Abstract
The introduction of consensus −35 (TTGACA) and −10 (TATAAT) hexamers and a TG motif into the Lactobacillus acidophilus ATCC 4356 wild-type slpA promoter resulted in significant improvements (4.3-, 4.1-, and 10.7-fold, respectively) in transcriptional activity in Lactobacillus fermentum BR11. In contrast, the same changes resulted in decreased transcription in Lactobacillus rhamnosus GG. The TG motif was shown to be important in the context of weak −35 and −10 hexamers (L. fermentum BR11) or a consensus −10 hexamer (L. rhamnosus GG). Thus, both strain- and context-dependent effects are critical factors influencing transcription in Lactobacillus.
The bacterial RNA polymerase (RNAP) major ς factor recognizes and binds to conserved hexamers located at positions −35 (TTGACA) and −10 (TATAAT) with respect to the transcription initiation site in most organisms examined to date. These promoter hexamers are most frequently separated by a spacer of 17 ± 1 nucleotides, which often harbors a TG motif upstream of the −10 hexamer. Mutations which result in increased identity to the consensus hexamers generally lead to an increase in promoter efficiency in both gram-negative (11, 13, 20) and gram-positive (4, 10, 19) bacteria. The presence of the TG motif appears to be of considerable importance in gram-positive organisms, where introduction or deletion of the motif can influence promoter activity substantially (9, 18, 19).
While extensive studies of promoter sequences in Escherichia coli have been conducted (5, 11), promoters of gram-positive bacteria, with the possible exception of Bacillus subtilis, remain largely uncharacterized. Indeed, mutational analysis of promoters in the industrially and medically important gram-positive organism Lactobacillus has not been previously reported. Surveys of Lactobacillus promoters have revealed significant conservation of bases in the −35 and −10 regions, with consensus sequences identical to those of other organisms (11a, 15). The TG motif is a conserved feature in 26% of all Lactobacillus promoters, with the conservation frequency varying markedly between species (11a). Data suggest that the −35 hexamer, −10 hexamer, and TG motifs are significant determinants of promoter strength in lactobacilli (11a). The potential exists, therefore, to improve expression of heterologous genes in lactobacilli by specifically targeting these core promoter elements to enhance transcription initiation.
In the present study, we introduced consensus −35 and −10 hexamer elements and a TG motif into the wild-type Lactobacillus acidophilus ATCC 4356 slpA promoter (2) and examined the effects on promoter activity in two strains of Lactobacillus, L. fermentum BR11 and L. rhamnosus GG.
The slpA wild-type promoter region (extending from position −286 to −20, with respect to the translation start codon [2]) was initially amplified from L. acidophilus ATCC 4356 genomic DNA by using the primer pair 5′-AAAAGGATCCTGCTTGTGGGGTAAGCGG-3′ and 5′-TTTTCTGCAGATATAAAAAAATGTAATAGGCC-3′. The promoter fragment was introduced into the BamHI/PstI sites of the multiple cloning site of plasmid pNZ272RBS−, a ribosome binding site-deficient derivative of the E. coli-Lactobacillus shuttle vector pNZ272 harboring the promoterless gusA reporter gene (14). The use of this plasmid, which carries defective gusA protein machinery, avoids the deleterious effects associated with expression of the β-glucuronidase enzyme in these strains of lactobacilli (11a). Seven slpA promoter derivatives (Table 1) were generated by ligation of a 5′-phosphorylated oligonucleotide harboring the mutant base(s) during PCR of the slpA fragment (primer sequences and templates used are available upon request). PCR and cycling conditions were as previously described (12), with the following modifications: 1.25 mM MgCl2, 50 pmol of each primer, 200 μM (each) deoxynucleoside triphosphate, 1 to 15 ng of template (genomic or plasmid) DNA, 1 to 3 U of Expand High Fidelity Taq DNA polymerase (Boehringer Mannheim), and a 55°C annealing temperature. The slpA derivative PCR products were purified, digested with BamHI/PstI, ligated to BamHI/PstI-digested pNZ272RBS−, and transformed into E. coli KW1 for selection of mutant clones. Plasmid constructs were sequenced to screen for incorporation of the desired base substitutions. Positive constructs were transformed into L. fermentum BR11 and L. rhamnosus GG essentially as previously described (16). Penicillin concentrations of 1 and 10 μg/ml were used for L. fermentum BR11 and L. rhamnosus GG, respectively. Plasmid constructs from lactobacilli were analyzed by restriction digestion and direct sequencing to ensure plasmid integrity and maintenance of the mutant bases within the slpA promoter sequence.
