Abstract
Herpes simplex virus DNA polymerase consists of a catalytic subunit, Pol, and a processivity subunit, UL42, that, unlike other established processivity factors, binds DNA directly. We used gel retardation and filter-binding assays to investigate how UL42 affects the polymerase-DNA interaction. The Pol/UL42 heterodimer bound more tightly to DNA in a primer-template configuration than to single-stranded DNA (ssDNA), while Pol alone bound more tightly to ssDNA than to DNA in a primer-template configuration. The affinity of Pol/UL42 for ssDNA was reduced severalfold relative to that of Pol, while the affinity of Pol/UL42 for primer-template DNA was increased ∼15-fold relative to that of Pol. The affinity of Pol/UL42 for circular double-stranded DNA (dsDNA) was reduced drastically relative to that of UL42, but the affinity of Pol/UL42 for short primer-templates was increased modestly relative to that of UL42. Pol/UL42 associated with primer-template DNA ∼2-fold faster than did Pol and dissociated ∼10-fold more slowly, resulting in a half-life of 2 h and a subnanomolar Kd. Despite such stable binding, rapid-quench analysis revealed that the rates of elongation of Pol/UL42 and Pol were essentially the same, ∼30 nucleotides/s. Taken together, these studies indicate that (i) Pol/UL42 is more likely than its subunits to associate with DNA in a primer-template configuration rather than nonspecifically to either ssDNA or dsDNA, and (ii) UL42 reduces the rate of dissociation from primer-template DNA but not the rate of elongation. Two models of polymerase-DNA interactions during replication that may explain these findings are presented.
During DNA replication, long-chain DNA synthesis on the leading strand requires processivity factors to overcome the tendency of DNA polymerase to dissociate from the template after each catalytic step. In most of the organisms studied to date, these processivity factors interact with the catalytic subunit of polymerase and with DNA to increase the time of association of the holoenzyme with the DNA template. The best-characterized processivity factors are the so-called “sliding clamps,” which include the Escherichia coli β subunit of DNA polymerase III, bacteriophage T4 gp45, and eukaryotic PCNA. Biochemical and crystallographic studies have shown that these factors do not bind directly to DNA but, rather, form multimeric rings around DNA, which permits them to slide along the template (15, 19, 30, 31, 47, 58). Under physiological conditions, the association of a sliding clamp with DNA and its cognate polymerase requires auxiliary proteins that serve as “clamp loaders” (27, 32, 33, 40, 50) and occurs at the primer-template junction (reviewed in reference 20, 27, 44, and 45). A second type of processivity factor is exemplified by thioredoxin, which, unlike the sliding clamps, associates with T7 DNA polymerase in the absence of other protein factors or ATP to form a heterodimer (22, 24, 39, 48). However, like sliding clamps, thioredoxin does not bind DNA directly (39). Thus, during DNA synthesis, the interactions of the sliding clamps and thioredoxin with DNA and their translocation appear to be passive and dependent on their association with their cognate polymerases.
For herpes simplex virus (HSV), the details of how processive DNA synthesis occurs have yet to be resolved. The replicative DNA polymerase consists of a heterodimer of two proteins (6, 14). One of these proteins is the catalytic subunit, Pol (also known as UL30), whose intrinsic activities include a 5′-3′ polymerase and a 3′-5′ exonuclease (11, 16, 17, 29, 35, 42, 56). To carry out these functions, Pol has double-stranded DNA (dsDNA)- and single-stranded DNA (ssDNA)-binding activities that also can be found within individual proteolytic fragments of the enzyme (56). Moreover, DNA binding causes a conformational change in Pol (57). However, this DNA binding is not sufficient for highly processive DNA synthesis (14, 21).
The second HSV polymerase subunit is UL42, which is a processivity factor (14). It differs from other established processivity factors in that it binds directly and stably to dsDNA, albeit without sequence specificity (36, 43). Unlike sliding clamps, the association of Pol and UL42 does not require additional factors and can occur in the absence of DNA. Both the DNA-binding and Pol-binding activities of UL42 appear to be required for its function as a processivity factor, since mutations that specifically affect either of these activities severely reduce long-chain DNA synthesis and in vivo replication (3, 10). Moreover, the affinity of Pol/UL42 for a hairpin primer-template is greater than that of Pol alone, and the footprint on the dsDNA region of this template is extended when UL42 is present (13). Thus, the available data support the hypothesis (14) that UL42 functions as a tether between Pol and DNA and that its DNA-binding activity is crucial for its function as a processivity factor. However, many of the specifics regarding how UL42 affects the interaction of polymerase with DNA are not known, nor is it known whether UL42, which has high affinity for DNA, affects the movement of the polymerase along the template.
In this study, we tested three hypotheses: (i) that UL42 increases the specificity of the holoenzyme for a primer-template configuration, (ii) that UL42 limits the rate of dissociation of the holoenzyme from the primer-template, and (iii) that the increase in processivity conferred by UL42 is achieved at the expense of a decrease in the rate of elongation, i.e., that the polymerase sacrifices speed for distance. To examine these hypotheses, bandshift and competition assays were used to determine the binding affinities of Pol, UL42, and the Pol/UL42 heterodimer on different templates in the presence or absence of magnesium. These assays allowed a comparison of the binding preferences of these proteins to different configurations of DNA. The association and dissociation rates of Pol and Pol/UL42 on DNA in a primer-template configuration were determined. Finally, the rates of elongation of Pol and Pol/UL42 were compared by using a rapid-quench technique. The results indicate that UL42 both increases the specificity of polymerase binding to primer-template DNA and decreases the rate of dissociation from primer-template DNA. However, despite the increased binding to DNA, UL42 does not retard the rate of elongation of polymerase.
MATERIALS AND METHODS
Proteins.
HSV type 1 (HSV-1) Pol, UL42, and the Pol/UL42 heterodimer from recombinant baculovirus-infected SF9 cells were purified as described previously (14, 35).
DNA templates.
To prepare an ∼100-bp dsDNA template with four-base 5′ ssDNA overhangs, an SmaI fragment of the UL42 gene from position 1259 to position 1348 (38) was cloned into pUC18 to generate pUCUL42-SF16. This plasmid was then digested with EcoRI and BamHI, and the ∼100-bp EcoRI-BamHI fragment was gel purified. An ssDNA template was prepared from this dsDNA template by heat denaturation, followed by immediate chilling on ice. An ∼30-bp dsDNA template with four-base 5′ ssDNA overhangs was generated by EcoRI and XbaI digestion of pGEM-3Zf(+) (Promega), followed by gel purification. M13mp18 positive-strand and replicative-form (RF) DNA templates were purchased from Pharmacia. Primer 2447 (New England Biolabs), 5′-CGCCAGGGTTTTCCCAGTCACGA, is complementary to nucleotides (nt) 6311 to 6353 of M13mp18. Primers and DNA fragments were 5′ end labeled by T4 nucleotide kinase with [γ-32P]ATP (6,000 Ci/mmol) by using standard procedures. Labeled reaction mixtures were phenol extracted. The labeled DNA primer was mixed with M13mp18 positive-strand template DNA at a molar ratio of 3:1 in the presence of 100 mM NaCl in a final volume of 100 μl. The mixture was heated to 95°C for 5 min and allowed to cool to room temperature over 30 min. To remove excess primer, the hybridized solution was spun through a Centricon-100 column (Amicon) and washed with 300 μl of Tris-EDTA (10:1) buffer (pH 7.5) in accordance with the manufacturer’s directions. The primed template was collected in ∼50 μl of Tris-EDTA.
Gel retardation assays.
