Abstract
Some retroviruses with an extended repeat (R) region encode the polyadenylation signal within the R region such that this signal is present at both the 5′ and 3′ ends of the viral transcript. This necessitates differential regulation to either repress recognition of the 5′ polyadenylation signal or enhance usage of the 3′ signal. The human immunodeficiency virus type 1 (HIV-1) genome encodes an inherently efficient polyadenylation signal within the 97-nucleotide R region. Polyadenylation at the 5′ HIV-1 polyadenylation site is inhibited by downstream splicing signals, and usage of the 3′ polyadenylation site is triggered by an upstream enhancer element. In this paper, we demonstrate that this on-off switch of the HIV-1 polyadenylation signal is controlled by a secondary RNA structure that occludes part of the AAUAAA hexamer motif, which we have termed the polyA hairpin. Opening the 5′ hairpin by mutation triggered premature polyadenylation and caused reduced synthesis of viral RNA, indicating that the RNA structure plays a pivotal role in repression of the 5′ polyadenylation site. Apparently, the same hairpin structure does not interfere with efficient usage of the 3′ polyadenylation site, which may be due to the presence of the upstream enhancer element. However, when the 3′ hairpin was further stabilized by mutation, we measured a complete loss of 3′ polyadenylation. Thus, the thermodynamic stability of the polyA hairpin is delicately balanced to allow nearly complete repression of the 5′ site yet efficient activation of the 3′ site. This is the first report of regulated polyadenylation that is mediated by RNA secondary structure. A similar hairpin motif that occludes the polyadenylation signal can be proposed for other lentiviruses and members of the spumaretroviruses, suggesting that this represents a more general gene expression strategy of complex retroviruses.
Reverse transcription of a retroviral genome produces a double-stranded DNA copy that is longer than the RNA template at both the 5′ and 3′ ends (40). This additional genetic information is generated in an intricate reverse transcription mechanism that includes strand transfers onto redundant sequences. All retroviral RNA genomes therefore contain a repeat (R) region that constitutes the extreme 5′ and 3′ ends of the viral transcript. The length of this R region varies significantly among retroviruses. It can be as short as 16 nucleotides in the mouse mammary tumor virus and as long as 228 nucleotides in the human T-cell leukemia virus type 1 (HTLV-1). At the 3′ end of the viral RNA, the AAUAAA polyadenylation signal is recognized by cellular enzymes that produce a polyadenylated RNA. Some retroviruses with an extended R region encode the polyadenylation signal within the R region such that it is present at both the 5′ and 3′ ends of the viral transcript. This necessitates differential regulation either to repress recognition of the 5′ polyadenylation signal or to enhance usage of the 3′ signal.
The human immunodeficiency virus type 1 (HIV-1) has been reported to have both regulatory features. Usage of the 3′ polyadenylation site is promoted by an upstream enhancer motif that is uniquely present at the 3′ end of viral transcripts (13, 20, 23, 41, 42). This upstream sequence element (USE) appears to stabilize binding of the cleavage polyadenylation specificity factor (CPSF) to the AAUAAA hexamer motif (24). Repression of the 5′ polyadenylation site is mediated by two mechanisms. First, the 5′ polyadenylation site becomes active when moved further downstream in the transcript (14, 44). Thus, the 5′ HIV-1 polyadenylation site is repressed because it is positioned too close to the transcription initiation site. A possible mechanistic explanation for this effect was recently provided by the observation that polyadenylation factors gain access to the nascent transcript through the RNA polymerase II complex (34). It is possible that the transcription complex engaged in synthesis of the HIV-1 leader transcript is not yet competent for polyadenylation. A second possible repression mechanism is that the 5′ polyadenylation site is negatively influenced by the major splice donor signal (SD) that is uniquely present downstream of the 5′ polyadenylation site (3, 4). Mutational inactivation of the SD site in full-length HIV-1 transcripts triggered usage of the 5′ polyadenylation site (3). This repression is mediated by binding of the U1 snRNP to the splice donor site, but it is currently unknown how this affects 5′ polyadenylation site usage (4). This example may be one of a growing number of cases in which the splicing machinery influences the process of polyadenylation (16).
Previous studies with reporter gene constructs indicated that the HIV-1 sequence represents an inherently efficient polyadenylation signal. Transfection studies with mutant HIV-1 polyadenylation signals indicated that the efficiency of this polyadenylation site can be down-modulated by stable RNA structure (30). When in vitro evolution techniques were used to select for functional variants of the HIV-1 polyadenylation site, sequences lacking stable secondary structure were obtained (27, 28). Notwithstanding the potential repressive effect of RNA structure, we previously reported that the natural HIV-1 polyadenylation site is embedded within a hairpin structure of intermediate stability, which we have termed the polyA hairpin (Fig. 1 and 2), which flanks the well-known trans-acting responsive (TAR) element (17). Although the sequence of this part of the viral genome varies significantly among different HIVs and simian immunodeficiency viruses (SIVs), all isolates can fold a similar polyA hairpin structure of comparable thermodynamic stability (9). The phylogenetic conservation suggested a critical role for this structured RNA motif in virus replication, and this was confirmed in studies with mutant viruses (18). Opening of the hairpin structure in both the 5′R and 3′R regions of the HIV-1 genome did severely affect virus replication. Through prolonged culturing of these mutant viruses, revertant viruses with improved replication capacity were obtained. Analysis of such phenotypic revertants revealed that additional mutations had been introduced into the sequences encoding the polyA hairpin. Although different mutations were observed in individual revertants, all nucleotide changes restored the hairpin conformation (10). These results demonstrate an absolute requirement for this structured RNA motif in virus replication, but they do not reveal the function of the 5′ or 3′ element. Previous studies indicated that the 5′ polyA hairpin contributes to packaging of the RNA genome into virus particles (18, 33), but the hairpin may play additional roles in the replication cycle.
FIG. 1.
