Abstract
Gene rgpG is required for biosynthesis of rhamnose-glucose polysaccharide (RGP) in Streptococcus mutans. Its deduced amino acid sequence had similarity to WecA, which initiates syntheses of enterobacterial common antigen and some O antigens in Escherichia coli. Gene rgpG complemented a wecA mutation of E. coli, suggesting that rgpG may function similarly in RGP synthesis.
Cell wall antigens of Streptococcus mutans are rhamnose-glucose polysaccharides (RGPs), which are composed of α1,2- and α1,3-linked rhamnan backbones and glucose side chains (8, 14). The biofunction of RGP is receiving increasing attention because of the facts that the serotype f-specific RGP induces the release of inflammatory cytokines (19) and also provokes nitric oxide production (9).
We recently cloned three loci that are related to RGP synthesis. Four rml genes (rmlA, rmlB, rmlC, and rmlD) that are directly involved in the synthesis of dTDP-l-rhamnose (22, 23) and a gene (gluA) encoding glucose-1-phosphate uridylyltransferase, which synthesizes UDP-d-glucose (28), were identified in these loci. The two nucleotide sugars were found to be immediate precursors for RGP synthesis (22, 23, 28). In addition, we found six rgp genes (rgpA, rgpB, rgpC, rgpD, rgpE, and rgpF) required for RGP synthesis in the region downstream from rmlD (27). Some of these rgp genes are probably involved in the transport and assembly of RGP.
Here, we describe the identification and characterization of an additional gene required for RGP synthesis, which is located in a new locus.
Isolation of an RGP-defective mutant of S. mutans.
A complete Sau3AI digest of the S. mutans Xc chromosome was ligated to BamHI- and BglII-digested pResEmBBN. pResEmBBN was produced by Shiroza and Kuramitsu in the process of constructing pResEmMCS11 (18) to prepare an S. mutans genomic library and is equivalent to pResEmMCS11, except that it lacks the restriction sites from XbaI to NotI in the multicloning site. S. mutans Xc (22) was randomly mutated by transformation with the S. mutans genomic library. Transformants were selected on tryptic soy agar plates containing 10 μg of erythromycin per ml. Both rml and rgp mutants of S. mutans, which are completely unable to synthesize RGP, have a characteristic colony morphology (22, 23, 27). The fact enables us to visually distinguish a mutant defective in RGP synthesis from other mutants.
Fifteen transformants that appeared to be RGP-defective mutants were selected after visually screening 9,000 transformants. Immunodiffusion analysis of their cell wall polysaccharide extracts with serotype c-specific rabbit antiserum (23) and a high-pressure liquid chromatography analysis (23) of sugar components in their cell wall fractions revealed that 7 of the 15 transformants actually lost serotype c-specific polysaccharides.
Southern blot analyses of XbaI- and BstEII-digested chromosomes of the seven transformants with defective RGP synthesis were performed using digoxigenin (DIG)-labeled PCR probes specific for rmlC and rmlD as previously described (22, 23). The results indicated that one of seven transformants did not have an insert in the known rgp or rml gene cluster. We designated this transformant Xc51.
Characterization of plasmid insertion point in Xc51.
Southern blotting with a DIG-labeled PCR probe specific for the Emr gene revealed that the probe hybridized with a 4.6-kb EcoRI fragment from Xc51 but not with any fragments in the wild-type strain Xc. A PCR probe specific for the Emr gene was prepared by PCR with a set of primers (5′-CTTAGAAGCAAACTTAAG-3′ and 5′-TTATTTCCTCCCGTTAAA-3′), using pResEmBBN as a template. Strain Xc was transformed with the chromosome DNA prepared from Xc51. Every Emr transformant showed the same colony morphology as that of Xc51. Five randomly selected transformants were further confirmed to be defective in RGP synthesis. Southern blot analysis with the specific probe for the Emr gene of these transformants gave identical results to that for Xc51. These findings indicated that integration of pResEmBBN in a specific locus on the Xc chromosome, which was different from the rml and rgp loci involved in RGP synthesis, produced a novel defect in RGP synthesis.
Cloning and sequencing of the region flanking the plasmid insertion in strain Xc51.
