Abstract
Several compounds that specifically inhibited replication of the H1 and H2 subtypes of influenza virus type A were identified by screening a chemical library for antiviral activity. In single-cycle infections, the compounds inhibited virus-specific protein synthesis when added before or immediately after infection but were ineffective when added 30 min later, suggesting that an uncoating step was blocked. Sequencing of hemagglutinin (HA) genes of several independent mutant viruses resistant to the compounds revealed single amino acid changes that clustered in the stem region of the HA trimer in and near the HA2 fusion peptide. One of the compounds, an N-substituted piperidine, could be docked in a pocket in this region by computer-assisted molecular modeling. This compound blocked the fusogenic activity of HA, as evidenced by its inhibition of low-pH-induced cell-cell fusion in infected cell monolayers. An analog which was more effective than the parent compound in inhibiting virus replication was synthesized. It was also more effective in blocking other manifestations of the low-pH-induced conformational change in HA, including virus inactivation, virus-induced hemolysis of erythrocytes, and susceptibility of the HA to proteolytic degradation. Both compounds inhibited viral protein synthesis and replication more effectively in cells infected with a virus mutated in its M2 protein than with wild-type virus. The possible functional relationship between M2 and HA suggested by these results is discussed.
Infection of cells by influenza virus is initiated by binding of the viral hemagglutinin (HA) to sialic acid-containing receptors on the surface of the cell. HA anchored in the viral membrane is a trimer composed of identical monomers, each composed of two disulfide-linked polypeptides, HA1 and HA2, generated by proteolytic cleavage of the primary translation product, HA0. Following binding, the virus is internalized by endocytosis. Within the low-pH (5.0 to 5.5) environment of the endosome, the HA undergoes a conformational rearrangement which releases the hydrophobic NH2-terminal amino acid residues of HA2 from their buried position within the molecule at the interface of the HA trimer. This fusion peptide is then inserted into the endosomal membrane, which, after further multistep conformational changes in the protein and the target lipids, results in fusion of the viral and endosomal membranes and formation of a fusion pore (reviewed in references 9, 43, 48, 50, 51, 59–61, 63, 64). Concomitant with this process, the viral membrane-bound M2 protein acts as an ion channel (19, 35, 52, 56) for the uptake of protons into the interior of the virion, which results in the dissociation of the M1 protein from the viral ribonucleoproteins (RNPs) (6, 32). Following completion of the fusion process, the RNPs are released into the cytoplasm, where, free of bound M1, they are able to enter the nucleus and initiate mRNA synthesis (21, 32, 42).
Most of what is known about the conformational changes that accompany fusion has been determined from studies of the HA from the X31 strain of influenza A virus, an H3 subtype. The crystallographic structures of most of the molecule in its neutral-pH form (65) and of a soluble fragment in the low-pH form (7) have been solved for this strain. These and other studies (8, 13, 47) have demonstrated extensive rearrangement of HA2 residues at low pH with respect to their relative orientation as well as coil-coil formation, loop-to-helix transitions, and helix-to-loop transitions. Different HA subtypes also undergo the conformational change and subsequent fusion reaction but do so at different pHs and temperatures (18, 25, 38). Recent accounts have identified several compounds that inhibit viral infectivity by blocking the low-pH-induced conformation change of HA. In one report, computer-assisted modeling was used to identify a group of benzo- and hydroquinones that bind to and stabilize the native form of X31 HA, resulting in inhibition of viral infectivity (2, 22). In other reports, a quinolizidine-linked benzamide was shown to block the conformational change of H1 and H2, but not H3, subtypes of HA (29, 30).
Here we describe several compounds that inhibit infectivity of H1, H2, and to a lesser extent H3 subtypes. Studies in which virus or purified HA was exposed to low pH demonstrated that the compounds act by blocking the conformational change in the HA. Consistent with this mechanism of action, the compounds also blocked virus-induced hemolysis of erythrocytes (RBCs) and fusion of infected cells at low pH. Resistant mutants which had amino acid changes in and topologically near the fusion peptide of HA2 were obtained. Computer modeling predicted a potential binding site for the compounds in this region. Differential effects of the compounds on wild-type (wt) and mutant viruses suggest a possible functional relationship between the viral HA and M2 proteins.
MATERIALS AND METHODS
Cells and virus stocks.
MDCK cells were obtained from the American Type Culture Collection (ATCC). MDBK cells and the WSN strain of influenza A virus were provided by Robert Krug. Cells were grown in Dulbecco’s modified Eagle medium (DMEM; Cellgro; Mediatech) containing glutamine, penicillin, and streptomycin (complete DMEM) supplemented with 10% fetal calf serum (FCS; Gibco, BRL). All other influenza A and B strains were obtained from the ATCC. Stocks of ATCC viruses were prepared by infecting MDCK cells at a multiplicity of infection (MOI) of 0.002 to 0.01 and incubating them for 2 days at 37°C under 5% CO2 in complete DMEM containing 1 μg of trypsin (Worthington) per ml. The stocks were expanded by infecting 11-day-old embryonated eggs with 2 × 103 to 20 × 103 PFU/egg and collecting the allantoic fluid after 2 days at 37°C (no CO2). Stocks of WSN virus were grown in MDBK cells in the presence of complete DMEM–2% calf serum (Gibco) and 5% CO2.
Automated ELISAs.
Growth of WSN virus in MDBK cells after 18 h in the presence of test compounds was monitored by enzyme-linked immunosorbent assay (ELISA) in microtiter plates (4 × 104 cells/well, MOI of 0.01), using a primary monoclonal antibody specific for influenza A nucleoprotein (Biodesign, Kennebunk, Maine). The assay was developed with β-galactosidase-linked secondary antibody (American Qualex, La Mirada, Calif.) and the fluorogenic substrate 4-methyl-umbelliferyl-β-d-galactoside (Sigma, St. Louis, Mo.).
Plaque assays.
Serial dilutions of virus in phosphate-buffered saline (PBS) containing Ca2+, Mg2+, and 0.2% bovine serum albumin (BSA) were used to infect 106 MDCK cells plated in six-well, 35-mm-diameter tissue culture dishes for 1 h at 22°C. After removal of virus, cells were overlaid with 2 ml of 0.6% agarose containing 1× modified Eagle medium (Gibco), glutamine, penicillin, streptomycin, and 1 μg of trypsin per ml. Plaques developed after 2 days at 37°C and were fixed by treatment with 10% trichloroacetic acid for 10 min followed by 10 min in 0.5% crystal violet in 80% methanol–PBS.
[35S]methionine labeling of proteins synthesized in virus-infected cells.
MDCK or MDBK cells (2 × 105 cells/well, plated in 24-well dishes) were infected at an MOI of 1 to 10, as specified, for 1 h at 0°C. After removal of virus, 0.5 ml of complete DMEM (without serum) was added and the cells were incubated for various times at 37°C with or without inhibitory compounds. The media was then removed and replaced with 0.2 ml of methionine-free modified Eagle medium-glutamine containing 10 μCi of [35S]methionine. After 30 min at 37°C, the labeled medium was removed and the cells were lysed in 150 μl of Laemmli buffer (26).
SDS-PAGE.
The standard Tris-glycine buffer system (26) was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels contained 13% acrylamide and 0.1% bisacrylamide (20 by 20 cm) and were electrophoresed either overnight at 50 V or for 5 h at 35 mA. Gels were fixed in 10% acetic acid–40% methanol, immersed in 1 M sodium salicylate for 15 min, dried, and exposed to X-ray film at −70°C.
Selection of mutant FM viruses resistant to CL 61917, CL 62554, and amantadine.
Influenza A/FM/47 virus at an MOI of 0.1 was used to infect 106 MDCK cells in 35-mm-diameter dishes in the presence of either CL 61917 at 2 μg/ml, CL 62554 at 4 μg/ml, or amantadine at 0.2 μg/ml. After 2 days, lysates were used to reinfect fresh cells in the presence of the compounds at 4, 8, or 0.4 μg/ml, respectively. After another 2 days, the infection was repeated with the new lysates in the presence of the compounds at 50, 100, or 10 μg/ml, respectively. Lysates from this infection were used to produce viral plaques. One plaque from each infection was picked and again plaque purified. Virus stocks grown in both MDCK cells and eggs were prepared from these plaques. Compounds were maintained at 50, 100, or 10 μg/ml, respectively, throughout the growth of all viral stocks in both MDCK cells and eggs. Three independent CL 61917-resistant isolates (61917r-A, -B, and -C), two CL 62554-resistant isolates (62554r-A and -B), and one amantadine-resistant (Amr) virus was obtained; the Amr virus was subjected to a second round of selection, this time against CL 61917, to generate a mutant resistant to both compounds (Amr 61917r virus).
Cloning and sequencing of HA and M genes.