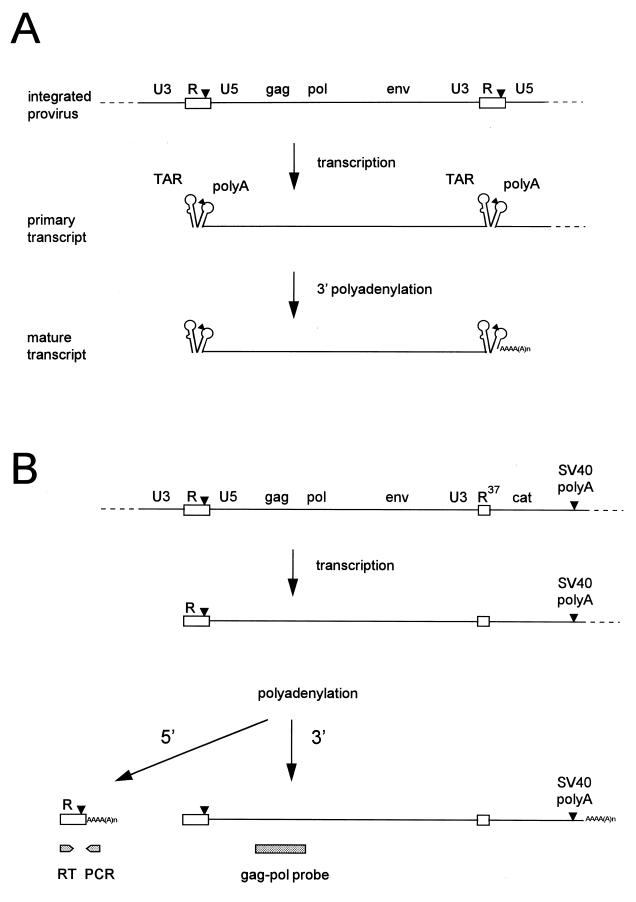
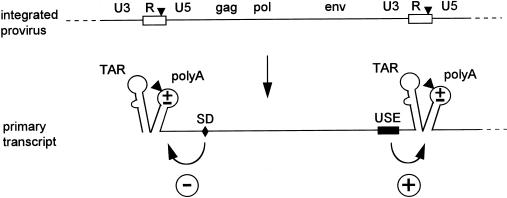
A tandem hairpin motif is present at both ends of HIV-1 genomic RNA. (A) A schematic of HIV-1 proviral DNA is shown at top with the LTRs subdivided into the U3, R, and U5 domains. The terminal R and U5 elements (R region is boxed) are also present in the primary transcript. This R-U5 region encodes the TAR and polyA hairpins, and details of the latter’s RNA structure are provided in Fig. 2. The polyA hairpin encompasses the polyadenylation signal (the AAUAAA hexamer motif, indicated by a triangle). This motif is apparently ignored in the 5′ context but is used efficiently at the 3′ end of the HIV-1 sequences. Cleavage and subsequent polyadenylation occur 19 nucleotides downstream of the hexamer. The resulting mature transcript represents the unspliced genomic HIV-1 RNA. (B) The pLAI-R37 construct used in this study contains a 3′LTR deletion that truncates the 3′R. The 3′ HIV-1 polyadenylation site is absent and replaced by the SV40 polyadenylation site (also marked as a triangle). The primary transcript and the products of 5′ and 3′ polyadenylation are shown. The full-length HIV-1 RNA was specifically detected by a gag-pol probe in dot blot assays (see Fig. 3). An RT-PCR protocol was used to specifically amplify the prematurely polyadenylated transcript form (see Fig. 6).
FIG. 2.
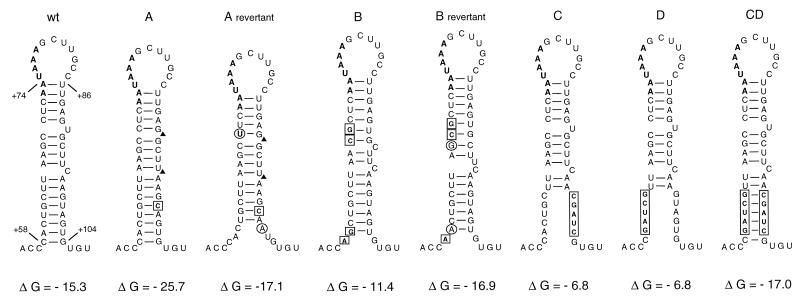
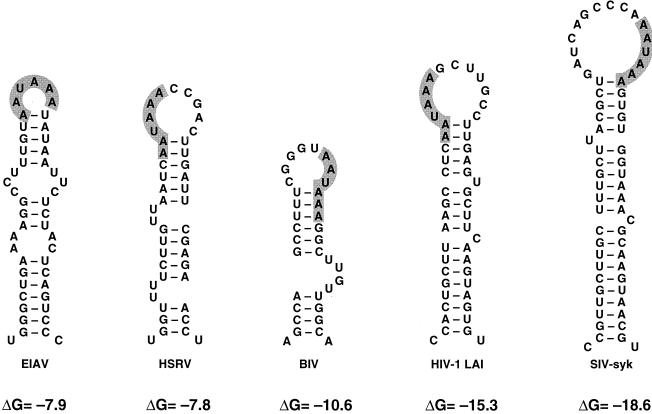
The wild-type and mutant polyA hairpins. The wild-type polyA hairpin conformation was established by several lines of evidence: RNA structure probing, phylogenetic comparison, and the analysis of mutants (reviewed in reference 7). The hairpin was mutated in different ways (nucleotide numbers refer to positions in the 5′ R-U5 of HIV-1 RNA). The polyadenylation signal, the AAUAAA hexamer, is indicated in boldface type. The thermodynamic stability (free energy, ΔG) of the RNA structures was calculated by the Zuker algorithm (46) and is indicated below the stems (in kilocalories per mole). In mutant A, the hairpin was stabilized by a single nucleotide substitution (boxed) and deletion (▴) of two bulges. The hairpin was destabilized in mutant B by four nucleotide substitutions (boxed). The hairpins of the A revertant and the B revertant were obtained upon prolonged culturing of the corresponding mutant viruses (clones A120-3 and B127-4 in reference 18). We circled the reversion-based mutations that restore the wild-type hairpin stability. In mutants C and D, destabilizing mutations (boxed) were introduced in the right- and left-hand side of the stem, respectively. The mutated segments of C and D were combined in the double mutant CD, which repairs the lower part of the stem.
In this study, we set out to test whether the HIV-1 polyA hairpin is involved in differential regulation of polyadenylation. For instance, the stem-loop structure may either stimulate polyadenylation in the 3′R context or inhibit polyadenylation in the 5′R context. We therefore analyzed the viral transcripts generated by HIV-1 genomes with mutant or revertant hairpin motifs at either the 5′ or 3′ end. We report that destabilization of the 5′ polyA hairpin triggers premature polyadenylation, suggesting that the wild-type structure is involved in occlusion of the 5′ polyadenylation site. In the 3′ context with the USE enhancer, the wild-type polyA hairpin does not interfere with efficient polyadenylation. However, 3′ polyadenylation can be efficiently inhibited by further stabilization of the hairpin. These results suggest that the role of the polyA hairpin is to create a regulatable polyadenylation site, which can be either repressed in the presence of silencers (5′ situation) or activated in the presence of an enhancer (3′ situation). The thermodynamic stability of the polyA hairpin needs to be fine-tuned to allow the on-off switching of polyadenylation.
MATERIALS AND METHODS
Cells and viruses.
C33A cervix carcinoma cells (ATCC HTB31) (5) were grown as a monolayer in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and minimal essential medium nonessential amino acids at 37°C and in 5% CO2. C33A cells were transfected by the calcium phosphate method. Cells were grown to 60% confluency in 24-well multidish plates. One microgram of DNA in 22 μl of water was mixed with 25 μl of 50 mM HEPES (pH 7.1), 250 mM NaCl, 1.5 mM Na2HPO4 and 3 μl of 2 M CaCl2, incubated at room temperature for 20 min, and subsequently added to the culture medium (1 ml). The culture medium was changed after 16 h.
CA-p24 levels.
Culture supernatant was heat inactivated (30 min at 56°C) in the presence of 0.05% Empigen-BB (Calbiochem, La Jolla, Calif.). CA-p24 concentration was determined by a twin-site enzyme-linked immunosorbent assay (ELISA) with D7320 (Biochrom, Berlin, Germany) as the capture antibody, alkaline phosphatase-conjugated anti-p24 monoclonal antibody (EH12-AP), and the AMPAK amplification system (Dako Diagnostics Ltd., ITK Diagnostics BV) as described previously (35, 36). Recombinant CA-p24 expressed in a baculovirus system was used as the reference standard.
