Abstract
We have recently identified a DNA-binding protein (DBP) from the baculovirus Bombyx mori nucleopolyhedrovirus (BmNPV) which can destabilize double-stranded DNA (V. S. Mikhailov, A. L. Mikhailova, M. Iwanaga, S. Gomi, and S. Maeda, J. Virol. 72:3107–3116, 1998). DBP was found to be an early gene product that was not present in budded or occlusion-derived virions. In order to characterize the localization of DBP during viral replication, BmNPV-infected BmN cells were examined by immunostaining and confocal microscopy with DBP antibodies. DBP first appeared as diffuse nuclear staining at 4 to 6 h postinfection (p.i.) and then localized to several specific foci within the nucleus at 6 to 8 h p.i. After the onset of viral DNA replication at around 8 h p.i., these foci began to enlarge and eventually occupied more than half of the nucleus by 14 h p.i. After the termination of viral DNA replication at about 20 h p.i., the DBP-stained regions appeared to break down into approximately 100 small foci within the nucleus. At 8 h p.i., the distribution of DBP as well as that of IE-1 or LEF-3 (two proteins involved in baculovirus DNA replication) overlapped well with that of DNA replication sites labeled with bromodeoxyuridine incorporation. Double-staining experiments with IE-1 and DBP or IE-1 and LEF-3 further confirmed that, between 8 and 14 h p.i., the distribution of IE-1 and LEF-3 overlapped with that of DBP. However, IE-1 localized to the specific foci prior to DBP or LEF-3 at 4 h p.i. In the presence of aphidicolin, an inhibitor of DNA synthesis, immature foci containing IE-1, LEF-3, and DBP were observed by 8 h p.i. However, the subsequent enlargement of these foci was completely suppressed, suggesting that the enlargement depended upon viral DNA replication. At 4 h p.i., the number of IE-1 foci correlated with the multiplicity of infection (MOI) between 0.4 and 10. At higher MOIs (e.g., 50), the number of foci plateaued at around 15. These results suggested that there are about 15 preexisting sites per nucleus which are associated with the initiation of viral DNA replication and assembly of viral DNA replication factories.
Bombyx mori nucleopolyhedrovirus (BmNPV; Baculoviridae) has been widely used for the expression of foreign genes in B. mori cell culture and larvae (29). BmNPV has a 128,413-bp-long circular double-stranded DNA (dsDNA) genome which encodes 136 potential genes (GenBank accession no. L33180). The organization of the BmNPV genome closely resembles that of Autographa californica nucleopolyhedrovirus (AcNPV) (32), which is the most extensively studied baculovirus. Although baculoviruses, including BmNPV and AcNPV, efficiently replicate in the nuclei of susceptible arthropod cells, the dynamics and mechanism of DNA replication within the infected cell are poorly understood.
We have recently purified and characterized two DNA-binding proteins, DBP and LEF-3, from nuclear lysates of BmNPV-infected BmN cells (36). DBP contains 317 amino acids and is encoded by BmNPV open reading frame 16 (ORF16), which is a homolog of AcNPV ORF25 (96% homology) (36). DBP preferentially binds single-stranded DNA (ssDNA) at least 1 order of magnitude higher than it binds dsDNA and unwinds partial DNA duplexes in vitro. The DNA binding site of DBP is about 30 nucleotides per protein monomer on the basis of exonuclease assays. BmNPV LEF-3 shows 92% amino acid sequence identity to AcNPV LEF-3, which may function as an ssDNA-binding protein (SSB) (14, 17). BmNPV LEF-3 also specifically binds ssDNA but cannot unwind DNA duplexes in vitro (36).
Genes from baculoviruses AcNPV (21, 28) and Orgyia pseudotsugata nucleopolyhedrovirus (OpNPV) (2, 4, 5) involved in DNA replication have been identified by transient replication assays. Kool et al. (21) reported that the products of six AcNPV genes (ie-1, lef-1, lef-2, lef-3, dnapol, and p143) are essential for replication of plasmid DNA from a baculovirus origin of replication (21). It is likely that the core replication machinery is formed by a putative DNA polymerase (dnapol) (3, 8, 35, 42), putative DNA helicase (p143) (3, 23, 25, 30), and a primase complex encoded by lef-1 and lef-2 (13). The other two essential gene products (LEF-3 and IE-1) are characterized as DNA-binding proteins. As previously mentioned, LEF-3 was predicted to be an SSB (14, 17). IE-1 is a well-studied transactivator of immediate-early genes (see reference 15) and possibly an origin-binding protein during DNA replication (9, 24).
Virus-expressed SSBs have been reported to form part of the DNA replication complex of herpes simplex virus type 1 (10). Antibodies raised against these SSBs have also been successfully used to label the herpes simplex virus type 1 replication complex. Although DBP functions as a typical SSB in in vitro assays (36), DBP homolog of AcNPV and OpNPV were not found to be essential or stimulatory for plasmid DNA replication by transient replication assays (5, 21). This apparent discrepancy prompted us to examine the relationship between DBP and DNA replication by using DBP-specific antibodies. In this report, we describe the temporal and spatial localizations of DBP, especially with reference to viral DNA replication sites. We show here that at 8 h postinfection (p.i.) DBP localized to specific foci where viral DNA replication was also observed, indicating that DBP is involved in viral DNA synthesis. The temporal and spatial distributions of IE-1 and LEF-3, both of which are essential for DNA replication, were then compared with those of DBP. Furthermore, analysis of the relationship between the number of IE-1 foci formed and multiplicity of infection (MOI) suggested that nuclear domains are found in insect cells which may be homologous to nuclear domain 10 (ND10) of mammalian cells.
MATERIALS AND METHODS
Cell line and viral infection.
BmN (BmN-4) cells were maintained in TC-100 medium (Funakoshi Co., Tokyo, Japan) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (29). The BmNPV wild-type isolate T3 (31) was propagated on BmN cells. The BmN cells were plated onto 22- by 22-mm coverslips (no. 1; Matsunami, Tokyo, Japan) and allowed to adhere for several hours or overnight prior to infection. The cell number on each coverslip was estimated to be 3 × 105 to 6 × 105. The BmN cells were infected with BmNPV at an MOI of 10 (29) unless otherwise stated. In all experiments, time zero was defined as the point at which fresh medium was added following the 1-h virus adsorption period. When appropriate, aphidicolin (a gift from S. Ikegami, Hiroshima University [18]) was added to the culture medium (20 μM) at time zero, in order to suppress host and viral DNA synthesis, and was present until fixation.
Polyclonal antibody production.
