Abstract
A central feature of the adeno-associated virus (AAV) latent life cycle is persistence in the form of both integrated and episomal genomes. However, the molecular processes associated with episomal long-term persistence of AAV genomes are only poorly understood. To investigate these mechanisms, we have utilized a recombinant AAV (rAAV) shuttle vector to identify circular AAV intermediates from transduced HeLa cells and primary fibroblasts. The unique structural features exhibited by these transduction intermediates included circularized monomer and dimer virus genomes in a head-to-tail array, with associated specific base pair alterations in the 5′ viral D sequence. In HeLa cells, the abundance and stability of AAV circular intermediates were augmented by adenovirus expressing the E2a gene product. In the absence of E2a, adenovirus expressing the E4 open reading frame 6 gene product decreased the abundance of AAV circular intermediates, favoring instead the linear replication form monomer (Rfm) and dimer (Rfd) structures. In summary, the formation of AAV circular intermediates appears to represent a new pathway for AAV genome conversion, which is consistent with the head-to-tail concatemerization associated with latent-phase persistence of rAAV. A better understanding of this pathway may increase the utility of rAAV vectors for gene therapy.
Adeno-associated virus (AAV) is a nonpathogenic parvovirus with a single-stranded DNA (ssDNA) genome of 4,680 nucleotides. Productive infection requires helper function supplied by a second coinfecting virus, such as adenovirus or herpes virus. In the absence of a helper virus, AAV establishes a latent infection by integrating into the host genome (3). Recombinant AAV (rAAV) has recently been recognized as an extremely attractive vehicle for gene delivery (18). rAAV vectors have been developed by substituting a therapeutic minigene for all virus open reading frames (ORFs) while retaining the cis elements contained in two inverted terminal repeats (ITRs) (25). Following transduction, rAAV genomes can persist as episomes (1, 6, 9), or, alternatively, they can integrate into the cellular genome (3, 5, 16). However, little is known about the mechanisms enabling rAAV vectors to persist in vivo or the identity of cellular factors which may modulate the efficiency of transduction and persistence. Although transduction of rAAV has been demonstrated in vitro in cell culture (18) and in vivo in various organs (2, 4, 10, 12, 26, 27), the mechanisms of rAAV-mediated transduction remain unclear.
The integration of wild-type AAV (wtAAV) is site specific and occurs on chromosome 19 at the AAVS1 loci through a Rep-facilitated mechanism (14, 24). In contrast to wtAAV, the mechanism for latent-phase persistence of rAAV is less clear. rAAV integration into the host genome is not site specific, because of deletion of the AAV Rep gene (21). Analyses of integrated provirus structures of both wtAAV and rAAV have demonstrated that head-to-tail genomes are the predominant structural forms. The formation of these structures could potentially be mediated either through linear concatemerization or through circularization of the virus genomes. Circular intermediate forms have previously been demonstrated for retroviruses (19), which share certain common structural features with AAV, including terminal repeats at the ends of their genomes. Furthermore, recent studies of junctional sequences of integrated wtAAV in an Epstein-Barr virus system have also raised the possibility of circular preintegration intermediates for AAV (14).
To test the hypothesis that circular intermediates are part of the AAV transduction process, we infected HeLa cells with an rAAV shuttle vector containing the ampicillin resistance gene and a bacterial origin of replication. rAAV circular intermediates were then isolated from infected cells and characterized. Coinfection with recombinant adenovirus at high multiplicities of infection (MOIs) led to augmentation of circular intermediates in rAAV-infected cells. Reconstitution experiments demonstrated that the E2a adenovirus protein, but not E4 ORF6, was capable of modulating circular intermediate formation. These studies support the hypothesis that two independent pathways for rAAV genome conversion exist which involve both linear replication form and circular intermediates.
MATERIALS AND METHODS
Construction of rAAV shuttle vector.
