Abstract
We have studied the transcription of the glnB and glnA genes in Rhodospirillum rubrum with firefly luciferase as a reporter enzyme. Under NH4+ and N2 conditions, glnBA was cotranscribed from a weak and a strong promoter. In nitrogen-fixing cultures, activity of the latter was highly enhanced by NtrC, but transcription from both promoters occurred under both conditions. There is no promoter controlling transcription of glnA alone, supporting our proposal that the glnA mRNA is produced by processing.
Rhodospirillum rubrum, a photosynthetic purple free-living bacterium, is capable of fixing nitrogen anaerobically in N-free medium. As in other diazotrophs, the ammonium produced by nitrogenase is assimilated via the glutamine synthetase-glutamate synthase pathway in R. rubrum (2).
The PII protein, a homotrimer encoded by glnB, plays a key role in controlling nitrogen metabolism in enteric bacteria. PII can be either unmodified or uridylylated, with different regulatory properties in the control of glutamine synthetase activity and transcriptional regulation involving NtrC, which in the phosphorylated form acts as a transcriptional activator. Under nitrogen-limited conditions, PII is uridylylated by a bifunctional enzyme, the uridylyltransferase, encoded by glnD. Conversely, under conditions of nitrogen excess, the uridyl-removing activity of GlnD dominates and the unmodified form of PII is produced. This form of PII stimulates the adenylylation activity of another bifunctional enzyme, adenylyltransferase, the product of glnE, which leads to the adenylylation of glutamine synthetase. PII also binds to NtrB, yet another bifunctional enzyme, which then acts as a phosphatase, catalyzing the hydrolysis of phosphate from NtrC-P and thereby inactivating transcription from NtrC-P-dependent promoters. The uridylylated form of PII-UMP is not able to bind to NtrB, which then acts as a kinase catalyzing the phosphorylation of NtrC, leading to the activation of transcription from NtrC-P-dependent promoters. PII-UMP also stimulates the deadenylylating activity of adenylyltransferase, causing deadenylylation (activation) of glutamine synthetase (8, 14, 19, 21). In R. rubrum, glutamine synthetase is not only adenylylated but also ADP-ribosylated (29), although the effect of this modification on activity has not been demonstrated.
As in R. rubrum, the glnB gene has been identified upstream of glnA in Rhodobacter capsulatus, Rhodobacter sphaeroides, Azospirillum brasilense, Rhizobium leguminosarum, Bradyrhizobium japonicum, and Azorhizobium caulinodans, but the regulation of the glnBA operon varies among these bacteria (3–6, 12, 15, 20, 25, 31). The glnB-like gene of Herbaspirillum seropedicae is not clustered with glnA and its expression is constitutive and independent of NtrC, as it is in Escherichia coli and Klebsiella pneumoniae (1).
In R. rubrum, the glnB gene is cotranscribed with glnA from a putative ς54-dependent promoter (glnBp2) and from a proposed ς70-dependent promoter (glnBp1), overlapping a possible NtrC binding site (9). However, in addition to the glnBA transcript we have demonstrated a glnA mRNA and proposed that this is due to specific mRNA processing. We have used firefly luciferase as a reporter enzyme to further analyze the transcriptional regulation of the glnB and glnA genes of R. rubrum and to provide additional evidence for processing of the glnBA mRNA.