Rabbit polyclonal DBP antiserum was prepared with a DBP–six-histidine fusion protein (N-terminal six-histidine tag) overexpressed in Escherichia coli according to the manufacturer’s instructions (Novagen, Madison, Wis.). The coding region of dbp (BmNPV ORF16) (36) was amplified from BmNPV genomic DNAs by PCR with two primers, 5′-GGCATATGGCAACTAAACGCAA-3′ and 5′-GGGGATCCGCAAGACATTTTGAC-3′, which generated appropriate sites for cloning. The PCR-amplified DNA fragment was digested with NdeI and BamHI and subcloned into the NdeI and BamHI sites of the expression vector pET-28a(+). The resulting plasmid was transformed into E. coli BL21(DE3)pLysS. Transformed E. coli cells were harvested 12 h after induction with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). The overexpressed recombinant DBP (rDBP) was purified by chromatography under denaturing conditions on the His-Bind metal chelation resin (Novagen). The purified His-tagged rDBP was subcutaneously injected into a rabbit with complete Freund’s adjuvant for the initial injection and incomplete Freund’s adjuvant for subsequent injections (2 to 3 weeks between injections). Rabbit antiserum was collected 1 week after the third injection and tested by enzyme-linked immunosorbent assay and Western blot analyses.
PAGE and Western blotting.
Virus- or mock-infected BmN cells (106 cells) were collected by centrifugation (3,500 × g, 3 min) and extracted in 100 μl of SLB buffer containing 20 mM dithiothreitol, 2% sodium dodecyl sulfate (SDS), 10% glycerol, 62.5 mM Tris-HCl (pH 6.8), and 0.03% bromophenol blue. The extracts (5 μl; 5 × 104-cell equivalent) were then mixed with 8.5 μl of T-SLB containing 1 part of 2 M Tris base and 9 parts of SLB buffer and boiled for 3 min at 100°C. After addition of 1.5 μl of 1 M iodoacetamide, resulting mixtures were analyzed by SDS–11% polyacrylamide gel electrophoresis (PAGE) as described by Laemmli (22). Gels were electrophoretically transferred to Clear-Blot membrane P by using a semidry blot apparatus (Atto) according to the manufacturer’s guidelines. Western blots were probed with a 1:3,000 dilution of rabbit anti-DBP serum or rabbit polyclonal antiserum to AcNPV LEF-3 (a generous gift from G. F. Rohrmann, Oregon State University [14]). The resulting membranes were washed and incubated with a 1:5,000 dilution of goat anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (ICN Pharmaceuticals) and visualized with an ECL detection system (Amersham). In order to quantify the amounts of DBP and LEF-3 in infected cells, Western blotting of proteins from BmNPV-infected BmN cells at 14 h p.i. was performed in parallel with known amounts of purified DBP and LEF-3. The blot obtained after SDS-PAGE was probed first with antiserum against DBP and then with antiserum against LEF-3. Quantitative estimates of DBP and LEF-3 were based on the assumption that the efficiency of transfer was the same for equivalent amounts of protein from the purified fraction and from the infected cell extract.
Immunohistochemistry and confocal microscopy.
Virus- or mock-infected BmN cells were fixed for 10 min with 2% formalin in phosphate-buffered saline (PBS), washed three times with PBS, and permeabilized for 2 min in cold acetone (−20°C). The cells were rehydrated with PBS, blocked with 1% FBS in PBS for 1 h, and then subjected to antibody treatments. Antigen localization was determined after incubation of the cells with rabbit anti-DBP serum (1:50 dilution with 1% FBS in PBS), guinea pig anti-IE-1 serum (1:100 dilution; a generous gift from H. Bando, Hokkaido University), or rabbit anti-LEF-3 serum (1:50 dilution [14]) for 1 h at room temperature. Anti-IE-1 serum was prepared from guinea pigs by using maltose-binding protein–IE-1 fusion protein overexpressed in E. coli and was preabsorbed with maltose-binding protein prior to use (6a). Anti-DBP serum was preabsorbed with uninfected BmN cells. After the incubation with the appropriate primary antibody, cells were washed four times (5 min per wash) with PBS and then treated with the appropriate secondary antibody, fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (1:1,000 dilution; Cappel, Aurora, Ohio) or FITC-conjugated goat anti-guinea pig IgG (1:200 dilution; Cappel), for 1 h at room temperature. After five washes with PBS (5 min per wash), the cells were mounted with the Slow Fade light antifade kit (Molecular Probes, Eugene, Oreg.). For the double-staining experiments, Cy-5-conjugated goat anti-rabbit IgG (1:200 dilution; Amersham Life Sciences) was used in place of the FITC-conjugated goat anti-rabbit IgG. The stained cells were analyzed with a laser confocal microscope (Leica TCS NT equipped with an Ar-Kr laser; Leica, Heidelberg, Germany).
In order to measure DNA synthesis, virus- or mock-infected BmN cells were pulse-labeled with the thymidine analog 5-bromo-2′-deoxyuridine (BrdU) (10 μM) (Calbiochem, La Jolla, Calif.) for 30 min prior to fixation (10). The immunofluorescence assay was performed as described above, except that, after permeabilization by acetone, the BrdU-labeled cells were treated with 4 N HCl for 10 min to expose the incorporated BrdU residues. The primary anti-BrdU monoclonal antibody (1:40 dilution) was purchased from Calbiochem, and the secondary rhodamine red X-conjugated goat anti-mouse IgG (1:200 dilution) was purchased from Molecular Probes.
For the double-stained samples, only one light source was used for any single-channel recording to avoid cross talk between channels: 488-nm light for FITC, 568-nm light for rhodamine red X, and 647-nm light for Cy-5. To obtain merged figures, single-channel images were digitally superimposed with PhotoShop 4.0 software. Variability of the progression of the infection cycle in any given experiment was relatively small (see Fig. 2) but present. The number of cells studied in each sample exceeded 100, and the experiments were repeated at least twice. At 20 or 26 h p.i., cells undergoing secondary infection were sometimes observed, but these cells were less than 10% of the population and were easily distinguishable.
FIG. 2.
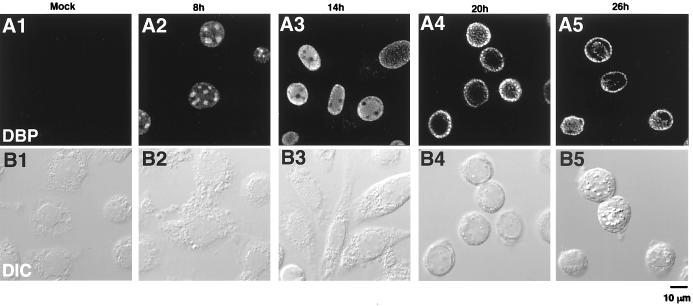
Distribution of DBP in BmNPV-infected BmN cells. (A) DBP immunofluorescence images. (B) Differential interface contrast images of the same fields as in panel A. BmN cells were mock infected (column 1) or infected with BmNPV for 8, 14, 20, or 26 h (columns 2 to 5, respectively) and then fixed with 2% formalin for 10 min, permeabilized with cold acetone for 2 min, and incubated with anti-DBP antibody followed by FITC-conjugated goat anti-rabbit IgG. The bar indicates 10 μm.
RESULTS
Analysis of DBP expression.