In one set of experiments, 1 fmol of labeled (10,000 cpm/fmol) ds or ss template DNA derived from an ∼100-bp fragment was incubated at room temperature with various amounts of Pol/UL42, Pol, or UL42 for 10 min in a 20-μl volume in buffer C (10 mM HEPES-KOH [pH 7.9], 5% glycerol, 0.25 mM dithiothreitol [DTT], 0.1 mM EDTA, 10 mM KCl, 100 μg of bovine serum albumin per ml). MgCl2 at 2 mM was included in the appropriate reaction mixtures. For determination of binding constants, a range of concentrations of a labeled ∼30 bp ds template with four-base 5′ ss overhangs (1, 2, 4, 8, and 16 nM) was titrated with either 2 × 10−9 M Pol or UL42 or 0.75 × 10−9 M Pol/UL42. Mixtures of protein and DNA were incubated for 10 min at room temperature. Following addition of 2 μl of loading buffer (10 mM HEPES-KOH [pH 7.9], 25% Ficoll 100, 0.1% bromphenol blue, 0.1% xylene cyanol), bound DNA and free DNA were resolved by fractionation on a 5% (for templates derived from the ∼100-bp fragment) or 10% (for the ∼30-bp template) native polyacrylamide gel. Gels were dried and autoradiographed, and the data were quantified by densitometry of the resulting autoradiograms.
Filter-binding assays.
Reaction conditions were identical to those used for the gel retardation assays with 2 mM MgCl2 included in the reaction mixtures, labeled template DNA derived from the ∼100-bp fragment, and various concentrations of unlabeled M13mp18 DNA included as indicated. At the end of the incubation, mixtures were diluted with 1 ml of buffer D (10 mM HEPES-KOH [pH 7.9], 0.25 mM DTT, 0.1 mM EDTA) for ss template DNA and buffer D plus 2 mM MgCl2 for ds template DNA. The diluted mixture was immediately filtered through alkali-washed nitrocellulose filters (pore-size, 0.45 μm). Filters were washed and dried, and radioactivity was measured by liquid scintillation counting.
Determination of dissociation constants.
Apparent dissociation constants from the filter-binding assays using the dsDNA and ssDNA templates based on the ∼100-bp fragment were calculated by using saturation isotherm analysis. (The values are apparent because the number of sites bound per DNA molecule cannot be determined.) The fraction of filter-bound DNA was plotted against the protein concentration. Apparent Kd = [P][D]/[PD], where apparent Kd is the apparent dissociation constant and [P], [D], and [PD] are the concentrations of free protein, free DNA, and the protein-DNA complex, respectively. The protein concentration that led to half saturation is referred to as K1/2. When half of the input DNA is bound, [D] = [PD]; thus, apparent Kd = [P] = K1/2. This is only true if [D] ≪ Kd, and the assays were performed under those conditions.
Dissociation constants were also calculated via Scatchard analysis by using data from gel retardation assays with the shorter template under conditions under which only one protein bound per DNA molecule. The ratio of [bound DNA]/[free DNA] was plotted as a function of [bound DNA]. The slope of the line equals −1/Kd.
Determination of on and off rates.
For measurement of kon, 10−7 M Pol or 5 × 10−8 M Pol/UL42 was incubated with the 33-bp fragment at 10−9 M in 100 μl at room temperature. Aliquots (10 μl) were removed at the indicated times, and binding was quenched by the addition of a 1,000-fold excess of competitor DNA (in 1 μl). The competitor used was a hairpin oligonucleotide, HP96 (50), purchased from Genosys (The Woodlands, Tex.). At the end of the time course, the samples were analyzed on a native acrylamide gel. Dried gels were autoradiographed, and the relative amounts of bound DNA and free DNA were determined by densitometry. 1/([P0] − [D0]) ln {[D0][P0] − [PD]/[P0]([D0] − [PD])} was plotted as a function of time, where [P0] and [D0] are the concentrations of free protein and free DNA at time zero and [PD] is the concentration of the protein-DNA complex at time t. kon corresponds to the slope of the straight line fitted to the data (r2 > 0.97 for both plots).
For measurement of koff, the mixture was first incubated for 10 min on ice in 100 μl. At time zero, a 1,000-fold competitor excess was added (in 10 μl) and the mixture was incubated at room temperature. At the indicated times, 11-μl aliquots were removed and immediately analyzed on a native polyacrylamide gel. Dried gels were autoradiographed, and the relative concentrations of bound DNA and free DNA were determined by densitometry. koff was determined by plotting ln ([PD]/[PD0]) versus time, where [PD0] and [PD] are the protein-DNA complex concentrations at times zero and t. The slope of the straight line fitted to the data (r2 > 0.97 for both plots) is equal to −koff. Half-lives of the complexes can be calculated by t1/2 = −(ln 0.5)/koff.
Rapid-quench analysis.
Stop flow reactions were performed on Kintek Instrument Rapid Quench Flow-3 (26) at 37°C. Reaction cocktails in one syringe contained, per 20 μl of reaction mixture, 50 fmol of singly primed M13 template DNA; 800 or 200 fmol of Pol or Pol/UL42, respectively; and 120 μM dCTP in buffer A [100 or 50 mM (NH4)SO4, 20 mM Tris-Cl (pH 7.5), 0.1 mM EDTA, 0.5 mM DTT, 4% glycerol, 40 μg of bovine serum albumin per ml]. Reactions were initiated by mixing an equal volume of this cocktail with 120 μM (each) dATP, dGTP, and TTP and 6 mM MgCl2 in buffer A. Reactions were terminated by a constant volume (60 μl) of quench solution containing 1% sodium dodecyl sulfate, 20 mM EDTA, and 20 mg of salmon sperm DNA per ml. Reaction times were 5, 20, 100, 500, and 1,000 ms. At least three determinations were made per time point in each of two separate experiments. Reaction products were ethanol precipitated in the presence of 0.3 M sodium acetate, resuspended in 5 or 10 μl of formamide dye solution, and fractionated on an 8 or 12% urea gel. The dried gel was exposed to a PhosphorImager screen. ImageQuant software was used to compare the intensity and frequency of each size product. Weighted values were used to estimate the number of nucleotides extended at each time point. Data were fitted to a simple straight line with an r2 value of >0.99.
RESULTS
Binding of Pol/UL42 to DNA in the absence or presence of magnesium.
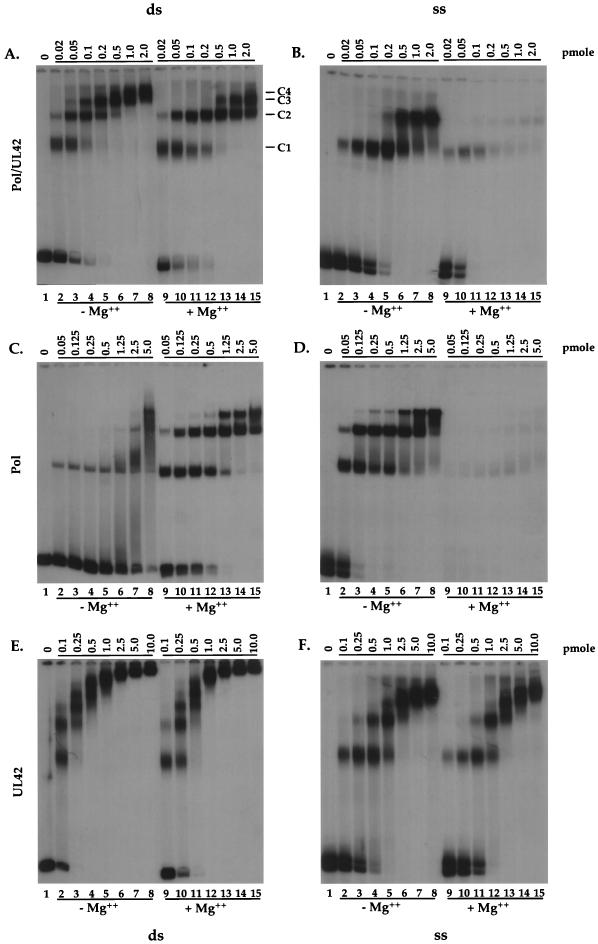
We wished to examine the binding of HSV DNA polymerase and each of its subunits to DNA templates in various configurations. Our working hypothesis was that the holoenzyme might exhibit affinities for certain configurations of DNA (e.g., dsDNA versus ssDNA) that differ from those of Pol alone. In our initial experiments, a gel retardation assay was employed to determine the DNA-binding activities of Pol/UL42, Pol, and UL42 to two kinds of radiolabeled DNA templates. The first was an ∼100-bp fragment that was dsDNA for most of its length but whose 5′ ends were four-base ssDNA overhangs. Thus, a protein could bind to this template via interactions either with dsDNA or with the ends of the DNA which are in a primer-template configuration. For the sake of brevity, we will refer to this as the ds template. The second template was the same fragment that had been heat denatured and was thus entirely ssDNA. Because (see below) Pol/UL42 and Pol degraded ssDNA in the presence of magnesium, we compared the binding of the various proteins to the different templates in the absence or presence of magnesium. In these assays, increasing concentrations of protein were titrated against a fixed amount of either of these templates. In Fig. 1, the panels on the left are binding assays with the dsDNA template, while those on the right were performed with the ssDNA template. Figure 2 presents a lower range of concentrations of Pol/UL42 binding to the ds template only. Within each panel, lanes 2 to 8 represent reactions incubated in the absence of magnesium, while the reactions in lanes 9 to 15 were incubated in the presence of 2 mM MgCl2.