The bacterial strains and plasmids used in this study are listed in Table 1. R. rubrum S1MJ is a spontaneous streptomycin-resistant strain, identical to the wild-type S1 in all respects studied (8a). E. coli DH5α was used for plasmid transformation and E. coli S17-1 was used for plasmid mobilization by conjugation into R. rubrum. E. coli strains were grown in Luria-Bertani medium (26). R. rubrum strains were grown anaerobically either with ammonium as a N source (N+) or with N2 (N−) at 30°C (24). A red filter (cutoff at 610 nm) was used to minimize tetracycline phototoxicity (16). Where required, antibiotics were added to the growth medium (final concentrations are in micrograms per milliliter): tetracycline, 15 for E. coli and 3 for R. rubrum; streptomycin, 200 for S1MJ and 100 for the ntrBC mutant UR381 of R. rubrum (30); kanamycin, 20 for UR381; and ampicillin, 50 for E. coli.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− Δ Φ80d lacZΔM15 endA1 recA1 hsdR17(rm− mK+) supE44 thi-1 gyrA96 Δ(lacZYA-argF)U169 | 7 |
| S17-1 | Smr Spr Tra+; pro thi hsdR recA; chromosomal integration of RP4-2-Tc::Mu-Kan::Tn7 | 27 |
| R. rubrum | ||
| S1MJ | Smr; a spontaneous strain of S1 | This laboratory |
| UR381 | Kmr Smr; Δ ntrBC1::Kan | 30 |
| Plasmids | ||
| pRKD418 | Tcr Tpr; cloning vector | 17 |
| pGEM-luc | Apr; source for luc | Promega |
| pJJ303 | Apr; pGEM-luc derivative containing tac promoter upstream of luc | 22 |
| pJOM | Apr; pGEM3zf(+) derivative containing glnB, glnA, and flanking regions | 9 |
| pJCN100 | Apr; pJJ303 derivative containing glnB and glnA | This work |
| pSNC208 | Tcr Tpr;pRKD418 digested by BamHI-XhoI and ligated with BamHI-XhoI luc from pGEM-luc | This work |
| pRKD419 | Tcr Tpr; pRKD418 with a complete BgIII-BamHI digestion and religation | This work |
| pSNC109 | Tcr Tpr; pRKD419 digested by NsiI-XhoI and ligated with NsiI-XhoI luc from pGEM-luc | This work |
| pSNCBP1 | Tcr Tpr; pSNC109 derivative containing glnBp1 | This work |
| pSNCB1 | Tcr Tpr; pSNC109 derivative containing glnBp1 and glnBp2 | This work |
| pSNCB2 | Tcr Tpr; pSNC109 derivative containing glnBp2 | This work |
| pSNCB1A | Tcr Tpr; pSNC109 derivative containing glnBp1 and glnBp2, glnB and upstream of glnA | This work |
| pSNCB2A | Tcr Tpr; pSNC109 derivative containing glnBp2, glnB and upstream of glnA, | This work |
| pSNCA1+ | Tcr Tpr; pSNC208 derivative containing glnB and upstream of glnA | This work |
| pSNCA1− | Tcr Tpr; same as pSNCA1+ but with insertion in opposite direction | This work |
| pSNCglnA | Tcr Tpr; pSNC109 derivative containing 189-bp 3′-end of glnB and upstream of glnA | This work |
| pSNCref | Tcr Tpr; pSNC109 derivative containing 53 bp upstream of glnA | This work |
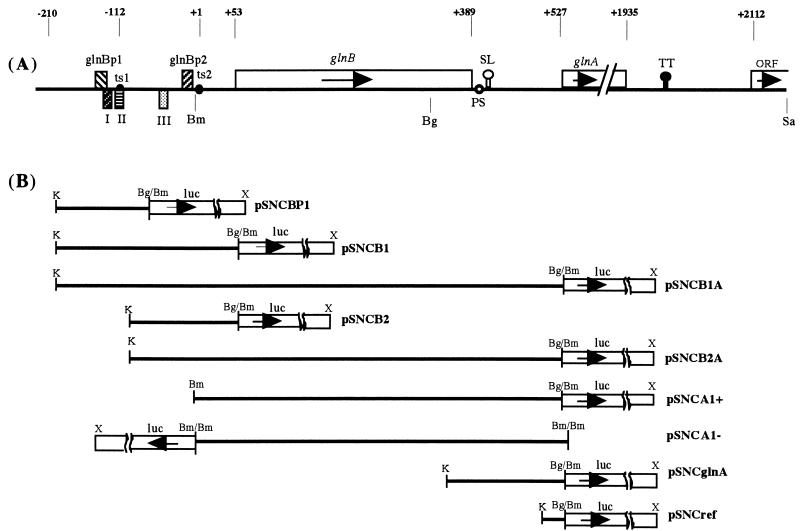
Plasmid purification, cloning, and transformation and restriction enzyme digestion were performed according to standard methods (26). Six PCR primers (28- to 33-mers) were synthesized, based on the nucleotide sequence of the glnBA operon of R. rubrum (9), including a KpnI, BglII, or BamHI restriction site near the 5′ end. The same ribosome binding site and similar numbers of nucleotides between the ribosome binding site and the coding start site of luc in the amplified DNA as those in chromosomal glnB and glnA were designed. With pJOM (Fig. 1A) as a template, DNA fragments containing the possible promoter region and/or glnB were amplified by PCR. After each amplification, DNA was digested by KpnI and BamHI or BglII and inserted upstream of luc (encoding firefly luciferase) in a broad-host-range promoterless plasmid, pSNC109 or pSNC208 (Table 1; Fig. 1B). The constructs were verified by restriction enzyme digestion analysis.