DNA constructs.
A derivative of the full-length molecular HIV-1 clone pLAI (37) was used to produce wild-type and 5′ long terminal repeat (LTR)-mutated viruses. In this vector, pLAI-R37, the 3′LTR was truncated at the SacI site (12). Because this results in deletion of the 3′ HIV-1 polyadenylation site, sequences encompassing the chloramphenicol acetyltransferase gene and a simian virus 40 (SV40) polyadenylation site were placed downstream of the truncated 3′LTR (Fig. 1). The polyA hairpin sequence was modified by oligonucleotide-directed in vitro mutagenesis on the Blue-5′LTR subclone as previously described (18). All mutations were verified by sequence analysis. The mutated 5′LTR was subsequently introduced into the pLAI-R37 vector as a XbaI-ClaI fragment. Nucleotide numbers presented here refer to the position on the genomic HIV-1 RNA transcript, with +1 being the capped G residue.
We also constructed pLAI variants with the mutated polyA hairpin sequences in both the 5′ and 3′ ends of the viral genome. The respective Blue-5′LTR subclones were used as templates in a standard PCR with the sense primer in the R region (AD-R1, positions +7 to +29) and the antisense primer LAI-3′A (proviral DNA positions 9746 to 9767). The latter primer was designed to amplify the exact LTR region and introduced a flanking AatII restriction site. The PCR fragments and the Blue-3′LTR subclone (32) were double digested with SacI-AatII to replace the 3′LTR region. The mutant 3′LTR was subsequently introduced as a XhoI-BglI fragment in the corresponding 5′-mutated pLAI-R37 constructs to create the 5′-3′ double mutants.
Isolation of virion and cellular viral RNA.
At 2 or 3 days after transfection of C33A cells, the culture medium was centrifuged at 2,750 × g for 5 min to remove cells. Viral RNA was isolated from 300 μl of the virus-containing supernatant by incubation with 500 μg of proteinase K per ml in the presence of 1% sodium dodecyl sulfate (SDS), 2.5 mM EDTA, and 1 μg of glycogen (as a carrier to increase the efficiency of subsequent precipitations) at 37°C for 30 min and extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1). The RNA was precipitated with 0.3 M sodium acetate (pH 5.2) and 70% ethanol at −20°C, pelleted by centrifugation (20 min at 16,000 × g), washed with 70% ethanol, and dried.
Cells were washed with phosphate-buffered saline, and total cellular RNA was isolated by the acid guanidinium thiocyanate-phenol-chloroform method (15). Ten micrograms of glycogen was added to the isolated RNA to increase the efficiency of subsequent ethanol precipitations.
To remove any contaminating DNA from the virion and cellular viral RNA, the RNA was resuspended in 100 μl of 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM MgCl2, and 1 mM dithioerythritol and incubated with 5 U of DNase I (RNase free; Boehringer Mannheim) at 37°C for 30 min. After extraction with phenol-chloroform-isoamyl alcohol (25:24:1), the RNA was precipitated with 0.3 M sodium acetate and 70% ethanol. The RNA was pelleted at 16,000 × g for 20 min, washed with 70% ethanol, and dried. Pellets were resuspended in 50 μl of water and stored at −20°C.
Quantification of HIV-1 RNA.
Virion and cellular viral RNA was spotted onto nitrocellulose membranes (BA-S 85; Schleicher and Schuell) by a slot blot manifold and hybridized with a 32P-labeled HIV-1 gag-pol probe (PvuII fragment of pLAI, positions +691 to +2881), as previously described (18). Hybridization signals were quantitated with a PhosphorImager (Molecular Dynamics). To verify the absence of contaminating DNA, a duplicate RNA sample was incubated with 0.5 N NaOH at 55°C for 30 min prior to slot blotting. This resulted in a complete loss of the hybridization signals, indicating that the observed hybridization signals corresponded exclusively to genomic RNA.
Reverse transcriptase PCR (RT-PCR) protocol.
RNA isolated from transfected C33A cells was used as template for first-strand cDNA synthesis with oligonucleotide G(T)19 as primer. RNA (4 μl) was mixed with 100 ng of G(T)19 primer in 10 μl (final volume) of 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol, and deoxynucleoside triphosphates (dNTPs) (dATP, dCTP, dGTP, and dTTP; 3 mM each). The primer was annealed onto the RNA by incubation at 85°C for 2 min and at 65°C for 10 min and then cooled to room temperature in approximately 30 min. Five units of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) in 10 μl of 50 mM Tris-HCl (pH 8.5)–8 mM MgCl2–30 mM KCl–1 mM dithiothreitol was added, and the mixture was incubated at 42°C for 30 min.
To amplify the cDNA products of HIV-1 transcripts that were polyadenylated at the 5′ site, the cDNA was used as template for PCR with a 5′ primer identical to HIV-1 R sequences (AD-R1, positions +7 to +29) and G(T)19 as 3′ primer. Four microliters of cDNA was mixed with 100 ng of AD-R1, 100 ng of G(T)19 and 1 U of Taq polymerase (AmpliTaq, Perkin-Elmer) in 50 μl (final volume) of 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.2 mM each dNTP and 100 μg of bovine serum albumin. The mixture was heated at 95°C for 5 min, and then 35 PCR cycles were performed (1 min at 95°C, 1 min at 55°C, and 20 s at 72°C), followed by a 10-min incubation at 72°C and a rapid cooling down to 4°C. PCR products were analyzed by agarose gel electrophoresis and Southern blotted onto a nylon membrane (Zeta probe; Bio-Rad). To quantitate the PCR products, the filters were hybridized with a 32P-labeled anti-TAR oligonucleotide (sequence complementary to positions +27 to +56) in 0.5 M sodium phosphate (pH 7.2), 7% SDS, 1 mM EDTA, and 50 μg of salmon testis DNA per ml at 60°C for 16 h. Membranes were washed in 40 mM sodium phosphate (pH 7.2)–1% SDS at 60°C three times for 5 min and once for 15 min, and then for 5 min in 40 mM sodium phosphate (pH 7.2) at room temperature. Hybridization signals were quantitated with a PhosphorImager (Molecular Dynamics).
RNase protection assay.
For the production of 32P-labeled antisense transcripts, plasmids containing wild-type or mutant LTR sequences were used. In the wild-type construct, the XbaI-ClaI fragment of pLAI, encompassing the complete 5′LTR untranslated leader and 5′ part of the gag gene, had been cloned in pBluescriptII KS+ (Blue-5′LTR in the study described in reference 8). Mutant and revertant polyA hairpins had been introduced in this plasmid (18). Plasmid DNA was digested with XbaI, and 0.5 μg of linearized template was transcribed in vitro with 10 U of T3 RNA polymerase (Boehringer) at 37°C for 1 h, in 10 μl of transcription buffer (40 mM Tris-HCl [pH 8.0], 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, 0.1 mM UTP) containing 40 μCi of [α-32P]UTP (3,000 Ci/mmol) and 10 U of RNase inhibitor (Boehringer). After incubation with 5 U of DNase I (RNase free) at 37°C for 15 min, the RNA product was size separated on a 4% polyacrylamide–7 M urea sequencing gel. The gel fragment containing the full-length transcript was isolated and incubated with 400 μl of 0.3 M sodium acetate (pH 5.2) and 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1) at 37°C for 16 h. After centrifugation at 16,000 × g for 5 min, the upper phase was transferred to a new tube, and the eluted RNA transcript was precipitated by the addition of 3 volumes of ethanol. The RNA was pelleted at 16,000 × g for 20 min and resuspended in 200 μl of 10 mM Tris-HCl (pH 8.0)–1 mM EDTA. After extraction with phenol and phenol-chloroform-isoamylalcohol (25:24:1), the RNA was precipitated with 0.3 M sodium acetate and 70% ethanol, washed with 70% ethanol, and dried. The riboprobe was resuspended in 80 μl of hybridization buffer {80% formamide, 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] [pH 6.4], 0.4 M NaCl, 1 mM EDTA}.
