Abstract
NS1, the major nonstructural parvovirus protein of the minute virus of mice, is a multifunctional protein responsible for several aspects of viral replication. NS1 transactivates the P38 promoter (used to express the structural proteins), as well as its own strong promoter, P4. To study the mechanism of activation and to map regions of NS1 responsible for transactivation, NS1 and various deletions of NS1 were cloned in frame with the GAL4DB and cotransfected into COS-7 and LA9 cells with a synthetic GAL4-responsive reporter plasmid. These studies showed NS1 can directly activate transcription through its 129 carboxyl-terminal amino acid residues. Any deletion from this region of the C terminus, even as few as 8 amino acids, completely abolishes transactivation. A yeast two-hybrid system used to identify protein-protein interactions demonstrated that NS1 is able to dimerize when expressed in yeast cells. However, only an almost complete NS11–638 bait was able to interact with the full-length NS1. A two-hybrid screen identified a HeLa cell cDNA clone (NS1-associated protein 1 [NSAP1]) that interacts with NS11–276 and NS11–638. An additional sequence was predicted from human EST (expressed sequence tag) data, and the cDNA was estimated to be at least 2,221 bp long, potentially encoding a 562-amino-acid protein product. A polyclonal antibody raised to a synthetic peptide within NSAP1 recognizes an ∼65-kDa cellular protein. This NSAP1 cDNA has not previously been characterized, but the predicted protein sequence is 80% identical to the recently identified heterogeneous nuclear ribonucleoprotein (hnRNP) R (W. Hassfeld et al., Nucleic Acids Res. 26:439–445, 1998). NSAP1 contains four ribonucleoprotein domains, as well as a highly repetitive C-terminal region. A closely related mouse cDNA (deduced from murine EST data) encodes a protein with only a single amino acid residue change from the human protein. NSAP1 is predicted to be a 65-kDa polynucleotide binding protein, and it likely functions in the regulation of splicing and/or transport of mRNAs from the nucleus.
Minute virus of mice (MVM) is an autonomously replicating parvovirus. MVM has a small, single-stranded, negative-sense DNA genome of 5,149 nucleotides (nt) with nonidentical terminal palindromic hairpins. Replication is dependent on its major nonstructural protein, NS1, a multifunctional 83-kDa nuclear phosphoprotein. NS1 supplied in trans allows replication of MVM minigenomes which include only the viral hairpins and an essential cis-acting internal replication sequence but exclude virtually all of the coding sequence (52). Most mutations, insertions, or deletions of NS1 destroy the ability of MVM to replicate.
The NS1 polypeptide has ATPase, DNA binding, helicase, site-specific endonuclease, and transcriptional activation activities. Nuclear localization was found to depend on a triple lysine sequence, including amino acid (aa) residues 214 to 216 (39), and a nucleoside triphosphate (NTP)-binding motif (aa 394 to 486) was identified through homology to the simian virus 40 (SV40) T antigen and the papillomavirus E1 protein (2). Point mutations of conserved amino acid residues in this domain are still able to bind NTPs, although some at a reduced level, while mutations in this region had various effects on ATPase activity. In contrast, all mutations in the conserved NTP binding domain abolish the helicase activity of NS1 (26, 57). Resolution of viral replicative forms (RFs) requires the site-specific endonuclease (nickase) activity of NS1, and this nickase activity is thought to be encoded in a conserved rolling-circle replication motif (25). The dimer bridge RF is nicked by NS1, leaving NS1 covalently attached to the 5′ end of the single-stranded genome, but the precise mechanism of resolution remains unclear (11, 12, 34).
NS1 activates transcription from both its own strong P4 promoter (16) and the otherwise weak P38 promoter used to express the structural genes (15, 45), supporting the observed temporal expression of viral proteins with an “early” promoter expressing the nonstructural genes and a “late” promoter expressing the structural genes (10). Deletions within either terminal of NS1 abolished DNA replication, transcriptional activation, and cytotoxicity (30, 50).
NS1 has been shown in coprecipitation studies to bind directly or via a host cell protein to a (ACCA)2–3 DNA repeat (13) which is present in both viral promoters, as well as throughout the viral genome. A transactivation responsive element (tar) was previously identified in the P38 promoter by deletional analysis (46), and NS1 has been shown to bind specifically to the ACCA repeats within this element (8). The consensus repeat is also present in the P4 promoter. The NS1 activation of both P4 and P38 requires the TATA box, an SP1 site, and possibly the NS1 binding sites (20, 36, 42, 46). NS1 has also been shown to interact directly with SP1 in vitro, through coimmunoprecipitation studies, and in vivo by using a two-hybrid test (29, 36) which may be sufficient to localize NS1 to the promoter in the absence of the ‘tar.’
In this study, the transactivation region of NS1 was determined by fusing regions of NS1 to a GAL4 DNA binding domain and nuclear localization signal (22, 47). Since parvovirus replication is absolutely dependent on the host cell due to the small size of the genomes and the limited number of virally encoded proteins and since NS1 is a large, multifunctional protein, it seemed reasonable to assume that it functions in concert with host cell proteins. Furthermore, the highly purified NS1 appears to lose some of its biochemical activities, including site-specific nicking (12, 34) and DNA binding (9) activities. At the outset of these studies we presumed that NS1 acts with the help of host cell cofactors or as part of cellular complexes to exert its many functions. Hence, we decided to use the two-hybrid system (21) to investigate the homooligomerization properties of NS1 and to identify novel host proteins that interact with this polypeptide. The present study shows that a cellular heterogeneous nuclear ribonucleoprotein (hnRNP)-like protein (possibly hnRNP R2) interacts with the N-terminal region of NS1.
MATERIALS AND METHODS
Cell lines.
COS-7 cells (19) were grown in Dulbecco modified Eagle medium (DMEM; GIBCO-BRL) supplemented with 10% fetal bovine serum (FBS). An A9 variant of mouse L cells (LA9) was grown in DMEM with 5% FBS (33). Cell lines were transfected by the DEAE-dextran method (35).
Plasmids.
