Abstract
Studies by R. Lin et al. (J. Bacteriol. 174:1948–1955, 1992) suggested that the Escherichia coli leu operon might be a member of the Lrp regulon. Their results were obtained with a leucine auxotroph; in leucine prototrophs grown in a medium lacking leucine, there was little difference in leu operon expression between lrp+ and lrp strains. Furthermore, when leuP-lacZ transcriptional fusions that lacked the leu attenuator were used, expression from the leu promoter varied less than twofold between lrp+ and lrp strains, irrespective of whether or not excess leucine was added to the medium. The simplest explanation of the observations of Lin et al. is that the known elevated leucine transport capacity of lrp strains (S. A. Haney et al., J. Bacteriol. 174:108–115, 1992) leads to very high intracellular levels of leucine for strains grown with leucine, resulting in the superattenuation of leu operon expression.
Lrp (leucine-responsive regulatory protein) controls the expression of a number of operons in Escherichia coli involved in amino acid biosynthesis and degradation, the transport of amino acids, and one carbon metabolism (for a review, see references 4, 5, and 16). In general, anabolic target genes are positively regulated and catabolic genes are negatively regulated by Lrp. It is for these reasons that Lrp has been considered to be a global regulator of metabolism in E. coli, playing an especially important role when cells make transitions between rich nutritional conditions and lean conditions in which they must synthesize most of their building blocks from simple carbon sources and salts.
Another interesting feature of Lrp is that its mode of action is sometimes but not always affected by elevated levels of the amino acid leucine. Altogether, six different patterns of regulation by Lrp have been recognized, depending upon whether Lrp acts negatively or positively and upon the way in which leucine affects expression. For those cases in which Lrp acts positively as an activator, leucine sometimes overcomes the effect of Lrp (thus causing reduced expression), sometimes potentiates the effect of Lrp, and sometimes has no effect on Lrp-mediated activation. Similarly, for cases in which Lrp acts negatively, there are examples in which leucine overcomes the effect, is required for the effect, or has no effect upon Lrp-mediated repression.
The leuPABCD operon of E. coli is known to be regulated by a transcription attenuation mechanism (8, 25). Some work by Lin et al. suggested that the leu operon may also be controlled by Lrp (14). Among E. coli strains containing placMu9 insertions that Lin et al. isolated, several had insertions within the leu operon, including strain CP55 with an insertion in leuB (14, 22). For strain CP55 [Φ(leuB-lacZ)] grown with excess leucine, β-galactosidase levels were more than 10-fold higher than those in an isogenic strain containing an inactive lrp gene, suggesting that Lrp acts positively on leu operon expression (14). Furthermore, exogenous leucine increases the growth rate of strains lacking a functional lrp gene (2, 4), leading some to conclude that leucine synthesis in such strains is impaired (2, 16). If Lrp is an activator of leu operon expression, then strains lacking Lrp might be impaired in leucine biosynthesis and thus exhibit a partial leucine requirement (2, 16).
Here we investigate in more detail the role that Lrp plays in regulating expression of the leu operon and the question of whether leucine synthesis is indeed impaired in lrp-containing strains. We found that mutations in Lrp affect leu operon expression to a limited extent, but only indirectly, and that leucine synthesis is likely not impaired in a strain lacking functional Lrp.
We repeated some of the experiments reported by Lin et al. (14) and observed, as they did, a more than 10-fold-lower level of β-galactosidase activity in strain CP55 (lrp+) than in an isogenic lrp strain that we created (Tables 1 and 2). Similar results were obtained with another isogenic set of strains derived from strain P90C to which we transferred the leuB::placMu9 allele (Tables 1 and 2).
TABLE 1.