Total RNA (10 μl) isolated from transfected C33A cells was lyophilized, resuspended in 10 μl of hybridization buffer, and mixed with 20 μl of riboprobe (6 × 106 cpm). This mix was overlaid with 2 drops of paraffin oil, heated at 85°C for 5 min, and incubated at 56°C for 16 h. After cooling to room temperature, 700 μl of RNase digestion buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 5 mM EDTA, 40 μg of RNase A/ml) was added, and single-stranded RNA was degraded at 30°C for 1 h. The mix was incubated with 0.1 mg of proteinase K/ml and 0.5% SDS at 37°C for 1 h and then extracted with phenol-chloroform-isoamyl alcohol (25:24:1). Protected RNA fragments were precipitated with 3 volumes of ethanol in the presence of 10 μg of glycogen. After centrifugation at 16,000 × g for 15 min, the pellet was resuspended in formamide loading buffer and analyzed on a 6% polyacrylamide–7 M urea sequencing gel. The gel was analyzed with a PhosphorImager (Molecular Dynamics).
RESULTS
The polyA hairpin mutants.
We recently demonstrated that stabilization and destabilization of the polyA hairpin (Fig. 2, mutants A and B) significantly impaired virus replication (18). Upon prolonged culturing of these mutants, phenotypic revertants with improved replication capacity were obtained. These revertants had acquired additional mutations that restored the thermodynamic stability of the polyA hairpin (Fig. 2, A and B revertants). The pivotal role of this hairpin structure in virus replication was also demonstrated by another set of mutants in which destabilizing mutations had been introduced in the right- or left-hand side of the base-paired stem (mutants C and D, respectively) and a double mutant CD that combined these mutations, allowing the formation of new base pairs. Whereas the destabilized mutants C and D showed a severe replication defect, the double mutant replicated more rapidly than the two individual virus mutants (18).
Destabilization of the 5′ polyA hairpin reduces the level of cellular HIV-1 RNA.
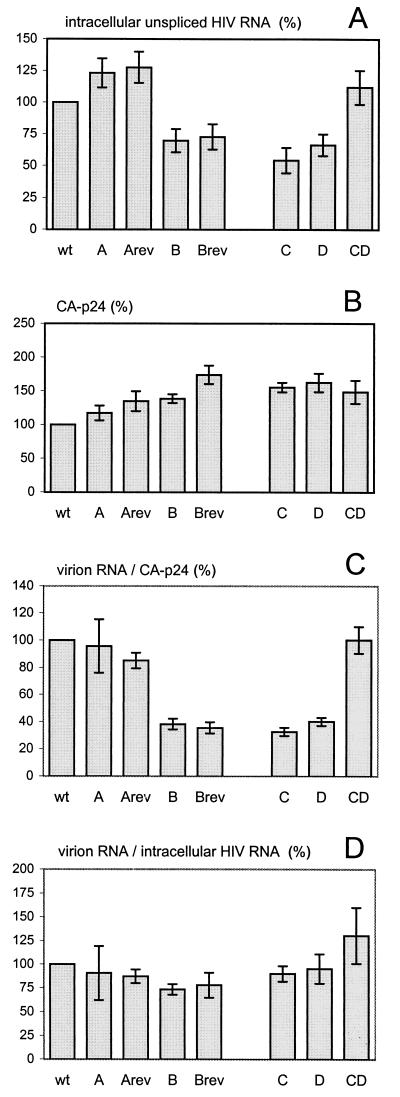
To identify the role of the 5′ polyA hairpin in HIV-1 replication, the mutant and revertant polyA hairpin sequences were introduced into a derivative of the full-length proviral clone pLAI. In this plasmid, pLAI-R37, the 3′LTR region is truncated, and the polyA hairpin sequence is present only in the complete 5′LTR (Fig. 1B). This construct was used to distinguish among the 5′ and 3′ terminal HIV-1 sequences in RNA probing experiments (see below). An SV40 polyadenylation site was placed downstream of the truncated 3′LTR to ensure the production of polyadenylated viral transcripts. Wild-type, mutant, and revertant constructs were transfected into C33A cells (human cervix carcinoma cells that do not express the CD4 receptor), resulting in the transient production of viral RNA and protein and the assembly of virions. At 2 days after transfection, cellular RNA was isolated, and the level of unspliced HIV-1 RNA was determined by dot blot analysis with a gag-pol probe (Fig. 3A). For the destabilized mutants (B, C, and D), the level of viral RNA was reduced to approximately 60% of the wild-type level. The HIV-1 RNA level was not affected for the stabilized mutant A and constructs with a hairpin of approximately wild-type stability (the double mutant CD and the A revertant but not the B revertant). Apparently, destabilization of the 5′ polyA hairpin reduced the level of viral RNA in the cell. The production of viral proteins was measured in the culture supernatant by CA-p24 ELISA (Fig. 3B). Despite the differences in the level of intracellular viral RNA, we measured similar CA-p24 levels for the wild-type, mutant, and revertant constructs, which is in agreement with previous Western blotting experiments with cellular extracts (18). There may be differences in the translatability of the wild-type and mutant HIV-1 transcripts, but this issue was not further addressed in this study.
FIG. 3.
Gene expression and virus production by HIV-1 genomes with a mutated 5′ polyA hairpin. C33A cells were transfected with the wild-type and mutant proviral constructs and cultured for 2 days. (A) The intracellular HIV-1 RNA content was measured with a gag-pol probe that specifically detects unspliced transcripts (illustrated in Fig. 1B). (B) The CA-p24 level was measured in the culture supernatant by ELISA. (C) The genomic RNA content of virions was determined by slot blot analysis and compared with the CA-p24 values. (D) The RNA packaging efficiency was calculated as the ratio of virion RNA to intracellular HIV-1 RNA. All parameters were arbitrarily set at 100% for the wild-type (wt) construct. The standard error of the mean was calculated for four to six independent transfections. For panels C and D, we first calculated the ratio per independent transfection and then calculated the mean value and standard error of the mean. Abbreviations: A, mutant A; Arev, A revertant; B, mutant B; Brev, B revertant; C, mutant C; D, mutant D; CD, double mutant CD.
The effect of the polyA hairpin on RNA packaging.