All plasmids were propagated in Escherichia coli DH5α (Life Technologies) or SURE (Stratagene) strains. Bacteria were transformed by electroporation (Bio-Rad). Plasmids for the transcription transactivation test have been described previously (47). pSG424 encodes the GAL4 DNA-binding domain (GAL4DB) from the SV40 early promoter. This vector was used to create GAL4DB-NS1 constructs. pG5BCAT contains five tandem GAL4 specific 17-mers upstream of a TATA box directing the expression of a CAT gene.
Yeast plasmids required for the two-hybrid selection method were generously provided by the Brent laboratory (21), and detailed information is available from the Massachusetts General Hospital Molecular Biology Internet Gopher server (http://xanadu.mgh.harvard.edu/brentlabweb/).
pLexA (pEG202) baits were constructed from pEG202 by cloning the desired sequence in frame with LexA (202 aa, including the DNA binding and dimerization domains). LexA-NS1 junctions were sequenced by using the Sequenase kit (USB). Fusions are expressed from the strong constitutive ADH1 promoter.
The lexAop-lacZ reporter pLexAop-lacZ (pSH18-34) was created by inserting four lexA operators (lexAop) (8 LexA dimer binding sites) into a GAL1-lacZ reporter gene with glucose- and galactose-responsive elements deleted.
pLexA-GAL4TA (pSH17-4) is a positive control encoding the DNA binding LexA1–87 fused to the activator GAL474–881 expressed from the ADH1 promoter.
pJG4-5 is a vector used for the HeLa-acid cDNA library of fusion proteins. The HeLa cDNA clones are fused to a sequence encoding the acidic E. coli B42 activator, the SV40 nuclear localization signal, and the hemagglutinin epitope. The HeLa-acid fusion proteins are expressed from the galactose-induced GAL1 promoter. This promoter is repressed by glucose.
pGAL4TA-NS1 (pPCNS1) is a plasmid that encodes a GAL4TA-NS1 fusion expressed from the constitutive ADC1 promoter. The plasmid was created by cloning the entire coding sequence of NS1 in frame into pPC86 (7).
Testing of baits.
Before the baits can be used in the two-hybrid system, they must be tested to ensure that they are expressed, are transcriptionally inert, and can bind to the lexAop in the yeast nucleus. A Western blot was performed to confirm that each bait plasmid expressed a LexA-NS1 fusion. A mouse α-LexA monoclonal antibody (MAb; Clontech) was used to detect each hybrid. LexA fusions of approximately the expected size for each construct were observed. CE10 α-NS1 MAb (58) identified bands in LexA-NS11–638 and LexA-NS1386–638 constructs (data not shown). All NS1-containing baits had a deletion of the C-terminal transcriptional activation domain as the C-terminal region acts as a transactivator in yeast cells (data not shown). All LexA-NS1 fusions were transcriptionally inert. LexA-NS11–638 induced very low expression of the reporter genes, which was visible if the plates were left for several additional days at 30°C or several weeks at 4°C. To ensure that the baits were not transcriptionally inert due to exclusion from the nucleus, they were tested for their ability to repress a GAL1 promoter by binding to an inserted lexAop. All of the baits had equal or greater repression activity than the LexA-bicoid positive control, confirming that they are expressed, transported to the nucleus, bound to the lexAop, and transcriptionally inert.
Transformation of cDNA library into EGY48(pLexAop-lacZ/pLexA-NS11–276).
Yeast cells were grown in the appropriate selection medium with a 2% glucose carbon source unless otherwise noted. Strain EGY48 (ura3 his3 trp1 lexAop-leu2) carrying pLexAop-lacZ/pLexA-NS1–276 was transformed by using the lithium acetate method (48) with the HeLa-acid library in pJG4-5. A total of 1.5 × 105 colonies were pooled, and titers of the EGY48(pLexAop-lacZ/pLexA-NS1–276/pJG4.5) HeLa-acid library were determined at 2 × 105 CFU/μl.
Screening of HeLa-acid library for interactors.
The EGY48(pLexAop-lacZ/pLexA-NS1–276/pJG4.5) HeLa-acid library was plated onto galactose-raffinose Ura− His− Trp− Leu− agar at 105 CFU/100-mm plate and incubated at 30°C for 5 days. Colonies were transferred to galactose-raffinose Ura− His− Trp− X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates and incubated for 3 days at 30°C. The plates were monitored every 12 h, and colonies that turned blue were streaked onto glucose Ura− His− Trp− plates. The dependence of the blue color and growth phenotypes on the HeLa-acid library clone was tested by repeating the selection and screening but with glucose instead of galactose-raffinose as the carbon source. The original tests on the galactose-raffinose plates were also repeated. Library plasmids were rescued from yeast cells into bacteria (27), and all of the tests were repeated with EGY48 transformed with the original plasmids.
RESULTS AND DISCUSSION
NS1 C-terminal activation domain.
GAL4DB-NS1 fusions were assayed for their ability to activate transcription of a synthetic GAL4-responsive promoter (47) following transfection into COS-7 and LA9 cells (Fig. 1). Correct expression and localization of the fusions was confirmed by immunofluorescence (data not shown).
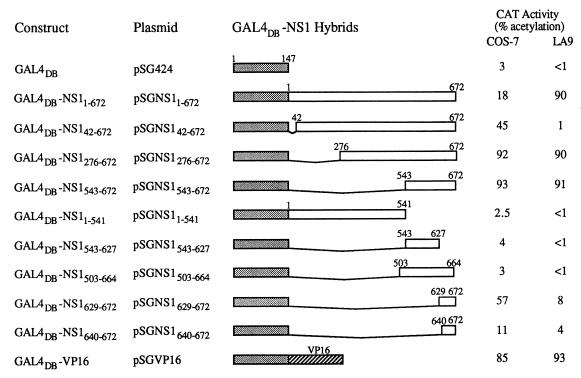
FIG. 1.