Five milliliters of lysate from 5 × 106 MDCK cells infected with wt or mutant virus was clarified by centrifugation at 2,000 × g for 10 min. The supernatants were centrifuged at 40,000 rpm in a Beckman SW50.1 rotor for 90 min at 4°C. The pelleted virus was extracted with RNAzol B (Cinna/Biotecx), and the RNA was precipitated with ethanol, dried, and dissolved in 100 μl of water. Two microliters of RNA was annealed to an oligonucleotide complementary to the 3′ end of the HA gene (viral RNA sense) and which also contained several nonviral restriction sites: CCGGGATCCTCTTCGAGCAAAAGCAGGGGAAAAT. cDNA was prepared according to the instructions of the manufacturer (Gibco, BRL), using murine leukemia virus reverse transcriptase. An aliquot of the reaction was used as template for PCR amplification using Vent DNA polymerase (New England Biolabs), the above oligonucleotide, and a second oligonucleotide complementary to the 5′ end of the HA viral RNA that also contained a T7 RNA polymerase promoter and additional restriction sites: AACTGCAGAAGCTTTAATACGACTCACTATAAGTAGAAACAAGGGTGTTTTTCCTTATATT. The amplified 1.8-kb PCR product was purified by agarose gel electrophoresis and electroelution onto DEAE-acetate membrane (Whatman). The recovered DNA was digested with BamHI and HindIII and cloned into similarly digested pUC 19/SP6 (pUC 19 containing an SP6 polymerase promoter) or pUC 119. Alternatively, it was inserted by blunt-end cloning into SmaI-digested pUC 118. Sequencing of double- and single-stranded DNA was performed with T7 DNA polymerase (United States Biochemicals). Mutant and wt M genes were cloned into the SphI and EcoRI sites of pUC 19/SP6 by using a similar approach. The 3′ oligonucleotide used was CGGAATTCTCTTCGAGCAAAAGCAGGTAGATATTG, and the 5′ oligonucleotide was GCATGCCGCATGCTAATACGACTCACTATAAGTAGAAACAAGGTAG.
Molecular modeling of HA structure. (i) Model building.
A homology model of the HA of FM virus was constructed based on the X-ray data available for the HA of the X31 (Aichi) strain of influenza A virus (65). Sequence alignment was done by using the algorithm of Smith and Waterman (45). Forty randomizations were performed to ensure that the homology was not spurious. Homology model building of the HA2 was done with WHATIF (55), which replaces the side chains of the template protein with those of the target protein. The resulting model was subjected to energy minimization using CHARMM (5), keeping all backbone atoms fixed and allowing the side chains to move.
(ii) Ligand docking.
The resulting model was then used to determine possible modes of interaction between the protein and inhibitor molecules. Shapesearch methodology (12) was used to identify a potential binding site on the protein which can then dock the inhibitor molecule by selecting from a pool of predetermined molecular conformations. Since many of the mutated residues in the viruses selected for resistance to the inhibitor were clustered in the same general area of HA2 close to (and including) the fusion peptide, this area was selected as a probable region of interaction with the inhibitor. A square grid consisting of points 1.5 Å apart was constructed in the available space in the center core of the protein; 20 grid points were retained for the analysis. The molecules were then docked by a pairwise graph-matching algorithm in which a minimum of four atom-grid correspondences must be made. The highest-scoring docking is presented. Ligand docking was also performed with the HA of X31 virus.
Virus-induced cell-cell fusion.
Confluent CV-1 cells in 35-mm-diameter dishes were infected with wt or mutant FM viruses at an MOI of 1. After incubation for 9 h at 37°C in complete DMEM–trypsin (1 μg/ml), the medium was replaced with prewarmed DMEM adjusted either to pH 5.0 with dilute acetic acid or left unchanged (pH 7.2), containing CL 61917 at 50 μg/ml where indicated. After incubation at 37°C for 10 min, the medium was removed and replaced with complete DMEM (pH 7.2)–2% FCS with or without CL 61917. After 2 h of incubation at 37°C, the medium was removed, and the cells were washed with PBS, then fixed with methanol-acetone (1:1) at −20°C for 5 min, and subjected to Giemsa staining.
Virus-induced hemolysis of RBCs.
Aliquots of 10 to 100 μl of undiluted virus stock (stocks varied from 2 × 106 to 2 × 108 PFU/ml) were mixed at 0°C with 400 μl of guinea pig RBCs diluted to 1% in a solution of PBS diluted 1:4 with normal saline (0.2× PBS). After 30 min, CL compounds (dissolved in dimethyl sulfoxide) were added and incubation was continued for 5 min at 0°C. The final dimethyl sulfoxide concentration was 0.5%. Then 50 μl of 0.4 M 2-[N-morpholino]ethanesulfonic acid (Sigma), pH 5.0, was added, and the mixtures were incubated at 37°C for 45 min. Controls included mixtures in which either virus or pH 5 buffer was replaced with an equal volume of 0.2× PBS. After centrifugation at 10,000 rpm for 30 s, the optical density at 540 nm (OD540) of the supernatant was determined. Control values were subtracted. Generally, the amount of virus used produced an OD540 of between 0.5 and 1.0. With some viruses, the HA titer was low and an OD540 of no more than 0.3 could be produced, even with 100 μl of virus. The IC50 was defined as that concentration of compound necessary to reduce the OD540 by 50%. To determine the pH of hemolysis of the various mutant viruses, the pH 5.0 buffer was replaced by buffers at pH 5.2, 5.4, 5.6, 5.8, 6.0, and 6.2.
Low-pH inactivation of influenza virus.
One million PFU of virus (100 μl) was incubated with or without CL compounds for 10 min at 22°C; 1.0 ml of DMEM (pH 5.0) with or without CL compounds was added, and incubation continued at 37°C for 15 min. Controls were incubated in neutral-pH DMEM. Virus was then serially diluted in PBS–0.2% BSA to ∼100 PFU/ml, and 0.5 ml was used to infect MDCK cells for plaque assay.
Purification of [35S]methionine-labeled HA from infected cells.
MDCK cells (2 × 106) were infected with either wt or 61917r-A FM virus at an MOI of 5. After incubation for 6 h at 37°C in complete DMEM, the medium was removed and replaced with methionine-free MEM-glutamine to which was added 1 mCi of [35S]methionine. After incubation for an additional 18 h, the cells were collected by centrifugation and frozen at −70°C. The cells were thawed and resuspended in 2 ml of buffer containing 2.5 mM HEPES (pH 7.5) and 150 mM NaCl. To enhance conversion of HA0 to HA1 plus HA2, trypsin was added to 10 μg/ml and the cells were incubated at 37°C. After 1 h, the cells were centrifuged and extracted in 2 ml of HEPES-NaCl buffer containing 0.5% Triton X-100 and soybean trypsin inhibitor (Sigma) at 25 μg/ml. The extract was centrifuged, and the supernatant applied to a column containing 0.3 ml of Ricinus communis lectin-agarose (Agglutinin RCA120; Sigma) equilibrated in HEPES-NaCl buffer. After washing of the column with this buffer, the HA was eluted with the same buffer containing 0.2 M d-(+)-galactose.
Proteinase K digestion of purified [35S]methionine-labeled HA.
Aliquots of 20 μl of purified HA were diluted to 50 μl with HEPES-NaCl buffer. Various amounts of inhibitor compounds were added as indicated; 1 μl of 3 M acetic acid-acetate buffer (pH 5.3) was added, which lowered the pH to 5.0. After incubation for 15 min at 37°C, the mixture was neutralized with 1 μl of 2 M Tris base. Proteinase K (Sigma) was added to 20 μg/ml, and the mixture was incubated at 37°C for another 30 min. BSA (100 μg) was added as carrier, and the proteins were precipitated with ice-cold 10% trichloroacetic acid. After centrifugation, the pellets were washed with acetone, dissolved in Laemmli buffer, and analyzed by SDS-PAGE.
Synthesis of CL 385319.
CL 385319 was prepared by reacting 1-(2-aminoethyl)piperidine (Aldrich) with 5-fluoro-3-trifluoromethylbenzoyl chloride (Lancaster) in methylene chloride at room temperature and collecting the hydrochloride product by filtration.
3H labeling of CL 61917.
Random tritiation of CL 61917 (10 Ci/mmol, 2 mCi/ml) was performed by Sibtech, Inc., Tenafly, N.J.