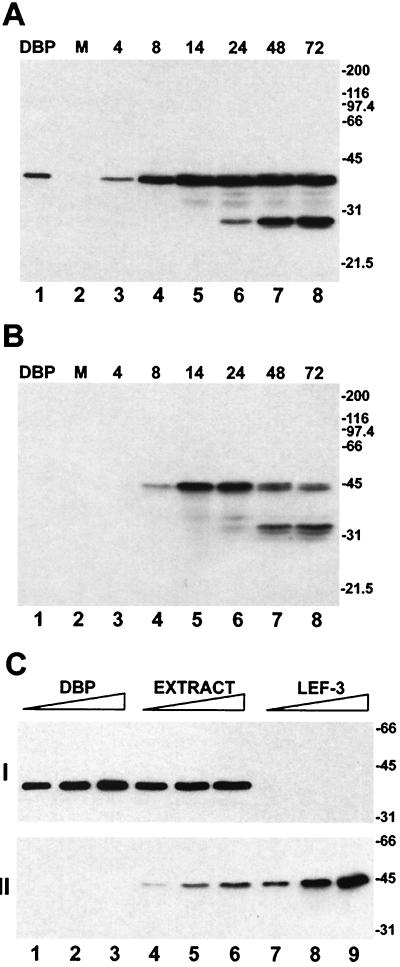
The expression of DBP in BmNPV-infected BmN cells was characterized by Western blotting with polyclonal antibodies raised against His-tagged DBP (Fig. 1A). The antibodies specifically recognized a protein with an apparent molecular mass of 38 kDa which migrated at the same position as purified DBP (36). DBP expression was initially detected in the cell extracts at 4 h p.i. The amount of DBP increased until 14 h p.i. and then remained at relatively steady levels until 72 h p.i. The time course of DBP expression in infected cells was consistent with that of an early viral product. The appearance of an immunoreactive polypeptide of about 28 kDa at 24 h p.i. indicated that proteolytic cleavage of DBP occurred at late stages in infection (Fig. 1A). DBP was not found among structural proteins from budded or occlusion-derived virions by Western blotting (data not shown).
FIG. 1.
Western blot analysis of DBP and LEF-3 in BmNPV-infected BmN cells. (A and B) Time course of the accumulation of DBP and LEF-3 in BmNPV-infected cells. Extracts (5 × 104 cells) were prepared from mock-infected cells (lanes 2) or from BmNPV-infected cells collected at the indicated times p.i. (lanes 3 to 8) as described in Materials and Methods. The cell extracts were separated by SDS–11% PAGE followed by Western blotting with antiserum against DBP (A) or LEF-3 (B). Purified DBP (10 ng) was analyzed for lanes 1. (C) Quantitative determination of DBP and LEF-3 in BmNPV-infected BmN cells. The extract was prepared from infected cells collected at 14 h p.i. and analyzed by SDS–11% PAGE in amounts equivalent to 0.8 × 104 cells (lane 4), 2 × 104 cells (lane 5), or 4 × 104 cells (lane 6). The purified proteins analyzed in the same gel were taken in the following amounts: DBP, 20 ng (lane 1), 50 ng (lane 2), and 100 ng (lane 3); LEF-3, 20 ng (lane 7), 50 ng (lane 8), and 100 ng (lane 9). The proteins were detected by Western blotting with antiserum against DBP (I) and then antiserum against LEF-3 (II). The migration of molecular size markers (sizes in kilodaltons) is shown to the right of each blot.
With antibodies against the AcNPV DNA-binding protein LEF-3 (14), Western blot analysis showed that LEF-3 expression can be detected in BmNPV-infected BmN cells at 8 h p.i. (Fig. 1B). The amount of LEF-3 increased between 8 and 14 h p.i. At 24 h p.i., LEF-3 appeared to be degraded, resulting predominantly in a 33-kDa polypeptide. The time course of LEF-3 expression in BmNPV-infected BmN cells was similar to that described earlier for AcNPV-infected Sf cells (17). There was no apparent cross-reactivity between the antibodies against LEF-3 and those against DBP.
Quantitative Western blot analysis showed that in BmNPV-infected BmN cells DBP was more abundant than LEF-3 (Fig. 1C). At 14 h p.i., there were approximately 7 × 107 DBP molecules per cell, whereas there were only approximately 1 × 107 LEF-3 molecules per cell.
Temporal changes in the nuclear localization of DBP during the infection cycle.
In order to visualize the distribution of DBP, BmNPV-infected BmN cells were immunostained with anti-DBP antibodies and scanned with a laser confocal microscope (Fig. 2). In mock-infected cells, very weak nonspecific staining was sometimes observed in the cytoplasm. However, this staining was far below the detection threshold of the parameters used. In contrast, BmNPV-infected cells showed a distinctively bright DBP immunostaining pattern, which was detected only in the nucleus (Fig. 2). At an early stage of infection (4 to 6 h p.i.), DBP antibodies weakly stained the entire nucleoplasm. At 8 h p.i., DBP appeared to accumulate into several (3 to 10) small, circular foci within the nucleus (Fig. 2A2). The sizes of these foci appeared to substantially differ in each cell, presumably due to differences in the progression of the infection cycle. These foci dramatically increased in size after 8 h p.i. (see Fig. 5B2 and B3) and by 14 h p.i. reached a maximum size (approximately 5 to 10 μm in diameter), occupying more than half of the nucleoplasm (Fig. 2A3). At 14 h p.i., the nuclear membrane was obviously stained, and by 20 h p.i., the heavily stained regions appeared to break down into numerous small foci within the nucleus and particularly near the nuclear membrane (Fig. 2A4). At 26 h p.i., these foci were localized mainly near the nuclear membrane and within an uncharacterized structure at the center of the nucleus (Fig. 2A5). Polyhedra were also observed in about 10% of the cells at this time (Fig. 2B5).
Temporal changes in the sites of viral DNA synthesis.
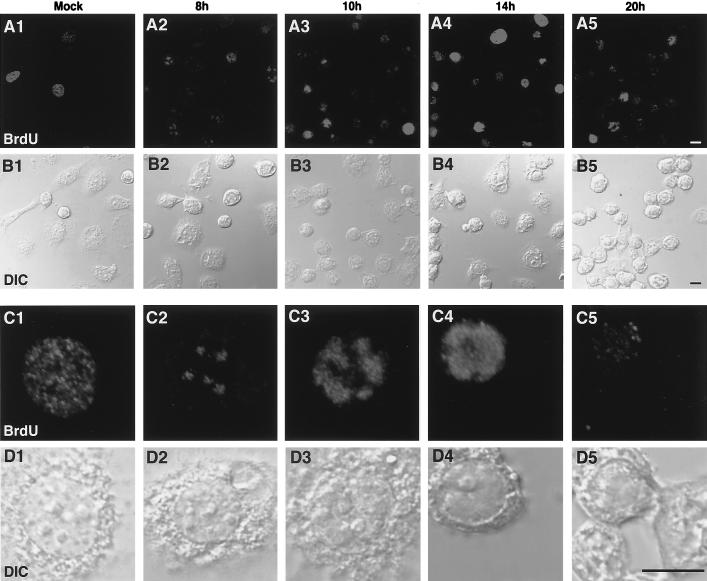
DBP has been shown elsewhere to strongly bind ssDNA and unwind dsDNA in vitro (36). These characteristics suggest that DBP is involved as an SSB during viral DNA replication. In order to examine the temporal progression of viral DNA replication, mock- or BmNPV-infected BmN cells were pulse-labeled with BrdU followed by immunofluorescence detection with an anti-BrdU antibody (10, 37). Numerous fine granular spots were stained by the anti-BrdU antibody in the nuclei of mock-infected cells, indicating the presence of host cell DNA synthesis (Fig. 3, column 1). This pattern was similar to that observed in other cultured cells (10, 37). In general, BrdU staining indicated that 10 to 30% of the mock-infected cells were in the S phase when measured between several hours and 1 day after plating. Similar patterns of BrdU staining were observed at 4 or 6 h p.i. (data not shown). Infection-specific BrdU staining was initially observed at 8 h p.i., as several large (1 to 2 μm in diameter) foci within a nucleus (Fig. 3, column 2). Although the staining intensity was variable, similar patterns were observed in almost half of the cells at 8 h p.i. At 10 h p.i., the intensity, size, and percentage of the BrdU foci increased (Fig. 3, column 3). At 14 h p.i., more than 70% of cells showed heavy BrdU staining which occupied more than half of the nucleoplasm (Fig. 3, column 4), suggesting a high rate of viral DNA replication. At 20 h p.i., the number of cells showing bright immunofluorescence dramatically decreased (Fig. 3, column 5).
