to that of gluthathione reductase. No N-terminal signal peptide is present in this protein, although it is attached to the periplasmic surface of the cytoplasmic membrane (12). In the C-terminal region, no significant homology exists between SQR and dihydrolipoamide dehydrogenase, making the function of the C terminus as a signal for translocation unlikely. Possibly, a surface-exposed pattern is necessary for translocation of FAD-binding proteins that are attached to the periplasmic surface of the cytoplasmic membrane by a so-far-unknown mechanism. Nevertheless, the C terminus of SQR is essential for translocation, and it is also required for activity, as shown with mutant 22/11. In this mutant, a truncated sqr gene [sqr(−75) in Fig. 1] which lacks the 3′-end 75 bp is present. From its position in the genome, this gene should be expressed similarly to sqr in the wild type. However, neither SQR activity nor SQR was found in membranes from strain 22/11. In addition, strain F14 transformed with pPSAP389, which expressed an SQR::PhoA fusion protein lacking the C-terminal 38 amino acid residues of SQR, could not grow autotrophically on sulfide. More detailed examinations will be necessary to understand the specific function of the C terminus for both activity and translocation of SQR.
ACKNOWLEDGMENTS
We are indebted to Y. Shahak, E. Padan, and M. Bronstein for intense discussion. We thank R. A. Siddiqui for discussion about phosphatase fusion experiments and C. Dahl for providing plasmid pPHU233.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. Ha 852/10).
Footnotes
Dedicated to A. Trebst on the occasion of his 70th birthday.
REFERENCES
- 1.Alberti M, Burke D H, Hearst J E. Structure and sequence of the photosynthesis gene cluster. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1083–1106. [Google Scholar]
- 2.Arieli B, Shahak Y, Taglicht D, Hauska G, Padan E. Purification and characterization of sulfide-quinone reductase (SQR), a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica. J Biol Chem. 1994;268:5705–5711. [PubMed] [Google Scholar]
- 3.Baccarini-Melandri A, Melandri B A. Partial resolution of the photophosphorylating system of Rhodopseudomonas capsulatus. Methods Enzymol. 1972;23:556–561. [Google Scholar]
- 4.Berks B C. A common export pathway for proteins binding complex redox factors. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 phages transducing. J Mol Biol. 1975;96:307–331. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 7.Brune D C. Sulfur compounds as photosynthetic electron donors. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer; 1995. pp. 847–870. [Google Scholar]
- 8.Cohen Y, Jorgensen B B, Padan E, Shilo M. Sulfide-dependent anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. Nature. 1975;257:489–492. doi: 10.1128/jb.123.3.855-861.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 10.Elhai J, Wolk C P. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 1990;9:3379–3388. doi: 10.1002/j.1460-2075.1990.tb07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engels A, Pistorius E K. Characterization of a gene encoding dihydrolipoamide dehydrogenase of the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology. 1997;143:3543–3553. doi: 10.1099/00221287-143-11-3543. [DOI] [PubMed] [Google Scholar]
- 12.Engels A, Kahmann U, Ruppel H G, Pistorius E K. Isolation, partial characterization and localization of a dihydrolipoamide dehydrogenase from the cyanobacterium Synechocystis sp. strain PCC 6803. Biochim Biophys Acta. 1997;1340:33–44. doi: 10.1016/s0167-4838(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, C. G. 1998. Physiology and genetics of bacterial sulfur oxidation. Adv. Microb. Physiol. ••:236–289. [DOI] [PubMed]
- 14.Grant S, Jersee J, Bloom F, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen T A, van Gemerden H. Sulfide utilization by purple nonsulfur bacteria. Arch Microbiol. 1972;86:49–56. doi: 10.1007/BF00412399. [DOI] [PubMed] [Google Scholar]
- 16.Imhoff J F. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer; 1995. pp. 1–15. [Google Scholar]
- 17.Irgens R L. Thioacetamide as a source of hydrogen sulfide for colony growth of purple sulfur bacteria. Curr Microbiol. 1983;8:183–186. [Google Scholar]