Bacterial strains of E. coli used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| P90C | ara thi Δlac-pro | 20 |
| CP55 | Δlac ΔilvA leuB::placMu9 | E. Newman |
| CV1008 | P90C ilvIH::Mu dI 1734 lrp-35::Tn10 | 18 |
| CV1216 | P90C lrp-35::Tn10 | This laboratory |
| CV1512 | CP55 lrp-35::Tn10 | This study |
| CV1513 | P90C leuB::placMu9 | This study |
| CV1514 | CV1513 lrp-35::Tn10 | This study |
| CV1517 | P90C λ Pleu att-lacZ | This study |
| CV1518 | CV1517 lrp-35::Tn10 | This study |
| CV1520 | P90C λ Pleu-lacZ | This study |
| CV1521 | CV1520 lrp-35::Tn10 | This study |
| CV1543 | P90C λ PilvIH Pleu att-lacZ | This study |
| CV1544 | CV1543 lrp-35::Tn10 | This study |
| CV1547 | P90C λ PilvIH Pleu-lacZ | This study |
| CV1548 | CV1547 lrp-35::Tn10 | This study |
| CV1549 | P90C λ P(−)ilvIH Pleu-lacZ | This study |
| CV1550 | CV1549 lrp-35::Tn10 | This study |
Strains CV1216, CV1512, CV1514, CV1518, CV1521, CV1544, CV1548, and CV1550 were created by transduction with P1 phage grown on strain CV1008, with selection for growth in the presence of 15 μg of tetracycline per ml. Similarly, strain CV1513 was formed with phage grown on strain CP55 and with 50 μg of kanamycin per ml. Strain CV1517 was constructed by amplifying region −247 to +230 from the leu operon of E. coli by PCR and cloning into plasmid pRS415 upstream of the lacZ gene (20). After transferring the leuP-lacZ fusion to λ phage by homologous recombination (20), single lysogens were identified as described earlier (24). Strains CV1520, CV1543, CV1547, and CV1549 were constructed similarly and contained the following: CV1520, 265 bp (position −247 to +18 from the leu operon); CV1543, 2,147 bp (from +50 downstream of the ilvIH promoter to +230 downstream from the leu promoter); CV1547, 1,927 bp (from position +50 downstream of the ilvIH promoter to +18 downstream from the leu promoter); CV1549, 1,840 bp (from −37 upstream of the ilvIH promoter to +18 downstream from the leu promoter).
TABLE 2.
Specific activity of β-galactosidase in lrp+ and lrp strains having a leuPAB′-lacZ translational fusiona
| Strain | lrp allele | Sp. act. of β-galactosidaseb |
|---|---|---|
| CP55 | lrp+ | 334 ± 46 |
| CV1512 | lrp-35::Tn10 | 19.5 ± 2.4 |
| CV1513 | lrp+ | 166 ± 11 |
| CV1514 | lrp-35::Tn10 | 16.2 ± 0.6 |
Strains were grown at 37°C with shaking in SSA minimal salts (11) containing 0.2% glucose; 50 μg (each) of isoleucine, valine, leucine, and proline per ml; and 5 μg of thiamine per ml. Cultures were grown overnight and diluted to an A550 of 0.01, and samples were taken at different times in the log phase for assay.
The results of Lin et al. (14) and the results shown in Table 2 were obtained with strains that were leucine auxotrophs and therefore the effect of lrp mutations could only be tested for these strains grown with excess leucine or under conditions of leucine limitation. To determine whether an lrp::Tn10 allele affects leu operon expression in cells grown in a minimal medium without excess or limited leucine, we measured the specific activity of the leuB gene product, β-isopropylmalate (β-IPM) dehydrogenase, in a strain having a wild-type leu operon. The results (Table 3) show that a null mutation in lrp had only a small effect upon leuB expression in cells grown in a minimal medium in the absence of leucine. It may be noted that Lin et al. found only a 2.6-fold difference in reporter gene expression between strain CP55 [Φ(leuB-lacZ)] and an isogenic strain carrying an lrp null allele when the two strains were grown to the point where they had depleted their supply of leucine (14). Therefore, a null mutation in lrp has a relatively small effect upon leu operon expression when the intracellular leucine concentration is either undisturbed (our results) or limiting for growth (14).
TABLE 3.
Specific activity of β-IPM dehydrogenase in lrp+ and lrp strainsa
| Strainb | lrp allele | Doubling time (min) in medium
|
Sp. act. of β-IPM dehydrogenaseb
|
||
|---|---|---|---|---|---|
| Without Leu | With Leu | Without Leu | With Leu | ||
| P90C | lrp+ | 57 | 67 | 2.0 ± 0.2 (3) | 0.2 ± 0.05 (3) |
| CV1216 | lrp-35::Tn10 | 82 | 62 | 1.5 ± 0.03 (3) | 0.07 ± 0.03 (3) |
Strains were grown in minimal glucose medium containing 50 μg (each) of isoleucine, valine, and proline per ml and 5 μg of thiamine per ml in the presence or absence of 100 μg of leucine per ml. Cultures were grown overnight, diluted to an A550 of 0.01, and samples were taken at different times in the log phase for assay.