FIG. 3.
Distribution of DNA replication sites in BmNPV-infected BmN cells. (A and C) BrdU immunofluorescence images. (B and D) Differential interface contrast images of the same fields as in panels A and C, respectively. In panels C and D, higher magnifications of specific cells in panels A and B are shown. Note that infection-specific BrdU incorporation appeared as unique foci at 8 h p.i. The size and intensity of these foci increased until 14 h p.i. BrdU incorporation then diminished at 20 h p.i. BmN cells were mock infected (column 1) or infected with BmNPV for 8, 10, 14, or 20 h (columns 2 to 5, respectively). Cells were labeled with 10 μM BrdU for 30 min prior to fixation in 2% formalin. The fixed cells were permeabilized with acetone and treated with 4 N HCl for 10 min to expose incorporated BrdU residues. The cells were then incubated with anti-BrdU antibody followed by rhodamine red X-conjugated goat anti-mouse IgG. Bars, 10 μm.
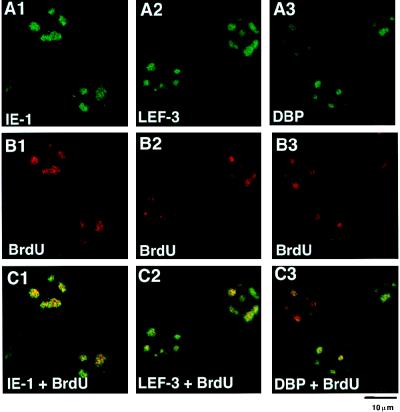
The unique pattern of DNA synthesis observed in BmNPV-infected cells was different from that resulting from host DNA replication and was likely due to viral DNA synthesis. To confirm this possibility, we performed double-staining experiments in which BrdU incorporation as well as IE-1, LEF-3, and DBP localization was determined. If the unique BrdU incorporation patterns were truly related to the sites of viral DNA replication, then early gene products that are involved in DNA replication were predicted to accumulate at these foci. Figure 4 shows that at 8 h p.i. the staining of IE-1 and LEF-3, gene products essential for baculovirus DNA synthesis (21), as well as that of DBP, strongly overlapped with the BrdU foci.
FIG. 4.
Double staining of BmNPV-infected BmN cells with BrdU and IE-1, LEF-3, or DBP. (A) Immunofluorescence images of BmNPV-infected BmN cells at 8 h p.i. with antibodies against IE-1 (A1), LEF-3 (A2), or DBP (A3). For IE-1 staining, FITC-conjugated goat anti-guinea pig IgG was used. For DBP and LEF-3 staining, FITC-conjugated goat anti-rabbit IgG was used. (B) BrdU immunofluorescence images of the same cells as in panel A. For BrdU staining, rhodamine red X-conjugated goat anti-mouse IgG was used. (C) Panels A and B merged.
These results indicated that viral DNA replication initiated at 8 h p.i. or earlier in conjunction with the accumulation of viral replicative proteins into specific foci. The time course of viral DNA replication based on BrdU incorporation at specific sites was comparable to that obtained by dot hybridization assay of BmNPV-infected BmN cells (16) and AcNPV-infected Sf-9 cells (40).
Colocalization of IE-1 and LEF-3 and DBP.
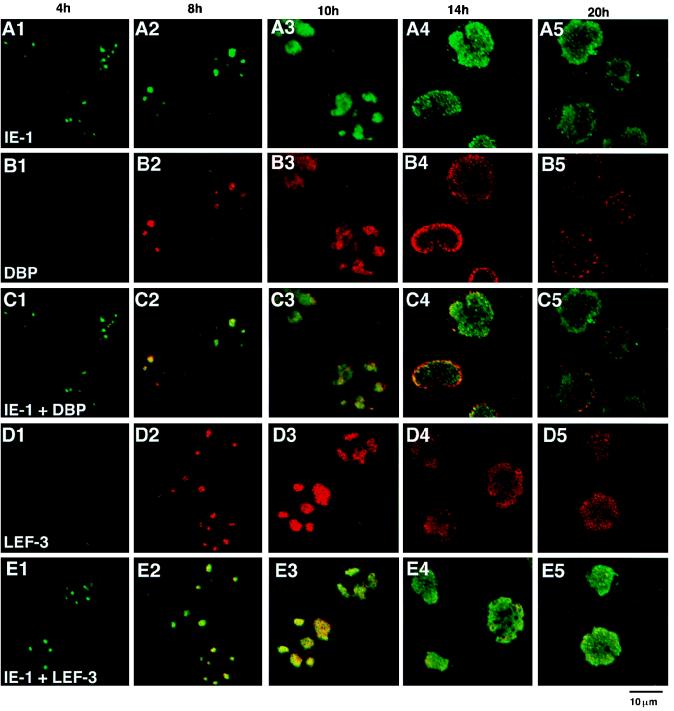
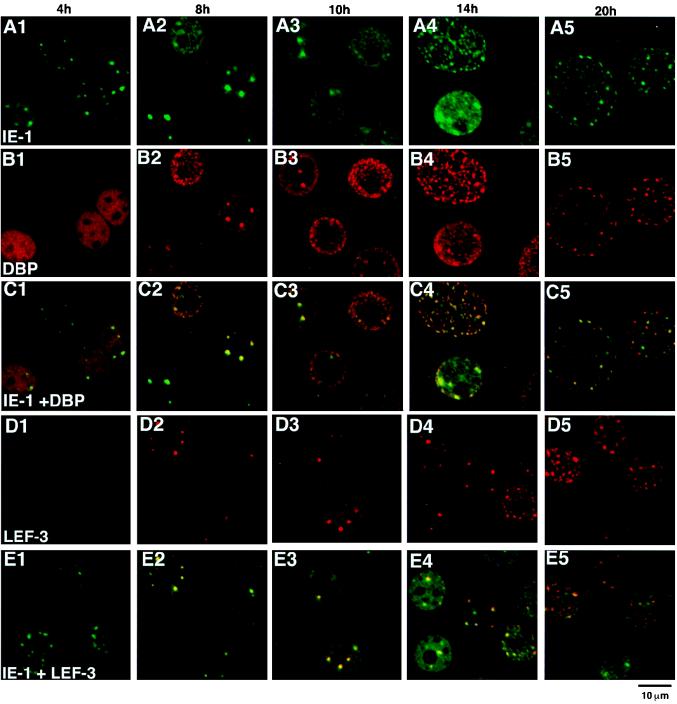
In order to address the question of whether the DBP antibody-stained regions always corresponded to those of IE-1 and LEF-3, double-staining experiments were carried out (Fig. 5). Between 8 and 10 h p.i., a period of rapid expansion of viral DNA replication sites, DBP and IE-1 (Fig. 5C2 and C3) and IE-1 and LEF-3 (Fig. 5E2 and E3) were found to colocalize. Therefore, all three of these proteins colocalized to the sites of viral DNA replication (Fig. 4). The distribution of these three proteins also overlapped at 14 h p.i., although DBP tended to accumulate closer to the nuclear membrane than did IE-1 at this time (Fig. 5, column 4). These results are consistent with the putative function of IE-1 and LEF-3 during viral DNA replication in infected cells and indicated that DBP is also associated with the viral DNA replication centers. In sharp contrast, the colocalization of IE-1, DBP, and LEF-3 was not observed at 4 h p.i., i.e., prior to the initiation of viral DNA replication. At 4 h p.i., IE-1 clearly localized at several foci (Fig. 5A1), whereas DBP stained weakly and appeared randomly throughout the nucleoplasm (Fig. 5B1), and LEF-3 was not detected (Fig. 5D1). LEF-3 immunostaining was initially observed at 6 h p.i. (data not shown). These results suggested that IE-1 initially localized to specific foci, followed by DBP and LEF-3, possibly resulting in the formation of a complex with IE-1 or with viral DNA. At later stages of infection, i.e., 20 h p.i., DBP appeared not to colocalize as strongly with IE-1 (Fig. 5C5).