- 18.Kabak H R. Bacterial membranes. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- 19.Kaneko T, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 20.Klenk H P, et al. The complete genome sequence of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 21.Klug G, Drews G. Construction of a gene bank of Rhodopseudomonas capsulata using a broad host range DNA cloning system. Arch Microbiol. 1984;139:319–332. doi: 10.1007/BF00408373. [DOI] [PubMed] [Google Scholar]
- 22.Klughammer C, Hager C, Padan E, Schütz M, Schreiber U, Shahak Y, Hauska G. Reduction of cytochromes with menaquinol and sulfide in membranes from green sulfur bacteria. Photosynth Res. 1995;43:27–34. doi: 10.1007/BF00029459. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 25.Manoil C, Mekalanos J J, Beckwith J. Alkaline phosphatase fusions: sensors of subcellular location. J Bacteriol. 1990;172:515–518. doi: 10.1128/jb.172.2.515-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Research Council. Subcommittee on Medical and Biologic Effects on Environmental Pollutants, Division of Medical Science, Assembly of Life Science, Hydrogen sulfide. Baltimore, Md: University Park Press; 1979. [Google Scholar]
- 27.Pattaragulwanit K, Brune C D, Trüper H G, Dahl C. Molecular genetic evidence for extracytoplasmic localization of sulfur globules in Chromatium vinosum. Arch Microbiol. 1998;169:434–444. doi: 10.1007/s002030050594. [DOI] [PubMed] [Google Scholar]
- 28.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 29.Pronk J T, Meulenberg R, Hazeu W, Bos B, Kuenen J G. Oxidation of reduced sulfur compounds by acidophilic thiobacilli. FEMS Microbiol Rev. 1990;75:293–306. [Google Scholar]
- 30.Reinartz M, Tschäpe J, Brüser T, Trüper H G, Dahl C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol. 1998;170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 31.Schütz M, Klughammer C, Griesbeck C, Quentmeier A, Friedrich C G, Hauska G. Sulfide-quinone reductase activity in membranes of the chemotrophic bacterium Paracoccus denitrificans GB17. Arch Microbiol. 1998;170:353–360. [Google Scholar]
- 32.Schütz M, Shahak Y, Padan E, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: purification, cloning and expression. J Biol Chem. 1997;272:9890–9894. doi: 10.1074/jbc.272.15.9890. [DOI] [PubMed] [Google Scholar]
- 33.Shahak Y, Klughammer C, Schreiber U, Padan E, Herrmann I, Hauska G. Sulfide-quinone and sulfide-cytochrome reduction in Rhodobacter capsulatus. Photosynth Res. 1994;39:175–181. doi: 10.1007/BF00029384. [DOI] [PubMed] [Google Scholar]
- 34.Shahak Y, Schütz M, Bronstein M, Griesbeck C, Hauska G, Padan E. Sulfide-dependent anoxygenic photosynthesis in prokaryotes: sulfide-quinone reductase (SQR), the initial step. In: Peschek G A, Löffelhardt W, Schmetterer G, editors. Proceedings of the 9th International Symposium on Phototrophic Procaryotes (Vienna 1997). New York, N.Y: Plenum Press; 1997. pp. 217–228. [Google Scholar]
- 35.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–98. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 36.Tal R, Wong H C, Calhoon R, Gelfand D, Fear A L, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J Bacteriol. 1998;180:4416–4425. doi: 10.1128/jb.180.17.4416-4425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trüper H G, Schlegel H G. Sulphur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii. Antonie Leeuwenhoek. 1964;30:225–281. doi: 10.1007/BF02046728. [DOI] [PubMed] [Google Scholar]
- 39.Van Gemerden H. The sulfide affinity of phototrophic bacteria in relation to the location of elemental sulfur. Arch Microbiol. 1984;139:289–294. [Google Scholar]
- 40.Van Visser J, de Jong G A H, Robertson L A, Kuenen J G. A novel membrane-bound flavocytochrome c sulfide dehydrogenase from the colourless sulfur bacterium Thiobacillus sp. W5. Arch Microbiol. 1997;167:295–301. doi: 10.1007/s002030050447. [DOI] [PubMed] [Google Scholar]
- 41.Vlcek C, Paces V, Maltsev N, Paces J, Haselkorn R, Fonstein M. Sequence of a 189-kb segment of the chromosome of Rhodobacter capsulatus SB1003. Proc Natl Acad Sci USA. 1997;94:9384–9388. doi: 10.1073/pnas.94.17.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver P F, Wall J D, Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–221. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]