Activation domain of NS1. GAL4DB-NS1 fusion constructs are shown with the plasmid name and a bar representation. All plasmids were derived from pSG424 (47). Expression and localization of the GAL4DB-NS1 fusions was confirmed by indirect immunofluorescence with the CE10 primary MAb (58). Constructs pSGNS1629–672 and pSGNS1640–672 were fully sequenced, since they do not contain the CE10 epitope. Each construct was tested in COS-7 and LA9 cells for its ability to stimulate a synthetic promoter containing the UASG upstream of a CAT reporter gene (pG5BCAT). At 48 h posttransfection, cells were harvested and assayed for CAT activity. CAT reactions were incubated for 1 h at 37°C and run on a thin-layer chromatography plate. To quantitate acetylation, the spots were excised, placed in Aquasol, and counted in a Beckman scintillation counter (18). Percentages given represent the total amount of acetylated chloramphenicol divided by the amount of input chloramphenicol. All values represent an average of at least three separate experiments and have an error of <10%. The shaded bars represent GAL41–147, the open bars represent segments of NS1, and the slashed bar represents the VP16 activation domain (the 78 C-terminal amino acid residues of HSV VP16).
An activation domain was identified at the C-terminal end of NS1 (24). The present study was done concurrently with that of Legendre and Rommelaere (31) and has also been confirmed by Krady and Ward (29), who have further mapped the activation domain to the 88 C-terminal amino acid (aa) residues. In our study, GAL4DB-NS1543–672 was the smallest construct to have full activity, and smaller C-terminal regions had reduced activity (GAL4DB-NS1629–672 and GAL4DB-NS1640–672). Even an 8-aa deletion at the C-terminal end, GAL4DB-NS1503–664, abolished activation. In COS-7 cells, GAL4DB-NS11–672 stimulated chloramphenicol acetyltransferase (CAT) expression to a relatively low level (18% acetylation), but a deletion of the first 275 N-terminal residues, GAL4DB-NS1276–672, activated expression of CAT to maximal levels (92% acetylated). A smaller N-terminal deletion, GAL4DB-NS142–672, increased CAT activity to 45% acetylation and half-maximal activity. In general the activities of the contructs in LA9 cells were similar to those observed with COS-7 cells with the following exceptions. In LA9 cells, GAL4DB-NS142–672 had almost no activity (1% acetylation) compared to half-maximal activity (45% acetylation) in COS-7 cells, and the most significant difference between the two cell lines was that GAL4DB-NS11–672 had full activity in LA9 cells compared with 18% activity in COS-7 cells.
These results suggest that in this assay the N-terminal region of NS1 can exert a negative effect on transactivation. Since the N terminus of NS1 is required for activation of P38, it is possible that this region is responsible for the DNA binding activity, either directly or through interaction with a host cell protein. Also, this region could be binding a host cell factor responsible for another function.
NS1 is able to dimerize.
NS1 was tested for self-association in the two-hybrid system to determine whether it is able to dimerize as was suggested by the cotransportation studies (39). Baits (Fig. 2) were tested against GAL4TA-NS1. The only clone to give a positive result in the dimerization assay was a construct containing residues 1 to 638 of NS1 [EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS11–638) (Fig. 3)]. This LexA-NS1 fusion was strongly positive for both the Leu+ phenotype (growth on a Leu− plate) and the LacZ+ phenotype (blue colonies on an X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] plate).
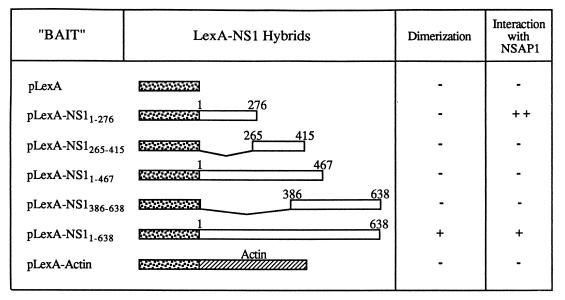
FIG. 2.
Baits tested in the two-hybrid system. Baits were tested by the two-hybrid system (21) for their ability to dimerize or to interact with NSAP1. A “+” indicates an interaction, and a “−” indicates no interaction. A “++” indicates a stronger interaction result than “+.” The dimerization was shown by an interaction between the bait and the entire NS1 coding sequence fused to the GAL4 activation domain (GAL4TA-NS1). The bait LexA-NS11–276 was used to trap NSAP1 from a two-hybrid screen of an acid activator-tagged HeLa cell cDNA library. All baits were then tested for interaction with NSAP1. The LexA constructs were created by cloning segments of NS1 in frame into pEG202 (21). The resulting fusions are shown with the stippled bar representing LexA, the open bar representing NS1, and the slashed bar representing actin. LexA-Actin was used as a negative control for the interactions.
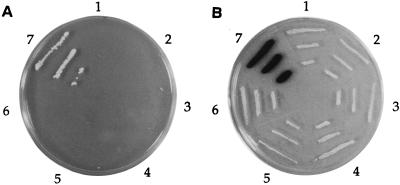
FIG. 3.
Dimerization activity of NS1. Baits were tested against a GAL4TA-NS1 fusion by using the two-hybrid system to determine if they were able to dimerize. The pEG202 (21)-based bait plasmids containing segments of NS1 and the plasmid encoding the GAL4TA-NS1 fusion (derived from pPC86 [7]) were transformed into the yeast EGY48 strain containing the lexAop-lacZ reporter plasmid (pSH18-34). Each fusion was tested for growth on a glucose Ura− His− Trp− Leu− plate (A) by stimulation of the integrated lexAop-LEU2 and for blue color on a galactose-raffinose Ura− His− Trp− X-Gal plate (B) by stimulation of lexAop-lacZ. Plate sections: 1, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-Actin) (negative control); 2, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA) (negative control); 3, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS11–276); 4, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS1265–415); 5, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS11–467); 6, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS1386–638); and 7, EGY48(pLexAop-lacZ/pGAL4TA-NS1/pLexA-NS11–638).