FIG. 1.
Binding of the HSV polymerase holoenzyme and subunits to different DNA configurations. The indicated amounts of Pol/UL42 (A and B), Pol (C and D), and UL42 (E and F) were incubated for 10 min with 1 fmol of a 5′-end-labeled ds template with ends in a primer-template configuration (ds; A, C, and E) or ss template (ss; B, D, and F) in the absence (lanes 2 to 8) or presence (lanes 9 to 15) of 2 mM magnesium chloride. In each panel, lane 1 is free template DNA that was not incubated with protein. Complexes and free DNA were fractionated on native polyacrylamide gels, which were autoradiographed.
FIG. 2.
Two binding sites for Pol/UL42 on the ds template at low protein-DNA ratios. The indicated amounts of Pol/UL42 were incubated with 1 fmol of a 5′-end-labeled ds template in the absence (lanes 2 to 8) or presence (lanes 9 to 15) of magnesium, as in Fig. 1. Lane 1 is free template DNA that was not incubated with protein. Complexes and free DNA were fractionated on native polyacrylamide gels, which were autoradiographed.
Incubation of Pol/UL42 with the ds template in the absence of magnesium resulted in four distinct complexes, which we term C1 to C4 (Fig. 1A, lanes 2 to 8). When lower concentrations of Pol/UL42 were used, only three complexes were observed (Fig. 2). The highest-mobility complex, C1, was formed when only 5 fmol of Pol/UL42 was present (Fig. 2, lane 1), and the next-highest-mobility complex (C2) appeared with 20 fmol of Pol/UL42 (Fig. 1, lane 2; Fig. 2, lane 4). Starting with 40 fmol of Pol/UL42, a lower-mobility complex (C3) was observed while the amount of C1 correspondingly diminished (Fig. 1A, lanes 4 to 5; Fig. 2, lane 6 to 8). With greater than 500 fmol of protein, the amount of C2 diminished as well, accompanied by the appearance of a fourth complex, C4 (Fig. 1A, lanes 6 to 8). The changes in the pattern of complexes formed with increasing amounts of protein added suggested that C1 contained a single Pol/UL42 heterodimer and C2, C3, and C4 contained two, three, and four heterodimers, respectively.
The complexes observed between Pol/UL42 and the ds template in the presence of magnesium (Fig. 1A and 2, lanes 9 to 15) differed quantitatively from those observed in the absence of divalent cation. Incubation of Pol/UL42 and the ds template resulted primarily in the formation of only two complexes, C1 and C2, (Fig. 2, lanes 9 to 15), even when as much as 200 fmol of protein was added (Fig. 1A, lanes 9 to 12). A complex with lower mobility (C3) was formed only when over 500 fmol of protein was included in the incubation mixture, and it was accompanied by a decrease in the amount of C1 (Fig. 1A, lanes 13 to 15). This amount is 10-fold higher than that required to form C3 in the absence of magnesium (compare lanes 3 and 13). Little C4 was observed, even at 2,000 fmol of protein, and the amount of C2 remaining was only slightly diminished compared to that which remained after incubation in the absence of magnesium (Fig. 1A, lanes 8 and 15).
Two distinct complexes were observed when the Pol/UL42 heterodimer was incubated with the ss template (Fig. 1B). In the absence of magnesium, only a single complex was predominant at those protein concentrations that yielded two complexes with the ds template (compare lanes 2 to 4 in Fig. 1A and B). As more protein was included in the incubation (Fig. 1B, lanes 5 to 8), an additional complex was formed, concomitant with a decrease in the amount of the faster migrating complex. When magnesium was included in the incubation mixture, complexes were present in much lower abundance, evidently due to the intrinsic exonuclease activity of HSV Pol, which is more active against ssDNA than against dsDNA (35, 56). Nevertheless, overexposure of the autoradiograph showed that the pattern and mobilities of shifted complexes were similar to those observed in the absence of magnesium. This was confirmed by incubating Pol/UL42 and ssDNA at 4°C, a temperature at which the exonuclease activity was limited (55).
Binding of Pol to DNA.
Results of gel shift assays of DNA with Pol in the absence of UL42 are shown in Fig. 1C and D. The effect of magnesium on binding of Pol to the ds template was even more pronounced than that observed for Pol/UL42 (Fig. 1C). In the absence of magnesium, Pol did not bind very efficiently, in that saturation was never achieved, even with the largest amounts of proteins used (lanes 7 and 8), as evidenced by the free (unbound) DNA. Moreover, a second, higher-mobility complex was formed only with greater than 2,500 fmol of protein, 50-fold more than was necessary when magnesium was present (compare Fig. 1, lanes 7 and 9). When Pol was allowed to bind the ds template in the presence of magnesium, three specific complexes were formed. As was true with Pol/UL42, the two highest-mobility complexes appeared at low protein concentrations (Fig. 1, lanes 9 to 11). The third complex was formed when protein amounts were greater than 250 fmol (Fig. 1, lanes 11 to 15). This was accompanied by a decrease in the amount of the complex with the greatest mobility. Regardless, comparison of the binding of Pol to the ds template with the binding of Pol/UL42 to this template suggested that Pol/UL42 bound with higher affinity than did Pol alone, as only 10 to 20 fmol of Pol/UL42 was required to shift more than half of the DNA (e.g., Fig. 1A, lanes 2 and 9, and Fig. 2, lanes 4 and 10), while more than 50 fmol of Pol was required to shift more than half of the DNA (Fig. 1C, lanes 3 and 10).
In contrast to binding to the ds template, specific complexes were readily formed between Pol and ssDNA in the absence of magnesium (compare Fig. 1C and D, lanes 2 to 8). The amount of complex observed in the presence of magnesium was reduced as expected from the 3′-5′ exonuclease activity of Pol. Again, incubation at 4°C instead of room temperature severely limited the exonuclease activity (55). Under these conditions, the patterns of Pol binding to ssDNA were virtually identical in the presence and absence of magnesium (55). For ss template DNA, comparisons between the binding of Pol and Pol/UL42 (Fig. 1B and D) suggested that Pol bound with higher affinity than did Pol/UL42.
Binding of UL42 to DNA.
Complex formation between UL42 and DNA in the presence or absence of magnesium was very similar (compare lanes 2 to 8 to lanes 9 to 15 in Fig. 1E or F). At the lowest protein concentrations, at least two complexes were formed between UL42 and the ds template (Fig. 1E, lanes 2, 3, 9, and 10). Over the range of protein concentrations used, six different complexes could be observed (54). At the highest concentrations of protein used, the complexes migrated slowly in the gel (Fig. 1E, lanes 5 to 8 and 12 to 15). As expected (36, 43), UL42 bound the ds template more readily than ssDNA, with free ds template disappearing at lower protein concentrations than free ssDNA (Fig. 1E and F, lanes 2 to 4 and 9 to 11). Furthermore, fewer complexes were observed when ssDNA was incubated with the same amounts of protein compared with the dsDNA template (compare Fig. 1E and F).