RESULTS
From an automated screen of compounds tested for growth inhibition against a panel of viruses, two compounds that specifically inhibited replication of the WSN strain of influenza A virus, an H1N1 subtype, emerged. CL 61917 (Fig. 1a) inhibited viral replication with an IC50 of 2 μg/ml (6 μM) as determined by either an 18-h growth assay (where growth was quantitated by ELISA) or by a plaque reduction assay. CL 62554 (Fig. 1b) was somewhat less effective, with an IC50 of 12 μg/ml (25 μM). When tested against other influenza A virus strains in plaque reduction assays, CL 61917 had similar inhibitory activities against H1 and H2 subtypes; it was much less effective against H3 subtypes and virtually ineffective against an influenza B virus (Table 1). The 50% cytotoxic concentration for both compounds against MDCK cells was about 250 μg/ml.
FIG. 1.
Inhibitors of influenza A virus replication.
TABLE 1.
Inhibition of influenza virus replication by CL compounds
| Compound | IC50 (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/WSN/33b | A/FM/47b | A/Japan/305/57c | A/Japan/170/62c | A/Hong Kong/8/68d | A/Victoria/3/75d | B/GL | FM mutant
|
||||
| Amr | Amr 61917r | 61917r-A | 62554r-A | ||||||||
| CL 61917 | 2 | 1.5 | 0.4 | 1 | 25 | 25 | 75 | 0.8 | 25 | 75 | 50 |
| CL 62554 | 12 | 7 | ND | ND | ND | ND | ND | 4 | ND | 75 | 50 |
| CL 385319 | 0.1 | 0.3 | ND | 0.7 | 25 | 25 | 50 | 0.15 | 2 | 50 | ND |
| Amantadine | ND | 0.05 | ND | ND | ND | ND | ND | ND | ND | 0.05 | 0.05 |
Determined by plaque reduction assays. For WSN virus, we also performed ELISAs, which gave IC50s identical to those obtained by plaque assay. ND, not done.
H1N1 strain.
H2N2 strain.
H3N2 strain.
Effects of CL 61917 and CL 62554 on WSN virus-specific protein synthesis.
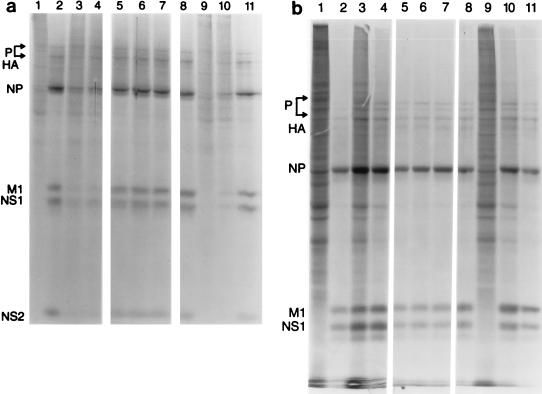
Time-of-addition experiments were done to identify the viral process(es) inhibited by the compounds. When added 1 h before infection (not shown) or immediately after removal of the infecting virus and then maintained in the media throughout the course of the experiment, CL 61917 and CL 62554 inhibited virus-specific protein synthesis occurring between 2.5 and 3 h postinfection (p.i.) (Fig. 2a, lanes 4 and 6). In contrast, when the compounds were added at 30 min p.i., no effect on viral protein synthesis was seen at any subsequent time point (Fig. 2b, lanes 4 to 7). These results suggest that both compounds act early in the viral life cycle, at a step subsequent to binding of the virus to the cell but probably before the onset of viral mRNA and protein synthesis (21, 49). Under the conditions tested, the CL compounds differ in the ability to block this early viral function because by 5 h p.i., protein synthesis had recovered to control levels in infected cells treated with CL 62554, whereas only partial recovery was seen in cells treated with the more potent CL 61917 (Fig. 2a; compare lanes 5 and 7 with lane 3).
FIG. 2.
Effects of inhibitory compounds on protein synthesis in MDBK cells infected with WSN virus. Cells were infected at an MOI of 1. Compounds (CL 61917 at 25 μg/ml; CL 62554 at 50 μg/ml) were added immediately after removal of the infecting virus but before incubation at 37°C (a) and after 30 min incubation at 37°C (b). Cells were labeled with [35S]methionine for 30 min. Lanes 2, 4, and 6, 2.5 h p.i.; lanes 3, 5, and 7, 5 h p.i. Lane 1, mock infected; lanes 2 and 3, infected, no compounds added; lanes 4 and 5, CL 61917 added; lanes 6 and 7, CL 62554 added. Lysates were analyzed by SDS-PAGE. None of the compounds had any effect on host-cell protein synthesis in mock-infected cells (not shown).
Generation of mutant viruses (FM strain) resistant to CL 61917 and CL 62554.
Two viral proteins, M2 and HA, are directly involved in early events in viral infection. To determine whether either of these proteins is the target of CL 61917 and CL 62554, mutants resistant to these compounds and to amantadine (which targets the M2 protein [19, 42]) were selected from the wt population by growth in the presence of compound. Because WSN virus is highly resistant to amantadine (28, 31), the FM strain of influenza A virus (H1N1 subtype) was used instead for all the mutant selections. Three mutant viruses resistant to CL 61917 (61917r-A, -B, and -C), two mutant viruses resistant to CL 62554 (62554r-A and -B, which had identical mutations [see below]), and an Amr virus were obtained. In addition, an Amr 61917r virus was isolated by growing the Amr mutant in the presence of CL 61917. Several of these viruses were assayed for sensitivity to the various inhibitors by a plaque reduction assay. The 61917r-A mutant was found to be cross-resistant to CL 62554, and the 62554r-A mutant was cross-resistant to CL 61917 (Table 1). These results suggest that both compounds target the same or a functionally related site on the same viral protein. Both mutants were as sensitive to amantadine as was wt virus. The Amr virus was somewhat more sensitive to both compounds than was wt FM virus. The Amr 61917r double mutant was more sensitive to CL 61917 than were the 61917r-A and 62554r-A viruses.
Effects of CL compounds and amantadine on wt and mutant FM virus-specific protein synthesis.
The properties of mutant and wt viruses were further characterized by monitoring virus-specific protein synthesis in MDCK cells at various times after infection (Fig. 3). Virus-specific protein synthesis in wt FM virus-infected cells is first detected at about 4 to 4.5 h p.i. (not shown). Both CL 61917 and CL 62554 significantly inhibited wt viral protein synthesis at 5 to 5.5 h p.i. (Fig. 3a, lanes 3 and 4). Viral protein synthesis in cells infected with 61917r-A virus was not inhibited by either CL compound at 5 to 5.5 h p.i., as expected (Fig. 3a, lanes 6 and 7). In cells infected with the Amr virus, protein synthesis was inhibited strongly by CL 61917, slightly less by CL 62554, and not at all by amantadine (Fig. 3a, lanes 9 to 11). At 14 to 14.5 h p.i., only CL 61917 continued to inhibit protein synthesis and did so only in cells infected with Amr virus (Fig. 3b, lane 9), suggesting that this mutant is particularly sensitive to inhibition by this compound. Neither of the compounds had any effect on protein synthesis in cells infected with influenza B virus (GL strain [data not shown]).
FIG. 3.
Effects of inhibitory compounds on protein synthesis in MDCK cells infected with wt and mutant FM viruses. Cells were mock infected (lane 1) or infected at an MOI of 10. Compounds (CL 61917 at 30 μg/ml; CL 62554 at 30 μg/ml; amantadine hydrochloride at 10 μg/ml) were added immediately after removal of virus but before incubation at 37°C. (a) Lanes 2 to 4, wt virus; lanes 5 to 7, 61917r-A virus; lanes 8 to 11, Amr virus. Lanes 3, 6, and 9, CL 61917 added; lanes 4, 7, and 10, CL 62554 added; lane 11, amantadine added. Cells were labeled with [35S]methionine for 30 min at 5 h p.i. (b) Like panel a except that cells were labeled at 14 h p.i.
Cloning and sequencing of HA and M genes.
To determine the precise mutations responsible for resistance to CL 61917, the HA genes of all 61917r and 62554r mutants as well as the HA and M genes of Amr and wt viruses were amplified by reverse transcription-PCR, cloned into pUC vectors, and sequenced. Five unique, single-base HA mutations that resulted in changes in the amino acid sequence were obtained (Table 2). Four of the mutations were in HA2, and one was near the NH2 terminus of HA1. In addition, all genes had several additional changes in amino acid sequence compared to the published sequence; these changes were, however, identical in the wt and mutant HAs. Whereas each of the mutations in the 61917r viruses was unique, the two 62554r virus isolates had the same mutation in HA. As expected, the Amr virus had a wt HA sequence and contained a mutation in the transmembrane region of the M2 protein.
TABLE 2.