These results demonstrated that NS1 self-associates in yeast cells and supports a previous observation that mutant NS1 with a deleted nuclear localization signal can comigrate to the nucleus with wild-type NS1 (39). These results are also in agreement with studies that showed small deletions over a large portion of NS1 (aa 221 to 529) reduce its oligomerization activity (43). Further work has shown that a single peptide (aa 258 to 275) is able to block coimmunoprecipitation, helicase activity, and NS1-dependent DNA replication (43). These observations are not surprising since many helicases (56) and transcriptional activators (e.g., p53 [53] and human immunodeficiency virus [HIV] Tat [3]) function as multiunit complexes. Also, SV40 T antigen with functional and limited sequence homology to NS1 is fully active as a double hexamer (37, 55).
These observations suggest that the oligomerization function requires either a large portion of the NS1 or tertiary structure which is disrupted by the larger deletions or multiple disperse domains. Functional oligomerization by the SV40 T antigen requires ATP binding (37, 44), and the NTP binding domain of NS1 (aa 394 to 486) is presumed to be required for its oligomerization. Therefore, oligomerization of NS1 requires at least the NS1 oligomerization domain, probably the NTP binding domain, and possibly other unidentified regions of NS1. It seems likely that NS1 functions in vivo as a multimer to unwind DNA, activate transcription, bind DNA, and effect viral replication.
Product of a partial HeLa cDNA clone interacts with NS1.
LexA-NS11–276 was used as a bait in a two-hybrid screen of an acid activation-tagged HeLa cDNA library (HeLa-acid cDNA library). A library plasmid from a clone that was Leu+ and LacZ+ on galactose-raffinose but Leu− and LacZ− on glucose was isolated and transformed into E. coli SURE cells. The HeLa cDNA-acid hybrid is expressed from the glucose-repressed GAL1 promoter (pJG4-5) and is therefore not expressed when grown on glucose media. The positive clone was called B4, and the isolated HeLa-acid plasmid was designated pJG4-5B4. pJG4-5B4 was retransformed into EGY48(pLexAop-lacZ/pLexA-NS11–276), and the other NS1 baits (Fig. 2) were also tested against pJG4-5B4 (Fig. 4). All of the other baits were negative for interaction with pJG4-5B4 except for pLexA-NS11–638. LexA-NS11–276 has a “stronger” phenotype than LexA-NS11–638, turning blue more quickly on an X-Gal plate and growing more quickly on Leu− selective medium. In a two-hybrid system, this indicates a weaker interaction with pJG4-5B4 for pLexA-NS11–638 than for pLexA-NS11–276. An unexpected result was that pLexA-NS11–467 did not give a positive interaction result. This clone contains the entire sequence of pLexA-NS11–276 but may produce a product with the interacting region masked. The B4 clone was renamed NS1-associated protein 1 (NSAP1).
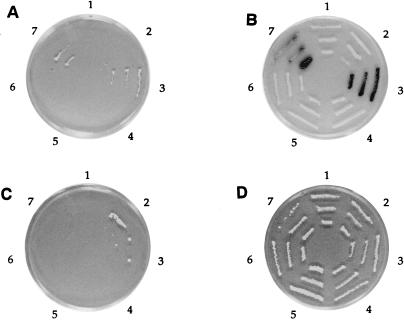
FIG. 4.
Interaction of NSAP1 with NS1. A partial cDNA clone that interacted with LexA-NS11–276 was isolated from an acid-tagged cDNA library by using the two-hybrid system. The plasmid DNA (pJG4-5B4) was rescued from the yeast cells and retransformed into yeast EGY48 with the lexAop-lacZ reporter plasmid (pSH18-34) and each of the baits. The pEG202-based plasmids contain segments of NS1. Each fusion was tested for growth on a galactose-raffinose Ura− His− Trp− Leu− plate (A) by stimulation of the integrated lexAop-LEU2 and for blue color on a galactose-raffinose Ura− His− Trp− X-Gal plate (B) by stimulation of lexAop-lacZ. The dependence of these phenotypes on expression of the HeLa cDNA clone from the GAL1 promoter was tested by repeating the tests on glucose Ura− His− Trp− Leu− and Ura− His− Trp− X-Gal plates (C and D, respectively). Plate sections: 1, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-Actin) (negative control); 2, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA) (negative control); 3, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-NS11–276); 4, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-NS1265–415); 5, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-NS11–467); 6, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-NS1386–638); and 7, EGY48(pLexAop-lacZ/pJG4-5B4/pLexA-NS11–638).
An in vitro interaction between NSAP1 and NS1 has not been demonstrated. This may be due to the complex nature of the interaction, which may require a cofactor, specific cellular conditions, or assembly of a larger complex. It is also possible that the in vitro interaction assays are much less sensitive than the two-hybrid results (17, 43).
Sequence analysis of NSAP1 cDNA.