The EcoRI-digested chromosome of Xc51 was self-ligated. Escherichia coli DH5 was transformed with this DNA, and transformants were isolated on Luria-Bertani agar plates containing 200 μg of erythromycin per ml. All plasmids isolated from transformants were identical (4.6 kb) in size. One of them was designated pKU51. The plasmid labeled with DIG-dUTP via random primer labeling hybridized with the 4.6-kb EcoRI fragment in Xc51 and a 2.6-kb EcoRI fragment in Xc. The 2.6-kb EcoRI-fragment of Xc was cloned in pBluescript SKII+ by colony hybridization. The resulting plasmid, pBluescript SKII+ carrying the 2.6-kb EcoRI-fragment, was designated pKU52 and the nucleotide sequence of the insert was determined with a 373 STRETCH automated sequencer (Applied Biosystems, Inc., Foster City, Calif.) as described previously (26).
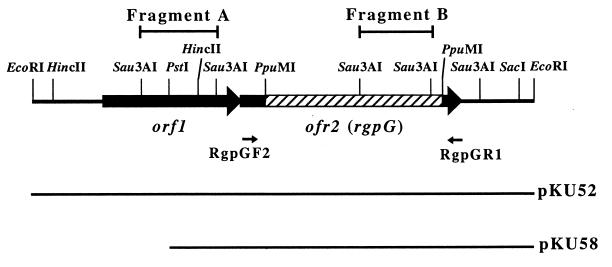
The nucleotide sequence analysis revealed the presence of two open reading frames (ORFs), as shown in Fig. 1. They were oriented in the same direction as the plasmid lacZ gene. No ORF on the opposite strand of this region was more than 250 bp in length. A possible Shine-Dalgarno sequence was identified just upstream from the potential initiation codons of orf1. A consensus −10 and −35 E. coli promoter-like sequence (TTGAAA-N17-TATAAT; positions 302 to 330) for orf1 and an inverted repeat structure (positions 2257 to 2274) with a free energy of −16.8 kcal/mol followed by a polyT sequence, which may act as a transcriptional terminator for orf2, were recognized. On the other hand, there was no typical ribosomal binding motif for orf2.
FIG. 1.
Restriction map of the EcoRI 2.6-kb insert fragment cloned in pKU52. The large arrows indicate the locations of the two ORFs. The hatched bar shows the 0.9-kb PpuMI fragment that was replaced with the Emr gene in Xc52. The small arrows labelled RgpGF2 and RgpGR1 show the positions of primers RgpGF2, 5′-GATAAGCTAGATGATACCTT-3′ (complementary to positions 1078 to 1097), and RgpGR1, 5′-AAATTACTTTTTCTTCTTAC-3′ (complementary to positions 2244 to 2225), respectively. The locations of the inserted fragments are indicated in the lower portion of the diagram.
The nucleotide database was searched for orf1 and orf2 using the program FASTA (13) on the DDBJ server at the National Institute of Genetics, Mishima, Japan. The amino acid sequence deduced from orf1 showed 25.5% identity with that from the mecA gene of Bacillus subtilis, which is involved in the negative regulation of genetic competence (7). The amino acid sequence deduced from orf2 showed 19.1% identity with that from the wecA (rfe) gene, involved in initiating the syntheses of enterobacterial common antigen (ECA) and O8 and O9a antigens in E. coli (5, 10, 15).
The insert fragment of the integration plasmid should locate within the target gene to disrupt the gene by Campbell-type recombination. Two Sau3AI fragments (fragments A and B) located within orf1 and orf2 (Fig. 1) are candidates as the insert fragment in the integration plasmid that produced Xc51. However, pKU51 possessed a unique PstI site. This suggests that the Sau3AI fragment within orf2 (fragment B) must be the insert fragment in the integration plasmid that produced Xc51, because the insert fragment must be duplicated when it is recovered in pKU51. Based on these findings, we hypothesize that the disruption of orf2 caused the RGP-defective phenotype of S. mutans.
Insertional inactivation of orf2.
The 0.9-kb PpuMI fragment within orf2 on pKU52 was replaced with the 1.0-kb Emr gene (18). The resultant plasmid was linearized by EcoRI digestion and used to transform strain Xc. Four randomly selected Emr transformants were examined by immunodiffusion analysis, and all were ascertained to be defective in RGP synthesis. An appropriate disruption of orf2 in these transformants was confirmed by PCR amplification of the 1.2-kb region spanning orf2 with primers RgpGF2 and RgpGR1 (Fig. 1) and the subsequent HincII digestion of this fragment, because there is a HincII site within the Emr gene but none in orf2. One of the transformants was designated Xc52.
