Abstract
A lysozyme-osmotic shock method is described for fractionation of Alcaligenes faecalis which uses glucose to adjust osmotic strength and multiple osmotic shocks. During phenylethylamine-dependent growth, aromatic amine dehydrogenase, azurin, and a single cytochrome c were localized in the periplasm. Their induction patterns are different from those for the related quinoprotein methylamine dehydrogenase and its associated redox proteins.
Aromatic amine dehydrogenase (AADH) from Alcaligenes faecalis is induced during growth on phenylethylamine (19). It catalyzes the oxidative deamination of primary amines (12) and subsequent electron transfer to azurin, a type I blue copper protein (13). AADH is the second enzyme (7) known to contain the tryptophan tryptophylquinone (TTQ) cofactor (17). The other TTQ enzyme is a methylamine dehydrogenase (MADH), which has been isolated from various sources (8, 11, 15, 16, 20, 22). MADH is localized in the periplasmic space and donates electrons to periplasmic c-type cytochromes via another copper protein, amicyanin (9, 10). With a newly developed fractionation method, AADH, azurin, and a single c-type cytochrome were localized in the periplasm of A. faecalis. Their induction patterns were also characterized and compared with those of MADH and the analogous periplasmic redox proteins with which it interacts.
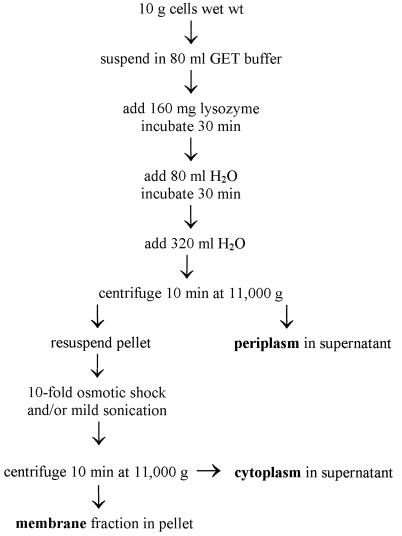
A. faecalis was cultured aerobically at 30°C with β-phenylethylamine as the sole carbon source (7). It was not previously possible to efficiently fractionate A. faecalis with conventional lysozyme-osmotic shock methods (18, 23) that generate spheroplasts, using sucrose to adjust osmotic strength. We hypothesized that the problem was that the exclusion size of the outer membrane porin of A. faecalis is too small to allow easy diffusion of sucrose (14). Use of glucose rather than sucrose allowed efficient fractionation of the cells. Glycine and NaCl were also tested as osmotic agents and found to be better than sucrose but not as good as glucose. Furthermore, whereas a single osmotic shock is typically used, with A. faecalis use of a second osmotic shock increased the release of the periplasm from about 15 to 75%. This allowed us to localize the redox proteins that are associated with phenylethylamine-dependent growth. A flowchart describing the new modified fractionation method is shown in Fig. 1. Purifications of AADH (7) and azurin (5) from the periplasmic extract were as described previously.
FIG. 1.
Flowchart for optimal fractionation of A. faecalis. All procedures were performed at 30°C. Cells were resuspended in 0.5 M glucose plus 1 mM EDTA in 0.2 M Tris-HCl, pH 7.4 (GET buffer), to a concentration of 1 g (wet weight) of cells per 8 ml of buffer. Lysozyme, which was first suspended in water, was then added to a concentration 1 mg/ml. Phenylmethylsulfonyl fluoride (a protease inhibitor) was added to 100 μg/ml. Under conditions when some release of cytoplasmic components was suspected, 20 mM MgCl2 plus 10 μg of DNAse I per ml were added the supernatant to reduce viscosity due to release of DNA. Disruption of spheroplasts that were separated from the periplasm was accomplished by a 10-fold osmotic shock followed by mild ultrasonication (30 W of power for 10 min).
A large majority of the total AADH and azurin was found in the periplasmic fraction (Table 1). Malate dehydrogenase, a cytoplasmic marker, was essentially absent from the osmotic shock supernatant and found primarily in the soluble fraction after disruption of spheroplasts. The single soluble cytochrome c that was identified (discussed later) was also localized in the periplasm.