Inhibition of hemolysis and pH 5 inactivation of wt and mutant FM viruses by CL 61917 and CL 385319a
| Virus | pH50 | IC50 (μg/ml)b
|
|||
|---|---|---|---|---|---|
| Hemolysis
|
pH 5 inactivation
|
||||
| CL 61917 | CL 385319 | CL 61917 | CL 385319 | ||
| FM (wt) | 5.82 | >50 | 12 | >100 | 2.5 |
| Mutants | |||||
| 61917r-A (HA2-N50D) | 6.12 | ≫50 | ≫50 | ≫500 | ≫500 |
| 61917r-B (HA2-Y34H) | ND | >50 | >50 | ND | ND |
| 61917r-C (HA1-L37F) | ND | 20 | 20 | ND | ND |
| 62554r-A and -B (HA2-F110S) | 5.76 | ≫50 | ≫50 | ND | ND |
| Amr (M2-S31N) | 5.82 | >50 | 12 | >100 | 1.25 |
| Amr 61917 (M2-S31N, HA2-F3L) | 5.90 | 12 | 4 | 25 | 1.25 |
Numbering of amino acids in HA1 begins with the initiator methionine counted as residue 1. Amino acid 1 of HA2 is the glycine residue C terminal to the cleavage site at arginine 344. Our clone of wt FM HA contained several differences, including HA1-S340P and HA2-W47G, plus a silent change of G to A at nucleotide 542, from the sequence available in GenBank (accession no. IVU02085).
>, inhibition was 20 to 40% at the indicated concentration; ≫, inhibition was 10% or less at the indicated concentration; ND, not done.
Computer-assisted molecular modeling.
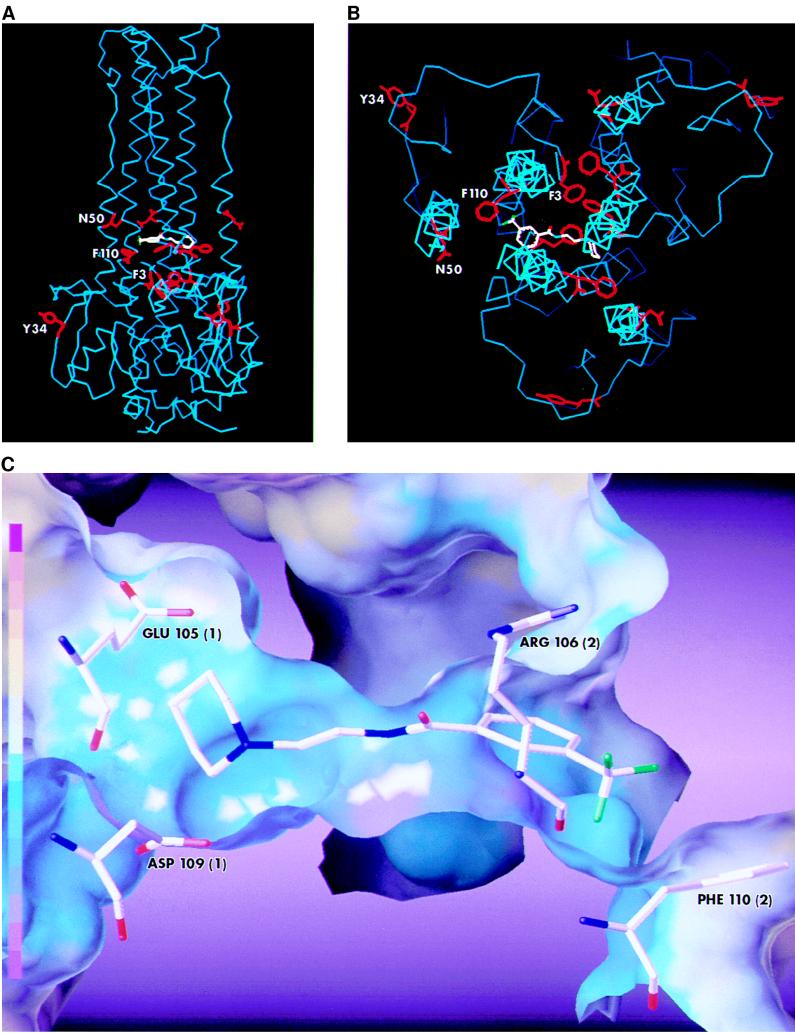
The neutral-pH structure of the HA of the X31 strain of influenza virus, an H3 subtype, has been solved to atomic resolution (65). If the HA of FM virus has a structure similar to that of the X31 virus, all five HA mutations (N50D, F110L, F3L, and Y34H in HA2 and L37F in HA1) in FM virus that render it resistant to CL 61917 would map to the stem of the HA trimer. Based on the >50% sequence identity between HA2 of the X31 and FM strains and the known crystal structure the former, a homology model of FM HA2 was constructed by computer-assisted molecular modeling. Using Shapesearch methodology (12), a docking site for CL 61917 was found in the middle of the stem region of the FM HA (Fig. 4A and B) near the HA2 fusion peptide that in X31 HA is known to undergo significant rearrangement during low-pH-induced fusion (7). The docking site is surrounded by three (Phe 3, Asn 50, and Phe 110) of the four HA2 amino acid residues that are altered in the resistant mutants. If the HA1 chains in the FM HA are arranged in the same way as they are in X31 HA (65), then Leu 37 in FM HA1 would also be situated very near the putative docking site (not shown). Close inspection of the binding site indicates that the compound interacts with the HA through both polar and hydrophobic forces (Fig. 4C). Glu 105 and Asp 109 from one of the monomer chains of HA2 can participate in a charge-charge interaction with the piperidine nitrogen, while Arg 106 from a second HA2 chain can hydrogen bond to the amide carbonyl oxygen of the compound. In addition, Phe 110 from the second chain forms part of a hydrophobic pocket into which the trifluorophenyl group is oriented. A similar docking attempt performed with the X31 HA yielded a significantly poorer fit, consistent with the fact that CL 61917 is a much poorer inhibitor of H3 viral subtypes than of H1 viruses (data not shown). The weaker binding may be ascribed to the fact that the interacting amino acid residues found in FM HA are absent in X31 HA; instead, X31 HA contains Gln 105, His 106, and Leu 110. In addition, four of the five amino acids whose mutation render FM HA resistant to the compound are also different in X31 HA.
FIG. 4.
Computer-generated model of the docking of CL 61917 in the stem of the FM HA. For purposes of clarity, only the backbones (colored blue) and selected side chains (colored red) of the three HA2 polypeptide chains in the HA trimer are depicted. (A) Side view; (B) view down the vertical axis (parts of the polypeptide backbones have been deleted for clarity). Because of the significant sequence differences between the X31 HA1 and FM HA1 polypeptide chains, modeling of the latter was not done. The side chains are those of the four amino acid residues whose mutation result in resistant virus. The labels indicate their position in one of the three polypeptides. CL 61917 (colored pink) is positioned between the three polypeptide chains below N50 and above F110. (C) Interactions between CL 61917 and selected residues in the putative binding site. Two acidic residues, Glu 105 and Asp 109, from one of the HA2 monomers (numbered arbitrarily as 1 in parentheses) are positioned to form a charge-charge interaction with the piperidine nitrogen. Arg 106 from another monomer (numbered arbitrarily as 2) is positioned so as to be able to hydrogen bond with the amide carbonyl oxygen. The trifluoromethylphenyl group is in a hydrophobic pocket which is partially formed by Phe 110 (from monomer 2). A solvent-accessible surface is shown around the binding pocket. The surface is color coded by electrostatic potential. The color coding scheme is shown by a color ramp at the left, with red the most positive and violet the most negative. The surface and rendering were done with the Sybyl software distributed by Tripos Associates, St. Louis, Mo.
Inhibition of cell-cell fusion by CL 61917.