FIG. 1.
Genetic mapping of the glnBA region of R. rubrum and construction of plasmids carrying promoters glnB and/or glnA. (A) The putative ς70-dependent (glnBp1) and ς54-dependent (glnBp2) promoters and NtrC binding sites (I, II, and III) are shown by rectangles. The arrows indicate the direction of transcription. Transcription start sites of glnB and glnA are marked ts1 and ts2. The point of origin +1 in the numbering above the map corresponds to ts2. A stem-loop structure (SL) in the intergenic region of glnB and glnA and a potential transcriptional terminator (TT) downstream of glnA are shown as open- and closed-circle pins, respectively. A putative processing site of the glnBA transcript is designated PS. (B) Relevant DNA fragments amplified by PCR and inserted upstream of the firefly luc gene in the broad-host-range promoterless plasmid pSNC109 or pSNC208. Restriction enzymes sites: Bg, BglII; Bm, BamHI; K, KpnI; X, XhoI; Sa, SacI.
Plasmid transfer from E. coli S17-1 into R. rubrum was essentially carried out according to the mating procedure of Liang et al. (13), but tetracycline and streptomycin or tetracycline, streptomycin, and kanamycin were used to select transconjugated R. rubrum. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting of PII, glutamine synthetase, and luciferase were essentially performed as described previously (10). An affinity-purified luciferase antibody (Promega) was used to probe the luciferase. The amount of protein was estimated by laser densitometry with a Molecular Dynamics Personal Densitometer. Luciferase activity was determined in cell extracts by a luciferase assay system (Promega). Glutamine synthetase activity was measured by the transferase assay (28), except that 0.1 M Tris-HCl buffer (pH 7.6) was used. Protein concentration was measured by the Bio-Rad protein assay.
In a previous report, evidence was provided for the transcription of the glnBA operon from two promoters, glnBp1 and glnBp2, with similarity to the consensus sequences of ς70- and ς54-dependent promoters, respectively, whose activity was dependent on N status (9). However, unlike what was observed for other N-regulated promoters (18), transcription from glnBp2 under nitrogen-sufficient conditions could also be demonstrated and, in an ntrBC mutant of R. rubrum (30), a glnBp2 transcript was still produced (9). Another central issue is the origin of a glnA mRNA, as we were not able to identify a third promoter controlling the transcription of glnA alone. To address these questions, a reporter system was constructed with firefly luciferase as the reporter enzyme. In R. rubrum strains containing the plasmids (Table 1; Fig. 1B), PII and luciferase were expressed separately from plasmids carrying luc and/or glnB; no truncated forms of PII or luciferase were present, as demonstrated by SDS-PAGE and Western blotting (data not shown).