TABLE 1.
Localization of proteins in A. faecalis
| Cell fractiona | % Total recovery (n = 4)
|
|||
|---|---|---|---|---|
| AADHb | Azurinc | Cytochromed | Malate dehydrogenasee | |
| Periplasm | 73.9 ± 2.5 | 76.5 ± 3.1 | 100 | 7.5 ± 0.9 |
| Cytoplasm | 24.3 ± 2.1 | 24.5 ± 0.3 | 0 | 89.8 ± 3.1 |
| Membrane | 1.6 ± 0.2 | NDf | 0 | 2.6 ± 0.1 |
Cells were fractionated as described in Fig. 1.
The steady-state assay for AADH activity was performed as described previously (12).
The amount of purified azurin was quantitated using the extinction coefficient ɛ600 = 4,000 M−1 cm−1 (13).
Cytochrome c levels were quantitated by performing densitometry of the heme-stained proteins after SDS-PAGE.
The assay of malate dehydrogenase was performed as described previously (1).
ND, it was not possible to determine what azurin if any remains associated with the membrane fraction as it is a soluble protein that can be detected only spectrophotometrically.
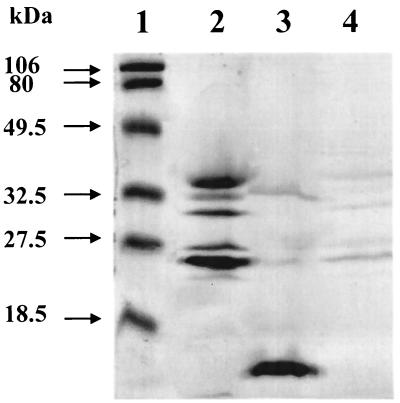
Whereas cytoplasmic dehydrogenases typically transfer electrons to the respiratory chain via NADH, periplasmic dehydrogenases often transfer electrons to the respiratory chain via c-type cytochromes. Heme staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of cell fractions of phenylethylamine-grown A. faecalis revealed several heme-containing proteins (Fig. 2). Reducing agents were included in the sample buffer because they substantially diminish the peroxidase activity of noncovalent hemoproteins, making this strain reasonably specific for c-type cytochromes (6). One 13-kDa cytochrome is clearly localized in the periplasm. The other heme-staining species are primarily associated with the membrane. We previously identified two cytochromes in whole-cell sonicated extracts of A. faecalis that could accept electrons from AADH via azurin in vitro (5). These appear to correspond to the 13- and 25-kDa cytochromes. The latter was most likely released from the membrane into the soluble fraction in the earlier study by the prolonged sonication which was employed to completely disrupt cells.
FIG. 2.
Localization of c-type cytochromes of A. faecalis. Samples were subjected to SDS-PAGE on a 15% polyacrylamide gel. An approximately equivalent amount of each cell fraction on a per-cell-weight basis was loaded onto the gel. The gel was specifically stained for heme-containing proteins as previously described (6). Lane 1, Bio-Rad prestained molecular weight markers; lane 2, membrane fraction; lane 3, periplasmic fraction; lane 4, cytoplasmic fraction.
Nearly 100% of the 13-kDa cytochrome appears in the periplasmic fraction, whereas only 74% of the AADH activity is present (Table 1, Fig. 2). This could be due to the relative size of the two proteins, 13 kDa for the cytochrome and 125 kDa for AADH. If only partial degradation of the cell wall occurs during lysozyme treatment, the smaller protein may be more readily released. Interestingly, only 77% of azurin (13.4 kDa) is released. This may mean that AADH and azurin are associated in the periplasm prior to release from the cell.
To identify which of these proteins are specifically induced by phenylethylamine, cells were also grown on Luria-Bertani (LB) medium and fractionated. No AADH activity was detected in these cells. Azurin was produced by these cells, but the level of azurin was only 28% of that produced by phenylethylamine-grown cells. Levels of the 13-kDa periplasmic cytochrome and the membrane-associated cytochromes were similar in cells grown on LB and on phenylethylamine (data not shown).