When the total amount of hexosamine in the purified cell wall preparation of Xc52 was determined by using the colorimetric method of Strominger et al. (21), the hexosamine content in the cell wall of Xc52 (163 ± 13 μg per mg [dry weight] of the purified cell wall preparation) was not very different from those in the cell walls of Xc and the purified cell wall preparations of its rml mutants (23). It seems unlikely that rgpG is involved in peptidoglycan synthesis.
Complementation analysis of rgpG.
We constructed a new E. coli-streptococcus shuttle plasmid, pKU55, containing a tetracycline resistance marker, the tetM916 gene derived from pLN2 (2), which functions both in E. coli and in streptococci, and the pUC and pC194 replicons for maintenance in E. coli and in streptococci, respectively, which are derived from pTH10 (4). Expression of rgpG was ensured by the promoter sequence in the 0.35-kb upstream region of the Emr gene from pAM77 (3). The 0.35-kb region was amplified by PCR with primers EmF2, 5′-AGAGAGTCTAGAGAAGCAAACTTAAGAGTGTG-3′ (XbaI site underlined), and EmR2, 5′-GTGTGTCTGCAGTTTCGTCGTTAAATGCC-3′ (PstI site underlined), using pResEmBBN as a template. The PCR fragment was digested with XbaI and PstI and ligated to the XbaI and PstI sites of pKU52, producing pKU54, which contains the streptococcal promoter sequence at the 5′ terminus of rgpG. The fragment containing both promoter sequence and rgpG in the same direction was excised from pKU54 by digestion with XbaI and HindIII and ligated to the XbaI and HindIII sites of pKU55; the resulting plasmid was designated pKU56.
Direct transformation of the rgpG mutant with pKU56 was abandoned because we tried unsuccessfully to transform Xc52 with shuttle plasmid pKU55 and also with a tetracycline resistance marker on the chromosomal DNA from strain GS5DD (2). The surface structure of cells with normal RGP synthesis might be critical to the genetic competence of S. mutans. As an alternative, wild-type strain Xc was initially transformed with pKU56 and subsequently transformed with EcoRI-digested pKU53. Three transformants which were randomly selected retained their serotype c antigenicity and had normal rhamnose and glucose contents in cell wall preparations, suggesting that the rgpG gene located on the shuttle plasmid complemented the inactivated chromosomal rgpG. The result ruled out a possible polar effect of the pResBBN insertion and proved a direct effect of the disruption of rgpG on the phenotype of Xc52.
Functional analysis of rgpG.
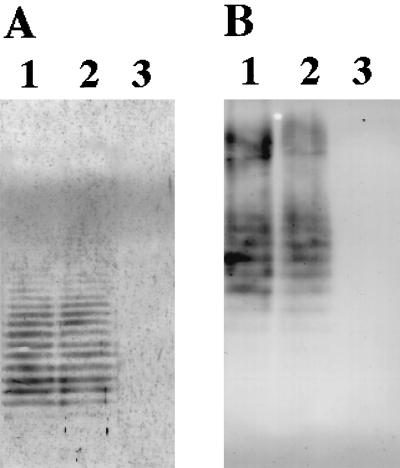
We examined whether rgpG complements wecA. A plasmid, pKU58, which has only rgpG on the insert fragment, was constructed by PstI digestion of pKU52 and self-ligation for removal of orf1 (Fig. 1). The plasmid was introduced into E. coli 21548, which is a wecA-defective mutant of strain K-12 (10). Strain 21548 was kindly provided by P. D. Rick, Department of Microbiology, Uniformed Services University of Health Sciences, Bethesda, Md. ECA and O9a antigen production in E. coli strains were detected by immunoblotting analysis with anti-ECA serum and anti-O9a monoclonal antibody (MAb F719), respectively, as previously described (5, 12), because the wecA gene is known to be involved in the syntheses of ECA (10) and O9a antigen (5). Strain 21548 transformed with pKU58 and its parental strain AB1133 produced ECA, whereas strain 21548 transformed with pBluescript SKII+ did not (Fig. 2A). Furthermore, O9a antigen production was observed in strain 21548 transformed with both pKU58 and pNKB26 and in strain AB1133 transformed with only pNKB26 but not in 21548 transformed with pNKB26 (Fig. 2B). These findings indicate that rgpG complemented the wecA-deficient phenotype of E. coli, suggesting a functional similarity between the gene products of rgpG and wecA.
FIG. 2.