The results shown in Table 2 indicate that in wild-type (S1MJ) strains containing plasmids with the glnBp2 promoter included, alone or together with glnBp1, there was a 3- to 17-fold increase in luciferase activity when cells were grown under N− conditions, compared to N+ conditions. The highest activity was obtained with plasmids containing both promoters (pSNCB1 and pSNCB1A), indicating that the entire glnBp1 and glnBp2 regions are required for maximal transcription. In the absence of either one or the other, expression was significantly lower than with both. Expression from glnBp2 alone was higher than from glnBp1 alone, and the former was required for N regulation (compare pSNCB2 with pSNCBP1). There are three potential NtrC binding sequences upstream of glnBp2 (Fig. 1) which could account for the observed N regulation, confirmed by the absence of N regulation in the ntrBC mutant strain (9, 30). The observation that significant NtrC-dependent transcription occurs from glnBp2 even without two of the potential NtrC binding sequences is interesting and might indicate that NtrC can activate transcription from solution, as was recently shown (23), or that NtrC can bind to another sequence less like the consensus NtrC binding sequence. Other possibilities could be that there are alternate activators in R. rubrum or that transcription by another RNA polymerase can occur from glnBp2 or from a sequence overlapping this region. In this context, it is interesting that the C normally found at position −12 in ς54-dependent promoters is an A in glnBp2 in R. rubrum (19).
TABLE 2.
Luciferase activity in R. rubrum cultures carrying plasmids with inserts upstream of luca
| Plasmid | Features upstream of luc | Activity of strain in medium
|
|||
|---|---|---|---|---|---|
| S1MJ
|
UR381
|
||||
| N− | N+ | N− | N+ | ||
| pSNCBP1 | glnBp1 | 769 | 509 | 947 | 741 |
| pSNCB1 | glnBp1 and glnBp2 | 21,007 | 1,231 | 2,291 | 1,950 |
| pSNCB1A | glnBp1, glnBp2, and glnB | 9,302 | 1,508 | 2,543 | 2,170 |
| pSNCB2 | glnBp2 | 4,070 | 1,042 | 1,418 | 1,140 |
| pSNCB2A | glnBp2 and glnB | 3,494 | 1,169 | 1,662 | 1,302 |
| pSNCA1+ | glnB | 194 | 168 | 171 | 159 |
| pSNCA1− | glnB, reversed direction | 150 | 138 | 140 | 110 |
| pSNCglnA | 3′ of Bg in glnB | 165 | 141 | 150 | 135 |
| pSNCref | 53 bp upstream of glnA | 179 | 163 | 123 | 110 |
| pSNC109 | No promoter | 140 | 128 | 127 | 125 |
| pSNC208 | No promoter | 135 | 141 | 124 | 131 |
R. rubrum S1MJ and UR381 (ntrBC−) were anaerobically grown in NH44-containing (N+) or N-free (N−) medium. Luciferase activity was measured in the cell extract as quanta seconds−1 nanograms of protein−1. The data shown are the averages of three independent assays. The standard deviation was less than 15% of the average.
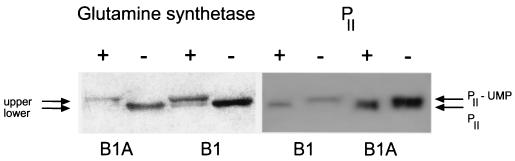
When the glnB gene was included in the insert upstream of luc (pSNCB1A and pSNCB2A), a lower level of transcription than that with the corresponding plasmid without glnB (pSNCB1 and pSNCB2) was obtained. We suggest that the reason for this effect is an accumulation of PII and thereby a higher level of the unmodified form. This would lead to an increase in the phosphatase activity of NtrB and thus a lower level of phosphorylated NtrC, i.e., decreased activation of transcription. This proposal is supported by the results shown in Fig. 2, where an increase in the total amount of both PII (compare B1 and B1A; Fig. 2, right) and the unmodified form (compare − lanes in B1 and B1A; Fig. 2, right) is demonstrated in the strain containing pSNCB1A (the separation of two bands in the B1A + lane is clearly seen with a shorter exposure of the film, but then the B1 bands are barely detectable). Furthermore, the amount of glutamine synthetase was significantly lower in that strain (compare B1A and BA; Fig. 2, left), indicating that transcription from the glnBp1-glnBp2 region of the chromosomal copy of the glnBA operon is regulated in the same way as the one in the plasmid. These results are supported by measurements of glutamine synthetase activity (Table 3). The strains carrying plasmids with the glnB gene show about 50% activity compared to those without glnB.