TABLE 1.
slpA promoter base substitution mutants and relative transcriptional activities in Lactobacillus strains
| Plasmid | Sequence | Relative gusA transcript levela
|
|
|---|---|---|---|
| L. fermentum BR11 | L. rhamnosus GG | ||
| −35 TG −10 | |||
| pNZslpb | TTGCTATTTCTTGAAGAGGTTAGTACAAT | 100 | 100 (450) |
| pNZS10 | -------------------------T--- | 412* | 7.5** (34) |
| pNZS35 | ---AC------------------------ | 426** | 33** (149) |
| pNZS1035 | ---AC--------------------T--- | 312 | 100 (450) |
| pNZTG | ---------------------G------- | 1,072** | 15** (68) |
| pNZT10 | ---------------------G---T--- | 500** | 16** (72) |
| pNZT35 | ---AC----------------G------- | 116 | 16** (72) |
| pNZT1035 | ---AC----------------G---T--- | 131 | 212** (954) |
Represented as percent wild-type activity (values in parentheses represent L. rhamnosus GG transcript levels normalized to those of L. fermentum BR11). Significance levels (Student’s t test) are as follows: *, P < 0.05; **, P < 0.01. Values in boldface type are maximal gusA mRNA levels in each strain.
Wild type.
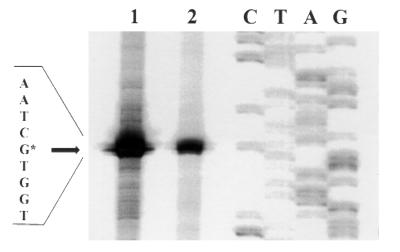
The activity of each promoter in lactobacilli was determined directly by a quantitative Northern blot procedure. Total RNA was extracted from Lactobacillus clones (14), electrophoresed through 1.5% agarose formaldehyde gels, and transferred to positively charged nylon membranes (Boehringer Mannheim) by capillary transfer (17). Membranes were subsequently probed with a digoxigenin (DIG)-labelled 895-bp gusA PCR fragment (produced from linearized pNZ272RBS− with the primer pair 5′-AATGTCACTAACCTGCCC-3′ and 5′-GGGTTGGGGTTTCTACAGGACGTA-3′) to determine specific transcript levels and with a DIG-labelled species-specific 212-bp 16S rRNA PCR fragment (produced from genomic DNA with primer pair 5′-AGCAGCCGCGGTAATAC-3′ and 5′-CTTGCGCACTGGTGTTC-3′) to standardize for gel loading and transfer. Hybridization signals were quantified via highly standardized optical scanning (Scanjet 4C; Hewlett Packard) and densitometry (Imagequant; Molecular Dynamics). Relative transcript levels were corrected for relative plasmid copy numbers (14) within each recombinant strain to eliminate gene dosage effects. Student’s t test was used to define significant differences between means of at least three independent determinations for each Lactobacillus clone. Primer extension analysis of transcripts was done essentially as described by Platteeuw et al. (14), except that oligonucleotide GUS-AS was labelled with [γ-33P]ATP via T4 polynucleotide kinase (Boehringer Mannheim) prior to the annealing reaction. Transcription was initiated from the start site published for the natural host L. acidophilus ATCC 4356 (2) in both L. fermentum BR11 and L. rhamnosus GG (Fig. 1). The slpA wild-type promoter was utilized with 4.5-fold-higher efficiency in L. rhamnosus GG than in L. fermentum BR11. Mutants analyzed by primer extension indicated that the transcription start site remained unchanged following substitution of bases within the promoter region (data not shown). The slpA promoter is not known to be subject to regulation in vivo. The second promoter structure postulated by Boot et al. (3), located upstream of the slpA promoter, was not active in either Lactobacillus strain examined in the present study (data not shown). Thus, the changes in transcript levels described in this work were deemed to be due to the direct influence of the slpA promoter sequence on transcription initiation.