The virion RNA level was measured by dot blot analysis with the gag-pol probe and compared with the amount of CA-p24 (Fig. 3C). This virion RNA content was reduced significantly for the destabilized mutants B, C, and D, and the B revertant, whereas a wild-type level was observed for the A mutant and revertant and the CD double mutant. This finding is consistent with published results and was previously interpreted to indicate a defect in the process of RNA packaging (18, 33). However, because the opening of the polyA hairpin also affected the intracellular level of HIV-1 RNA (Fig. 3A), the virion RNA content may not be an appropriate measure of the RNA packaging efficiency. Therefore, we calculated the ratio of virion RNA to intracellular HIV-1 RNA to determine the packaging efficiency (Fig. 3D). An approximately wild-type level of RNA packaging is now apparent for all mutant and revertant constructs. These results suggest that the process of RNA packaging is not directly affected by the opening of the polyA hairpin. It appears that the RNA packaging efficiency is sensitive to changes in the intracellular concentration of HIV-1 RNA.
The polyA hairpin is involved in repression of 5′ polyadenylation.
Destabilization of the 5′ polyA hairpin reduced the intracellular level of HIV-1 RNA (Fig. 3A). Several mechanistic explanations can be proposed. Similar to the function of the TAR hairpin, the polyA hairpin may be required for efficient transcription from the upstream LTR promoter. Alternatively, opening of the 5′-terminal hairpin may reduce the stability of the viral RNA. Because we measured only unspliced RNA, the possibility that mutation of the polyA hairpin led to an increase in the splicing efficiency cannot be excluded either. Finally, the polyadenylation site located within the 5′ polyA hairpin may be activated by destabilization of the structure, resulting in premature polyadenylation and the production of extremely short HIV-1 transcripts.
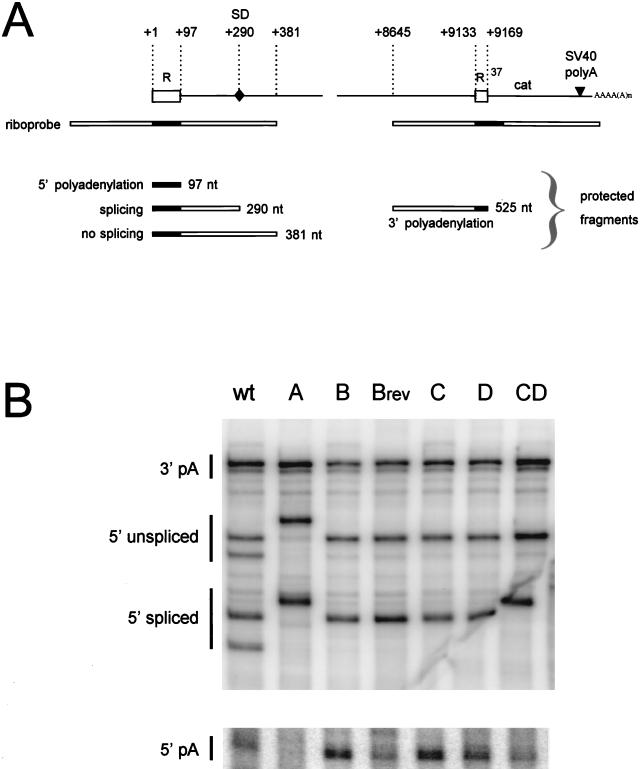
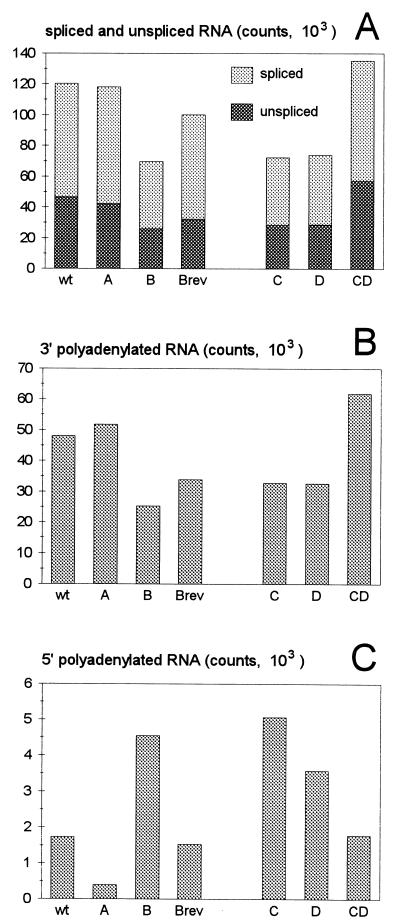
To study the effect of the hairpin mutations on viral RNA production, total cellular RNA was isolated from transfected C33A cells and analyzed by RNAse protection assays. HIV-1 RNA was hybridized to a riboprobe complementary to the U3, R, and U5 sequences (Fig. 4A). Upon digestion with RNase A, protected fragments resulting from 5′ polyadenylated RNAs and from spliced and unspliced 3′ polyadenylated RNAs were detected (Fig. 4B) and quantitated (Fig. 5). For this quantitative comparison of the different protected fragments, all signals were corrected for the number of labeled nucleotides incorporated in the respective probe fragments. For the destabilized mutants B, C, and D, the intracellular level of HIV-1 RNA, both the spliced and unspliced transcripts, was reduced to approximately 60% of the wild-type level (Fig. 5A). The RNA level was partially restored for the B revertant and was fully restored for the double mutant CD. Similar results were obtained when HIV-1 RNA was quantitated by the probe fragment that is protected by the 3′-terminal HIV-1 sequences just upstream of the cat gene and the SV40-based polyadenylation site (Fig. 5B). Stabilization of the 5′ hairpin in mutant A did not affect the level of HIV-1 RNA, as measured by the combined 5′ signals (spliced and unspliced), as well as the 3′ signal. The RNase protection results confirm the results of the dot blot analysis (Fig. 3A) and demonstrate that destabilization of the 5′ polyA hairpin causes a reduction in the amount of viral RNA. The results also indicate that mutation of the polyA hairpin does not affect the splicing efficiency of the primary transcript because the ratio of spliced to unspliced RNAs was not significantly altered for these constructs (Fig. 5A), with the possible exception of the B revertant (see below).
FIG. 4.
Analysis of 5′ and 3′ polyadenylated HIV-1 RNA by RNase protection. (A) Shown is the unspliced, genomic HIV-1 RNA that is polyadenylated at the 3′ end. The two R regions are blocked, and nucleotide numbers refer to positions on the HIV-1 RNA. The position of the major splice donor site (SD, position 290) is indicated. The riboprobe used in the RNase protection assay and the expected protected fragments are indicated (the R region is marked black in the riboprobe and the protected fragments). Premature 5′ polyadenylation will result in the protection of a 97-nucleotide probe fragment. Read-through transcripts protect fragments at both the 5′ and 3′ ends. At the 5′ side, two fragments of 381 or 290 nucleotides are protected by the unspliced and spliced HIV-1 RNAs, respectively. At the 3′ side, a 525-nucleotide fragment is protected by both the unspliced and spliced transcripts. (B) Individual 32P-labeled RNA probes were hybridized to total RNA from cells transfected with the corresponding HIV-1 plasmids (indicated at the top of the panel). The samples were treated with RNase and subjected to denaturing polyacrylamide gel electrophoresis. Control samples from mock-transfected cells and mock-treated RNAs did not produce the signals that are indicated (not shown). Several RNA transcripts of distinct length were used as size markers (not shown). The somewhat aberrant migration of the 5′ spliced signal of the CD mutant was caused by folding of the gel during drying. The doublet bands observed for the 5′ spliced and unspliced signals of the wild-type (wt) construct were not observed in parallel probing experiments with RNase T1. Larger RNase digestion products were observed for the riboprobe of mutant A, which is most likely caused by stable secondary RNA structure in this antisense RNA. In fact, this probe even resisted complete digestion in the absence of HIV-1 RNA (not shown). The data were quantitated by PhosphorImager analysis and corrected for the differences in probe length (amount of incorporated labeled nucleotides), and these results are presented in Fig. 5. Abbreviations are as defined in the legend for Fig. 3.