FIG. 2.
Accumulation and modification of PII and glutamine synthetase in R. rubrum. Extracts from cells grown under N+ or N− conditions were subjected to SDS-PAGE and Western blotting. B1 denotes strain S1MJ, carrying pSNCB1, and B1A denotes strain S1MJ, carrying pSNCB1A. The left panel shows glutamine synthetase and the right panel shows PII. Upper, adenylylated form of glutamine synthetase, lower, unmodified form of glutamine synthetase.
TABLE 3.
Glutamine synthetase activity of R. rubrum S1MJ and the ntrBC mutant UR381 harboring plasmids with different inserts upstream of luca
| Plasmid | Activity of strain or mutant in medium
|
|||
|---|---|---|---|---|
| S1MJ
|
UR381
|
|||
| N− | N+ | N− | N+ | |
| None | 4.4 | 1.0 | 0.7 | 0.6 |
| pSNCB1 | 4.5 | 0.9 | 0.9 | 0.7 |
| pSNCB1A | 2.7 | 0.8 | 1.0 | 0.9 |
| pSNCB2 | 4.6 | 1.1 | 1.0 | 1.0 |
| pSNCB2A | 2.8 | 1.0 | 0.9 | 0.8 |
| pSNCA1+ | 4.6 | 1.1 | 0.9 | 0.8 |
| pSNCA1− | 4.3 | 1.0 | 1.1 | 0.9 |
| pSNCglnA | 4.7 | 1.1 | 0.9 | 0.7 |
| pSNCref | 4.8 | 1.3 | 1.1 | 0.9 |
R. rubrum cultures were grown in N-free (N−) medium or NH4+-containing (N+) medium, and glutamine synthetase activity was measured in cell extracts as γ-glutamyltransferase activity (in micromoles minutes−1 milligrams of protein−1). The results shown are the average of two different experiments.
Although maximal transcription of the glnBA operon occurs under N− conditions, requiring NtrC and the region containing the putative NtrC binding sequence(s) close to glnBp1, there was still significant transcription in UR381 (the ntrBC mutant), containing the same plasmids as the S1MJ strains (Table 2), which again shows that transcription from glnBp2 is not absolutely dependent on NtrC. It should be emphasized that NtrC is not required for nif transcription in R. rubrum, as shown by Zhang et al. (30).
Transcription from the glnBp1 promoter was also investigated by inserting this promoter directly upstream of the luc gene (pSCNBP1). As shown in Table 2, transcription was rather low, indicating that even under N+ conditions transcription from glnBp2 plays a more important role. It should be noted that transcription from glnBp1 was above background, as defined by the luciferase activity in strains containing plasmids with luc but no promoter (pSNC109 and pSNC208). The possible physiological role of glnBp1 is to provide a basic (low) level of glnBA products, whereas activation from glnBp2 occurs when an increase in glutamine synthetase is required. In the light of the results reported here, the mechanism(s) of such activation may involve more than the Ntr system.
An important goal of this investigation was to establish the origin of the glnA mRNA. Four plasmids (pSNCA1+, SNCA1−, pSNCglnA, and pSNCref) containing different parts of the glnBA operon between glnBp2 and the start of glnA (Fig. 1B) were constructed to address this issue. As shown in Table 2, neither of the strains containing these plasmids showed luciferase activity that was significantly higher than background. We believe that this clearly shows the absence of a complete promoter 3′ of the BamHI site in glnB and therefore that the glnA mRNA results from processing of the glnBA transcript.