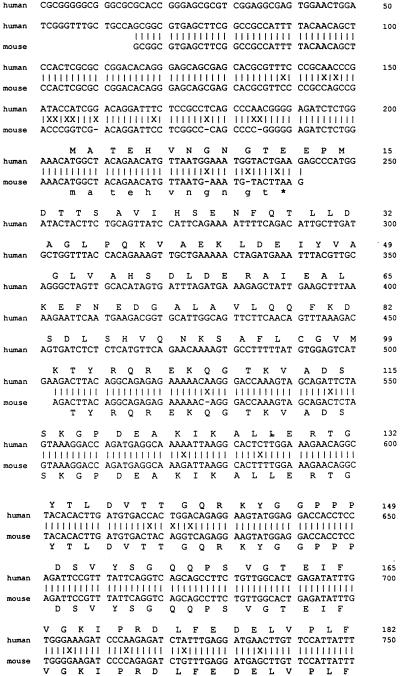
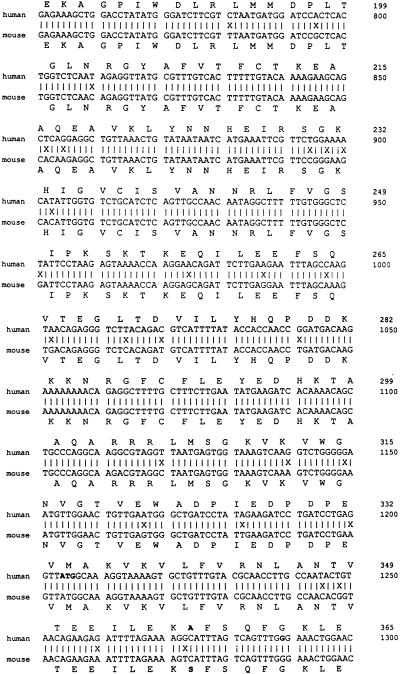
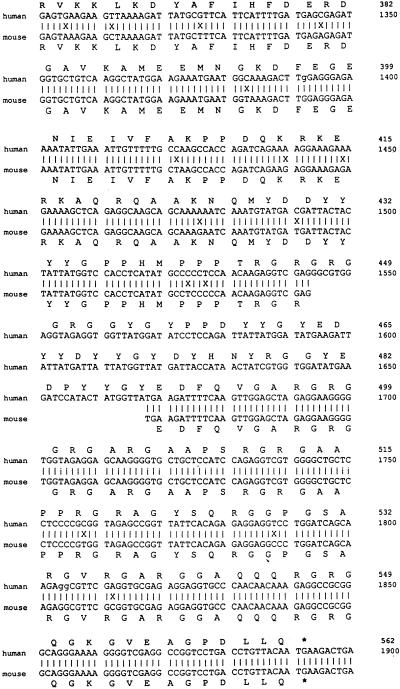
The nucleotide sequence of the partial HeLa cDNA clone was determined and found to contain a 932-bp insert (Fig. 5). A BLAST search (1) of EST (expressed sequence tag) databases (4) allowed PCR amplification of a slightly longer cDNA, providing a further 85 bp (1204 to 1288), including an upstream ATG codon which also matched the EST sequences. An additional 1,203 bp was predicted from hESTs, and the total 2,221 bp represents the minimum cDNA length. The cDNA sequence includes a strong Kozak consensus ATG (nt 205) (28), polyadenylation signal (AATAAA, nt 2176), and poly(A) tail (nt 2204). The 2,221-bp sequence may include the entire coding sequence, as no matching clones extending further 5′ are present in the dbEST. (These EST sequences were derived from a wide variety of tissue types). From the predicted cDNA sequence a protein of 562 aa residues would be produced with a molecular size of approximately 60 to 70 kDa (Fig. 5).
FIG. 5.
Human and mouse NSAP1 sequences. The partial cDNA clone isolated by the yeast interaction trap extends from nucleotide position 1289 (G) to 2221. An additional 85 bp (bp 1204 to 1288 from ATG) were sequenced from a larger cDNA clone isolated through amplification by using primers based on the predicted 5′ sequence. In total, 1,027 bp were sequenced from primers on the library plasmid and primers made from internal sequence. The first 1,204 bp are predicted sequence from hEST database clones. The mouse sequence shown was derived entirely from overlapping mEST sequences. Not all regions of the mouse cDNA are represented in the mEST data at this time. The predicted amino acid sequence is shown for both the human (above) and the mouse (below) clones. An asterisk indicates a stop codon. The boldface amino acid residues mark a difference between the human and mouse sequences. Lowercase letters in the DNA sequence indicate bases determined from EST sequences not seen when sequenced manually. The “=” sign indicates a single base deletion between the human and mouse sequences.
Mouse homologue of NSAP1.
The sequence of mouse NSAP1 was predicted from EST data and is aligned with human NSAP1 (Fig. 5). Mouse EST data for NSAP1 are incomplete; several regions show no corresponding mouse sequence. EST databases have concentrated on human cDNAs, with attention only recently shifting to include other species. The 5′ mouse sequence includes an in-frame stop codon (TAA) just upstream of an early ATG codon (bp 247), and the mouse sequence appears to deviate from the human sequence upstream of the stop codon. Only a single mouse EST is present in this region, and it contains the most mismatches compared with the predicted human sequence of any mouse EST.
DNA sequences (human compared with mouse) were found to be at least 95% identical overall, and only a single base-pair change within the putative coding region affected the protein sequence (Ala-357 to Ser, bp 1273), indicating that NSAP1 is a highly conserved protein whose sequence is likely functionally constrained.
NSAP1 is transcribed in several mouse tissues.
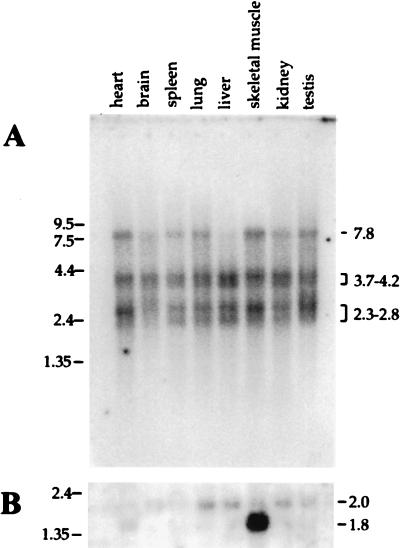
To determine the size of the transcript for NSAP1 and to determine its expression in various tissue types, a Northern blot was performed on mRNA from several mouse tissues with an NSAP1 probe (Fig. 6). Multiple bands visible in every tissue type confirm the EST data that NSAP1 is widely expressed. There appear to be up to five bands per lane, ranging in size from 2.3 to 7.8 knt. A single band at about 7.8 knt is most prominent in heart and skeletal muscle tissue and is least prominent in the liver. The other bands can be divided into two groups (3.7 to 4.2 and 2.3 to 2.8 knt), each consisting of at least two bands. The groupings are based on size and on constant relative intensities of each set of bands. The smallest band corresponds in length to the minimum length of NSAP1 (Fig. 5). Multiple bands within a set could represent the secondary structure of a partially denatured transcript. Also, the larger bands may represent more complete mRNAs, alternatively spliced mRNAs, or unprocessed or partially processed hnRNA, or they may represent transcripts from related but distinct genes. However, a more stringent washing of the blot did not reduce or affect the relative intensity of any bands (data not shown). Although NSAP1 was isolated from a human HeLa cell cDNA library, a nearly identical cDNA is expressed in many mouse tissues that are permissive for MVM replication.