FIG. 5.
Double staining of BmNPV-infected BmN cells with IE-1 and DBP or IE-1 and LEF-3. (A) IE-1 immunofluorescence images. (B) DBP immunofluorescence images of the same fields as in panel A. (C) Panels A and B merged. (D) LEF-3 immunofluorescence images. (E) Panel D and the corresponding IE-1 immunofluorescence images (not shown) merged. The time p.i. is shown at the top of each column. For IE-1 staining, FITC-conjugated goat anti-guinea pig IgG was used. For DBP and LEF-3 staining, Cy-5-conjugated goat anti-rabbit IgG was used.
Spatial localization.
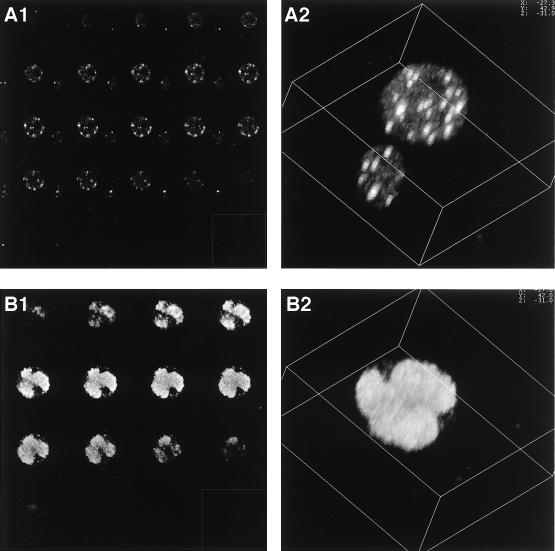
Figures 2, 3, 4, and 5 clearly show the temporal dynamics of the putative viral DNA replication centers and the association of DBP and other replication proteins. These figures, however, are the results of single optical sections taken by the laser confocal microscope; thus, the three-dimensional (3D) localization and structure are not clearly defined. Therefore, 3D images were made by computer-generated reconstitution of the optical sections (Fig. 6). IE-1-specific antibodies were used for this analysis, since IE-1 gave the strongest fluorescence signal (compared to DBP and LEF-3) at early stages of infection. Figure 6A2 shows the 3D image of an IE-1-stained cell at 4 h p.i. that was generated by combining 24 optical sections (Fig. 6A1). The 3D image revealed IE-1 foci which were usually less than 1 μm in diameter and arbitrarily distributed within the nucleus. Association with the nuclear membrane or nucleoli was not clearly observed. At 14 h p.i., IE-1 appeared to form a few partially fused egg-shaped masses, which occupied most of the nucleus (Fig. 6B2).
FIG. 6.
3D images of IE-1 distribution. (A) IE-1 immunofluorescence images taken at 4 h p.i. Twenty-four optical sections (A1) were used to reconstitute the 3D image of IE-1 foci observed at 4 h p.i. by using Leica 3D software. Even though the pinhole size was reduced to 80% of the optimum value for the lens (water immersion type PLAPO63) that was used, substantial blurring was observed along the z axis. However, the number of foci could still be accurately estimated. The distribution of the foci did not correlate with visible structures such as the nuclear membrane or nucleoli. (B) IE-1 fluorescence images taken at 14 h p.i. To reconstitute the 3D image of IE-1 foci at 14 h p.i. (B2), 16 optical sections (B1) were used.
MOI and distribution of IE-1.
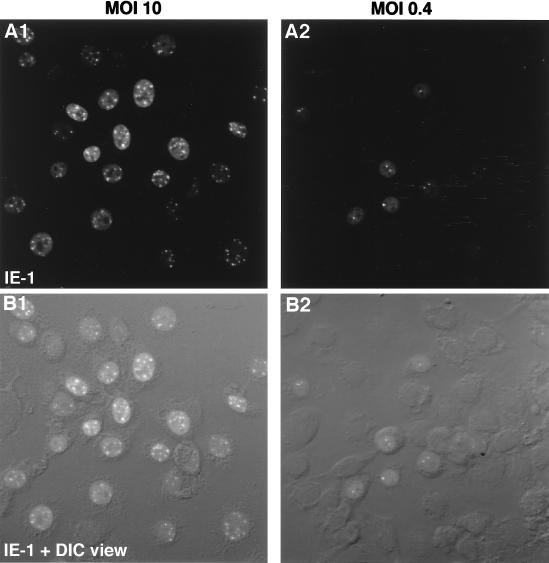
In the course of the trials to form the 3D images, it was found that the number of IE-1 foci at 4 h p.i. was accurately countable and in general less than 20. In addition, lowering the MOI appeared to result in a decrease in the number of foci. Therefore, the relationship between the number of foci and MOI was further characterized. Although 3D reconstitution gave an accurate count of the number of foci (Fig. 6A2), this procedure was time-consuming. Therefore, for these experiments, eight sections covering the entire nucleus were scanned at relatively low magnification, and then these sections were combined and projected onto one plane (Fig. 7). This procedure potentially underestimates the true number of foci by superimposing foci present at the same z axis; however, preliminary trials indicated that the small size and the limited number of foci minimized this type of error (data not shown). Figure 7 shows representative BmN cells following inoculation at MOIs of 0.4 and 10. Of 42 cells inoculated at an MOI of 0.4, three, two, and one focus were observed in one, three, and five cells, respectively (Fig. 7A2 and B2). No staining was observed in 33 cells, indicating that 80% of cells were not infected in this photograph. At an MOI of 10, an average of about 10 foci per cell was observed in 28 of 30 cells (Fig. 7A1 and B1).