To summarize the results of these experiments, Pol/UL42 bound and formed lower-mobility complexes on the ds template (whose ends are in a primer-template configuration) at lower concentrations of protein than did Pol alone, regardless of the presence or absence of magnesium. In the presence of magnesium, both Pol/UL42 and Pol formed the two highest-mobility complexes on this template at much lower protein concentrations than were required to form the third complex, suggesting that there were two preferred binding sites per template. Pol formed lower-mobility complexes on ssDNA at lower concentrations than did Pol/UL42. UL42 bound and formed multiple lower-mobility complexes on the dsDNA template at lower concentrations than it did on ssDNA.
Competition experiments to test the importance of specific binding to ends of the ds template.
Given that the mobility shift patterns of Pol/UL42 and Pol on the ds template suggested two preferred binding sites per template, we hypothesized that Pol and Pol/UL42 would bind preferentially to the primer-template ends of the ds template. This would explain why at low concentrations of protein, the C1 (one end bound) and C2 (two ends bound) complexes were observed, while additional complexes resulting from binding to internal ds regions were only observed at high concentrations of protein. To test this hypothesis, filter-binding experiments were performed by using the same radiolabeled ds template that was used in the gel retardation assays and using M13mp18 DNA as a competitor. The competitor DNA was either circular dsDNA or ssDNA or linearized (with EcoRI) dsDNA or ssDNA. Binding could thus occur at both ends (primer-template configuration) of the linearized competitor DNAs or at internal sites, whereas only internal binding would be permitted with the circular templates. Pol/UL42, Pol, or UL42 was incubated with the 32P-labeled templates in the presence of various concentrations of competitor DNA, and the amounts of labeled DNA bound to nitrocellulose filters by virtue of being bound to protein were measured. The concentrations of competitor DNA that reduced filter-bound radioactivity by 50% (IC50) were determined. The lower the IC50, the more efficiently bound is that conformation of competitor DNA. It should be noted that because the proteins could bind to internal sites (albeit with various efficiencies), the IC50s obtained were low relative to the Kd values obtained with shorter templates (see below) because M13mp18 is large (7.2 kb) and thus has many internal binding sites per molecule.
Linearized M13mp18 dsDNA, with an IC50 of 20 pM, was the most efficient competitor for Pol/UL42 binding to ds template DNA, being 15- to 20-fold more efficient than linear or circular ssDNA and 3- to 4-fold more efficient than circular dsDNA (Table 1). In contrast, circular and linear M13mp18 ssDNAs were the most efficient competitors for the binding of the Pol subunit to ds template DNA, having an IC50 of 90 pM (Table 1). Nevertheless, linearized M13mp18 dsDNA was again more effective at competing for Pol binding than was circular dsDNA, having a fourfold lower IC50 (Table 1). Additional competition experiments showed that relatively short dsDNA fragments with 3′ overhangs or blunt ends were less efficient competitors for Pol binding than dsDNA fragments with 5′ overhangs (55). Moreover, Strick and Knopf (46) have reported that Pol/UL42 from HSV-infected cells binds preferentially to dsDNA with 5′ overhangs over dsDNA with blunt ends. These observations were consistent with our hypothesis that Pol/UL42 and Pol interact preferentially with the primer-template configurations at the ends of the ds template. Therefore, the two higher-mobility complexes observed in the right-hand halves of Fig. 1A and C most likely represent binding to the primer-template ends of the dsDNA fragment, while additional complexes were formed by nonspecific binding to internal binding sites.
TABLE 1.
Competition efficiencies of circular and linearized ssDNA and dsDNAa
| Protein and competitor DNAc | Mean IC50 (nM)b ± SD
|
|
|---|---|---|
| ssDNA | dsDNA | |
| Pol/UL42 | ||
| Circular | 0.4 ± 0.006 | 0.07 ± 0.005 |
| Linear | 0.3 ± 0.05 | 0.02 ± 0.003 |
| Pol | ||
| Circular | 0.09 ± 0.006 | 2 ± 0.12 |
| Linear | 0.09 ± 0.02 | 0.5 ± 0.006 |
| UL42 | ||
| Circular | 0.5 ± 0 | 0.01 ± 0.0006 |
| Linear | 0.5 ± 0.01 | 0.01 ± 0.0006 |
A 15-fmol sample of a radiolabeled ds template (∼100 bp) was incubated with 0.5 pmol of Pol/UL42, Pol, or UL42 and various amounts of competitor DNA in 10-μl reaction mixtures in the presence of magnesium as detailed in Materials and Methods. The amount of radioactivity that bound to filters was used to deduce the IC50 of each competitor.
Data from three separate experiments are presented.
M13mp18 DNA was used as the competitor. The circular ssDNA and dsDNA used were positive-strand and RFI M13mp18, respectively. Linearized dsDNA was generated by restriction of RFI M13mp18 with EcoRI. Linearized ssDNA was obtained by heat denaturation of this restricted DNA.
In contrast to Pol and Pol/UL42, UL42 binding to the ds template was competed by linearized and circular M13mp18 dsDNAs with equal efficiency (Table 1). Either of the ssDNA competitors was 50-fold less efficient by comparison. Thus, unlike Pol/UL42 and Pol, UL42 exhibited a preference not for linearized DNA but only for dsDNA over ssDNA.
Determination of relative apparent affinities.
We then used the filter-binding assay to determine relative affinities of HSV polymerase and its subunits for the ds template whose ends are in a primer-template configuration and for the ss template. Increasing amounts of protein were added to 1 fmol of 5′-end-labeled template DNA in the presence of magnesium for the ds template and in the absence of magnesium for the ss template. The amount of filter-bound radioactivity was measured to determine the fraction of bound DNA, which was plotted against protein concentration. With these experimental conditions, in which the concentration of DNA was very low, we could then apply a saturation isotherm analysis to calculate apparent Kds from the concentration of protein that led to half saturation (Table 2). The values obtained were apparent rather than absolute because the assay cannot distinguish between DNAs with one or more sites bound.
TABLE 2.
Apparent binding affinities of Pol/UL42, Pol, and UL42a
| Protein | Apparent dissociation constant (M)
|
|
|---|---|---|
| ss template | ds template | |
| Pol/UL42 | 1.6 × 10−9 | 0.33 × 10−9 |
| Pol | 0.5 × 10−9 | 4.6 × 10−9 |
| UL42 | 1.1 × 10−8 | 2.0 × 10−9 |
Apparent dissociation constants correspond to protein concentrations that led to half-maximal occupation of binding sites, as determined in filter-binding assays. Assays with ss and ds templates were performed in the absence and presence of MgCl2, respectively.
By this assay, Pol/UL42 exhibited a high apparent affinity (0.33 nM) for the ds template that was fivefold higher than that for the ss template (Table 2). Based on our analysis of the gel retardation experiments and competition experiments, this higher affinity for the ds template was due to binding to the ends, which are in a primer-template configuration. In contrast with Pol/UL42, Pol bound the ss template with 10-fold greater apparent affinity (0.5 nM) than the ds template (Table 2), even though, like Pol/UL42, its binding to the ds template was via binding to the primer-template ends. Additionally, the apparent affinity of Pol/UL42 for the primer-template configuration was 10- to 20-fold greater than that of Pol, while the apparent affinity of Pol for the ss template was ∼3-fold greater than that of Pol/UL42 (Table 2). Thus, not only did Pol/UL42 bind more tightly to primer-template DNA than did Pol, as previously shown by using hairpin primer-template DNA (13), but it bound less tightly to ssDNA than did Pol. UL42 exhibited ∼20-fold greater apparent affinity for dsDNA (2 nM) than for ssDNA (Table 2). The differences in apparent Kd between the ds and ss templates correlated well with the differences in IC50 between the linearized M13mp18 dsDNA and ssDNA in the competition assay (Table 1).
Determination of Kd for a ds template with primer-template ends by Scatchard analysis.