Immunoblots with anti-ECA serum (A) and monoclonal antibody against O9 antigen (B). Shown is complementation analysis of the E. coli wecA mutant (21548) with rgpG. ECA or lipopolysaccharide was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by immunoblotting with the respective antiserum. (A) Lane 1, strain AB1133; lane 2, strain 21548 transformed with pKU58; lane 3, strain 21548 transformed with pBluescript SKII+. (B) Lane 1, strain AB1133; lane 2, strain 21548 transformed with pKU58 and pNKB26; lane 3, strain 21548 transformed with pNKB26.
There was no typical preceding ribosomal binding motif for rgpG. Several potential start codons in the 5′-terminal part of rgpG in addition to the TTG codon at position 1075 were expected. Therefore, truncated PCR fragments were amplified and ligated into pBluescript SKII+. Transformants of strain 21548 with the ligated DNA were obtained, and their ECA production was checked by Western blot analysis with anti-ECA serum as described above. The transformant with the plasmid containing the largest rgpG fragment that covers the TTG codon at position 1075 produced ECA, whereas all the other transformants with plasmids containing smaller PCR fragments did not. The fidelity of the PCR amplification was confirmed by sequencing the insert of the plasmids. This complementation study suggested that translation of the largest coding frame is essential for the enzyme activity of the rgpG gene product. This study clearly showed that the TTG at position 1075 is the initiation codon for rgpG translation.
To date, three distinct pathways have been proposed for the initiation of cell surface polysaccharide synthesis in some bacteria. The enzyme encoded by wecA is an N-acetylglucosamine-1-phosphate transferase that forms lipid I (undecaprenyl pyrophosphoryl N-acetylglucosamine). In gram-negative bacteria, this enzyme is involved in the first step of the biosyntheses of ECA and several lipopolysaccharide O antigens, even when the mature polysaccharides do not contain N-acetylglucosamine residues (24). The second pathway, that for the biosynthesis of the heteropolysaccharide O antigen in Salmonella enterica, is initiated by the enzyme encoded by wbaP, which catalyzes the transfer of galactose-1-phosphate to a lipid carrier. A similar type of initiation of polysaccharide synthesis is speculated to occur in the capsular polysaccharide production systems of group B streptococci (17) and Streptococcus pneumoniae (6, 11). Finally, group A streptococci are not believed to utilize a lipid intermediate in the production of their hyaluronic acid capsule (20). In addition, it was recently reported that no lipid-linked intermediates are detected in the synthesis of polysaccharide intercellular adhesin in Staphylococcus epidermidis (1).
Functional similarity between RgpG and WecA hinted that transferring N-acetylglucosamine-1-phosphate to a lipid carrier may be the first step in RGP synthesis, even though repeating units of RGP do not contain N-acetylglucosamine. It is interesting that a gene highly homologous (47.3% identity) to rgpG was found in a locus involved in polysaccharide biosynthesis in Enterococcus faecalis (25). However, the gene (orfde3) is not required for antigenic polysaccharide biosynthesis, and its biological function remains to be determined. The present study is the first report of functional characterization of a gene homologous to wecA in gram-positive bacteria, although intermediates of teichuronic acid biosynthesis in Micrococcus luteus have earlier been shown to contain undecaprenyl pyrophosphoryl N-acetylglucosamine structure (3a).
Mechanisms of cell surface polysaccharide synthesis are more poorly characterized in gram-positive bacteria than in gram-negative bacteria (16). The lipid carrier involved in the synthesis of the cell surface polysaccharides of gram-positive bacteria, including S. mutans RGP, and the way these polysaccharides are linked to the cell wall still remain to be clarified. Further characterization of this enzyme is important to resolve the polysaccharide synthetic pathway in gram-positive bacteria.
Nucleotide sequence accession number.
The 2,614-bp nucleotide sequence described in this paper has been submitted to the EMBL/GenBank/DDBJ data bank under accession no. AB022909.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Developmental Scientific Research [(B)09470474] (Y.Y.) from the Ministry of Education, Science, Sports and Culture of Japan and the Kyushu University Interdisciplinary Programs in Education and Projects in Research Development.