FIG. 7.
Effect of MOI on the number of IE-1 foci at 4 h p.i. (A1 and B1) MOI of 10. (A2 and B2) MOI of 0.4. BmN cells were fixed at 4 h p.i. and subjected to immunofluorescence visualization. Primary antibody, anti IE-1; secondary antibody, FITC-conjugated goat anti-guinea pig IgG. To evaluate the number of IE-1 foci within the entire nuclei, eight consecutive fluorescence sections of cell nuclei were combined and projected onto a single plane by using Leica extended-focus software. (A) IE-1 immunofluorescence images. (B) Differential interface contrast images and the corresponding IE-1 immunofluorescence images from panel A merged.
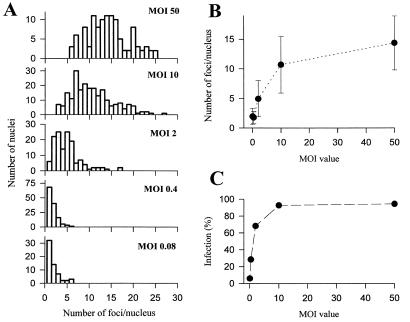
Quantitative analysis of the number of foci following inoculation at MOIs of 0.08, 0.4, 2, 10, and 50 is presented in Fig. 8. A large variance in number of foci per nucleus, ranging from 5 to 25, was observed at higher MOIs (MOIs of 10 and 50) (Fig. 8A). However, at MOIs of 0.4, 2, and 10, the average number of the foci per nucleus correlated well with the MOI value (Fig. 8A and B). Furthermore, increasing the MOI from 10 to 50 resulted in only a 35% increase in the average number of foci from 10.7 ± 4.8 (n = 228) at an MOI of 10 to 14.4 ± 4.6 (n = 110) at an MOI of 50, suggesting that the maximum number of foci per nucleus was around 15. This saturability characteristic was not due to incomplete infection at an MOI of 10, since the percentage of infected cells was around 94% (Fig. 8C). When the MOI was decreased to 0.08, the distribution histogram was indistinguishable from that for an MOI of 0.4 (Fig. 8A and B), while the percentage of infected cells decreased in response to the decrease in MOI (Fig. 8C). This indicated that when the viruses were adequately diluted, the number of foci per infected cell nucleus converged to one. It was also observed that larger nuclei contained more foci, whereas recently divided cells with smaller nuclei contained fewer foci, suggesting that the maximum number of foci per nucleus partly depended upon the nuclear volume. The saturability characteristics at high MOIs and dependence on nuclear volume suggested that the maximum number of foci was restricted by an intrinsic parameter of the infected cell. On the other hand, the fact that lowering the MOI caused convergence of the number of foci per infected cell to one suggested that the number of viral genomes which were able to enter the nucleus also had a critical influence on the formation of foci.
FIG. 8.
Quantitative analyses of the relationship between MOI and the number of IE-1 foci at 4 h p.i. (A) Distribution histogram. BmN cells infected with BmNPV at the indicated MOI were fixed at 4 h p.i. and subjected to immunofluorescence visualization. Primary antibody, anti IE-1; secondary antibody, FITC-conjugated goat anti-guinea pig IgG. Only infected cells containing nuclei with IE-1 foci (staining) were taken for the analysis. At least five images in each case were analyzed. Abscissas indicate the numbers of foci per nucleus (bin size = 1), and ordinates indicate the numbers of nuclei with corresponding numbers of foci per nucleus. (B) Relationship between MOI and the number of IE-1 foci per nucleus. (C) Relationship between MOI and infection percentage [(number of positively stained cells/number of total cells)] × 100].
Localization of DBP, LEF-3, and IE-1 in the absence of DNA synthesis.
If the association of DBP, LEF-3, and IE-1 is intrinsically important for viral DNA synthesis, the observed changes in the localization of these proteins (Fig. 5) may be disturbed by inhibiting viral DNA synthesis. In order to study the localization of DBP, LEF-3, and IE-1 in the absence of viral DNA replication, BmNPV-infected BmN cells were cultured in the presence of aphidicolin, an inhibitor of nuclear DNA replication in eukaryotic cells (18). Aphidicolin has been shown to efficiently block BmNPV DNA polymerase (35). In mock- and BmNPV-infected cells, the addition of 20 μM aphidicolin to the culture medium completely suppressed the incorporation of BrdU (data not shown).
At 4 h p.i., the presence of aphidicolin did not affect the formation of IE-1 foci (Fig. 9A1). At 8 h p.i., the foci formed by IE-1 and LEF-3 (Fig. 9E2) were indistinguishable from the IE-1–LEF-3 foci formed in the absence of aphidicolin (Fig. 5E2). DBP also began to colocalize with IE-1 (Fig. 9C2). These data suggested that aphidicolin treatment did not block the colocalization of DBP, LEF-3, and IE-1 at early times postinfection (i.e., at 8 h p.i. and earlier). However, the colocalization of DBP with IE-1 was not as efficient as that observed in the absence of aphidicolin (compare Fig. 9C2 with Fig. 5C2), and relatively high nucleoplasm and nuclear membrane staining was observed. In some cells (e.g., the uppermost cell in Fig. 9C2), sporadic nuclear staining began to be superimposed on and to obscure the IE-1–DBP foci.
At 10 and 14 h p.i., the formation of the DBP, LEF-3, and IE-1 foci differed depending upon the presence or absence of aphidicolin in the culture medium. In contrast to cells grown in the absence of aphidicolin (Fig. 5, columns 3 and 4), in which the foci formed by DBP, LEF-3, and IE-1 enlarged, the staining patterns of these three proteins became sporadic in the presence of aphidicolin (Fig. 9, columns 3 and 4). Deterioration of the DBP foci was most prominent. These results suggested that the enlargement of these foci was associated with DNA replication.
FIG. 9.
Distribution of DBP, LEF-3, and IE-1 in the presence of aphidicolin. Aphidicolin (20 μM) was added to the culture medium at time zero to inhibit viral and cellular DNA replication. (A) IE-1 immunofluorescence images. (B) DBP immunofluorescence images of the same fields as in panel A. (C) Panels A and B merged. (D) LEF-3 immunofluorescence images. (E) Panel D and the corresponding IE-1 immunofluorescence images (not shown) merged. The time p.i. is shown at the top of each column. For IE-1 staining, FITC-conjugated goat anti-guinea pig IgG was used. For DBP or LEF-3 staining, Cy-5-conjugated goat anti-rabbit IgG was used.