FIG. 5.
Premature polyadenylation is triggered by opening of the 5′ polyA hairpin. These results were derived from the RNase protection experiment shown in Fig. 4. Abbreviations are as defined in the legend for Fig. 3.
Enhanced 5′ polyadenylation is one possible explanation for the decline in the amount of full-length and spliced viral transcripts. The RNase protection assay allowed us to directly measure such prematurely polyadenylated transcript forms. Indeed, we measured an approximately threefold increase of this signal for the destabilized mutants B, C, and D compared with the wild-type construct (Fig. 5C), and the wild-type level of 5′ polyadenylation was restored in constructs that repair the hairpin stability (the B revertant and the CD double mutant). The 5′ polyadenylation efficiency of the A mutant with the excessively stable hairpin was reduced approximately fourfold compared with that of the wild type.
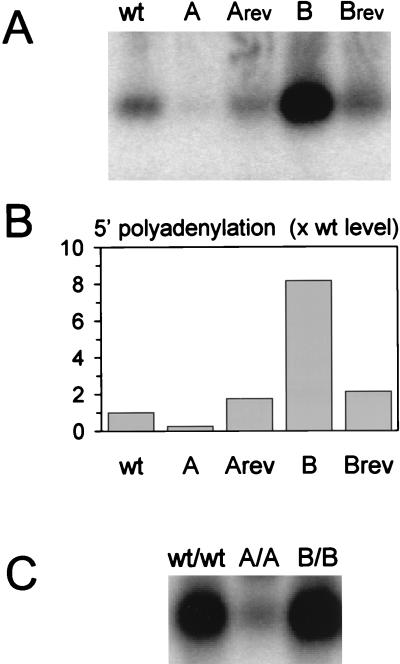
The RNase protection assay indicated that a small fraction of the wild-type HIV-1 transcripts are polyadenylated prematurely at the 5′ polyadenylation site. Because it turned out to be technically difficult to accurately measure this low level of the extremely short transcript (Fig. 4B), we also designed a sensitive RT-PCR protocol to specifically amplify 5′ polyadenylated HIV-1 RNA. First-strand cDNA synthesis was primed with a G(T)19 oligonucleotide that can anneal to poly(A) tails. HIV-1 specific cDNAs were amplified with a 5′ primer on the basis of the TAR sequence and the same G(T)19 oligonucleotide as 3′ primer (Fig. 1B). Prematurely (5′) polyadenylated HIV-1 transcripts will produce a 109-bp RT-PCR product that was visualized by Southern blotting and hybridization with a 32P-labeled TAR region probe. A small amount of 5′ polyadenylation product was detected for the wild-type HIV-1 construct (Fig. 6A). This signal was markedly reduced for the stabilized mutant A and restored to the wild-type level in the A revertant. An eightfold increase of 5′ polyadenylation was measured for the B mutant, and this effect was largely reversed in the B revertant. These results are quantitated in Fig. 6B, with the relative 5′ polyadenylation efficiency of the wild type set at the arbitrary value of 1. The combined results of the RNase protection and RT-PCR assays demonstrate that polyadenylation at the 5′ site is triggered by destabilization of the polyA hairpin and reduced below the wild-type level by further stabilization of this structure.
FIG. 6.
Analysis of the prematurely polyadenylated HIV-1 transcript by RT-PCR. (A) The HIV-1 plasmids with a wild-type or mutated 5′ polyA hairpin (indicated on top of the panel) were transiently transfected into C33A cells. Total cellular RNA was extracted, and HIV-1 sequences were amplified by a RT-PCR protocol (illustrated in Fig. 1B). The products were separated on an agarose gel, blotted, and probed with a TAR region probe. No PCR products were obtained in an RT-minus control reaction, indicating that the input plasmid DNA sequences were not amplified. The 5′ polyadenylated HIV-1 RNA yields a short cDNA product. Please note that read-through transcripts will be polyadenylated at the SV40 polyadenylation site. This RNA is expected to produce a much longer PCR product that includes the cat gene. This product is not detected with the TAR probe because of incomplete complementarity. Quantitation of the PCR products was performed by PhosphorImager analysis, and the results are presented in panel B. (B) The 5′ polyadenylation efficiency of wild-type HIV-1 RNA was arbitrarily set at the value 1. Abbreviations are as in the legend for Fig. 3. (C) Viral plasmids were constructed with the wild-type (wt) or mutant A or B hairpin in both LTRs. C33A cells were transfected with the individual plasmids (indicated on top of the panel), and total cellular RNA was isolated and analyzed by RT-PCR as described for panel A.
As one would expect, we measured an inverse correlation between the efficiency of premature polyadenylation at the 5′ polyadenylation site and the level of intracellular HIV-1 RNA for all constructs except the B revertant. Although this revertant effectively repressed the premature polyadenylation that was observed for the B mutant (Fig. 6), the HIV-1 RNA level was only partially restored (Fig. 3A and 5A). We do not fully understand the behavior of the B revertant. Inspection of the RNA data (Fig. 5A) suggests that the amount of spliced HIV-1 RNA is actually restored, unlike the level of full-length HIV-1 RNA (Fig. 5A). This result may suggest that the process of splicing is affected in this revertant, but this issue was not further addressed.
Polyadenylation at the 3′ end is suppressed by further stabilization of the polyA hairpin.
An intriguing observation is that the 5′ polyadenylation site of the wild-type HIV-1 genome is used at a low frequency. Thus, production of full-length viral transcripts can be optimized slightly by complete repression of the 5′ polyadenylation site. This may be the case for the stabilized mutant A, which showed no premature polyadenylation (Fig. 5C and 6). It can be argued that viral replication can be improved by stabilization of the 5′ polyA hairpin. However, because the sequence of the 5′R element will be dominantly inherited in both LTRs of the viral progeny after a single round of reverse transcription (31), it is anticipated that such an improvement of viral RNA expression will be overshadowed by a severe polyadenylation defect caused by the same RNA structure in the 3′R. To directly test this, we made new HIV-1 constructs with two complete LTRs encoding the wild-type, A, or B mutant polyA hairpin. We will refer to these double mutants as wt/wt, A/A, and B/B.