FIG. 6.
Mouse multiple tissue Northern blot. A Northern blot containing approximately 2 μg of poly(A)+ RNA per lane from different BALB/c mouse tissues (Clontech) was probed with a 32P randomly primed NSAP1 (bp 1204 to 1929) cDNA probe (2.6 × 107 cpm/50 ng of DNA) (A) and a 32P randomly primed β-actin control in 5 ml of fresh ExpressHyb (Clontech) (B). Blots were washed at room temperature several times with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.05% sodium dodecyl sulfate (SDS) for 1 h at room temperature in 2× SSC–0.05% SDS and twice for 20 min at 50°C in 0.1× SSC–0.1% SDS. The blots were exposed for 12.5 h (A) and 24 h (B) on a phosphor screen and read on a Molecular Dynamics PhosphorImager. More-stringent washes did not alter the appearance of any of the bands. The positions of RNA size standards (in kilonucleotides) are indicated on the left. The sizes of the resulting bands (in kilonucleotides) are shown on the right. Note that there are two forms of actin mRNA in heart and skeletal muscle (2 kbp and 1.6 to 1.8 kbp) (40).
NSAP1 encodes an ∼65-kDa protein.
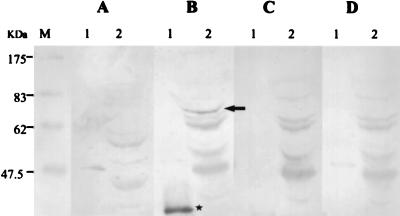
In order to determine if a cellular protein corresponding to the predicted size of NSAP1 (562 aa, 60 to 70 kDa) is expressed, we generated a rabbit polyclonal antibody to a synthetic peptide within the NSAP1 protein. Western blot analysis of mouse LA9 cell extract detected an ∼65-kDa protein (Fig. 7). A positive control of bacterially expressed His6X-partial NSAP1 (aa 334 to 562) produced the expected band of ∼40 kDa. Both immune antibody reactions were blocked by preincubating the serum with the specific NSAP1 synthetic peptide. This result demonstrates that a specific protein product of the expected size is produced in LA9 cells.
FIG. 7.
Expression of NSAP1 in LA9 cells. Western blot analysis was performed on nickel column-purified His6X-partial NSAP1 (aa 334 to 562) (lanes 1) and LA9 cell lysate after separation by SDS–12% polyacrylamide gel electrophoresis and transfer to polyvinylidene difluoride membrane (Millipore). A rabbit polyclonal antibody was raised to a synthetic peptide within NSAP1 (aa 397 to 416, EGENIEIVFAKPPDQKRKER). Part A shows a Western blot negative control with prebleed serum, while the others were developed with NSAP1 antiserum. Part B had no competition; in part C the serum was preincubated with 1 μg of synthetic NSAP1 peptide per ml and in part D the serum was preincubated with 100 μg of synthetic NSAP1 peptide per ml. An arrow marks a strong 65-kDa band, and a star marks a band corresponding to the partial NSAP1 product. M indicates the protein size standards (NEB).
NSAP1 and its putative function.
NSAP1 contains four tandem ribonucleoprotein (RNP) domains, together comprising all but the first 35 and last 154 aa residues. The RNP domain is common in proteins which bind pre-mRNA, mRNA, pre-rRNA, and small nuclear RNA. The first RNP repeat seems to encode only a partial RNP motif but may have a conserved structure and function. Alternatively, the modified RNP motif might be a component of a domain that is functionally distinct from the other RNP domains. A bacterial cold-shock protein uses a highly conserved portion of the RNP motif to bind single-stranded DNA (49).
The cDNA sequence of the 3′ terminus of NSAP1 is repetitive and encodes a highly repeated protein sequence. One short tyrosine-rich repeat (DYYGYE consensus) and longer glycine- and arginine-rich repeats (RGAAXXRGR and GRGRGGRGXRG consensus) were identified. The regions enriched for glycine and arginine have seven Arg-Gly-Gly (RGG) repeats found in RNA binding proteins, often in association with other RNA binding motifs (6).
Recently, a protein that is very similar to NSAP1 (80% identities, 88% positives, 2% gaps) has been characterized (23) (Fig. 8). The 82-kDa protein (633 aa) was identified as being the antigen for autoantibodies present in the serum of a patient suffering from an autoimmune disease. This protein was named heterogeneous nuclear RNP R (hnRNP R) since it precipitated with hnRNP complexes. NSAP1 and hnRNP R have similar structural organizations. The N-terminal sequences of NSAP1 and hnRNP R match, which suggests that the predicted sequence of NSAP1 is complete; however, hnRNP R has a glutamine- and an asparagine-rich 63-aa residue C-terminal extension that is not present in NSAP1. The C-terminal end of NSAP1 is likely not truncated since our original cDNA clone contains a poly(A) tract. NSAP1 may be an hnRNP and, if this is verified, it would become known as hnRNP R2.
FIG. 8.
Amino acid sequence comparison of hnRNP R and NSAP1. The amino acid sequence alignment of NSAP1 and hnRNP R is shown with an amino acid symbol indicating an identity (80% or 449/562), with a “+” indicating a conservative change (8% or 48/562) and a “−” indicating a gap (2% or 14/562). All motifs observed in hnRNP R (23) are present in NSAP1 except for the glutamine- and asparagine-rich C terminus. The region identified with the overbar is the peptide used to generate a polyclonal antibody to NSAP1.
All hnRNP proteins have a general affinity for RNA, as well as single-stranded DNA, and appear to bind in a transcript-specific manner (54). hnRNP K binds poly(C) RNA but also binds to single- and double-stranded DNA, and hnRNP K binds the CT element upstream of the c-myc P1 promoter and increases gene expression (6).