Assays were performed as described previously (19), except that cells were broken by sonication; 2,4-dinitrophenylhydrazones were extracted into toluene and then aqueous sodium carbonate as described by Stieglitz and Calvo (21), except that 1.5 ml of Na2CO3 and 0.25 ml of KOH were used and A540 was determined. Specific activity is expressed as micromoles per hour per milligram of total protein. Values in parentheses represent the number of samples analyzed. Each sample was assayed in duplicate. A repetition of this experiment gave similar results.
For the experiments described in the paragraph above, cells were also grown in the presence of leucine. For each strain, the addition of leucine to the medium resulted in reduced expression of the operon, a result expected because of the known control of this operon by a leucine-dependent transcription attenuation mechanism (8, 25). Under this condition of leucine excess, there was a difference in β-IPM dehydrogenase specific activity in lrp+ and lrp strains, but it was only about 3-fold, rather than the 10-fold difference observed with strains containing leuB-lac translational fusions. However, β-IPM dehydrogenase assays (Table 3) are not as sensitive as β-galactosidase assays (Table 2), and the β-IPM dehydrogenase values shown in Table 3 for strain CV1216 were at the limits of detection.
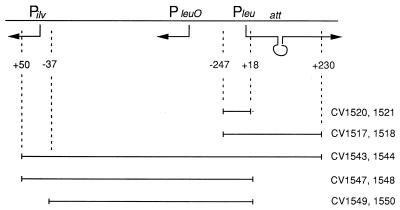
Any effects of an lrp mutation upon leu operon expression could be upon initiation of transcription or upon attenuation of transcription. In an attempt to distinguish between these possibilities, two sets of strains were prepared, each having a wild-type leu operon at its normal location in the chromosome and a leu promoter-lacZ transcriptional fusion in single copy at the phage lambda attachment site. One set of strains contained the leu promoter without the attenuator (position −247 to +18) directly attached to lacZ (strains CV1520 lrp+ and CV1521 lrp-35::Tn10), whereas the second set contained the leu promoter and attenuator (−247 to +230) attached to lacZ (strains CV1517 lrp+ and CV1518 lrp-35::Tn10) (Fig. 1). For constructs having the leu promoter without the attenuator, there was little or no difference in β-galactosidase specific activity between lrp+ and lrp strains grown in minimal medium with or without leucine (Table 4). These results suggest that the leu promoter is not regulated by Lrp, either directly or indirectly. For strains with constructs having both the leu promoter and attenuator and grown in the absence of exogenous leucine, an lrp null allele led to an approximately fourfold-higher level of reporter gene expression, a surprising result that will be discussed later. For the same strains, growth in the presence of leucine resulted in a substantial reduction in reporter gene expression, as expected for a system under attenuation control (23) (Table 4). However, the extent of the repression was about fivefold higher for the strain containing the lrp null allele than for the lrp+ strain, a result reminiscent of the original finding of Lin et al. (Table 2 and reference 14). Taken together, the results shown in Table 4 strongly suggest that any effects that Lrp might have on leu operon expression are not due to effects on transcription initiation but rather on some subsequent process such as transcription attenuation.
FIG. 1.
Schematic representation of constructs used in this study.
TABLE 4.
Specific activity of β-galactosidase in lrp+ and lrp strains having leuP-lacZ transcriptional fusions at the λ attachment sitea
| Strain | lrp allele | leu attenuatorb | Sp. act. of β-galactosidasec in medium
|
Ratio of sp. act. (without Leu/with Leu) | |
|---|---|---|---|---|---|
| Without Leu | With Leu | ||||
| CV1520 | lrp+ | Absent | 1,870 ± 425 | 2,780 ± 243 | 0.67 |
| CV1521 | lrp-35::Tn10 | Absent | 2,020 ± 240 | 1,960 ± 293 | 1.0 |
| CV1517 | lrp+ | Present | 432 ± 110 | 80.7 ± 10.4 | 5.5 |
| CV1518 | lrp-35::Tn10 | Present | 1,950 ± 518 | 64.0 ± 8.9 | 30 |
Culture conditions as in legend to Table 3.