FIG. 1.
Primer extension analysis of lactobacilli harboring the wild-type slpA promoter. Lane 1, L. rhamnosus GG; lane 2, L. fermentum BR11; lanes C, T, A, and G, sequencing reactions. Transcription was initiated from the start site published for the natural host L. acidophilus ATCC 4356 (2), with 4.5-fold-higher efficiency in L. rhamnosus GG than in L. fermentum BR11.
The effect of introducing consensus −35 and −10 hexamers and a TG motif into the slpA promoter was clearly dependent on the Lactobacillus strain in which it was harbored and varied when the promoter context was changed by the presence of another mutation (Fig. 2, Table 1). In L. fermentum BR11, introduction of an individual consensus −35 hexamer, −10 hexamer, or TG motif into an otherwise wild-type slpA promoter resulted in significant increases in transcription (4.3-, 4.1-, and 10.7-fold, respectively). These results indicate that the wild-type slpA promoter sequence was not optimal and could be improved considerably in L. fermentum BR11. The presence of a consensus −35 hexamer enhanced transcription (compared to the wild-type promoter) to the same level as a consensus −10 hexamer in the present study (Table 1), indicating that the consensus identities at the −35 and −10 hexamers were of equivalent importance in the slpA promoter in L. fermentum BR11. Interestingly, transcription directed from the slpA promoter in L. fermentum BR11 was not amenable to further enhancement by the introduction of multiple consensus motifs. gusA transcript levels produced from the slpA promoter derivative harboring concurrent consensus hexamer motifs (in pNZS1035) were not significantly different from levels produced from either of the derivatives harboring individual consensus hexamers (in pNZS35 and pNZS10) in L. fermentum BR11 (Table 1). This observation suggests that excessive RNAP-promoter contact in the hexamer regions may have resulted in the disruption of subsequent steps of transcription initiation in L. fermentum BR11, sufficient to counteract the additional interaction at the hexamer motifs. Similar hypotheses have been postulated for some consensus promoters of E. coli (5, 7).
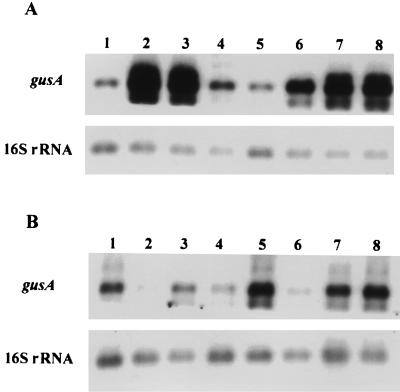
FIG. 2.
Northern blot analysis of RNA isolated from L. fermentum BR11 (A) and L. rhamnosus GG (B) harboring the slpA wild-type and derivative promoter constructs. Membranes were hybridized with DIG-labelled gusA and 16S rRNA probes as described in Materials and Methods. Lanes 1, pNZslp (wild type); lanes 2, pNZTG; lanes 3, pNZT10; lanes 4, pNZT35; lanes 5, pNZT1035; lanes 6, pNZS10; lanes 7, pNZS35; lanes 8, pNZS1035. Signals were quantified by densitometry and normalized for relative plasmid copy number. The relative transcriptional activities determined are given in Table 1.