DISCUSSION
Functional role of DBP and assembly of virus replication factories.
We have previously shown that DBP strongly and preferentially binds to ssDNA and has the ability to destabilize partial DNA duplexes in vitro (36), all of which are known to be characteristics of SSBs (34). Since SSBs are essential components of viral DNA replication systems (10), DBP should colocalize with viral replication centers and hence other proteins necessary for viral DNA replication. In the present study, this assumption was tested, with antibodies raised against rDBP. The findings provided evidence that DBP colocalized to the infection-specific BrdU incorporation (Fig. 4). Furthermore, during active viral DNA replication periods (8 to 14 h p.i.), the DBP antibody-stained areas overlapped with the IE-1 and LEF-3 antibody-stained areas (Fig. 5), both of which were essential for plasmid replication in AcNPV transient replication assays (21). Aphidicolin also completely inhibited the enlargement of the foci formed by DBP as well as that of foci formed by IE-1 and LEF-3 (Fig. 9). These data are consistent with the putative role that DBP plays in viral DNA replication.
Interestingly, the dbp homolog in AcNPV, ORF25, is not one of nine viral genes previously reported as necessary or stimulatory for plasmid DNA replication from an AcNPV origin of replication (21). The requirement of DNA replication for late viral gene expression is well recognized in many systems (11) including AcNPV (28), while ORF25 is not listed as a late expression factor (lef) gene (28, 41). Therefore, the function of ORF25 in DNA replication and/or successive late or very late gene expression is not clear. Obviously, there are complexities arising from host cell dependence on the essentiality of lef genes (27). The absolute necessity of dnapol is also controversial on the basis of transient replication assays (4, 21, 28). Since similar studies have not been performed for BmNPV, it is difficult to determine whether factors essential for transient DNA replication are similar between BmNPV and AcNPV. On the other hand, additional factors besides those essential for the transient replication of plasmids may be required for the replication of the entire virus genome in vivo.
Although colocalization of DBP, IE-1, and LEF-3 was observed during DNA replication, such colocalization was not found either before the onset of DNA replication or after the termination of DNA replication. At 4 h p.i., only IE-1 localized into discrete foci, suggesting that IE-1 initiates assembly of viral replication factories. This is not surprising, since IE-1 binds the homologous regions of viral DNA and homologous regions may function as sites for DNA-binding proteins (26). However, the onset of viral DNA replication was delayed until 8 h p.i., at which time other proteins such as DBP and LEF-3 colocalized with IE-1. This suggests that baculovirus DNA replication requires the proper assembly of a multiprotein complex.
When aphidicolin was added to the culture medium, the association of LEF-3 and DBP with the IE-1 foci was unaffected at early stages of infection. This is consistent with the fact that aphidicolin blocks only DNA synthesis and late or very late gene expression, not early gene expression (39). However, the colocalization of DBP and IE-1 was somewhat weak, and fairly strong background nuclear staining was also observed. Furthermore, the staining patterns of these three DNA-binding proteins became sporadic at later stages of infection. If binding sites for these proteins are provided by nascent viral DNA, the presence of aphidicolin should inhibit the synthesis of nascent DNAs and they would have no place to accumulate.
At 14 h p.i., the distribution of DBP was more diffuse than that of IE-1 and LEF-3 in the absence of aphidicolin (Fig. 5C4). If DBP is an essential part of the BmNPV replication factory and tightly binds to viral DNA, the dispersion of DBP at 14 h p.i. was somewhat unexpected because viral DNA replication as detected by BrdU incorporation was still very active in most cells at this time (Fig. 3, column 4). Since DBP is very abundant in infected cells at 14 h p.i., excessive production of DBP may account for this dispersion. At 14 h p.i., an approximately seven times molar excess of DBP was present compared to LEF-3. The excess abundance of DBP is consistent with the proposed SSB function of this protein, since SSB should saturate all replication forks during intensive viral replication. On the other hand, the dispersal of DBP after 14 h p.i. may reflect its release from mature viral DNAs. This is consistent with the finding that DBP is not present in budded or occlusion-derived virions. The maturation process is also likely to be accompanied in part by proteolytic digestion of DBP, which was observed in infected cells after 14 h p.i. (Fig. 1A).
Preexisting nuclear sites.
Increasing the input of infectious virus from 10 to 50 per cell did not substantially increase the number of IE-1 foci at 4 h p.i. In fact, the number of IE-1 foci plateaued at around 15. Furthermore, the maximum number of foci appeared to partly depend on the nuclear volume. These features are reminiscent of the presence of specific and preexisting nuclear sites for virus replication (19). The association of the IE-1 foci with the nuclear membrane or nucleoli was not observed during the infection. During the S phase in uninfected cells (Fig. 3, column 1), hundreds of DNA replication sites exist in cell nuclei (38), making it unlikely that cellular DNA replication sites restrict the number of viral replication centers. Domains with a high concentration of the splicing factor SC35 reportedly have a frequency of 20 to 50 per nucleus (7), while the number of ND10s is about 10 to 15 in a wide variety of cell lines (6). ND10 has been estimated to be 0.2 to 0.5 μm in diameter (6). In BmN cells, the initial IE-1 sites were arbitrarily distributed and their sizes were usually less than 1 μm in diameter. Since fluorescence measurements can overestimate the size of the foci, the 1-μm-diameter size appears consistent with the predicted size of ND10. Although there are no reports that insect cells have nuclear structures similar to ND10, the periphery of ND10 is thought to be a favorable site to deposit viral DNA for many dsDNA viruses including herpes simplex virus type 1 (19, 33), adenovirus type 5 (12, 19), simian virus 40 (19), and human cytomegalovirus (1, 20).
If IE-1 initially localizes to specific nuclear domains and then recruits viral DNA to these sites, the number of IE-1 foci should be independent of MOI value. On the other hand, if viral DNAs initially associate with specific nuclear sites, with subsequent binding of nascent IE-1, lowering the MOI should decrease the number of IE-1 foci. The finding that lowering the MOI causes the number of foci to converge to one suggests that the number of viral genomes which enter the nucleus is crucial for the formation of the foci and supports the later case. If so, the sequence of events in the formation of the baculoviral DNA replication factory may be as follows: (i) viral DNAs uncoat, enter into the nucleus, and bind to saturable (specific) nuclear domains; (ii) early genes are transcribed by the host RNA polymerase II; (iii) IE-1 then binds the baculovirus replication origin(s) and forms the core of the DNA replication machinery; and (iv) LEF-3 and DBP as well as helicase-primase and polymerase are recruited.
ACKNOWLEDGMENTS
We are grateful to Ke-Qin Xin for help in antibody production, George F. Rohrmann for the gift of the antiserum against AcNPV LEF-3 and for critical reading of the manuscript, Hisanori Bando for the gift of the antiserum against BmNPV IE-1, Susumu Ikegami for the gift of aphidicolin, and Shizuo G. Kamita for critical reading of the manuscript. We also thank Masaaki Kurihara for help with the culture of BmN cells and viral inoculation.