The induction pattern of periplasmic redox proteins in A. faecalis in the presence of phenylethylamine is different from the induction pattern of periplasmic redox proteins in Paracoccus denitrificans in the presence of methylamine. Like AADH, MADH is synthesized by P. denitrificans only when the substrate amine is present as the sole carbon source (11). The synthesis of amicyanin is also observed only with methylamine present as the sole carbon source (9). MADH and amicyanin genes are known to be under the control of the same promoter (3, 21). In contrast, while azurin levels are greater during growth on phenylethylamine, it is also synthesized in its absence. This suggests that in A. faecalis either the genes for AADH and azurin are not under control of the same promoter or multiple genes for azurin are present, one constitutive and one inducible. In P. denitrificans, three periplasmic c-type cytochromes are present during growth on methylamine, a constitutive cytochrome c-550 and two inducible cytochromes, c-551i and c-553i (10). Cytochrome c-551i is the most efficient acceptor of electrons from MADH and amicyanin in vitro (10). These inducible cytochromes c do not react directly with the membrane-bound respiratory chain but transfer electrons to it via cytochrome c-550 (4). In A. faecalis, when AADH is induced by phenylethylamine, no additional cytochromes c are induced and the level of the periplasmic 13-kDa cytochrome is unchanged. Thus, in A. faecalis, phenylethylamine-derived electrons are likely donated from AADH and azurin to the respiratory chain via the single periplasmic cytochrome c. Its physiologic role may be like that of cytochrome c-550 in P. denitrificans (2), which mediates electron transfer to cytochrome oxidase from multiple periplasmic dehydrogenases.
In the past, the characterization of proteins of A. faecalis as periplasmic was primarily inferred from the presence of signal sequences in their genes. Sequences of the genes for AADH and these A. faecalis cytochromes have not been reported. The new modified lysozyme-osmotic shock method described in Fig. 1 allows efficient fractionation of A. faecalis and will now allow a detailed study of the physiologic redox reactions associated with the dissimilation of aromatic amines, as well as better characterization of other periplasmic proteins and metabolic processes.
Acknowledgments
We thank Siddhartha De for technical assistance with densitometry measurements.
This work was supported by NIH grant GM-41574.
REFERENCES
- 1.Alefounder P R, Ferguson S J. The location of dissimilatory nitrite reductase and the control of dissimilatory nitrate reductase by oxygen in Paracoccus denitrificans. Biochem J. 1980;192:231–240. doi: 10.1042/bj1920231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S C, Ferguson S J, Ludwig B, Page M D, Richter O-M H, van Spanning R J M. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol Mol Biol Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistosedov A Y, Boyd J, Mathews F S, Lidstrom M E. The genetic organization of the mau gene cluster of the facultative autotroph Paracoccus denitrificans. Biochem Biophys Res Commun. 1992;184:1181–1189. doi: 10.1016/s0006-291x(05)80007-5. [DOI] [PubMed] [Google Scholar]
- 4.Davidson V L, Kumar M A. Cytochrome c-550 mediates electron transfer from inducible periplasmic c-type cytochromes to the cytoplasmic membrane of Paracoccus denitrificans. FEBS Lett. 1989;245:271–273. doi: 10.1016/0014-5793(89)80235-2. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S L, Davidson V L, Hyun Y L, Wingfield P T. Spectroscopic evidence for a common electron transfer pathway for two tryptophan tryptophylquinone enzyme. J Biol Chem. 1995;270:4293–4298. doi: 10.1074/jbc.270.9.4293. [DOI] [PubMed] [Google Scholar]