In conclusion, we have shown that the transcriptional regulation of the glnBA operon in R. rubrum is, in some central aspects, different from that in R. capsulatus, the phototrophic bacterium for which the genetics of nitrogen fixation and ammonium assimilation have so far been best characterized. Transcription from the glnBp2 promoter region is dominant under both N+ and N− conditions, and although significantly enhanced by NtrC under N− conditions, it is not strictly dependent on this activator. This could indicate that either the ς54-dependent RNA polymerase is activated by other factors in addition to NtrC or there is additional polymerase(s) catalyzing transcription from the glnBp2 region. We have never been able to detect significant changes in the amount of PII in R. rubrum under conditions where glutamine synthetase increases, and we believe that this is due to the processing of the glnBA transcript, which results in an increased level of glnA mRNA and a rapid degradation of the glnB mRNA, which we never detected. It is reasonable to assume that the PII level remains unchanged, as there is no evidence for an increased level in its target proteins, NtrB and GlnE, in R. rubrum. It is also possible that specific mRNA processing is more common in phototrophs than has been reported so far. However, whether the processing is regulated in R. rubrum (as it is in the best-studied processing system in phototrophs, the puf genes in R. capsulatus [11]) remains to be established.
Acknowledgments
This work was supported by grants to S.N. from the Swedish Natural Science Research Council.
We are indebted to Gary P. Roberts for giving us the R. rubrum ntrBC mutant. Michael W. Mather and Janet Jansson are acknowledged for the gift of plasmids. We also thank Janet Jansson for valuable discussions.
REFERENCES
- 1.Benelli E M, Souza E M, Funayama S, Rigo L U, Pedrosa F O. Evidence for two possible glnB-type genes in Herbaspirillum seropedicae. J Bacteriol. 1997;179:4623–4626. doi: 10.1128/jb.179.14.4623-4626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlberg I, Nordlund S. Purification and partial characterization of glutamate synthase from Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem J. 1991;279:151–154. doi: 10.1042/bj2790151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiuarazzi M, Iaccarino M. Transcriptional analysis of the glnB-glnA region of Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1990;4:1727–1735. doi: 10.1111/j.1365-2958.1990.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 4.de Zamaroczy M, Delorme F, Elmerich C. Characterization of three different nitrogen-regulated promoter regions for the expression of glnB and glnA in Azospirillum brasilense. Mol Gen Genet. 1990;224:421–430. doi: 10.1007/BF00262437. [DOI] [PubMed] [Google Scholar]
- 5.de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster-Hartnett D, Kranz R G. The Rhodobacter capsulatus glnB gene is regulated by NtrC at tandem rpoN-independent promoters. J Bacteriol. 1994;176:5171–5176. doi: 10.1128/jb.176.16.5171-5176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D. Techniques for transformation of E. coli. In: Clover M, editor. DNA cloning. Vol. 1. Oxford, England: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 8.Holtel A, Merrick M. Identification of the Klebsiella pnumoniae glnB gene: nucleotide sequence of wild-type and mutant alleles. Mol Gen Genet. 1988;215:134–138. doi: 10.1007/BF00331314. [DOI] [PubMed] [Google Scholar]
- 8a.Johansson, M. Unpublished data.