In order to determine the absolute binding affinities of Pol, Pol/UL42, and UL42 for a ds template with primer-template ends, we performed a gel retardation assay using a short template—an ∼30-bp ds fragment with four-base 5′ ss overhangs. In contrast with the longer template used in Fig. 1 and 2 and Table 2, incubation of this short template at all of the protein concentrations used to calculate affinities resulted in only one complex that migrated more slowly than the unbound template (Fig. 3A). Thus, the affinities measured reflect one protein bound to one DNA molecule. (In other assays, at higher protein-DNA ratios, a maximum of two complexes were observed with UL42 [55].) Constant amounts of protein were titrated with increasing amounts of 5′-end-labeled DNA. After a 10-min incubation, the protein-DNA complexes were resolved on a nondenaturing gel. Densitometric measurements of free and complexed DNAs were plotted as bound versus bound/free in a Scatchard analysis. Kds were determined as −1/slope.
FIG. 3.
Determination of dissociation constants by Scatchard analysis. (A) The indicated amounts of a 5′-end-labeled 33-bp DNA fragment with ends in a primer-template configuration were incubated with 7.5 fmol of Pol/UL42 (lanes 1 to 5), 20 fmol of Pol (lanes 6 to 10), or 20 fmol of UL42 (lanes 11 to 15). Complex formation was analyzed by gel retardation assay as detailed in Materials and Methods. (B) The relative amounts of DNA bound in panel A were measured by densitometry and plotted against the amounts of DNA added. (C) Scatchard plot of gel retardation data from panels A and B. Slope, −1/Kd.
The affinity of Pol/UL42 for the short template was 0.75 nM, 15-fold higher than that of Pol alone (Table 3, column 2). This difference was consistent with that observed for the longer template (Tables 1 and 2). Interestingly, the binding constants of Pol and Pol/UL42 obtained in the filter-binding assay using the longer ds template, which is large enough to bind two proteins to its ends, were approximately half of those obtained with the short template (compare Tables 2 and 3). This suggests that the conditions used in measuring the binding constants with the larger ds template were such that both ends of the fragment could be equivalently occupied. UL42 bound the short template with a Kd of 1.4 nM (Table 3). This value is slightly lower than that obtained with the longer ds template (Table 2), which is consistent with the longer template binding, on average, >1 UL42 molecule at concentrations at which about half of the DNA molecules were bound (Fig. 1E). The value is modestly higher than that for Pol/UL42 on the short template (Table 2); thus, Pol/UL42 bound to this template with higher affinity than did either of its subunits, especially Pol, consistent with previous results obtained by Gottlieb and Challberg by using hairpin primer-template DNA (13).
TABLE 3.
Dissociation constants and kinetic parametersa
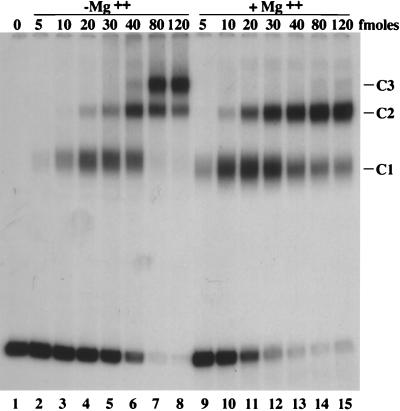
Determination of kon and koff.
We wished to determine whether the higher affinity of Pol/UL42 relative to Pol for primer-template DNA was due to more rapid association or diminished dissociation. Therefore, the kon and koff of Pol/UL42 and Pol for the short ds template were measured (Fig. 4). To measure kon, a saturating amount of enzyme was mixed with a reaction cocktail containing radiolabeled template DNA and incubated at room temperature. At appropriate times, an aliquot was removed and further binding of protein to labeled DNA was quenched with EDTA and an excess of a unlabeled competitor DNA. Each aliquot was analyzed on a nondenaturing polyacrylamide gel, the autoradiogram of which is shown in Fig. 4A. The amounts of bound DNA and free DNA were measured by densitometry of the resulting autoradiogram and plotted (Fig. 4B). koff was determined similarly, except that the enzyme and DNA were preincubated for 10 min at room temperature to allow binding. At time zero, unlabeled competitor DNA was added to the cocktail and the mixture was incubated at room temperature. At appropriate times, an aliquot was removed and immediately loaded onto a running gel, the autoradiogram of which is shown in Fig. 4C (the apparent decreasing mobility of the free DNA with time is due to the samples being loaded at different times on the running gel). The amounts of bound DNA and free DNA were assessed by densitometry (Fig. 4D). The plots in Fig. 4B and D were used to calculate kon and koff values (Table 3).
FIG. 4.
Determination of association and dissociation rates. (A) Gel retardation analysis of association rate. A 5′-end-labeled short ds template (10−9 M) was incubated with 5 × 10−8 fmol of Pol/UL42 (lanes 1 to 9) or 10−7 fmol of Pol (lanes 10 to 18) in a total volume of 100 μl. At the indicated times, 10 μl was removed and mixed with 2 μl of loading buffer and 1 μl of a 1,000-fold excess of unlabeled DNA to prevent any further association of protein with labeled DNA and analyzed on a native polyacrylamide gel. The samples from the time course using Pol/UL42 were applied to the gel first, electrophoresis was started at a low voltage, and then, after the time course using Pol was completed, those samples were applied to the gel. This accounts for the apparently slower mobility of free DNA in lanes 10 to 18. (B) Calculation of kon. The data from panel A were analyzed and plotted. To ensure that initial rates of association were measured, only data from the first 50 s were fitted to the plot. [P0] and [D0] are concentrations of free protein and free DNA, respectively, at time 0. [PD] is the protein-DNA complex concentration at a given time point. kon corresponds to the slope of the straight line fitted to the first 50 s of datum points (r2 > 0.97 for both plots). (C) Gel retardation analysis of dissociation rate. Experimental details are similar to those of panel A, except that Pol (lanes 1 to 7) or Pol/UL42 (lanes 8 to 14) and DNA were preincubated for 10 min. At time zero, to prevent any reassociation of protein and labeled DNA, a 1,000-fold molar excess of unlabeled DNA was added to the mixture. Aliquots of 11 μl were removed at the indicated times and immediately loaded onto a running polyacrylamide gel. (D) Calculation of koff. The data from panel C were analyzed and plotted. [PD0] and [PD] refer to the concentrations of bound species at time zero and at a given time point, respectively. The slope of the plot is equal to −koff.
The kon for Pol/UL42 was about twice that of Pol (Fig. 4 and Table 3), indicating that UL42 had only a modest effect in increasing the rate of association of polymerase with DNA. In contrast, the koff for Pol/UL42 was 10-fold lower than that for Pol, resulting in t1/2 of a 2 h for Pol/UL42 on the primer-template (Fig. 4 and Table 3). The Kd values calculated from the koff and kon values (Table 3, rightmost column) were severalfold lower than those determined by Scatchard analysis (Table 3, column 2). This may be due to overestimation of kon values as a result of manual quenching. Nevertheless, the two methods yielded very similar fold differences in affinity between Pol/UL42 and Pol. Thus, a decreased rate of Pol/UL42 dissociation from the primer-template is primarily responsible for the increased affinity of Pol/UL42 versus Pol.
Pol/UL42 and Pol exhibit similar rates of elongation.
We had previously shown that mutations that specifically and severely decrease the DNA-binding activity of UL42 severely reduce its ability to function as a processivity factor in vitro and to function in viral replication in vivo, strongly suggesting that DNA binding is required for UL42 function (3). Results in Table 3 show that UL42 stabilizes the interaction of the heterodimer with the primer-template largely by decreasing dissociation. A hypothesis that arises from these observations is that the direct and highly stable binding of DNA conferred on polymerase by UL42, while increasing processivity (macroscopic elongation), may slow the speed at which polymerase translocates (microscopic elongation). To address this question directly, the rates of elongation of Pol and Pol/UL42 on an M13mp18 template were determined. In order to measure elongation by Pol before it dissociated from the template, a stop-flow method was used to measure the rates at relatively short time intervals. This also allowed us to minimize the effects of secondary structure and sequence-dependent stops (1, 12, 53, 54). With that in mind, we chose a primer that yielded the fewest favored pause products within the time assessed (4).