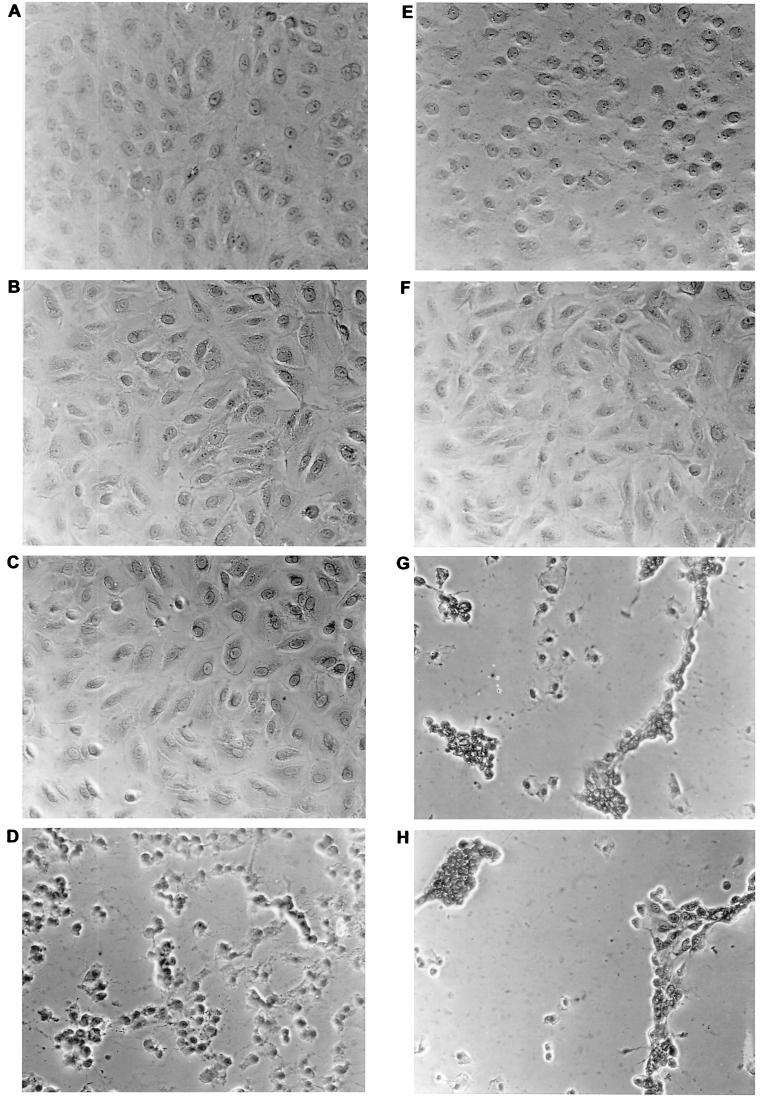
The region of the protein where the mutations cluster, together with the fact that CL 61917 inhibits viral protein synthesis when added immediately after infection but fails to inhibit when added 30 min after infection or later, suggested that the compound may act by inhibiting the fusogenic activity of HA. Fusion of viral and intracellular membranes occurs in the low-pH environment of the endosome and is a requirement for the release of the RNPs from the interior of the virus into the cytoplasm. Cell-to-cell fusion does not normally occur in influenza virus infection but can be induced by lowering the pH of the media (58). When wt virus-infected cells were incubated briefly at pH 5 and 37°C, extensive fusion and heterokaryon formation occurred (Fig. 5D). No fusion was observed with uninfected cells treated at pH 5 (Fig. 5B). In the presence of CL 61917, fusion and heterokaryon formation in virus-infected cells were completely blocked, although some morphological changes in the cell monolayer were observed (Fig. 5E). In contrast, fusion and heterokaryon formation in 61917r-A virus-infected cells could not be prevented by CL 61917 (Fig. 5H). At neutral pH, CL 61917 had no effect on the morphology of the cells (Fig. 5C). Amantadine at 10 μg/ml was completely ineffective in blocking fusion (not shown). The fact that the compound inhibits low-pH-induced fusogenic activity of HA bound to the outer cellular envelope strongly suggests that it also inhibits the same function of virion-bound HA when it is present within the endosome. In addition, binding of the compound to HA apparently occurs in a way that does not perturb the sialic acid-binding residues in the head of the HA trimer that are responsible for hemagglutination, because CL 61917 had no effect on the ability of virus to agglutinate RBCs (data not shown).
FIG. 5.
Inhibition of low-pH-induced cell-cell fusion of infected CV-1 cells by CL 61917. Cells were infected with either wt FM or 61917r-A virus at an MOI of 1. After 9 h, cells were incubated at 37°C with DMEM at pH 7 or pH 5 for 10 min, in the presence or absence of CL 61917 (50 μg/ml), and then incubated for 2 h at 37°C in complete DMEM (pH 7.2)–2% FCS either with or without CL 61917. (a) wt infected, pH 7; (b) uninfected, pH 5; (c) wt infected plus CL 61917, pH 7; (d) wt infected, pH 5; (e) wt infected plus CL 61917, pH 5; (f) 61917r-A infected, pH 7; (g) 61917r-A infected, pH 5; (h) 61917r-A infected plus CL 61917, pH 5.
Inhibition of viral replication and protein synthesis by CL 385319.
In an attempt to obtain compounds with improved antiviral efficacy, CL 385319, the 5-fluoro analog of CL 61917, was synthesized (Fig. 1c), and its IC50s against various H1, H2, and H3 viruses were determined by plaque reduction assays. The 50% cytotoxic concentration against MDCK cells was about 125 μg/ml. As was the case with CL 61917, CL 385319 was most effective against H1 and H2 viruses: for wt FM virus, the IC50 was about fivefold lower than that of CL 61917 (Table 1). Inspection of the computer-generated homology model indicated that the 5-fluoro group of CL 385319 can form an additional hydrogen bond with the N-terminal glycine of HA2, which may explain its greater potency (data not shown). 61917r-A virus was nearly as resistant to CL 385319 as it was to CL 61917. In addition, like CL 61917, CL 385319 inhibited the Amr virus somewhat more effectively than wt virus (Table 1). To confirm this apparent difference, CL 385319 and CL 61917 were tested for the ability to inhibit viral protein synthesis in cells infected with either wt or Amr virus. CL 385319 inhibited protein synthesis in Amr virus-infected cells more effectively (IC50 of 0.3 to 0.6 μg/ml) than in wt virus-infected cells (IC50 of 0.6 to 1.25 μg/ml). CL 61917 was also more effective against Amr virus-infected cells (IC50 of 1.25 to 2.5 μg/ml) than against wt virus-infected cells (IC50 of 2.5 to 5 μg/ml) (data not shown; IC50s estimated from the relative intensity of the viral protein bands in SDS-gels). These IC50s are only severalfold higher than the corresponding ones determined in multicycle infections by plaque reduction assay. CL 385319 did not inhibit viral protein synthesis in cells infected with 61917r-A virus (data not shown). In cells infected with the Amr 61917r double mutant, CL 61917 did not inhibit protein synthesis but CL 385319 inhibited strongly, consistent with the >10-fold-lower IC50 for the latter compound in plaque reduction assays (Table 1 and data not shown).
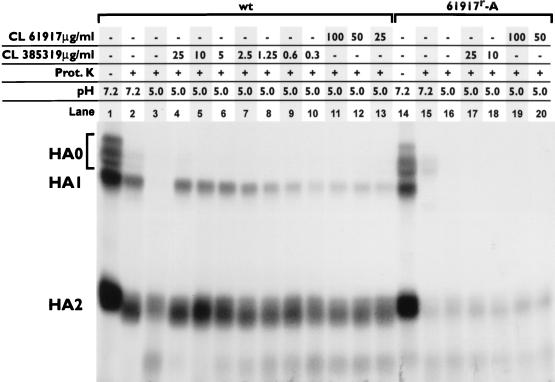
Proteinase K digestion.
Maturation of the HA protein is a multistep process. Following synthesis, the primary HA translation product HA0 is glycosylated in the endoplasmic reticulum, where it folds into its correct tertiary structure and then trimerizes. The trimers are transported through the Golgi apparatus, where further carbohydrate modifications occur, and are then exported to the cell membrane (reviewed in reference 40). Depending on the viral subtype and host cell, each monomer of trimeric HA0 is proteolytically cleaved into disulfide-linked HA1 and HA2 either intracellularly prior to export, on the cell surface by extracellular proteases, or at the stage of virus entry into target cells (3, 24). Following cleavage, the trimer reorganizes and becomes resistant to further proteolytic cleavage. If the protein is acidified, however, the ensuing conformational changes make the protein susceptible to extensive proteolytic degradation (44, 62). To determine whether the CL compounds could block this low-pH-induced sensitivity to proteolytic degradation, [35S]methionine-labeled HA from wt or 61917r-A virus-infected cells was purified by affinity chromatography, incubated at pH 5 with various amounts of CL 385319 or CL 61917, neutralized, and then digested with proteinase K. As purified from the infected cell extract, the protein contained substantial amounts of immature and incompletely processed HA0 (notwithstanding trypsin treatment during isolation) which was susceptible to proteinase K digestion even when the protein was maintained at neutral pH (Fig. 6; compare lanes 1 and 2). After incubation at pH 5, more extensive digestion of HA occurred: HA1 was completely and HA2 was partially digested by proteinase K (lane 3). In the presence of CL 385319 at concentrations of >5 μg/ml, nearly complete protection against this pH 5-promoted digestion of HA1 was attained, with 50% protection occurring at 1.25 to 2.5 μg/ml. CL 61917 was less effective, however, and only partially protected the protein, even at a concentration of 100 μg/ml (lane 11). The HA from 61917r-A virus-infected cells was much more sensitive to proteolytic digestion than wt HA, so that almost complete digestion of HA1 occurred even at neutral pH (lane 15), and neither compound protected the protein.
FIG. 6.
Inhibition of proteolytic digestion of purified HA by CL 61917 and CL 385319. Proteinase (Prot.) K digestion of HA purified from either wt or 61917r-A virus was performed as described in the text. The concentrations of the CL compounds are indicated, as is the pH to which the HA was exposed prior to proteolysis.