- 6.Francis R T, Becker R B. Specific indication of hemoproteins in polyacrylamide gels using a double-staining method. Anal Biochem. 1984;136:509–514. doi: 10.1016/0003-2697(84)90253-7. [DOI] [PubMed] [Google Scholar]
- 7.Govindaraj S, Eisenstein E, Jones L H, Sanders-Loehr J, Chistoserdov A Y, Davidson V L, Edwards S L. Aromatic amine dehydrogenase, a second tryptophan tryptophylquinone enzyme. J Bacteriol. 1994;176:2922–2929. doi: 10.1128/jb.176.10.2922-2929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haywood G W, Janschke N S, Large P J, Wallis J M. Properties and subunit structure of methylamine dehydrogenase from Thiobacillus A2 and Methylophilus methylotrophus. FEMS Microbiol Lett. 1982;15:79–82. [Google Scholar]
- 9.Husain M, Davidson V L. An inducible periplasmic blue copper protein from Paracoccus denitrificans. J Biol Chem. 1985;260:14626–14629. [PubMed] [Google Scholar]
- 10.Husain M, Davidson V L. Characterization of two inducible periplasmic c-type cytochromes from Paracoccus denitrificans. J Biol Chem. 1986;261:8577–8580. [PubMed] [Google Scholar]
- 11.Husain M, Davidson V L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987;169:1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyun Y L, Davidson V L. Mechanistic studies of aromatic amine dehydrogenase, a tryptophan tryptophylquinone enzyme. Biochemistry. 1995;34:816–823. doi: 10.1021/bi00003a015. [DOI] [PubMed] [Google Scholar]
- 13.Hyun Y L, Davidson V L. Electron transfer reactions between aromatic amine dehydrogenase and azurin. Biochemistry. 1995;34:12249–12254. doi: 10.1021/bi00038a020. [DOI] [PubMed] [Google Scholar]
- 14.Ishii J, Taiji N. Size of diffusion pore of Alcaligenes faecalis. Antimicrob Agents Chemother. 1988;32:378–384. doi: 10.1128/aac.32.3.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenny W C, McIntire W. Characterization of methylamine dehydrogenase from bacterium W3A1. Interaction with reductants and amino-containing compounds. Biochemistry. 1983;22:3858–3868. doi: 10.1021/bi00285a022. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto T, Hiroka B Y, Tobari J. Methylamine dehydrogenase of Pseudomonas sp. J. Isolation and properties of the subunits. Biochim Biophys Acta. 1978;522:303–310. doi: 10.1016/0005-2744(78)90064-5. [DOI] [PubMed] [Google Scholar]
- 17.McIntire W S, Wemmer D E, Chistoserdov A Y, Lindstrom M E. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991;252:817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- 18.Nossal N G, Heppel L A. The release of enzymes by osmotic shock from Escherichia coli in exponential phase. J Biol Chem. 1966;241:3055–3062. [PubMed] [Google Scholar]
- 19.Nozaki M. Aromatic amine dehydrogenase from Alcaligenes faecalis. Methods Enzymol. 1987;142:650–655. doi: 10.1016/s0076-6879(87)42077-6. [DOI] [PubMed] [Google Scholar]
- 20.Shirai S, Matsumoto T, Tobari J. Methylamine dehydrogenase of Pseudomonas AM1. A subunit enzyme. J Biochem. 1978;83:1599–1607. doi: 10.1093/oxfordjournals.jbchem.a132071. [DOI] [PubMed] [Google Scholar]
- 21.Van der Palen C J N M, Slotboom D, Jongejan L, Reijnders W N M, Harms N, Duine J A, Van Spanning R J M. Mutational analysis of mau genes involved in methylamine metabolism in Paracoccus denitrificans. Eur J Biochem. 1995;230:860–871. doi: 10.1111/j.1432-1033.1995.tb20629.x. [DOI] [PubMed] [Google Scholar]
- 22.Vellieux F M D, Frank J J, Swarte M B A, Groendijk H, Duine J A, Drenth J, Hol W G J. Purification, crystallization and preliminary X-ray investigation of quinoprotein methylamine dehydrogenase from Thiobacillus versutus. Eur J Biochem. 1986;154:383–386. doi: 10.1111/j.1432-1033.1986.tb09409.x. [DOI] [PubMed] [Google Scholar]
- 23.Witholt B, Boekhout M, Brock M, Kingma J, van Heerikhuizen H, de Leij L. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]