The TG motif significantly increased the wild-type slpA promoter activity in L. fermentum BR11 (Table 1), presumably by providing an additional RNAP contact point in the presence of the weaker (i.e., nonconsensus) hexamers (1). Furthermore, the TG motif alone conferred approximately threefold more strength than a “perfect” hexamer consensus sequence in L. fermentum BR11. In contrast, the TG motif displayed either no significant effect or a down-effect on transcription when introduced into a promoter which harbored consensus hexamers (cf. pNZS35 and pNZT35, pNZS10 and pNZT10, and pNZS1035 and pNZT1035) (Table 1). These data are consistent with observations in B. subtilis, where the TG motif is only of benefit in the context of weak −35 and −10 hexamers (18, 19).
The results with the modified promoter sequences in L. rhamnosus GG contrasted with those obtained for L. fermentum BR11. The introduction of individual consensus −35 and −10 hexamers and a TG motif all resulted in decreased transcription (3-, 13.3-, and 6.7-fold, respectively) compared to the wild-type slpA promoter in L. rhamnosus GG (Table 1). These data would suggest that the wild-type slpA promoter was operating near maximal strength in L. rhamnosus GG. Presumably, any increase in transcription initiation facilitated by the increased RNAP-promoter interaction at the consensus motifs in L. rhamnosus GG was exceeded by the significant disruption of later stages of transcription initiation (e.g., promoter clearance [7]).
The effect of introducing consensus hexamers on slpA-directed transcription in L. rhamnosus GG was clearly context dependent. Both the −35 and −10 consensus hexamers displayed up-effects on transcription when introduced into a promoter harboring a consensus hexamer at the alternate position, regardless of the status of the TG motif (Table 1). For example, introducing a consensus −10 hexamer to the promoter derivative harboring a consensus −35 hexamer resulted in a significant 3-fold (cf. pNZS35 and pNZS1035) or 13-fold (cf. pNZT35 and pNZT1035) increase in transcription (P < 0.01). Similarly, the addition of a consensus −35 hexamer to the promoter harboring a consensus −10 hexamer led to a 13-fold increase (P < 0.01) in relative gusA-specific transcript levels, regardless of the TG motif status (cf. pNZS10 and pNZS1035; cf. pNZT10 and pNZT1035). These results suggest that a consensus hexamer is able to compensate for tight binding at the alternate hexamer in L. rhamnosus GG. This presumably occurs by increasing the efficiency of another step in the transcription initiation process, such as providing optimal bending in the promoter spacer region or facilitating open complex formation.
Introduction of a TG motif upstream of the slpA −10 hexamer in an otherwise wild-type promoter resulted in a 6.7-fold decrease in a 6.7-fold decrease in promoter activity in L. rhamnosus GG (Table 1). Interestingly, however, the TG motif partially compensated (2.1-fold increase [P < 0.01]) for the down-effect of the −10 consensus hexamer in this strain, regardless of the status of the −35 hexamer (cf. pNZS10 and pNZT10; cf. pNZS1035 and pNZT1035). These results are in agreement with data obtained from gram-negative bacteria (6, 8) and suggest that the TG motif is able to enhance transcription in the context of a tightly bound −10 hexamer in L. rhamnosus GG, either by alleviating strong RNAP-promoter contacts in the vicinity of the −10 hexamer or by facilitating open complex formation. Interestingly, only the slpA promoter derivative harboring all three consensus motifs (in pNZT1035) led to a promoter activity which was significantly higher (2.1-fold) than the wild-type slpA promoter in L. rhamnosus GG. This observation is consistent with the hypotheses extended for the context-dependent changes in this strain. Comparative analysis suggested that L. rhamnosus GG and L. fermentum BR11 were capable of transcribing gusA mRNA to similar levels when the optimal slpA promoter derivative (in pNZT1035 and pNZTG, respectively) was provided (Table 1).
In the presence of a consensus −35 hexamer, the TG motif displayed a significant down-effect (2.1- to 3.4-fold [P < 0.05]) on transcription from the slpA promoter in both L. rhamnosus GG (cf. pNZS35 and pNZT35) and L. fermentum BR11 (cf. pNZS35 and pNZT35; cf. pNZS1035 and pNZT1035) (Table 1). Voskuil and Chambliss (18) postulated that in the presence of an efficient −35 hexamer, the TG motif may act by promoting or stabilizing a transcription initiation step. The data obtained in the present study suggest that the presence of both a consensus −35 hexamer and a TG motif may overstabilize transcription intermediates and thereby hinder later stages of the transcription process in Lactobacillus strains.