Inhibition of low-pH-induced viral inactivation and viral hemolysis of RBCs.
In the absence of target membranes with which to fuse, most HA subtypes are irreversibly inactivated by the conformational changes resulting from the exposure to low pH, rendering the virus uninfectious (18, 39, 46). In the presence of RBCs, fusion between the viral and cellular membranes occurs, resulting in hemolysis of the cells and release of their hemoglobin-colored contents to the surrounding solution (1). To determine the ability of the compounds to block these processes, wt and mutant FM viruses were incubated with the compounds at pH 5 either in the absence or in the presence of RBCs. The results are expressed as IC50 (the concentration of compound at which either loss of infectivity or hemolysis was inhibited by 50%) (Table 2). CL 385319 was quite effective in inhibiting low-pH inactivation of wt and Amr virus, with IC50s only about 10 times higher than those required to inhibit viral growth in plaque assays (Table 1). As in other assays, the compound was somewhat more effective against the Amr virus than against wt virus. The fact that the IC50 for protection of wt virus by CL 385319 against low-pH inactivation is very nearly the same as the IC50 estimated for the protection of purified wt HA against proteinase K digestion (Fig. 6) strongly suggests that regardless of whether the HA is inserted into the viral membrane or is free in solution, the binding site in the protein for the compound is accessible. Surprisingly, even at 100 μg/ml, CL 61917 was unable to inhibit inactivation of either wt or Amr virus. As expected, both compounds were also ineffective in inhibiting inactivation of 61917r-A virus. The Amr 61917r double mutant behaved anomalously; it was more sensitive to both compounds than wt virus.
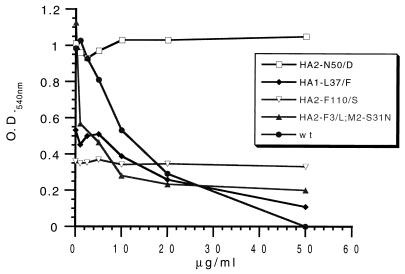
In hemolysis assays, we first determined the ability of wt and mutant FM viruses to hemolyze RBCs over a range of pHs, and these results are expressed in Table 2 as the pH at which hemolysis is 50% of maximal (pH50). In two of the mutants, the pH50 is higher than that of wt; in one mutant it is lower. The pH50 for the Amr virus was identical to that of wt virus. Changes in the pH of hemolysis are a hallmark of fusion mutants (11). CL 61917 and CL 385319 were then tested for their effects on hemolysis at pH 5.0. The inhibition profile of CL 385319 against several of the viruses is shown in Fig. 7, and the estimated IC50s for both compounds against these and additional viruses are summarized in Table 2. Both compounds exhibited identical IC50s against wt and Amr virus; this contrasts with the somewhat greater potency of these compounds against Amr virus than wt virus as measured in other assays. Three of the 61917r viruses (HA2-N50D, HA2-Y34H, and HA2-F110S) were resistant to high concentrations of both compounds, as would be expected. One of the 61917r viruses (HA1-L37F) however, was moderately sensitive. As in the inactivation assay, the double mutant was more sensitive to both compounds than wt virus.
FIG. 7.
Effect of CL 385319 on hemolysis of RBCs by wt and mutant FM viruses. Assays were performed as described in the text.
The IC50s of the compounds determined in both hemolysis and (to a lesser extent) viral inactivation assays are significantly higher (except for the double mutant) than those in multicycle growth assays or even in single-cycle protein synthesis assays. In a single-cycle infection, the virion-bound HA molecules present in the endosome shortly after infection are the same as those previously exposed to the external media and would be expected to be equally susceptible to the effects of an inhibitory compound. The fact that the IC50s differ suggests that the external environment to which either free HA or HA bound to the viral membrane is exposed in these in vitro assays may not fully reflect the endosomal environment in which fusion occurs in vivo. Possible explanations for the lower IC50s in vivo are (i) the HA within the endosome is more sensitive to inhibition because of some aspect of the intracellular environment, (ii) the inhibitors are metabolized within the cell to more active forms, and (iii) the inhibitors are selectively concentrated in the cell. In initial experiments to determine whether the latter hypothesis is correct, 3H-labeled CL 61917 was incubated with both infected and uninfected cells for various times and with various concentrations of unlabeled CL 61917. An approximate 100-fold selective concentration of the compound in both infected and uninfected cells was found (data not shown). Thus, it appears that viral replication is indeed inhibited by CL 61917 by virtue of its action against HA-driven fusion, but its potency is a function of its selective uptake by cells from the surrounding media.
DISCUSSION
We have found an N-substituted piperidine, CL 61917, that inhibits the infectivity of several H1, H2, and to a lesser extent H3 influenza A viruses. Inhibition appears to result from the compound’s ability to interfere with the fusogenic function of the viral HA. This was demonstrated by the ability of the compound, or its more potent analog CL 385319, to inhibit various manifestations of the fusogenic activity of a representative H1 virus, including low-pH-induced hemolysis of RBCs by virus, infected cell-cell fusion, and low-pH-induced inactivation of viral infectivity. In addition, inhibition of the low-pH-induced conformational change that is a prerequisite for fusion activity was directly demonstrated by the ability of the compound to protect purified HA against proteolytic digestion. Another compound, CL 62554, structurally unrelated to CL 61917, also inhibited viral replication. A mutant selected for resistance to this compound was also resistant to CL 61917. The mutation, F110S in HA2, is found in the same vicinity of the HA structure as the mutations in the viruses selected for resistance to CL 61917. At least one of these mutants, HA2-N50D, was also resistant to CL 62554. These results suggest that CL 62554 may bind at the same site as CL 61917 or at a nearby site, but we have not investigated this further.
Computer modeling of the HA of the FM virus subtype used in this study predicts a binding site for CL 61917 in the middle of the stem of the HA trimer in the vicinity of the buried fusion peptide. This site is fairly close to but not identical with the putative binding site of hydroquinone inhibitors in the X31 virus, a H3 subtype (2, 22). The likelihood that CL 61917 actually binds at this site is enhanced by the fact that four of five mutant viruses resistant to inhibition of infectivity by the compound have changes in amino acid residues that are near or surround the binding pocket. Because of its pivotal position, it is not unreasonable that occupation of this pocket by a small molecule could interfere with the structural rearrangements induced by low pH, either by disruption of the ionic and hydrophobic forces that maintain the protein in its prefusogenic state or by physically blocking the movements of the polypeptide chain during the conformational reorganization.
Alternative explanations for the effects of CL 61917 and CL 385319 on viral infectivity can be posited. Both compounds are weak bases. Other weak bases have been shown to inhibit influenza virus replication by raising the intraendosomal pH, thereby preventing the conformational change in HA (44). At high concentration, the compound norakin exerted its antiviral effect by raising the intraendosomal pH (34). Amantadine, another weak base, which at low concentration targets the M2 protein, can, at high concentration, also raise the intraendosomal pH (20). HA mutants that have a high pH of fusion can be selected when virus is grown at high norakin (36, 37) or amantadine (11) concentrations. One of the CL 61917-resistant mutants had a pH of fusion significantly higher than that of wt; the pHs of fusion of two other mutants, however, were only slightly above or below that of wt. While the possibility that the CL compounds raise the intraendosomal pH cannot be ruled out, this effect on viral replication, if any, is probably secondary to the demonstrated ability of the compounds (in particular CL 385319) to directly block the conformational change of the HA. In addition, if inhibition was due solely to an endosomal pH change, the compounds would be expected to be as effective against H3 viruses as against H1 and H2 viruses. Another formal possibility is that notwithstanding the computer-generated model, the compounds do not bind at the site proposed above but rather at some alternative site, whose ability to bind the compounds is reduced by mutations in amino acid residues elsewhere in the protein.