Strains CV1520 and CV1521 have region −247 to +18 from the leu operon (contains leu promoter but not attenuator) attached to a lacZ gene on phage λ in single copy at the λ attachment site (Fig. 1). Strains CV1517 and CV1518 are similar except that they have region −247 to +230 from the leu operon which contains both the leu promoter and attenuator.
Specific activity in Miller units. Values represent the means and standard deviations for multiple samples taken throughout the log growth phase from at least two independent experiments.
We return to the surprising result presented in Table 4 involving constructs having a leu promoter-attenuator–lacZ transcriptional fusion in single copy at the phage λ attachment site. For strains grown in the absence of leucine, reporter gene expression was about fourfold higher in an lrp strain than in an lrp+ strain. The sequence of the construct in the region of the promoter and attenuator was verified by sequencing, but out of concern that a mutation might have occurred during the transfer of leuP att-lacZ to phage lambda, we repeated these experiments with the plasmid-containing strains from which the single copy λ lysogens were prepared, normalizing specific activities for plasmid copy number (estimated by measuring β-lactamase activity) (9). Reporter gene specific activities were higher, as expected for plasmid-containing strains, but the pattern was similar, i.e., there was threefold-higher expression in an lrp strain than in an lrp+ strain (data not shown). In addition, in order to explore any possible effects of Lrp on translation, we cloned this same leu promoter-attenuator fragment (position −247 to +230) into plasmid pRS414 (20) creating a leuA′-lacZ translational fusion. This construct also showed a similar pattern of expression, being about sixfold higher in an lrp strain than in an lrp+ strain (data not shown).
The results in Table 3 (with the leu operon at its normal position in the chromosome) and Table 4 (with the leu promoter and attenuator located at the phage lambda attachment site) seem at odds in the case of cells grown in the absence of leucine, with the effect of an lrp allele being to reduce expression slightly in one case and increase expression in the other. We considered the possibility of this result being related to the long-range interaction between the leu-500 and ilvIH promoters described by M. Fang and H.-Y. Wu (6, 7). They demonstrated that transcription from the leu-500 promoter of Salmonella typhimurium is affected by transcription initiated at the ilvIH promoter, located 1.9 kb away. They postulate a promoter relay mechanism involving another gene, leuO, located between the leu and ilvIH operons. To determine whether similar long-range effects might be at play in the E. coli strains that we analyzed here, we prepared lacZ fusion constructs containing the leu promoter (with or without the attenuator) together with upstream DNA that did or did not include the ilvIH promoter (Fig. 1). For strains possessing the longest construct (upstream DNA including the ilvIH promoter and DNA downstream of the leu promoter, including the attenuator), there was little difference in reporter gene expression between lrp+ and lrp strains when cells were grown in either the absence or presence of leucine (Table 5; Fig. 1). For strains having similar constructs lacking the leu attenuator, reporter gene expression was higher as expected, but again, there were only modest differences (less than twofold) between lrp+ and lrp strains (Table 5; Fig. 1). Finally, to determine whether transcription from the ilvIH promoter had an effect upon leu promoter expression, we analyzed strains having constructs containing nearly the same amount of upstream DNA but lacking the ilvIH promoter. For these strains and conditions of growth, deleting the ilvIH promoter had no discernable effect (Table 5; Fig. 1). As with the shorter leu promoter construct (Table 4), we found only modest differences in reporter gene expression due to a mutation in lrp. Furthermore, comparison of the constructs shows that the upstream DNA containing the ilvIH promoter has no significant effect on expression from the leu promoter alone (Table 5). Taken as a whole, these results again suggest that Lrp has little effect on leu expression, except perhaps for cells grown with excess leucine. There was no hint that Lrp might decrease expression from the leu promoter, as was suggested from results with strains CV1517 and CV1518 (Table 4). These latter results we assume to be some sort of artifact, although we were unable to establish that in our experiments.
TABLE 5.