This investigation provides the first insight into the functional relevance of the −35 and −10 hexamers and the TG motif within promoters of Lactobacillus. The findings presented should facilitate the selection or development of promoter sequences for achieving high-level heterologous gene expression in these medically and industrially important organisms.
Acknowledgments
We thank W. de Vos (NIZO, The Netherlands) for generously donating plasmid pNZ272 and S. Mathews for technical advice and review of the manuscript.
This work was supported by research grant 941114 from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Barne K A, Bown J A, Busby S J W, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the ’extended −10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boot H J, Kolen C P A M, Pouwels P H. Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J Bacteriol. 1995;177:7222–7230. doi: 10.1128/jb.177.24.7222-7230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boot H J, Kolen C P A M, Andreadaki F J, Leer R J, Pouwels P H. The Lactobacillus acidophilus S-layer protein gene expression site comprises two consensus promoter sequences, one of which directs transcription of stable mRNA. J Bacteriol. 1996;178:5388–5394. doi: 10.1128/jb.178.18.5388-5394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst D W, Betley M J. Promoter analysis of the staphylococcal enterotoxin A gene. J Biol Chem. 1994;269:1883–1888. [PubMed] [Google Scholar]
- 5.Bujard H, Brunner M, Deuschle U, Kammerer W, Knaus R. Structure-function relationship of Escherichia coli promoters. In: Reznikoff W S, Burgess R R, Dahlberg J E, Gross C A, Record M T Jr, Wickens M P, editors. RNA polymerase and the regulation of transcription. New York, N.Y: Elsevier; 1987. pp. 95–103. [Google Scholar]
- 6.Burns H D, Belyaeva T A, Busby S J W, Minchin S D. Temperature-dependence of open-complex formation at two Escherichia coli promoters with extended −10 sequences. Biochem J. 1996;317:305–311. doi: 10.1042/bj3170305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinger T, Behnke D, Bujard H, Gralla J D. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 8.Graña D, Gardella T, Susskind M M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henkin T M, Sonenshein A L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol Gen Genet. 1987;209:467–474. doi: 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- 10.Kenney T J, Churchward G. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol. 1996;178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandecki W, Goldman R A, Powell B S, Caruthers M H. lac up-promoter mutants with increased homology to the consensus promoter sequence. J Bacteriol. 1985;164:1353–1355. doi: 10.1128/jb.164.3.1353-1355.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.McCracken, A., L. M. Hafner, and P. Timms. Unpublished data.
- 12.Michael S F. Mutagenesis by incorporation of a phosphorylated oligo during PCR amplification. BioTechniques. 1994;16:410–412. [PubMed] [Google Scholar]
- 13.Moyle H, Waldberger C, Susskind M M. Hierarchies of base pair preferences in the P22 ant promoter. J Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pouwels P H, Leer R J. Genetics of lactobacilli: plasmids and gene expression. Antonie Leeuwenhoek. 1993;64:85–107. doi: 10.1007/BF00873020. [DOI] [PubMed] [Google Scholar]
- 16.Rush C M, Hafner L M, Timms P. Genetic modification of a vaginal strain of Lactobacillus fermentum and its maintenance within the reproductive tract after intravaginal administration. J Med Microbiol. 1994;41:272–278. doi: 10.1099/00222615-41-4-272. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Voskuil M I, Chambliss G H. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 1998;26:3584–3590. doi: 10.1093/nar/26.15.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voskuil M I, Voepel K, Chambliss G H. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol Microbiol. 1995;17:271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]
- 20.Youderian P, Bouvier S, Susskind M M. Sequence determinants of promoter activity. Cell. 1982;30:843–853. doi: 10.1016/0092-8674(82)90289-6. [DOI] [PubMed] [Google Scholar]