CL 61917 partially protects HA against low-pH-induced proteolytic digestion by proteinase K. However, it is unable to inhibit low-pH inactivation of wt or Amr virus, even at high concentration. This suggests that a critical concentration of functional HA molecules on the viral membrane is required to effect fusion. If the majority of the HA molecules have been rendered fusogenically inactive by low pH, the remaining fusogenically competent molecules (those protected by the compound) are insufficient to effect fusion in the endosome and initiate viral replication. Fusion may in fact require all, or virtually all, of the available membrane-bound HA. This is illustrated by the effect of CL 61917 on low-pH-induced fusion of virus-infected cells. Even though the compound is presumably capable of blocking only a fraction of the HA molecules on the cell surface from undergoing the low-pH-induced conformational change, it is still capable of blocking cell-cell fusion and heterokaryon formation. These results can be explained by the fact that fusion has been shown to require the cooperation of three or more HA trimers to form the fusion pore (10, 14); thus, even a small reduction in the density of functional HA molecules could seriously impair such a cooperative process. A much lower concentration of the compound is sufficient to block fusion and virus replication within infected cells, but this is presumably due to the apparent 100-fold selective uptake of the compound, resulting in very high intracellular concentrations. CL 61917 is also a weak inhibitor of virus-induced RBC hemolysis. The fact that some hemolysis occurs in the presence of the compound is not necessarily inconsistent with its effectiveness in blocking heterokaryon formation—these are two different morphological changes that may reflect vastly different levels of fusogenic activity by HA. In contrast, CL 385319 is able to block low-pH inactivation of virus and fully protects HA from proteolytic digestion. This greater potency is presumably due to its ability to bind more tightly to the HA; in computer-aided modeling studies, the 5-fluorophenyl group is in position to form an additional hydrogen bond with the NH2 group of the N-terminal glycine of HA2.
Unlike wt virus, the HA of 61917r-A virus is sensitive to proteinase K digestion even at neutral pH. This suggests that the N50D mutation destabilizes the protein, thereby making the polypeptide chains more accessible to the protease. This result is consistent with the fact that the pH50 of fusion of this virus is 0.2 pH unit higher than for wt virus (Table 2). Mutations that raise the pH of fusion have been shown to destabilize the HA (11, 50).
Two recent reports describe the properties of another compound, BMY-27709, that can also inhibit the fusogenic activity of influenza A virus HA (29, 30). The compound, like the CL compounds described here, is specific for H1 and H2 viral subtypes and does not inhibit influenza B virus replication. BMY-27709 is structurally similar to the CL compounds in that it is composed of a substituted benzamide linked to a nitrogen-containing heterocyclic ring structure, although the substitutions and linkages differ in nature. Computer-aided modeling of the binding of this compound to a homology model of the HA of WSN virus used in that study (which is 90% identical to the FM HA) demonstrated hydrophobic and ionic interactions remarkably similar to those described here between the CL compounds and FM HA, involving the same amino acid residues. In addition, several of the mutant viruses resistant to BMY-27709 had changes in the same amino acid residues as in the mutants described in this report (HA2-N50 and F-110). One of the mutations that rendered WSN virus resistant to BMY-27709, I19V in HA1 (amino acid 19 is equivalent to amino acid 36 in our numbering system, which includes the signal peptide and N-terminal methionine), illustrates the close functional similarity between the BMY and CL compounds. The I19V mutation in this virus results in adjacent amino acid residues (V19 and F20) that are identical to those in the FM mutant HA1-L37F (V36 and F37). Thus, although each compound selected for different mutations in different viral HAs, the fact that the resulting amino acid sequences in both HA1 polypeptides were the same suggests that the binding of both compounds is affected by this region of the HA. Another mutation, however, illustrates the need for caution in making generalizations about the effects of mutations in one virus on different viral subtypes. The mutation F110S in HA2, which was identical in both viruses, raised the pH of fusion of WSN HA from 5.65 to 5.95 but lowered that of FM HA from 5.82 to 5.76. As mentioned previously, mutations that raise the pH of fusion destabilize the HA with respect to pH (11, 50). Thus, the same mutation, even in similar viral subtypes, can result in important functional differences.
In assays that assessed the ability of the compounds to block virus-specific protein synthesis, the Amr virus (which contains a wt HA) was several times more sensitive than wt virus to both CL 61917 and CL 385319. The S31N mutation in M2, which renders the protein Amr, when present in the Rostock and Weybridge strains of influenza fowl plague virus, has been shown to reduce the ion channel activity of the protein (17, 23). The increased sensitivity of the Amr mutant to the compounds might be explained if this mutation also reduced the ion channel activity of the M2 protein of FM virus. M2 has been shown to raise the intraluminal pH of the trans-Golgi network through which the nascent HA is transported, thereby preventing acidification and inactivation of the HA (of some, but not all, subtypes) before it reaches the cell surface (33, 53, 54). In the endosome, the ion channel activity of M2 regulates the low-pH-induced dissociation of M1 from the viral RNPs. Because this process must be completed before formation of the fusion pore and exposure of the RNPs to the neutral pH of the cytoplasm, it is possible that the temporal order of M1-RNP dissociation and fusion pore formation are controlled by the same mechanism—M2 regulation of the rate and extent of intravirion acidification. If a fully functional M2 is required to optimize the rate of viral fusion with endosomes, it is possible that when M2 activity is suboptimal, the effects of compounds that interfere with HA activity are enhanced, requiring lower concentrations to effect the same response. This idea is supported by the fact that amantadine slows the rate of fusion of virus with liposomes (4, 57), directly implicating M2 as a facilitator of the fusion process, possibly by promoting a low-pH-induced weakening of an association of the HA with the M1 protein (15, 27, 67).
In the case of the Amr 61917r double mutant, sensitivity of the virus to inhibition of viral replication by the CL compounds was reduced about 15- to 30-fold compared to the Amr virus (Table 1). Conversely, sensitivity of the double mutant to both compounds in hemolysis and low-pH inactivation assays was equal to or enhanced compared to the Amr virus (Table 2). These apparently contradictory results are not easily reconciled. They suggest that when the virus is exposed to exogenously added pH 5 buffer, the F3L mutation somehow acts to enhance binding of the compound, whereas within the highly regulated endosomal environment the same mutation reduces the binding affinity. Further study of the effects of the compounds on the conformational change of the HA of this virus as a function of time, temperature, and pH may be instructive. It would also be useful to determine the effect of this mutation in a virus lacking the mutation in M2. Ultimately, clarification of the precise interactions between the inhibitors and wt and mutant HAs must await the outcome of cocrystallization studies.
Footnotes
This work is dedicated to the memory of Yakov “Yasha” Gluzman.
REFERENCES
- 1.Barrett T, Inglis S C. Growth, purification and titration of influenza viruses. In: Mahy B W J, editor. Virology: a practical approach. Oxford, England: IRL Press; 1985. pp. 119–150. [Google Scholar]
- 2.Bodian D L, Yamasaki R B, Buswell R L, Stearns J F, White J M, Kuntz I D. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 3.Boycott R, Klenk H-D, Ohuchi M. Cell tropism of influenza virus mediated by hemagglutinin activation at the stage of virus entry. Virology. 1994;203:313–319. doi: 10.1006/viro.1994.1489. [DOI] [PubMed] [Google Scholar]
- 4.Bron R, Kendal A P, Klenk H-D, Wilschut J. Role of the M2 protein in influenza virus membrane fusion: effects of amantadine and monensin on fusion kinetics. Virology. 1993;195:808–811. doi: 10.1006/viro.1993.1435. [DOI] [PubMed] [Google Scholar]
- 5.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 6.Bukrinskaya A G, Vorkunova N K, Kornilayeva G V, Narmanbetova R A, Vorkunova G K. Influenza virus uncoating and effects of rimantidine. J Gen Virol. 1982;60:49–59. doi: 10.1099/0022-1317-60-1-49. [DOI] [PubMed] [Google Scholar]
- 7.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 8.Carr C M, Kim P S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 9.Carr C M, Kim P S. Flu virus invasion; halfway there. Science. 1994;266:234–236. doi: 10.1126/science.7939658. [DOI] [PubMed] [Google Scholar]
- 10.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels R S, Downle J C, Hay A J, Knossow M, Skehel J J, Wang M L, Wiley D C. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 12.Desjarlais R L, Sheridan R P, Dixon J S, Kuntz I D, Venkataraghavan R. Docking flexible ligands to macromolecular receptors by molecular shape. J Med Chem. 1986;29:2149–2153. doi: 10.1021/jm00161a004. [DOI] [PubMed] [Google Scholar]