Specific activity of β-galactosidase in lrp+ and lrp strains having leuP-lacZ transcriptional fusions with upstream DNA at the λ attachment sitea
| Strain | lrp allele | PilvIHb | leu attenuatorb | Sp. act. of β-galactosidasec
|
Ratio of sp. act. (without Leu/with Leu) | |
|---|---|---|---|---|---|---|
| Without Leu | With Leu | |||||
| CV1543 | lrp+ | + | + | 2,500 ± 250 | 96 ± 6.4 | 26 |
| CV1544 | lrp-35::Tn10 | + | + | 2,890 ± 189 | 36 ± 2.3 | 79 |
| CV1547 | lrp+ | + | − | 5,380 ± 473 | 3,370 ± 136 | 1.6 |
| CV1548 | lrp-35::Tn10 | + | − | 3,120 ± 338 | 2,510 ± 246 | 1.2 |
| CV1549 | lrp+ | − | − | 5,370 ± 454 | 3,340 ± 143 | 1.6 |
| CV1550 | lrp-35::Tn10 | − | − | 3,110 ± 267 | 2,430 ± 278 | 1.3 |
Strains were grown as described in Table 3.
+, present; −, absent.
Specific activity is in Miller units. Values represent the means and standard deviations for multiple samples taken throughout log growth phase from at least two independent experiments. Similar results were obtained for cells grown to late log and early stationary phases of growth.
We return to the original observation of Lin et al. (14) that leu operon expression is reduced in an lrp strain grown with excess leucine. We considered the possibility that Lrp indirectly affects leu transcription attenuation by affecting intracellular concentrations of leucine or of leucyl-tRNA. If Lrp were a repressor of leuS (encodes leucyl-tRNA synthetase), then increased levels of leucyl-tRNA synthetase in an lrp-35::Tn10 strain could result in higher levels of leucyl-tRNA and lower levels of leu operon expression through additional attenuation. We tested this possibility by preparing constructs containing leuS-lacZ transcriptional fusions and measuring β-galactosidase levels in lrp+ and lrp strains containing these constructs. No differences in reporter gene expression between the two strains were found (data not shown).
We also explored the potential effects of Lrp upon the transport of leucine that might result in altered intracellular levels of leucine. livJ and livKHMGF, operons involved in transporting leucine into E. coli, are negatively controlled by Lrp, both as measured by transport assays and by levels of reporter gene activity (10). It should be noted that for some strains having lacZ insertions within liv genes, the phenotypes suggest that liv genes are positively controlled by Lrp (3, 14, 22). The discrepancy in results obtained with different fusion constructs is not understood, but for the analysis that follows, we assume that Lrp negatively affects liv expression because this conclusion is also supported by direct measurement of transport activity (10). In an lrp+ strain, exogenous leucine causes extensive repression of the two liv operons (3, 10, 14, 22). By contrast, an lrp strain has high constitutive levels of transport activity and therefore is expected to have high intracellular levels of leucine when grown in the presence of exogenous leucine. Thus, the reduced expression of the leucine operon that is observed in an lrp strain grown in the presence of leucine (14) (Table 2) may be due to superattenuation caused by high intracellular leucine concentrations. To test this idea, we repeated the experiment described in Table 2, sometimes supplementing the medium with leucine-containing dipeptides instead of leucine. In E. coli, dipeptides are transported primarily by the dipeptide permease, encoded by the dppABCDF operon (1, 17). As shown in Table 6, exogenous glycyl leucine and, to a lesser extent, alanyl leucine caused severe repression of leu operon expression in the lrp+ strain while the lrp strain showed low values under all three conditions. This result is most easily explained by assuming that dipeptide transport is not affected by Lrp, and that the dipeptide permease system has a high rate of transport of leucine-containing dipeptides, resulting in high intracellular leucine concentrations and superattenuation of leu operon transcription. By this view, superattenuation of leu transcription is a consequence of very high intracellular leucine concentrations, concentrations that can be achieved by the growth of a wild-type strain with exogenous leucine-containing dipeptides or by the growth of an lrp strain in the presence of leucine.
TABLE 6.
Specific activity of β-galactosidase in lrp+ and lrp strains having a leuPAB′-lacZ translational fusion and grown in media supplemented with dipeptidesa
| Strain | lrp allele | Sp. act. of β-galactosidaseb
|
||
|---|---|---|---|---|
| Leu | Gly Leu | Ala Leu | ||
| CP55 | lrp+ | 470 ± 50 | 31 ± 6 | 124 ± 40 |
| CV1512 | lrp-35::Tn10 | 30 ± 7 | 20 ± 3 | 51 ± 17 |
As in legend to Table 2, except that cultures were sometimes supplemented with leucine at 50 μg/ml (Leu), glycyl leucine at 75 μg/m; (Gly Leu) or alanyl leucine at 77 μg/ml (Ala Leu).