The RT-PCR procedure was used to analyze viral RNA isolated from transiently transfected cells. With these full-length proviral constructs, the RT-PCR protocol cannot distinguish among 5′ and 3′ polyadenylated RNAs. Thus, the amount of cDNA product represents the sum of both types of RNA. A prominent cDNA signal was obtained for the wt/wt construct (Fig. 6C). Because the wild-type genome was shown to inefficiently use the 5′ polyadenylation site, this signal represents essentially the 3′ polyadenylated viral transcripts. An approximately wild-type level of RT-PCR product was obtained for the B/B construct. Because opening of the 5′ hairpin triggered significant levels of premature polyadenylation, this RT-PCR product represents the sum of both 5′ and 3′ polyadenylation. When both hairpins were stabilized, as in mutant A/A, the RT-PCR signal was reduced dramatically. This result indicates that polyadenylation in the 3′ context is also efficiently inhibited by stable RNA structure. Thus, having a more stable hairpin in the 5′R may modestly improve the production of viral transcripts, but the same structure causes an overwhelming polyadenylation defect in the 3′R. This result is consistent with the severe replication defect described for the mutant virus with the stable A hairpin in both R regions (18).
DISCUSSION
We tested whether secondary RNA structure played a role in the differential usage of the polyadenylation signal that is present in both the 5′R and 3′R region of the HIV-1 genome. Several models of regulated HIV-1 polyadenylation site usage have been proposed, including the presence of 5′-specific silencer and 3′-specific enhancer elements (3, 13, 14, 20, 23, 24, 41–44). The presentation of the AAUAAA polyadenylation signal in a hairpin structure may provide additional regulatory possibilities. This polyA hairpin has been demonstrated to be essential for virus replication (18). In this study, we report that the hairpin is required for efficient repression of the 5′ polyadenylation site. The opening of the 5′ structure induced premature polyadenylation and reduced the amount of spliced and unspliced viral transcripts. Apparently, this repressive potential of RNA structure is overcome at the 3′ polyadenylation site in the presence of the USE enhancer. Polyadenylation at the 3′ site was inhibited when the stability of the 3′ hairpin was increased further by mutation. Thus, the stability of the polyA hairpin is fine-tuned to allow efficient repression in the 5′R yet full activation in the 3′R. This modulating role is illustrated in the model presented in Fig. 7.
FIG. 7.
A multifactorial model for differential HIV-1 polyadenylation. Shown are the proviral DNA and the primary HIV-1 transcript with the typical tandem hairpin motif at both the 5′ and 3′ ends. The polyadenylation signal AAUAAA (triangle) is located in the polyA hairpin. Results presented in this study indicate that regulated HIV-1 polyadenylation is made possible by the polyA hairpin structure, which down-modulates this potentially efficient polyadenylation site (indicated by the ± sign). In the presence of additional repressive signals in the 5′ context (indicated is inhibition through the SD site, but promoter proximity may also play a negative role), polyadenylation is reduced to a low level (approximately 10%; see Discussion). In the presence of the USE enhancer motif in the 3′ context, the full potential of the HIV-1 polyadenylation sequence is realized. This scheme does not indicate which step of polyadenylation is regulated. However, because the polyadenylation signal is occluded by the hairpin structure, it is likely that the initial binding of CPSF to the hexamer motif is blocked.
The HIV-1 polyadenylation signal has been demonstrated to represent an inherently efficient polyadenylation sequence in reporter constructs. In our model, the role of the polyA hairpin is to repress this efficiency. The suboptimal context of the structured HIV-1 polyadenylation site allows both repression of 5′ polyadenylation through the SD mechanism (3, 4) or through promoter proximity (14, 44) and activation of 3′ polyadenylation through the USE mechanism (13, 20, 23, 24, 41, 42). Thus, the idea that occlusion of the HIV-1 polyadenylation site by RNA structure plays a critical role in differential polyadenylation does not replace the existing models but rather provides a mechanistic explanation for 5′ down-regulation and 3′ up-regulation. Conceivably, rapid folding of the polyA hairpin structure on the nascent viral transcript will delay the recognition by polyadenylation factors, such that sufficient time is available for recognition of the major SD by the U1 snRNP (4). As the RNA commits itself to the splicing reaction, e.g., is transported into spliceosomes, it will become less accessible to the polyadenylation factors. The presence of the inhibitory RNA conformation also explains why the 3′ polyadenylation site requires the USE enhancer for full activity. Thus, a complex interplay of polyadenylation and splicing signals, repressive RNA structure, and either enhancer or silencer motifs are involved in regulated HIV-1 polyadenylation.
Interestingly, the TAR hairpin structure (Fig. 1) is also thought to perform a critical function as part of the nascent viral transcript, that is, binding of the viral transcriptional activator protein Tat (11, 21). It is therefore likely that the nucleotide sequence of both the TAR and polyA elements are optimized to allow rapid base pairing of the stem regions. Given the fact that these two hairpins are immediately adjacent to each other, it is possible that the two stem regions stack coaxially to further stabilize this structured RNA domain that plays such a critical role in enhancement of transcription (TAR hairpin) and repression of premature polyadenylation (polyA hairpin) (see also reference 7).
Destabilization of the 5′ polyA hairpin resulted in a reduction of the intracellular viral RNA levels to approximately 60 to 70% of the wild-type level (Fig. 3A and 5A). This reduction in viral RNA synthesis coincided with a three- to eightfold increase in 5′ polyadenylation efficiency, as measured by an RNase protection assay and RT-PCR, respectively. It is possible that the RT-PCR protocol overestimates the effect because the amplified fragment included the polyA hairpin, which may form a more efficient template for reverse transcription in the case of the mutant sequence that encodes a less stable hairpin structure. Assuming that the 30 to 40% reduction of viral RNA was caused by the increase in premature polyadenylation, it can be calculated that the 5′ polyadenylation efficiency of the wild-type construct is around 5 to 10%. This value is raised to approximately 30 to 40% upon destabilization of the 5′ hairpin, and reduced fourfold upon stabilization. We and others have reported previously that the 5′ polyA hairpin structure contributes to packaging of the viral RNA into virion particles (18, 33). However, our current analysis suggests that the reduced packaging is a direct consequence of the reduced level of intracellular HIV-1 RNA.
The mechanistic model of Fig. 7 predicts that the stability of the polyA hairpin structure is critical for differential regulation of polyadenylation. Indeed, the analysis of revertants that evolved from poorly replicating virus mutants with a stabilized or destabilized hairpin demonstrated a definite drift to a thermodynamic stability comparable to that of the wild-type structure (10). Further experimentation is under way to test which step of the polyadenylation mechanism is blocked. Because the AAUAAA hexamer motif itself is partially occluded by base pairing (Fig. 2), a likely possibility is that binding of CPSF to this sequence motif is affected, thus inhibiting the initial step of the polyadenylation reaction. We have some preliminary evidence from in vitro RNA-protein binding assays that binding of CPSF is indeed blocked by stable RNA structure (30a). Although this is the first report of occlusion of polyadenylation sequence elements by a naturally occurring RNA structure, such a masking effect of RNA structure is not without precedent in molecular biology. RNA structure has been implicated in regulation of gene expression at several levels, including the processes of splicing and translation (2, 22, 26, 29). For instance, a remarkably similar case of a translational control mechanism in the coliphage MS2 was presented. Here, the thermodynamic stability of a hairpin encompassing the ribosomal binding site is inversely correlated with the translational activity (19). Detailed studies indicated that ribosomes bind exclusively to the unfolded RNA, with the equilibrium between the folded and unfolded RNA being determined by the thermodynamic stability of the hairpin. In the case of HIV-1, the situation seems more complex because the USE enhancer forms an additional binding site for CPSF. Thus, the USE enhancer may form the actual entry site for CPSF. From that position near the 3′ polyadenylation site, CPSF may await melting of the polyA hairpin to gain access to the hexamer motif.