- 13.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 14.Ellens H, Bentz J, Mason D, Zhang F, White J M. Fusion of influenza-expressing fibroblasts with glycophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- 15.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghendon Y, Markushin S, Heider H, Melnikov S, Lotte V. Hemagglutinin of influenza A virus is a target for the antiviral effect of norakin. J Gen Virol. 1986;76:1115–1122. doi: 10.1099/0022-1317-67-6-1115. [DOI] [PubMed] [Google Scholar]
- 17.Grambas S, Bennett M S, Hay A J. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 18.Gutman O, Danieli T, White J M, Henis Y I. Effects of exposure to low pH on the lateral mobility of influenza hemagglutinin expressed at the cell surface: correlation between mobility inhibition and inactivation. Biochemistry. 1993;32:101–106. doi: 10.1021/bi00052a014. [DOI] [PubMed] [Google Scholar]
- 19.Hay A J. The action of adamantanamines against influenza A viruses: inhibition of the M2 ion channel protein. Semin Virol. 1992;3:21–30. [Google Scholar]
- 20.Hay A J, Walstenholme A J, Skehel J J, Smith M H. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman L R, Kuntz I D, White J M. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J Virol. 1997;71:8808–8820. doi: 10.1128/jvi.71.11.8808-8820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holsinger L J, Nichani D, Pinto L H, Lamb R A. Influenza A virus M2 protein: a structure-function analysis. J Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klenk H-D, Garten W. Activation cleavage of viral spike proteins by host proteases. In: Wimmer E, editor. Cellular receptors for animal viruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 241–280. [Google Scholar]
- 25.Korte T, Ludwig K, Krumbiegel M, Zirwer D, Damaschun G, Herrmann A. Transient changes of the conformation of hemagglutinin of influenza virus at low pH detected by time-resolved circular dichroism spectroscopy. J Biol Chem. 1997;272:9764–9770. doi: 10.1074/jbc.272.15.9764. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lamb R A, Holsinger L J, Pinto L H. The influenza virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Cellular receptors for animal viruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 303–321. [Google Scholar]
- 28.Lubeck M D, Schulman J L, Palese P J. Susceptibility of influenza A viruses amantadine is influenced by the gene coding for M protein. Virology. 1978;28:710–716. doi: 10.1128/jvi.28.3.710-716.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo G-X, Torri A, Harte W E, Danetz S, Cianci C, Tiley L, Day S, Mullaney D, Yuo K-L, Ouellet C, Dextraze P, Meanwell N, Colonna R, Krystal M. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J Virol. 1997;71:4062–4070. doi: 10.1128/jvi.71.5.4062-4070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo G-X, Colonno R, Krystal M. Characterization of a hemagglutinin-specific inhibitor of influenza A virus. Virology. 1996;226:66–76. doi: 10.1006/viro.1996.0628. [DOI] [PubMed] [Google Scholar]
- 31.Markushin S, Ghiasi H, Sokolov N, Shilov A, Sinitsin B, Brown D, Klimov A, Nayak D. Nucleotide sequence of RNA segment 7 and the predicted amino sequence of M1 and M2 proteins of FPV/Weybridge (H7N7) and WSN (H1N1) influenza viruses. Virus Res. 1988;10:263–271. doi: 10.1016/0168-1702(88)90021-4. [DOI] [PubMed] [Google Scholar]
- 32.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 33.Ohuchi M, Cramer A, Vey M, Ohuchi R, Klenk H-D. Rescue of vector expressed fowl plague virus hemagglutinin in biologically active form by acidotrophic agents and coexpressed M2 protein. J Virol. 1994;68:920–926. doi: 10.1128/jvi.68.2.920-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott S, Wunderli-Allenspach H. Effect of the virostatic norakin (triperiden) on influenza activities. Antiviral Res. 1994;24:37–42. doi: 10.1016/0166-3542(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 35.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 36.Prösch S, Heider H, Schroeder C, Shilov A A, Sinitzyn B V, Blinov V M, Krüger D H, Frömmel C. Mapping mutations in influenza A virus resistant to norakin. FEBS Lett. 1990;267:19–21. doi: 10.1016/0014-5793(90)80277-p. [DOI] [PubMed] [Google Scholar]
- 37.Prösch S, Heider H, Schroeder C, Krüger D H. Mutations in the hemagglutinin gene associated with influenza virus resistance to norakin. Arch Virol. 1988;102:125–129. doi: 10.1007/BF01315569. [DOI] [PubMed] [Google Scholar]
- 38.Puri A, Booy F P, Doms R W, White J M, Blumenthal R. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J Virol. 1990;64:3824–3832. doi: 10.1128/jvi.64.8.3824-3832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramalho-Santos J, Nir S, Düsgünes N, Pato de Carvalho A, Pedroso de Lima M. A common mechanism for influenza virus fusion activity and inactivation. Biochemistry. 1993;32:2771–2779. doi: 10.1021/bi00062a006. [DOI] [PubMed] [Google Scholar]
- 40.Roth M G, Gething M-J, Sambrook J. Membrane insertion and intracellular transport of influenza virus glycoproteins. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 219–267. [Google Scholar]
- 41.Schroeder C, Heider H, Hegenscheid B, Schöffel M, Bubovich V I, Rosenthal H A. The anticholinergic anti-parkinson drug norakin selectively inhibits influenza virus replication. Antiviral Res Suppl. 1985;1:95–99. doi: 10.1016/s0166-3542(85)80014-0. [DOI] [PubMed] [Google Scholar]
- 42.Skehel J J. Influenza virus. Amantadine blocks the channel. Nature. 1992;358:110–111. doi: 10.1038/358110b0. [DOI] [PubMed] [Google Scholar]
- 43.Skehel J J, Steinhauer D, Wharton S A, Bullough P A, Hughson F M, Watowich S J, Wiley D. Receptor binding and membrane fusion by influenza hemagglutinin. In: Wimmer E, editor. Cellular receptors for animal viruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 187–193. [Google Scholar]
- 44.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith T F, Waterman M S. Comparison of biosequences. Adv Appl Math. 1981;2:482–489. [Google Scholar]
- 46.Stegmann T, Booy F P, Wilschut J. Effects of low pH on influenza virus. Activation and inactivation of the membrane fusion capacity of the hemagglutinin. J Biol Chem. 1987;262:17744–17749. [PubMed] [Google Scholar]
- 47.Stegmann T, Delfino J M, Richards R, Helenius A. The HA2 subunit of influenza hemagglutinin insets into the target membrane prior to fusion. J Biol Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 48.Stegmann T, Doms R W, Helenius A. Protein mediated membrane fusion. Annu Rev Biophys Biophys Chem. 1989;18:187–211. doi: 10.1146/annurev.bb.18.060189.001155. [DOI] [PubMed] [Google Scholar]
- 49.Stegmann T, Morselt W, Schloma J, Wilschut J. Fusion of influenza virus in an intracellular acidic compartment measured by fluorescence dequenching. Biochim Biophys Acta. 1987;904:165–170. doi: 10.1016/0005-2736(87)90100-3. [DOI] [PubMed] [Google Scholar]
- 50.Steinhauer D A, Sauter N K, Skehel J J, Wiley D C. Receptor binding and cell entry by influenza viruses. Semin Virol. 1992;3:91–100. [Google Scholar]
- 51.Stuart D. News and views: docking mission accomplished. Nature. 1994;371:19–20. doi: 10.1038/371019a0. [DOI] [PubMed] [Google Scholar]
- 52.Sugrue R J, Hay A J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeuchi K, Shaughnessy M A, Lamb R A. Influenza virus M2 protein ion channel activity is not required to maintain the equine-1 hemagglutinin in its native form in infected cells. Virology. 1994;202:1007–1011. doi: 10.1006/viro.1994.1428. [DOI] [PubMed] [Google Scholar]
- 54.Takeuchi K, Lamb R A. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J Virol. 1994;68:911–919. doi: 10.1128/jvi.68.2.911-919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vriend G, Sander C, DeFillipis V. Predicting local structural changes that result from point mutations. Protein Eng. 1994;7:1203–1208. doi: 10.1093/protein/7.10.1203. [DOI] [PubMed] [Google Scholar]
- 56.Wang C, Takeuchi K, Pinto L H, Lamb R A. The ion channel activity of the influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wharton S A, Belshe R B, Skehel J J, Hay A J. Role of virion M2 protein in influenza virus uncoating: specific reduction in the rate of membrane fusion between virus and liposomes by amantadine. J Gen Virol. 1994;75:945–948. doi: 10.1099/0022-1317-75-4-945. [DOI] [PubMed] [Google Scholar]
- 58.White J, Matlin K, Helenius A. Cell fusion by Semliki forest, influenza and vesicular stomatitus viruses. J Cell Biol. 1981;89:674–679. doi: 10.1083/jcb.89.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 60.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 61.White J M. Fusion of influenza virus in endosomes: role of the hemagglutinin. In: Wimmer E, editor. Cellular receptors for animal viruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 281–301. [Google Scholar]
- 62.White J M, Wilson I A. Anti-peptide antibodies detect steps in a protein conformation change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White J M, Hoffman L R, Arevalo J H, Wilson I. Attachment and entry of influenza virus into host cells. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1997. pp. 80–104. [Google Scholar]
- 64.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 65.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 66.Yu Y G, King D S, Shin Y-K. Insertion of a coiled-coil peptide from influenza virus hemagglutinin into membranes. Science. 1994;266:274–276. doi: 10.1126/science.7939662. [DOI] [PubMed] [Google Scholar]
- 67.Zebedee S L, Lamb R A. Growth restriction of influenza A virus by M2 protein antibody is genetically linked to the M1 protein. Proc Natl Acad Sci USA. 1989;86:1061–1065. doi: 10.1073/pnas.86.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]