Specific activity is in Miller units. Values represent the means and standard deviations for multiple samples taken throughout log growth phase from at least two independent experiments.
To summarize, we confirmed the original observation of Lin et al. that leu operon expression is extremely low in a strain having an lrp null allele (14), but found that this result is only seen in cells grown in the presence of excess leucine. For a prototrophic strain grown without excess leucine, an lrp null allele had little effect upon leu operon expression (Table 3). Furthermore, an lrp null allele had little effect upon leu promoter expression in strains having just the leu promoter fused to a lacZ reporter gene (Table 4 and 5). With our E. coli strains grown under our defined conditions, we did not find that the leu promoter was affected by transcription initiated almost 2 kb away at the ilvIH promoter, as has been suggested by Fang and Wu for the leu-500 promoter in S. typhimurium (6, 7). The effects of Lrp upon leu operon expression, originally observed by Lin et al. (14) and confirmed by us, must be related to events secondary to transcription initiation, likely either transcription attenuation at leu att or translation initiation at the beginning of structural genes. The simplest explanation of all of our results is that leucine transport capacity is elevated in a strain lacking Lrp (10) and that growth of such a strain in the presence of leucine causes high intracellular levels of leucine, which in turn cause very low levels of leu operon expression through the transcription attenuation mechanism. The results shown in Table 6 involving growth in media supplemented with leucine-containing dipeptides are consistent with this interpretation. The overall conclusion of these studies is that any effects of Lrp on leu operon expression are indirect.
While this overall conclusion seems justified, it must be noted that several aspects of our data are not readily explained. For example, in the presence of leucine and the leu attenuator, the ratio of leu::lacZ expression in lrp+ to that in lrp is more than 10 when the fusion is in leuB and all upstream DNA is present (Table 2), 2.7 when DNA stretching upstream to the ilvIH promoter is included (Table 5), and 1.3 when upstream DNA goes only to position −247 (Table 4). Or consider that for lrp+ strains grown in the absence of leucine, leu::lacZ expression was about threefold higher in constructs containing about 1,500 bp of upstream DNA than for those containing only 250 bp (Table 4 and 5). These comparisons suggest that there may be some long-distance effects of upstream sequences upon expression from the leu promoter.
Finally, the conclusion of previous studies that lrp strains grow slowly because they are starved for leucine (2, 16) needs to be evaluated in the context of our results. lrp strains do grow more slowly than isogenic lrp+ strains (Table 3) and the growth rate of lrp strains is increased by inclusion of leucine in the medium (Table 3), but lrp strains do not appear to be defective for leu operon expression (Table 3). In fact, if lrp strains were starved for leucine, the expected (though not observed) result is elevated leu operon expression caused by relief of transcription attenuation. The transcription attenuation mechanism is functional in lrp strains because Lin et al. showed in their original work that a leucine limitation imposed upon a lrp strain with a leu mutation resulted in elevated expression of the leu operon (14). The underlying basis for the slow growth of lrp strains and for leucine-mediated stimulation of growth remains unclear.
Acknowledgments
We thank E. Newman and M. Hartlein for strains, Alison Moskowitz for help with some of the experiments, and David Wilson for a careful reading of the manuscript.
This work was supported by NIH grant GM48861.