This work was supported, in part, by a Japan Society for Promotion of Science fellowship to V.S.M. and by the grants from the COE (Center for Excellence) program of the Science and Technology Agency and the Institute of Physical and Chemical Research (RIKEN) to S.M.
Footnotes
K.O. and V.S.M. dedicate this paper to the memory and achievements of Susumu Maeda, who died unexpectedly on 26 March 1998.
REFERENCES
- 1.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrens C H, Carlson C, Rohrmann G F. Identification, sequence, and transcriptional analysis of lef-3, a gene essential for Orgyia pseudotsugata baculovirus DNA replication. Virology. 1995;210:372–382. doi: 10.1006/viro.1995.1353. [DOI] [PubMed] [Google Scholar]
- 3.Ahrens C H, Rohrmann G F. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudotsugata. J Gen Virol. 1996;77:825–837. doi: 10.1099/0022-1317-77-5-825. [DOI] [PubMed] [Google Scholar]
- 4.Ahrens C H, Rohrmann G F. Identification of essential trans-acting regions required for DNA replication of the Orgyia pseudotsugata multinucleocapsid nuclear polyhedrosis virus: lef-1 is an essential replication gene. Virology. 1995;207:417–428. doi: 10.1006/viro.1995.1101. [DOI] [PubMed] [Google Scholar]
- 5.Ahrens C H, Rohrmann G F. Replication of Orgyia pseudotsugata baculovirus DNA: lef-2 and ie-1 are essential and ie-2, p34, and Op-iap are stimulatory genes. Virology. 1995;212:650–662. doi: 10.1006/viro.1995.1523. [DOI] [PubMed] [Google Scholar]
- 6.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bando, H., et al. Unpublished data.
- 7.Bregman D B, Du L, van der Zee S, Warren S L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaeychomsri S, Ikeda M, Kobayashi M. Nucleotide sequence and transcriptional analysis of the DNA polymerase gene of Bombyx mori nuclear polyhedrosis virus. Virology. 1995;206:435–447. doi: 10.1016/s0042-6822(95)80059-x. [DOI] [PubMed] [Google Scholar]
- 9.Choi J, Guarino L A. Expression of the IE1 transactivator of Autographa californica nuclear polyhedrosis virus during viral infection. Virology. 1995;209:99–107. doi: 10.1006/viro.1995.1234. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 11.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 12.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 13.Evans J T, Leisy D J, Rohrmann G F. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. J Virol. 1997;71:3114–3119. doi: 10.1128/jvi.71.4.3114-3119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans J T, Rohrmann G F. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J Virol. 1997;71:3574–3579. doi: 10.1128/jvi.71.5.3574-3579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friesen P D. Regulation of baculovirus early gene expression. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 141–170. [Google Scholar]
- 16.Gomi S, Zhou C E, Yih W, Majima K, Maeda S. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology. 1997;230:35–47. doi: 10.1006/viro.1997.8457. [DOI] [PubMed] [Google Scholar]
- 17.Hang X, Dong W, Guarino L A. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J Virol. 1995;69:3924–3928. doi: 10.1128/jvi.69.6.3924-3928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 19.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishov A M, Stenberg R M, Maul G G. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol. 1997;138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kool M, Ahrens C H, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Laufs S, Lu A, Arrell K, Carstens E B. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology. 1997;228:98–106. doi: 10.1006/viro.1996.8361. [DOI] [PubMed] [Google Scholar]
- 24.Leisy D J, Rasmussen C, Kim H T, Rohrmann G F. The Autographa californica nuclear polyhedrosis virus homologous region 1a: identical sequences are essential for DNA replication activity and transcriptional enhancer function. Virology. 1995;208:742–752. doi: 10.1006/viro.1995.1206. [DOI] [PubMed] [Google Scholar]
- 25.Lu A, Carstens E B. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1991;181:336–347. doi: 10.1016/0042-6822(91)90500-b. [DOI] [PubMed] [Google Scholar]
- 26.Lu A, Krell P J, Vlak J M, Rohrmann G F. Baculovirus DNA replication. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 171–191. [Google Scholar]
- 27.Lu A, Miller L K. Differential requirements for baculovirus late expression factor genes in two cell lines. J Virol. 1995;69:6265–6272. doi: 10.1128/jvi.69.10.6265-6272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda S. Expression of foreign genes in insects using baculovirus vectors. Annu Rev Entomol. 1989;34:351–372. doi: 10.1146/annurev.en.34.010189.002031. [DOI] [PubMed] [Google Scholar]
- 30.Maeda S, Kamita S G, Kondo A. Host range expansion of Autographa californica nuclear polyhedrosis virus (NPV) following recombination of a 0.6-kilobase-pair DNA fragment originating from Bombyx mori NPV. J Virol. 1993;67:6234–6238. doi: 10.1128/jvi.67.10.6234-6238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S, Kawai T, Obinata M, Fujiwara H, Horiuchi T, Saeki Y, Sato Y, Furusawa M. Production of human alpha-interferon in silkworm using a baculovirus vector. Nature. 1985;315:592–594. doi: 10.1038/315592a0. [DOI] [PubMed] [Google Scholar]
- 32.Maeda S, Majima K. Molecular cloning and physical mapping of the genome of Bombyx mori nuclear polyhedrosis virus. J Gen Virol. 1990;71:1851–1855. doi: 10.1099/0022-1317-71-8-1851. [DOI] [PubMed] [Google Scholar]
- 33.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 34.Meyer R R, Laine P S. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–380. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikhailov V S, Marlyev K A, Ataeva J O, Kullyev P K, Atrazhev A M. Characterization of 3′→5′ exonuclease associated with DNA polymerase of silkworm nuclear polyhedrosis virus. Nucleic Acids Res. 1986;14:3841–3857. doi: 10.1093/nar/14.9.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikhailov V S, Mikhailova A L, Iwanaga M, Gomi S, Maeda S. Bombyx mori nucleopolyhedrovirus encodes a DNA-binding protein capable of destabilizing duplex DNA. J Virol. 1998;72:3107–3116. doi: 10.1128/jvi.72.4.3107-3116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura H, Morita T, Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- 38.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ooi B G, Miller L K. Regulation of host RNA levels during baculovirus infection. Virology. 1988;166:515–523. doi: 10.1016/0042-6822(88)90522-3. [DOI] [PubMed] [Google Scholar]
- 40.Tjia S T, Carstens E B, Doerfler W. Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus II. Virology. 1979;99:399–409. doi: 10.1016/0042-6822(79)90018-7. [DOI] [PubMed] [Google Scholar]
- 41.Todd J W, Passarelli A L, Lu A, Miller L K. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J Virol. 1996;70:2307–2317. doi: 10.1128/jvi.70.4.2307-2317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomalski M D, Wu J G, Miller L K. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology. 1988;167:591–600. [PubMed] [Google Scholar]