There have been a few reports on cellular mRNAs that may control polyadenylation by RNA structure (25, 39, 45), and there is one other, well-documented retroviral system where RNA secondary structure plays a critical role in polyadenylation. The HTLV-1 uses an extensive RNA structure to guide the polyadenylation complex to a cleavage site that is located 274 nucleotides downstream of the AAUAAA hexamer signal (1, 6, 38). Here, the RNA structure facilitates efficient 3′ polyadenylation but is not involved in suppression of a 5′ polyadenylation site. In HTLV-1, the AAUAAA signal is encoded by the upstream U3 region and is therefore not present at the 5′ end of the viral transcript. This HTLV-1 case illustrates another retroviral strategy for a genome with identical 5′ and 3′ ends to undergo selective 3′ polyadenylation. The HTLV-1 strategy to place the AAUAAA signal in the U3 sequences is also used by retroviruses with a relatively short R region.
Likely candidates for a polyadenylation mechanism similar to that of HIV-1 are the retroviruses with a relatively extended R region that includes the AAUAAA signal. We screened several retroviral genomes that meet these criteria and remarkably, similar stem-loop structures could be drawn for retroviruses of the lentivirus and spumavirus groups. Some of these structures are shown in Fig. 8. We previously reported that the polyA hairpin is well conserved among HIVs and SIVs (9) despite a considerable divergence in nucleotide sequence of this part of the genome. As an example, we included the hairpin of the HIV-1LAI isolate and SIVsykes. A similar structure is predicted for the polyadenylation signal of other lentiviruses, such as the bovine immunodeficiency virus and the equine infectious anemia virus. The human spumaretrovirus was also able to base pair part of the AAUAAA signal in a hairpin structure (Fig. 8), and similar foldings were predicted for simian spumaviruses (not shown). There is considerable variation in the thermodynamic stability of these retroviral RNA structures, but stability is merely one of the many parameters that may control the efficiency of these polyadenylation sites. These variables include the AAUAAA signal and perhaps the flanking nucleotide sequences, the presence of enhancer or silencer elements, and the extent of base pairing of these sequences. These results suggest that regulation of polyadenylation by RNA structure is more widespread among the lentivirus and spumavirus groups and that this mechanism may represent a more common retroviral strategy.
FIG. 8.
Lentiviruses and spumaviruses have their polyadenylation signals encoded within a hairpin structure. All these viruses have a relatively extended R region encoding the AAUAAA hexamer motif (shaded in structure), which necessitates differential polyadenylation. Similar stem-loop structures were predicted for other HIV and SIV viruses (9) and also for simian spumaviruses (not shown). The free energies of the stem-loop structures (including the terminal stacks) were calculated with the Zuker algorithm (46), and the ΔG values are presented in kilocalories per mole. EIAV, equine infectious anemia virus; HSRV, human spumaretrovirus; BIV, bovine immunodeficiency virus.
ACKNOWLEDGMENTS
We thank Koen Verhoef and Bianca Klasens for critical reading of the manuscript, Bianca Klasens and Jeroen van Wamel for assistance in constructing the CD double mutant, and Wim van Est for artwork.
This work was supported in part by the Dutch Cancer Society (KWF), the Dutch AIDS Fund (AIDS Fonds), and the EC (grant 950675).
REFERENCES
- 1.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez C J, Romfo C M, Vanhoy R W, Porter G L, Wise J A. Mutational analysis of U1 function in Schizosaccharomyces pombe: pre-mRNAs differ in the extent and nature of their requirements for this snRNA in vivo. RNA. 1996;2:404–418. [PMC free article] [PubMed] [Google Scholar]
- 3.Ashe M P, Griffin P, James W, Proudfoot N J. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 4.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auersperg N. Long-term cultivation of hypodiploid human tumor cells. J Natl Cancer Inst. 1964;32:135–163. [PubMed] [Google Scholar]
- 6.Bar-Shira A, Panet A, Honigman A. An RNA secondary structure juxtaposes two remote genetic signals for human T-cell leukemia virus type 1 RNA 3′-end processing. J Virol. 1991;65:5165–5173. doi: 10.1128/jvi.65.10.5165-5173.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B, Klaver B. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 1993;21:5020–5024. doi: 10.1093/nar/21.22.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkhout B, Klaver B, Das A T. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 10.Berkhout B, Klaver B, Das A T. Forced evolution of a regulatory RNA helix in the HIV-1 genome. Nucleic Acids Res. 1997;25:940–947. doi: 10.1093/nar/25.5.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 12.Berkhout B, van Wamel J, Klaver B. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J Mol Biol. 1995;252:59–69. doi: 10.1006/jmbi.1994.0475. [DOI] [PubMed] [Google Scholar]
- 13.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherrington J, Ganem D. Regulation of polyadenylation in human immunodeficiency virus (HIV): contributions of promoter proximity and upstream sequences. EMBO J. 1992;11:1513–1524. doi: 10.1002/j.1460-2075.1992.tb05196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 17.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of the human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A T, Klaver B, Klasens B I F, van Wamel J L B, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Smit M H, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeZazzo J D, Kilpatrick J E, Imperiale M J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3′ end formation. Mol Cell Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activating-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng F J, Warner J R. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 23.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 25.Gimmi E R, Reff M E, Deckman I C. Alterations in the pre-mRNA topology of the bovine growth hormone polyadenylation region decrease poly(A) site efficiency. Nucleic Acids Res. 1989;17:6983–6998. doi: 10.1093/nar/17.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goguel V, Yang Y, Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol Cell Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graveley B R, Fleming E S, Gilmartin G M. Restoration of both structure and function to a defective poly(A) site by in vitro selection. J Biol Chem. 1996;271:33654–33663. doi: 10.1074/jbc.271.52.33654. [DOI] [PubMed] [Google Scholar]
- 28.Graveley B R, Fleming E S, Gilmartin G M. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 30.Klasens B I F, Das A T, Berkhout B. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 1998;26:1870–1876. doi: 10.1093/nar/26.8.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Klasens, B. I. F., et al. Submitted for publication.
- 31.Klaver B, Berkhout B. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res. 1994;22:137–144. doi: 10.1093/nar/22.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 35.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 37.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 38.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sittler A, Gallinaro H, Jacob M. The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in its loop. J Mol Biol. 1995;248:525–540. doi: 10.1006/jmbi.1995.0240. [DOI] [PubMed] [Google Scholar]
- 40.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 41.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weichs an der Glon C, Ashe M, Eggermont J, Proudfoot N J. Tat-dependent occlusion of the HIV poly(A) site. EMBO J. 1993;12:2119–2128. doi: 10.1002/j.1460-2075.1993.tb05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weichs an der Glon C, Monks J, Proudfoot N J. Occlusion of the HIV poly(A) site. Genes Dev. 1991;5:244–253. doi: 10.1101/gad.5.2.244. [DOI] [PubMed] [Google Scholar]
- 45.Woychik R P, Lyons R H, Post L, Rottman F M. Requirement for the 3′ flanking region of the bovine growth hormone gene for accurate polyadenylation. Proc Natl Acad Sci USA. 1984;81:3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]