Purified Pol or Pol/UL42 was preincubated with an M13mp18 template that had been annealed to a radiolabeled primer in Mg2+-free buffer containing the next nucleotide (dCTP). This permitted binding to primer-template DNA while limiting exonuclease activity. It has previously been reported that the polymerase activity of Pol and Pol/UL42 was dependent on the concentration of monovalent cations present in the assay, with Pol alone being more active at 50 mM (NH4)2SO4 and Pol/UL42 being more active at 100 mM (NH4)2SO4 (18). Thus, to maximize the chances of observing a difference in elongation rate between Pol and Pol/UL42, the buffer contained either 50 or 100 mM (NH4)2SO4. Reactions were initiated with the addition of the remaining deoxynucleoside triphosphates in 2× Mg2+ buffer and, using a rapid-quench flow apparatus, measurements were taken at time points of 5 through 1,000 ms.
Figure 5 represents the PhosphorImager analysis of products from 5-, 20-, 100-, 500-, and 1,000-ms reactions. As shown, the amounts of synthesis by Pol and Pol/UL42 at each time point were qualitatively similar at both 100 mM (Fig. 5A) and 50 mM (Fig. 5B) (NH4)2SO4. To ensure that UL42 functioned as a processivity factor under these conditions, similar reactions were performed for longer times and analyzed by using both alkaline agarose gels and denaturing polyacrylamide gels. In the reaction mixtures containing UL42, long products (>2 kb) appeared much earlier, the amount of intermediate products was minimal, and the fraction of primers utilized was much smaller than in the same reaction mixtures with Pol alone (4). Thus, UL42 was active as a processivity factor under these conditions. Also, at these longer time points, greater activity was observed with Pol/UL42 than with Pol at 100 mM (NH4)2SO4 and greater activity was observed with Pol than with Pol/UL42 at 50 mM (NH4)2SO4 (4), as previously reported (18).
FIG. 5.
Rapid-quench analysis of elongation rate. A primed M13mp18 ssDNA template was preincubated with dCTP and Pol or Pol/UL42, as indicated, in Mg2+-free buffer containing 100 mM (NH4)2SO4 (A) or 50 mM (NH4)2SO4 (B). Reactions were initiated by adding the remaining deoxynucleoside triphosphates and 2× Mg2+ buffer. Reaction times were 5 (lanes 1 and 6), 20 (lanes 2 and 7), 100 (lanes 3 and 8), 500 (lanes 4 and 9), and 1,000 (lanes 5 and 10) ms. Quenched reactions were processed as described in Materials and Methods. Products were fractionated on a 12% denaturing polyacrylamide gel. M13 only: primed template preincubated with Pol alone and dCTP at 50 mM (NH4)2SO4.
To calculate rates of elongation, densitometric plots for each time point were evaluated and the area under each peak was integrated. Figure 6A and B shows plots of the rates of elongation at 100 mM (NH4)2SO4, while Fig. 6C and D represents reactions at 50 mM (NH4)2SO4. The extension that represented the median or 75th percentile value of total radioactivity (excluding unextended primer) at each time point between 5 and 500 ms was used to plot the graphs in Fig. 6. The median value represented the average speed of the population. The 75th percentile was chosen to approximate the maximal elongation rate (too few molecules were synthesized at the actual maximal rate to permit analysis). The median value plots are shown on the left (Fig. 6A and C), and the 75th percentile plots are shown on the right (Fig. 6B and D). As shown, the data represented by the 5- to 500-ms time points indicate a linear rate of elongation within this time frame. The rates of elongation by Pol and Pol/UL42 were not meaningfully different at 50 or 100 mM (NH4)2SO4 (Fig. 5 and 6). Thus, despite the apparent requirement for DNA binding by UL42 for processivity (3) and despite the stable binding of Pol/UL42 to primer-template DNA (Table 3 and reference 13) and its slow dissociation (Fig. 4 and Table 3), UL42 did not slow the translocation of polymerase along the template.
FIG. 6.
Comparison of elongation rates of Pol and Pol/UL42. Plots were based on densitometric measurements of product bands from 5- to 500-ms reactions from data in Fig. 5, as detailed in Materials and Methods. Panels A and B, reactions performed at 100 mM (NH4)2SO4; panels C and D, reactions performed at 50 mM (NH4)2SO4. A and C represent median values; B and D represent 75th percentile values.
DISCUSSION
Critical interaction steps between a replicative DNA polymerase and its primer-template DNA include initial attachment and sustained association, during which catalysis and translocation occur. Studies reported here investigated aspects of these processes. Below, the results of these investigations are discussed and two models for DNA replication by HSV DNA polymerase are proposed.
Increased affinity of polymerase for primer-template DNA relative to Pol is due largely to a decreased dissociation rate.
Gottlieb and Challberg (13) showed that both Pol and Pol/UL42 protected a hairpin oligonucleotide that was in a primer-template configuration over the junction of the ds and ss regions. The Kd values for the oligonucleotide of Pol/UL42, Pol, and UL42 determined in that study were 0.78 × 10−9, 7.1 × 10−9, and 1.1 × 10−9, respectively. We used a ds template that could permit internal binding to entirely dsDNA or binding to primer-template configurations at the ends. We found that Pol and Pol/UL42 bound preferentially to the primer-template configuration at the ends of the template, with stronger binding by Pol/UL42, while UL42 showed no preference for ends, in agreement with the interpretations of Gottlieb and Challberg (13). Using a short template to permit measurements of Kd (Table 3) yielded values within 30% of those obtained by Gottlieb and Challberg for the hairpin oligonucleotide. Thus, confirming and extending the previous study (13), the Pol/UL42 heterodimer has higher affinity for primer-template DNA than does either of its subunits alone.
The 10- to 20-fold increase in Pol/UL42 affinity for primer-template DNA relative to Pol was mainly due to a substantially reduced rate of dissociation of the polymerase from the primer-template (Fig. 4). The effect on the association rate was less dramatic. The association rates of Pol and Pol/UL42 were well below the value of 108 to 109 that is expected for diffusion-controlled reactions (5, 23). This is consistent with the view that Pol and Pol/UL42 must approach the DNA in a particular orientation in order for productive and stable association to occur and/or that binding of Pol or Pol/UL42 is a two-step process (e.g., involving a conformational change in the enzyme [57]).
Decreased heterodimer binding to nonproductive templates.
Our studies permitted comparisons of binding to DNA in a primer-template configuration with binding to DNA in other configurations. As there can be many more of the latter binding sites than the former per replicating viral genome, it seems likely that minimizing nonproductive association with DNA that was either entirely ss or ds would increase the efficiency of DNA replication. It was thus interesting that Pol/UL42 bound DNA that was in a primer-template configuration more avidly than DNA in other configurations. Pol/UL42 bound ssDNA with much lower affinity than did Pol alone and bound DNA that was entirely ds with sevenfold lower affinity than did UL42. In contrast, Pol bound ssDNA more avidly than it did DNA in a primer-template configuration and UL42 bound DNA that was entirely ds as avidly as or more avidly than it did DNA in a primer-template configuration. Thus, without UL42, Pol would be more likely to associate nonproductively with ssDNA and without Pol, UL42 would be more likely to associate nonproductively with the more abundant binding sites that are entirely dsDNA. This reduction in nonspecific binding of Pol/UL42 compared to Pol alone or to UL42 alone could be a result of conformational changes in Pol and UL42 upon heterodimerization and/or due to regions in the individual subunits that contribute to nonspecific DNA binding becoming inaccessible. Regardless, detection of stimulation of Pol activity by UL42 in certain assays might not necessarily reflect increased processivity but might, instead, reflect fewer nonproductive associations with DNA.
HSV does not utilize auxiliary protein complexes to guide the polymerase to the primer-template junction, as do other organisms (27, 32, 33, 50). However, as recently reported, Pol and UL8, a component of the viral helicase-primase complex, can interact, at least in vitro (37). Such interactions between the primase and the DNA polymerase, combined with decreases in interaction of Pol/UL42 with DNA that is not in a primer-template configuration, might help guide polymerase to the primer-template junction. The observation that the kon of Pol/UL42 is approximately twofold greater than that of Pol alone also may abet this process.