Binding of hnRNPs is proposed to modulate RNA secondary structure and allow access to trans-acting factors to regulate splicing and mRNA export. hnRNP A1 has been shown to bind to the transcription-regulatory region of mouse hepatitis virus RNA and to regulate the expression of subgenomic mRNAs (32). Regulation of alternative splicing is an important step in establishing the steady-state levels of both VP1 and VP2/3 and also NS1 and NS2. Studies have shown that splicing of MVM transcripts is not controlled by viral trans acting factors but rather cis-acting sequences and host cell factors (41).
In addition to playing a role in RNA splicing, formation of various intermediary viral DNA structures may be facilitated by an hnRNP. Almost all annealing activities isolated from human cell nuclei copurify with known hnRNPs (54). NS1 may associate with DNA sequence near the 5′ internal replication sequence, where the DNA is thought to “open” and allow snapping back of the 5′ hairpins (5). At the bridge dimer, resolution models (12, 34) predict unstable hairpin formation, which likely requires the cooperation of viral and host factors, DNA strand separation, and reannealing into an alternate structure. These events presumably require several host cell proteins, and the hnRNPs are likely candidates.
A Gly-rich region interspersed with aromatic residues has been implicated in the protein-protein interaction and the annealing activities of hnRNP A1 (54). Interestingly, NSAP1 has a Gly-rich C terminus with many aromatic residues, and this region was present in the partial cDNA clone isolated through its interaction with NS11–276 in the two-hybrid system.
A cellular protein is required for the nickase activity to resolve the viral bridge dimer since purified NS1 is unable to nick DNA (12, 34). Resolution of the dimer bridge has some similarities to splicing, and host proteins required by MVM may be similar to splicing factors. Christensen et al. (9) have identified a novel 110-kDa cellular-site-specific DNA-binding factor that cooperates with NS1 in DNA binding in the dimer bridge. More recently, two proteins (96 and 79 kDa) have been identified; however, neither of these proteins appear to be the same as NSAP1.
NSAP1 might be a cofactor in the double-stranded DNA-binding activity of NS1. While NS1 has been shown to bind to the (ACCA)2–3 repeat, these experiments have been performed such that an associated protein may also be present in the assays (8, 13). A classic example of a viral activator using a host cell DNA binding protein is that of HSV activator VP16 and Oct-1, a DNA binding protein (51). NS1’s DNA binding activity extends from the double-stranded DNA (ACCA)2–3 to single-stranded DNA and RNA, and NS11–276 that interacts with NSAP1 also is responsible for DNA binding (38).
Finally, the expression of several hnRNPs is proliferation dependent, and if this is the case with NSAP1 this may contribute to the restriction of MVM to dividing cell populations.
Of interest is that others have also used the two-hybrid system to identify cellular interactors for the NS1 protein of the very closely related H-1 parvovirus (14). In that study a smaller protein, SGT (small glutamine-rich tetratricopeptide repeat containing protein), of 314 aa, was identified. It has one transcript (as shown by Northern blot), and transient expression of NS1 results in modification of SGT (possibly by phosphorylation). SGT is a novel protein, its function is unknown, and it is not related to NSAP1.
In summary, we have identified a cellular protein, NSAP1, which interacts in vivo with NS11–276. This protein is closely related but is not identical to the previously identified hnRNP R polypeptide, suggesting it has a function in regulating the splicing of transcripts and/or mRNA transport. Ongoing studies will hopefully elucidate the precise role of NSAP1 in MVM replication.
ACKNOWLEDGMENT
This work was supported by the MRC (Canada) (MA6728).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astell C R, Mol C D, Anderson W F. Structural and functional homology of parvovirus and papovavirus polypeptides. J Gen Virol. 1987;68:885–893. doi: 10.1099/0022-1317-68-3-885. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd H P, Fridell R A, Blair W S, Cullen B R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguski M S, Lowe T M J, Tolstoshev C M. dbEST—database for “expressed sequence tags.”. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 5.Brunstein J, Astell C R. Analysis of the internal replication sequences indicates that there are three elements required for efficient replication of minute virus of mice minigenomes. J Virol. 1997;71:9087–9095. doi: 10.1128/jvi.71.12.9087-9095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens K E, Pintel D J. The two transcription units of the autonomous parvovirus minute virus of mice are transcribed in a temporal order. J Virol. 1988;62:1448–1451. doi: 10.1128/jvi.62.4.1448-1451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore S F, Tattersall P. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J Virol. 1988;62:851–860. doi: 10.1128/jvi.62.3.851-860.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotmore S F, Christensen J, Nuesch J P F, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cziepluch C, Kordes E, Poirey R, Grewenig A, Rommelaere J, Jauniaux J-C. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein of parvovirus H1. J Virol. 1998;72:4149–4156. doi: 10.1128/jvi.72.5.4149-4156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doerig C, Hirt B, Beard P, Antonietti J-P. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J Gen Virol. 1988;69:2563–2573. doi: 10.1099/0022-1317-69-10-2563. [DOI] [PubMed] [Google Scholar]
- 16.Doerig C, Hirt B, Antonietti J-P, Beard P. Nonstructural protein of parvovirus B19 and minute virus of mice controls transcription. J Virol. 1990;64:387–396. doi: 10.1128/jvi.64.1.387-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gill G, Sadowski I, Ptashne M. Mutations that increase the activity of a transcriptional activator in yeast and mammalian cells. Proc Natl Acad Sci USA. 1990;87:2127–2131. doi: 10.1073/pnas.87.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 20.Gu M-L, Chen F-X, Rhode S L. Parvovirus H-1 P38 promoter requires the trans-activation region (tar), an SP1 site, and a TATA box for full activity. Virology. 1992;187:10–17. doi: 10.1016/0042-6822(92)90290-6. [DOI] [PubMed] [Google Scholar]
- 21.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 22.Harris C E, Astell C R. Transcriptional activation and SV40 T antigen responsive domains are present in NS1, the major nonstructural protein of minute virus of mice, abstr. P1–16. 5th International Workshop on Parvoviruses, Crystal River, Fla. 1993. [Google Scholar]