REFERENCES
- 1.Abouhamad W N, Manson M, Gibson M M, Higgins C F. Peptide transport and chemotaxis in Escherichia coli and Salmonella typhimurium: characterization of the dipeptide permease (Dpp) and the dipeptide-binding protein. Mol Microbiol. 1991;5:1035–1047. doi: 10.1111/j.1365-2958.1991.tb01876.x. [DOI] [PubMed] [Google Scholar]
- 2.Ambartsoumian G, D’Ari R, Lin R T, Newman E B. Altered amino acid metabolism in lrp mutants of E. coli K-12 and their derivatives. Microbiology. 1994;140:1737–1744. doi: 10.1099/13500872-140-7-1737. [DOI] [PubMed] [Google Scholar]
- 3.Bhagwat S P, Rice M R, Matthews R G, Blumenthal R M. Use of an inducible regulatory protein to identify members of a regulon: application to the regulon controlled by the leucine-responsive regulatory protein (Lrp) in Escherichia coli. J Bacteriol. 1997;179:6254–6263. doi: 10.1128/jb.179.20.6254-6263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo J M, Matthews R G. The leucine-responsive regulatory protein (Lrp), a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Ari R, Lin R T, Newman E B. The leucine-responsive regulatory protein: more than a regulator? Trends Biochem Sci. 1993;18:260–263. doi: 10.1016/0968-0004(93)90177-o. [DOI] [PubMed] [Google Scholar]
- 6.Fang M, Wu H-Y. A promoter relay mechanism for sequential gene activation. J Bacteriol. 1998;180:626–633. doi: 10.1128/jb.180.3.626-633.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang M, Wu H-Y. Suppression of leu-500 mutation in topA+ Salmonella typhimurium strains. J Biol Chem. 1998;273:29929–29934. doi: 10.1074/jbc.273.45.29929. [DOI] [PubMed] [Google Scholar]
- 8.Gemmill R M, Wessler S R, Keller E B, Calvo J M. The leucine operon of Salmonella typhimurium is controlled by an attenuation mechanism. Proc Natl Acad Sci USA. 1979;76:4941–4945. doi: 10.1073/pnas.76.10.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgiou G, Schuler M L, Wilson D B. Release of periplasmic enzymes and other physiological effects of β-lactamase overproduction in Escherichia coli. Biotechnol Bioeng. 1988;32:741–748. doi: 10.1002/bit.260320603. [DOI] [PubMed] [Google Scholar]
- 10.Haney S A, Platko J V, Oxender D L, Calvo J M. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J Bacteriol. 1992;174:108–115. doi: 10.1128/jb.174.1.108-115.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haughn G W, Squires C H, DeFelice M, Lago C T, Calvo J M. Unusual organization of the ilvIH promoter of Escherichia coli. J Bacteriol. 1985;163:186–198. doi: 10.1128/jb.163.1.186-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf J R, Levinthal M, Danchin A. The role of H-NS in one carbon metabolism. Biochimie. 1994;76:1063–1070. doi: 10.1016/0300-9084(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 13.Landgraf J R, Wu J, Calvo J M. The effect of nutrition and growth rate upon Lrp levels in Escherichia coli. J Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin R, D’Ari R, Newman E B. placMu insertions in genes of the leucine regulon: extension of the regulon to genes not regulated by leucine. J Bacteriol. 1992;174:1948–1955. doi: 10.1128/jb.174.6.1948-1955.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 16.Newman E B, Lin T T, D’Ari R. The leucine/Lrp regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1513–1525. [Google Scholar]
- 17.Olson E R, Dunyak D S, Jurss L M, Poorman R A. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J Bacteriol. 1991;173:234–244. doi: 10.1128/jb.173.1.234-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platko J V, Willins D A, Calvo J M. The ilvIH operon of Escherichia coli is positively regulated. J Bacteriol. 1990;172:4563–4570. doi: 10.1128/jb.172.8.4563-4570.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Searles L L, Calvo J M. Permeabilized cell and radiochemical assays for β-isopropylmalate dehydrogenase. Methods Enzymol. 1988;166:225–229. doi: 10.1016/s0076-6879(88)66029-0. [DOI] [PubMed] [Google Scholar]
- 20.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 21.Stieglitz B I, Calvo J M. Distribution of the isopropylmalate pathway to leucine among diverse bacteria. J Bacteriol. 1974;118:935–941. doi: 10.1128/jb.118.3.935-941.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchetina E, Newman E B. Identification of Lrp-regulated genes by inverse PCR and sequencing: regulation of two mal operons of Escherichia coli by leucine-responsive regulatory protein. J Bacteriol. 1995;177:2679–2683. doi: 10.1128/jb.177.10.2679-2683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 352–367. [Google Scholar]
- 24.Wang Q, Wu J, Friedberg D, Platko J, Calvo J M. Regulation of the Escherichia coli lrp gene. J Bacteriol. 1994;176:1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessler S R, Calvo J M. Control of leu operon expression in Escherichia coli by a transcription attenuation mechanism. J Mol Biol. 1981;149:579–597. doi: 10.1016/0022-2836(81)90348-x. [DOI] [PubMed] [Google Scholar]