Rate of fork movement.
The rate of DNA synthesis by HSV DNA polymerase purified from HSV-infected cells was originally estimated to be about 3 to 5 nt/s (42). Subsequent estimations using proteins purified from recombinant-baculovirus-infected insect cells gave rates of elongation for Pol/UL42 of 10 to 60 nt/s (14). Our results (26 to 33 nt/s), in agreement with the latter estimate and with the rate of fork movement of 50 nt/s in pseudorabies virus, another alphaherpesvirus (2), would be sufficient to sustain productive HSV infection at a rate of 10,000 copies per cell in 10 h (42). It is thus possible that the rate of fork movement in infected cells is dictated primarily by polymerase. Still, the rate of fork movement could be influenced by other replication factors. In simian virus 40 replication, for example, the helicase activity of T antigen (∼3 nt/s) appears to limit the rate of leading-strand synthesis in vitro and in vivo (41, 49). In the other direction, interaction of E. coli polymerase III DNA polymerase and helicase increases the rate of unwinding 10-fold (28). Since HSV Pol can interact with a helicase-primase subunit (37), it would be of interest to determine if HSV helicase activity is stimulated by Pol/UL42 or if the rate of unwinding limits fork movement instead.
UL42 is not a brake.
Embedded in the hypothesis that UL42 functions as a processivity factor by acting as a tether between Pol and DNA during DNA replication is a mechanistic paradox (14). Wouldn’t the stable association of UL42 and DNA decrease the rate of elongation of Pol? To address this paradox, we hypothesized that the increase in processivity afforded by UL42 would be achieved at the expense of a decrease in the rate of elongation. In other words, Pol alone would be like a sprinter, having high speed for short distances, while the Pol/UL42 complex would be like a marathoner, going farther by running more slowly. However, the microscopic elongation rates of Pol and Pol/UL42 were essentially the same. Although our results do not rule out the possibility that UL42 did reduce translocation speed but, at the same time, increased the rate of a catalytic step by the same amount, thereby preserving the overall elongation rate, the simplest interpretation is that UL42 does not brake elongation even though it “sticks” the polymerase to DNA.
How does Pol/UL42 rapidly translocate?
The apparent requirement for DNA binding by UL42 for processivity (3), combined with the very stable binding to primer-template DNA due to very slow dissociation conferred by UL42, implies a mechanism that is very different from that of the sliding clamps. Those processivity factors, which interact with DNA topologically rather than directly, appear to be designed to permit sliding and thus facile translocation. How, then, does Pol/UL42’s stable association with primer-template DNA permit rapid translocation? Two models for the translocation of Pol/UL42 along DNA are presented below.
Ratcheting.
In the ratcheting model, a catalytic step causes a conformational change such that HSV polymerase is no longer bound tightly to the primer terminus but can “scan” the DNA to arrive at its next preferred site. Such a conformational change has been advanced for T4 DNA polymerase, for which it was suggested that a conformer that binds specifically to primer-template junctions predominates during catalysis, while a conformer that binds less specifically to DNA via electrostatic interactions accomplishes the translocation step (12, 52). In one version of this model, Pol alone would also undergo a conformational change as it elongates. Interestingly, the C-terminal 12-kDa subdomain of Pol, which does not contain regions widely conserved among DNA polymerases, can bind DNA and UL42 independently from the rest of the molecule (56, 57). Certain mutations that affect this domain impair DNA polymerase activity and in vivo replication, even though they do not affect UL42 binding activity in vitro (8, 9). This C-terminal 12-kDa subdomain appears to bind preferentially to DNA ends (55); thus, it could conceivably serve as a lower-affinity, alternative primer-binding site that might be important during translocation.
In a second version of this model, Pol might transmit and coordinate a conformational change in UL42, such that UL42’s affinity for DNA would be temporarily decreased. In other words, the DNA-binding activity of UL42 would be moderated during the polymerization cycle in this model to allow the holoenzyme to advance.
(ii) Sitting.
In the sitting model (suggested to us by T. Kelly), Pol-associated UL42 itself does not translocate with respect to the DNA template but remains bound to the template upstream of the initial primer-binding site. Instead of Pol/UL42 moving along the DNA, the newly synthesized DNA loops and is extruded through the catalytic subunit. Indeed, replication factories, or focal sites of immobilized polymerizing complexes, have been proposed for replication of eukaryotic DNA (reviewed in reference 25). Lending credence to this model is evidence that HSV DNA replication appears to be confined to sites associated with the nucleoskeleton (7, 34, 51). Furthermore, this model predicts that the microscopic rate of elongation would be solely dependent on the chemistry of Pol, which is consistent with our results. However, the spatial arrangement of Pol/UL42 during catalysis would be rather different from that of other polymerases. Further studies to illuminate the molecular interactions between the enzyme subunits and DNA before, during, and after elongation would help distinguish between these two models.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank T. Kelly for suggesting the sitting model, A. Kuo for assistance with protein purification and binding experiments, M. Prahalad and C. Walsh for use of the rapid-quench flow apparatus and instruction and assistance, and B. Linder, J. Randell, and K. Grove for comments on versions of the manuscript.
This work was supported by grants from the NIH (AI19838, AI26077). K.W. was supported by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Bambara R A, Fay P J, Mallaber L M. Methods of analyzing processivity. Methods Enzymol. 1995;262:270–280. doi: 10.1016/0076-6879(95)62023-0. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Porat T, Blankenship M L, DeMarchi J M, Kaplan A S. Replication of herpesvirus DNA. III. Rate of DNA elongation. J Virol. 1977;22:734–741. doi: 10.1128/jvi.22.3.734-741.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow C S, Coen D M. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J Virol. 1995;69:6965–6971. doi: 10.1128/jvi.69.11.6965-6971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow, C. S., and D. M. Coen. 1997. Unpublished results.
- 5.Creighton T E. Proteins: structures and molecular properties. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1993. [Google Scholar]
- 6.Crute J J, Lehman I R. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5′-3′ exonuclease with ribonuclease H activity. J Biol Chem. 1989;264:19266–19270. [PubMed] [Google Scholar]
- 7.de Bruyn Kops A, Knipe D M. Preexisting nuclear architecture defines the intranuclear location of herpes virus DNA replication structures. J Virol. 1994;68:3512–3526. doi: 10.1128/jvi.68.6.3512-3526.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Digard, P., W. Bebrin, and D. M. Coen. 1993. Unpublished results.