- 23.Hassfeld W, Chan E K L, Mathison D A, Portman D, Dreyfuss G, Steiner G, Tan E M. Molecular definition of heterogeneous nuclear ribonucleoprotein R (hnRNP R) using autoimmune antibody: immunological relationship with hnRNP P. Nucleic Acids Res. 1998;26:439–445. doi: 10.1093/nar/26.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haut D D, Pintel D J. Intron definition is required for excision of the minute virus of mice small intron and definition of the upstream exon. J Virol. 1998;72:1834–1843. doi: 10.1128/jvi.72.3.1834-1843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicon from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jindal H K, Yong C B, Wilson G M, Tam P, Astell C R. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J Biol Chem. 1994;269:3283–3289. [PubMed] [Google Scholar]
- 27.Kaiser P, Auer B. Rapid shuttle plasmid preparation from yeast cells by transfer to E. coli. BioTechniques. 1993;14:552. [PubMed] [Google Scholar]
- 28.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 29.Krady J K, Ward D C. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol Cell Biol. 1995;15:524–533. doi: 10.1128/mcb.15.1.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legendre D, Rommelaere J. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J Virol. 1992;66:5705–5713. doi: 10.1128/jvi.66.10.5705-5713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legendre D, Rommelaere J. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J Virol. 1994;68:7974–7985. doi: 10.1128/jvi.68.12.7974-7985.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Littlefield J W. Three degrees of guanylic acid-inosinic acid pyrophosphorylase deficiency in mouse fibroblasts. Nature. 1964;203:1142–1144. doi: 10.1038/2031142a0. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q, Yong C B, Astell C R. In vitro resolution of the dimer bridge of the minute virus of mice (MVM) genome supports the modified rolling hairpin model for MVM replication. Virology. 1994;201:251–262. doi: 10.1006/viro.1994.1290. [DOI] [PubMed] [Google Scholar]
- 35.Lopata M A, Cleveland D W, Sollner-Webb B. High-level transient expression of a chloramphenicol acetyltransferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorson C, Pearson J, Burger L, Pintel D J. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology. 1998;240:326–337. doi: 10.1006/viro.1997.8940. [DOI] [PubMed] [Google Scholar]
- 37.Mastrangelo I A, Hough P V, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 38.Mouw M B, Pintel D J. Identification of an MVM nucleic acid binding domain and characterization of NS1-nucleic acid binding properties. VIIth International Parvovirus Workshop, Heidelberg, Germany. 1997. [Google Scholar]
- 39.Nuesch J P F, Tattersall P. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology. 1993;196:637–651. doi: 10.1006/viro.1993.1520. [DOI] [PubMed] [Google Scholar]
- 40.Pari G, Jardine K, McBurney M W. Multiple CArG boxes in the human cardiac actin gene promoter required for expression in embryonic cardiac muscle cells developing in vitro from embryonal carcinoma cells. Mol Cell Biol. 1991;11:4796–4803. doi: 10.1128/mcb.11.9.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pintel D J, Gersappe A, Haut D, Pearson J. Determinants that govern splicing of parvovirus pre-mRNAs. Semin Virol. 1995;6:283–290. [Google Scholar]
- 42.Pitluk Z W, Ward D C. Unusual Sp1-GC box interaction in a parvovirus promoter. J Virol. 1991;65:6661–6670. doi: 10.1128/jvi.65.12.6661-6670.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pujol A, Deleu L, Nuesch J P F, Czieluch C, Jauniaux J-C, Rommelaere J. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J Virol. 1997;71:7393–7403. doi: 10.1128/jvi.71.10.7393-7403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynisdottir I, Lorimer H E, Friedman P N, Wang E H, Prives C. Phosphorylation and active ATP hydrolysis are not required for SV40 T antigen hexamer formation. J Biol Chem. 1993;268:24647–24654. [PubMed] [Google Scholar]
- 45.Rhode S L. trans-activation of parvovirus P38 promoter by the 76K noncapsid protein. J Virol. 1985;55:886–889. doi: 10.1128/jvi.55.3.886-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhode S L, Richard S M. Characterization of the trans-activation-responsive element of the parvovirus H-1 P38 promoter. J Virol. 1987;61:2807–2815. doi: 10.1128/jvi.61.9.2807-2815.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiestl R H, Manivasakam P, Woods R A, Gietz R D. Introducing DNA into yeast by transformation. New York, N.Y: Academic Press, Inc.; 1993. pp. 79–85. [Google Scholar]
- 49.Schnuchel A, Wiltscheck R, Czisch M, Herrier M, Willimsky G, Graumann P, Marahiel M A, Holak T A. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature. 1993;364:169–171. doi: 10.1038/364169a0. [DOI] [PubMed] [Google Scholar]
- 50.Skiadopoulos M H, Salvino R, Leong W L, Faust E A. Characterization of linker insertion and point mutations in the NS-1 gene of minute virus of mice: effects on DNA replication and transcriptional activation functions of NS-1. Virology. 1992;188:122–134. doi: 10.1016/0042-6822(92)90741-7. [DOI] [PubMed] [Google Scholar]
- 51.Stern S D, Tanaka M, Herr W. The Oct-1 homeo domain directs formation of a multiprotein-DNA complex with the transactivator VP16. Nature. 1989;341:624–30. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 52.Tam P, Astell C R. Replication of minute virus of mice minigenomes: novel replication elements required for MVM DNA replication. Virology. 1993;193:812–824. doi: 10.1006/viro.1993.1190. [DOI] [PubMed] [Google Scholar]
- 53.Wang P, Reed M, Wang Y, Mayr G, Stenger J E, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: structure, oligomerization, and transformation. Mol Cell Biol. 1994;14:5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weighardt F, Biamonti G, Riva S. The roles of heterogenous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 55.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West S C. DNA helicases: new breeds of translocating motors and molecular pumps. Cell. 1996;86:177–180. doi: 10.1016/s0092-8674(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 57.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 58.Yeung D E, Brown G W, Tam P, Russnak R H, Wilson G, Clark-Lewis I, Astell C R. Monoclonal antibodies to the major nonstructural nuclear protein of minute virus of mice. Virology. 1991;181:35–45. doi: 10.1016/0042-6822(91)90467-p. [DOI] [PubMed] [Google Scholar]