- 9.Johansson M, Nordlund S. Transcription of the glnB and glnA genes in the photosynthetic bacterium Rhodospirillum rubrum. Microbiology. 1996;142:1265–1272. doi: 10.1099/13500872-142-5-1265. [DOI] [PubMed] [Google Scholar]
- 10.Johansson M, Nordlund S. Uridylylation of the PII protein in the photosynthetic bacterium Rhodospirillum rubrum. J Bacteriol. 1997;179:4190–4194. doi: 10.1128/jb.179.13.4190-4194.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klug G. Post-transcriptional control of photosynthesis gene expression. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1235–1244. [Google Scholar]
- 12.Kranz R G, Pace V M, Caldicott I M. Inactivation, sequence, and lacZ fusion analysis of a regulatory locus required for repression of nitrogen fixation genes in Rhodobacter capsulatus. J Bacteriol. 1990;172:53–62. doi: 10.1128/jb.172.1.53-62.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang J, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magasanik B. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochemie. 1989;71:1005–1012. doi: 10.1016/0300-9084(89)90104-1. [DOI] [PubMed] [Google Scholar]
- 15.Martin G B, Thomashow M F, Chelm B K. Bradyrhizobium japonicum glnB, a putative nitrogen-regulatory gene, is regulated by NtrC at tandem promoters. J Bacteriol. 1989;171:5638–5645. doi: 10.1128/jb.171.10.5638-5645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin J P, Colina J K, Logsdon N. Role of oxygen radicals in the phototoxicity of tetracycline towards Escherichia coli. J Bacteriol. 1987;169:2516–2522. doi: 10.1128/jb.169.6.2516-2522.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mather M W, McReynolds L M, Yu C-A. An enhanced broad-host-range vector for gram-negative bacteria: avoiding tetracycline phototoxicity. Gene. 1995;156:85–88. doi: 10.1016/0378-1119(95)00074-g. [DOI] [PubMed] [Google Scholar]
- 18.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 19.Merrick M J, Edwards R E. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel-Reydellet N, Desnoues N, Elmerich C, Kaminski P A. Characterization of Azorhizobium caulinodans glnB and glnA genes: involvement of the PII protein in symbiotic nitrogen fixation. J Bacteriol. 1997;179:3580–3587. doi: 10.1128/jb.179.11.3580-3587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minchin S D, Austin S, Dixon R A. Transcriptional activation of the Klebsiella pneumoniae nifLA promoter by NTRC is face-of-the-helix dependent and the activator stabilizes the interaction of sigma 54-RNA polymerase with the promoter. EMBO J. 1989;8:3491–3499. doi: 10.1002/j.1460-2075.1989.tb08514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Möller A, Gustafsson K, Jansson J K. Specific monitoring by PCR amplification and bioluminescence of firefly luciferase gene-tagged bacteria added to environmental samples. FEMS Microbiol Ecol. 1994;15:193–206. [Google Scholar]
- 23.North A K, Kustu S. Mutant forms of the enhancer-binding protein NtrC can activate transcription from solution. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 24.Ormerod J G, Ormerod K S, Gest H. Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch Biochem Biophys. 1961;94:449–463. doi: 10.1016/0003-9861(61)90073-x. [DOI] [PubMed] [Google Scholar]
- 25.Qian Y, Tabita F R. Expression of glnB and a glnB-like gene (glnK) in a ribulose bisphosphate carboxylase/oxygenase-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1998;180:4644–4649. doi: 10.1128/jb.180.17.4644-4649.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Simon R, Priefer U, Pühler A A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 28.Soliman A, Nordlund S. Purification and partial characterization of glutamine synthetase from the photosynthetic bacterium Rhodospirillum rubrum. Biochim Biophys Acta. 1989;994:138–141. doi: 10.1016/0167-4838(89)90152-0. [DOI] [PubMed] [Google Scholar]
- 29.Woehle D L, Lueddecke B A, Ludden P W. ATP-dependent and NAD-dependent modification of glutamine synthetase from Rhodospirillum rubrum in vitro. J Biol Chem. 1990;265:13741–13749. [PubMed] [Google Scholar]
- 30.Zhang Y, Cummings A D, Burris R H, Ludden P W, Roberts G P. Effect of an ntrBC mutation on the posttranslational regulation of nitrogenase activity in Rhodospirillum rubrum. J Bacteriol. 1995;177:5322–5326. doi: 10.1128/jb.177.18.5322-5326.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zinchenko V, Churin Y, Shestopalov V, Shestakov S. Nucleotide sequence and characterization of the Rhodobacter sphaeroides glnB and glnB genes. Microbiology. 1994;140:2143–2151. doi: 10.1099/13500872-140-8-2143. [DOI] [PubMed] [Google Scholar]