- 9.Digard P, Bebrin W R, Weisshart K, Coen D M. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J Virol. 1993;67:398–406. doi: 10.1128/jvi.67.1.398-406.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Digard P, Chow C S, Pirrit L, Coen D M. Functional analysis of the herpes simplex virus UL42 protein. J Virol. 1993;67:1159–1168. doi: 10.1128/jvi.67.3.1159-1168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorsky D I, Crumpacker C S. Expression of herpes simplex virus type 1 DNA polymerase gene by in vitro translation and effects of gene deletions on activity. J Virol. 1988;62:3224–3232. doi: 10.1128/jvi.62.9.3224-3232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairfield F R, Newport J W, Dolejsi M K, von Hippel P H. On the processivity of DNA replication. J Biomol Struct Dyn. 1983;1:715–727. doi: 10.1080/07391102.1983.10507477. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb J, Challberg M D. Interaction of herpes simplex virus type 1 DNA polymerase and UL42 accessory protein with a model primer template. J Virol. 1994;68:4937–4945. doi: 10.1128/jvi.68.8.4937-4945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb J, Marcy A I, Coen D M, Challberg M D. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J Virol. 1990;64:5976–5987. doi: 10.1128/jvi.64.12.5976-5987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulbis J M, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21 WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 16.Haffey M L, Stevens J T, Terry B J, Dorsky D I, Crumpacker D I, Wiestock S M, Rutechan W T, Field A K. Expression of herpes simplex virus type 1 DNA polymerase in Saccharomyces cerevisiae and detection of virus-specific enzyme activity in cell-free lysates. J Virol. 1988;62:4493–4498. doi: 10.1128/jvi.62.12.4493-4498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall J D, Orth K L, Claus-Walker D. Evidence that the nuclease activities associated with the herpes simplex type 1 DNA polymerase are due to the 3′-5′ exonuclease. J Virol. 1996;70:4816–4818. doi: 10.1128/jvi.70.7.4816-4818.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart G J, Boehme R E. The effect of the UL42 protein on the DNA polymerase activity of the catalytic subunit of the DNA polymerase encoded by herpes simplex virus type 1. FEBS Lett. 1992;305:97–100. doi: 10.1016/0014-5793(92)80872-e. [DOI] [PubMed] [Google Scholar]
- 19.Herendeen D R, Kassavetis G A, Geiduschek E P. A transcriptional enhancer whose function imposes a requirement that proteins track along DNA. Science. 1992;256:1298–1303. doi: 10.1126/science.1598572. [DOI] [PubMed] [Google Scholar]
- 20.Herendeen D R, Kelly T J. DNA polymerase III: running rings around the fork. Cell. 1996;84:5–8. doi: 10.1016/s0092-8674(00)80069-0. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez T R, Lehman I R. Functional interaction between the herpes simplex-1 DNA polymerase and UL42 protein. J Biol Chem. 1990;265:11227–11232. [PubMed] [Google Scholar]
- 22.Himawan J S, Richardson C C. Genetic analysis of the interaction between bacteriophage T7 DNA polymerase and Escherichia coli thioredoxin. Proc Natl Acad Sci USA. 1992;89:9774–9778. doi: 10.1073/pnas.89.20.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinkle D C, Chamberlin M J. Studies of the binding of Escherichia coli RNA polymerase to DNA. II. The kinetics of the binding reaction. J Mol Biol. 1972;70:187–195. doi: 10.1016/0022-2836(72)90532-3. [DOI] [PubMed] [Google Scholar]
- 24.Huber H E, Tabor S, Richardson C C. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 25.Hughes, T. A., A. Pombo, J. McManus, P. Hozak, D. A. Jackson, and P. R. Cook. 1995. On the structure of replication and transcription factories. J. Cell Sci. Suppl. 19:59–65. [DOI] [PubMed]
- 26.Johnson K A. Rapid kinetic analysis of mechanochemical adenosinetriphosphatases. Methods Enzymol. 1986;134:677–705. doi: 10.1016/0076-6879(86)34129-6. [DOI] [PubMed] [Google Scholar]
- 27.Kelman Z, O’Donnell M O. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Dallmann H G, McHenry C S, Marians K J. Coupling of a replicative polymerase and helicase: a t-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 29.Knopf K W. Properties of herpes simplex virus DNA polymerase and characterization of its associated exonuclease activity. Eur J Biochem. 1979;98:231–244. doi: 10.1111/j.1432-1033.1979.tb13181.x. [DOI] [PubMed] [Google Scholar]
- 30.Kong X P, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the β-subunit of E. coli DNA polymerase III holoenzyme: a sliding clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 31.Krishna T S R, Kong X P, Gary S, Burgers P M, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 32.Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. J Mol Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 33.Lee S, Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase δ, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukonis C J, Weller S K. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J Virol. 1997;71:2390–2399. doi: 10.1128/jvi.71.3.2390-2399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marcy A I, Olivo P D, Challberg M D, Coen D M. Enzymatic activities of overexpressed herpes simplex virus DNA polymerase purified from recombinant baculovirus-infected insect cells. Nucleic Acids Res. 1990;18:1207–1215. doi: 10.1093/nar/18.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsden H S, Campbell M E M, Haarr L, Frame M C, Parris D S, Murphy M, Hope R G, Muller M T, Preston C M. The 65,000-Mr DNA-binding and viron trans-inducing proteins of herpes simplex virus type 1. J Virol. 1987;61:2428–2437. doi: 10.1128/jvi.61.8.2428-2437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsden H S, McLean G W, Barnard E C, Francis G J, MacEachran K, Murphy M, McVey G, Cross A, Abbotts A P, Stow N D. The catalytic subunit of the DNA polymerase of herpes simplex virus type 1 interacts specifically with the C terminus of the UL8 component of the viral helicase-primase complex. J Virol. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 39.Modrich P, Richardson C C. Bacteriophage T7 deoxyribonucleic acid replication in vitro. Bacteriophage T7 DNA polymerase: an enzyme composed of phage- and host-specific subunits. J Biol Chem. 1975;250:5515–5522. [PubMed] [Google Scholar]
- 40.Munn M M, Alberts B M. The T4 DNA polymerase accessory proteins from an ATP-dependent complex on a primer-template junction. J Biol Chem. 1991;266:20024–20033. [PubMed] [Google Scholar]
- 41.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 42.O’Donnell M E, Elias P, Lehman I R. Processive replication of single-stranded DNA templates by the herpes simplex virus-induced DNA polymerase. J Biol Chem. 1987;262:4252–4259. [PubMed] [Google Scholar]
- 43.Powell K L, Purifoy D J M. DNA-binding proteins of cells infected with herpes simplex virus type 1 and type 2. Intervirology. 1976;7:225–239. doi: 10.1159/000149955. [DOI] [PubMed] [Google Scholar]
- 44.Sexton D J, Carver T E, Berdis A J, Benkovic S J. Protein-protein and protein-DNA interactions at the bacteriophage T4 DNA replication fork. J Biol Chem. 1996;271:28045–28051. doi: 10.1074/jbc.271.45.28045. [DOI] [PubMed] [Google Scholar]
- 45.Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 46.Strick R, Knopf C W. Improved band shift assay for the simultaneous analysis of protein-DNA interactions and enzymatic functions of DNA polymerases. FEBS Lett. 1992;300:141–144. doi: 10.1016/0014-5793(92)80182-g. [DOI] [PubMed] [Google Scholar]
- 47.Stukenberg P T, Studwell-Vaughn P S, O’Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 48.Tabor S, Huber H E, Richardson C C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 49.Tapper D P, Anderson S, DePamphilis M L. Maturation of replicating simian virus 40 DNA molecules in isolated nuclei by continued bidirectional replication to the normal termination region. Biochim Biophys Acta. 1979;565:84–97. doi: 10.1016/0005-2787(79)90084-4. [DOI] [PubMed] [Google Scholar]
- 50.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 51.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 52.von Hippel P H, Fairfield F R, Dolejsi M K. On the processivity of polymerases. Ann N Y Acad Sci. 1994;726:118–131. doi: 10.1111/j.1749-6632.1994.tb52803.x. [DOI] [PubMed] [Google Scholar]
- 53.Weaver D T, DePamphilis M L. The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J Mol Biol. 1984;180:961–986. doi: 10.1016/0022-2836(84)90266-3. [DOI] [PubMed] [Google Scholar]
- 54.Weisman-Shomer P, Dube D K, Perrino F W, Stokes K, Loeb L A, Fry M. Sequence specificity of pausing by DNA polymerases. Biochem Biophys Res Commun. 1989;164:1149–1156. doi: 10.1016/0006-291x(89)91789-0. [DOI] [PubMed] [Google Scholar]
- 55.Weisshart, K., and D. M. Coen. 1992. Unpublished results.
- 56.Weisshart K, Kuo A A, Hwang B C, Kumura K, Coen D M. Structural and functional organization of herpes simplex virus DNA polymerase investigated by limited proteolysis. J Biol Chem. 1994;269:22788–22796. [PubMed] [Google Scholar]
- 57.Weisshart K, Kuo A A, Painter G R, Wright L L, Furman P A, Coen D M. Conformational changes induced in herpes simplex virus DNA polymerase upon DNA binding. Proc Natl Acad Sci USA. 1993;90:1028–1032. doi: 10.1073/pnas.90.3.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao N, Turner J, Kelman Z, Stukenberg P T, Dean F, Shechter D, Pah Z, Hurwitz J, O’Donnell M. Cycling of the sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]