REFERENCES
- 1.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 2.Hanada N, Kuramitsu H K. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect Immun. 1989;57:2079–2085. doi: 10.1128/iai.57.7.2079-2085.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horinouchi S, Byeon W H, Weisblum B. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J Bacteriol. 1983;154:1252–1262. doi: 10.1128/jb.154.3.1252-1262.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Johnson G L, Hoger J H, Ratnayake J H, Anderson J S. Characterization of three intermediates in the biosynthesis of teichuronic acid of Micrococcus luteus. Arch Biochem Biophys. 1984;235:679–691. doi: 10.1016/0003-9861(84)90244-3. [DOI] [PubMed] [Google Scholar]
- 4.Kato C, Kuramitsu H K. Molecular basis for the association of glucosyltransferases with the cell surface of oral streptococci. FEMS Microbiol Lett. 1991;63:153–157. doi: 10.1111/j.1574-6968.1991.tb04521.x. [DOI] [PubMed] [Google Scholar]
- 5.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferase, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolkman M A, Morrison D A, Van Der Zeijst B A, Nuijten P J. The capsule polysaccharide synthesis locus of Streptococcus pneumoniae serotype 14: identification of the glycosyl transferase gene cps14E. J Bacteriol. 1996;178:3736–3741. doi: 10.1128/jb.178.13.3736-3741.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong L, Siranosian K J, Grossman A D, Dubnau D. Sequence and properties of mecA, a negative regulator of genetic competence in Bacillus subtilis. Mol Microbiol. 1993;9:365–373. doi: 10.1111/j.1365-2958.1993.tb01697.x. [DOI] [PubMed] [Google Scholar]
- 8.Linzer R, Reddy M S, Levine M J. Immunochemical aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. pp. 29–38. [Google Scholar]
- 9.Martin V, Kleschyov A L, Klein J-P, Beretz A. Induction of nitric oxide production by polyosides from the cell walls of Streptococcus mutans OMZ 175, a gram-positive bacterium, in the rat aorta. Infect Immun. 1997;65:2074–2079. doi: 10.1128/iai.65.6.2074-2079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 11.Morona J K, Morona R, Paton J C. Characterization of the locus encoding the Streptococcus pneumoniae type 19F capsular polysaccharide biosynthetic pathway. Mol Microbiol. 1997;23:751–763. doi: 10.1046/j.1365-2958.1997.2551624.x. [DOI] [PubMed] [Google Scholar]
- 12.Ohta M, Ina K, Kusuzaki K, Kido N, Arakawa Y, Kato N. Cloning and expression of the rfe-rff gene cluster of Escherichia coli. Mol Microbiol. 1991;5:1853–1862. doi: 10.1111/j.1365-2958.1991.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 13.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritchard D G, Gregory R L, Michalek S M, McGhee J R. Biochemical aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. pp. 39–49. [Google Scholar]
- 15.Rick P D, Hubbard G L, Baar K. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 17.Rubens C E, Heggen L M, Haft R F, Wessels M R. Identification of cpsD, a gene essential for type III capsule expression in group B streptococci. Mol Microbiol. 1993;8:843–855. doi: 10.1111/j.1365-2958.1993.tb01631.x. [DOI] [PubMed] [Google Scholar]
- 18.Shiroza T, Kuramitsu H K. Construction of a model secretion system for oral streptococci. Infect Immun. 1993;61:3745–3755. doi: 10.1128/iai.61.9.3745-3755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soell M, Lett E, Holveck F, Schöller M, Wachsmann D, Klein J-P. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-α release. J Immunol. 1995;154:851–860. [PubMed] [Google Scholar]
- 20.Stoolmiller A C, Dorfman A. The biosynthesis of hyaluronic acid by Streptococcus. J Biol Chem. 1969;244:236–246. [PubMed] [Google Scholar]
- 21.Strominger J L, Park J T, Thompson R E. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J Biol Chem. 1959;234:3263–3268. [PubMed] [Google Scholar]
- 22.Tsukioka Y, Yamashita Y, Nakano Y, Oho T, Koga T. Identification of a fourth gene concerned with dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol. 1997;179:4411–4414. doi: 10.1128/jb.179.13.4411-4414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, Murray B E, Weinstock G M. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita Y, Takehara T, Kuramitsu H K. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 1993;175:6220–6228. doi: 10.1128/jb.175.19.6220-6228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita Y, Tsukioka Y, Tomihisa K, Nakano Y, Koga T. Genes involved in cell wall localization and side chain formation of rhamnose-glucose polysaccharide in Streptococcus mutans. J Bacteriol. 1998;180:5803–5807. doi: 10.1128/jb.180.21.5803-5807.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita Y, Tsukioka Y, Nakano Y, Tomihisa K, Oho T, Koga T. Biological function of UDP-glucose synthesis in Streptococcus mutans. Microbiology. 1998;144:1235–1245. doi: 10.1099/00221287-144-5-1235. [DOI] [PubMed] [Google Scholar]