An rAAV shuttle vector (AV.GFP3ori) containing a green fluorescent protein (GFP) transgene cassette, a bacterial ampicillin resistance gene, and a bacterial origin of replication was generated from a cis-acting plasmid (pCisAV.GFP3ori) (6). Expression of the GFP gene was directed by the cytomegalovirus (CMV) promoter and enhancer and simian virus 40 polyadenylation sequences. pCisAV.GFP3ori was constructed with pSub201-derived ITR elements (25), which included 27 bp of 3′-polyadenylation sequences from wt AAV. The integrity of the ITR sequences was confirmed by restriction analysis with SmaI and PvuII and by sequencing. rAAV stocks were generated by cotransfection of 293 cells with pCisAV.GFP3ori and pRep/Cap, and coinfection by recombinant Ad.CMVlacZ (5). Following transfection of 40 150-mm-diameter plates, cells were collected at 42 h by centrifugation and resuspended in 12 ml of buffer (10 mM Tris [pH 8.0]). Virus was released from cells by three cycles of freeze-thawing and passage through a 25G needle six times. Cell lysates were then treated with 1.3 mg of DNase I per ml at 37°C for 30 min and 1% deoxycholate (final concentration in grams per milliliter) and 0.05% trypsin (final concentration in grams per milliliter) at 37°C for 30 min. Samples were then placed on ice for 10 min and centrifuged to remove large particulate material at 3,000 rpm for 30 min in a Beckman GS-6R centrifuge with GH-3.8 rotor. rAAV was purified by isopycnic density gradient centrifugation in CsCl (ρ = 1.4) in an SW55 rotor for 72 h at 35,000 rpm in a Beckman ultracentrifuge. Peak fractions of AAV were combined and repurified through two more rounds of CsCl centrifugation, followed by heating at 58°C for 60 min to inactivate all contaminant helper adenovirus. Typically, this preparation gave approximate AAV titers of 1012 DNA molecules/ml and 2.5 × 108 GFP-expressing units/ml. Recombinant virus titers were assessed by slot blotting and quantified against pCisAV.GFP3ori controls for DNA particles. Functional transducing units were quantified by GFP transgene expression in 293 cells. The absence of helper adenovirus was confirmed by histochemical staining of rAAV-infected 293 cells for β-galactosidase, and no recombinant adenovirus was found in 1010 particles of purified rAAV stocks. The absence of significant wtAAV contamination was confirmed by immunocytochemical staining of rAAV-adenovirus-coinfected 293 cells with anti-Rep antibodies. These studies, which had a sensitivity of 1 wtAAV particle in 1010 rAAV particles, demonstrated an absence of Rep staining compared to that in pRep/Cap plasmid-transfected controls.
Isolation and structural evaluation of AAV circular intermediates from HeLa cells.
HeLa cells were grown in 35-mm-diameter dishes in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were infected at 80% confluency in 2% FBS–DMEM with recombinant AV.GFP3ori (MOI = 1,000 particles/cell; 109 total particles/plate), and Hirt DNAs were isolated as described previously at 6, 12, 24, 48, and 72 h postinfection (6). In experiments analyzing the effects of adenovirus, plates were coinfected with either Ad.CMVlacZ (MOI = 5,000 particles/cell), Ad5.dl802 (MOI = 50, 500, and 5,000 particles/cell), or Ad5.dl1004 (MOI = 50, 500, and 5,000 particles/cell) in the presence of 2% FBS–DMEM. Zero-hour controls were generated by mixing 109 particles of AV.GFP3ori with cell lysates prior to Hirt DNA preparation. Hirt DNA isolated at each time point was used to transform Escherichia coli SURE cells (Stratagene, La Jolla, Calif.). Typically, 1/10 of the Hirt DNA preparation (total of 20 μl) was used to transform 40 μl of competent bacteria by electroporation. The resultant total number of bacterial colonies was quantified for each time point, and the structure of circular intermediates was evaluated for greater than 20 plasmid clones for each time point from two independent experiments. Structural determinations were based on restriction enzyme analysis with PstI, SphI, and AseI single and double digests together with Southern blotting against GFP, stuffer, and ITR probes. Similarly, studies were also performed with 293 cells and primary fibroblasts to evaluate the abundance and structure of AAV circular intermediates in the absence of helper adenovirus.
Control experiments transforming various forms of double-stranded and single-stranded AAV genomes with and without exogenous ligase were performed to rule out the artifactual generation of AAV circular intermediates in bacteria as previously described (6). Results from these studies suggest that linear ssDNA and double-stranded DNA (dsDNA) forms of rAAV genomes are unable to give rise to replication-competent plasmids in our rescue assay. However, following the addition of exogenous T4 ligase, dsDNA, but not ssDNA, linear rAAV genomes gave rise to the typical head-to-tail monomer circular form genomes isolated from Hirt DNA of rAAV-infected HeLa cells.
Evaluation of E2a and GFP gene expression in HeLa cells.
E2a gene expression was evaluated by immunofluorescent staining of HeLa cells superinfected with E1-deleted Ad.CMVlacZ (MOI = 0, 500, or 5,000 particles/cell). Briefly, cells were fixed in methanol at −20°C for 10 min followed by air drying. Cells were then incubated at room temperature with hybridoma supernatant against Ad5 72-kDa DNA binding protein (DBP) (23), followed by goat anti-mouse fluorescein isothiocyanate-conjugated antibody (5 μg/ml) for 30 min at room temperature. In studies evaluating augmentation of AAV GFP transgene expression by adenovirus, HeLa cells were harvested at 24 or 72 h postinfection by trypsinization, resuspended in 2% FBS–phosphate-buffered saline (PBS), and evaluated by fluorescence-activated cell sorter (FACS) analyses. Thresholds were set by using uninfected controls, and the percent and/or the average relative fluorescent intensity was determined by sorting of greater than 105 cells per experimental condition.
Sequence analysis of AAV circular intermediates.
Sequence analysis of the ITR array within circular intermediates was performed with primers EL118 (5′-CGGGGGTCGTTGGGCGGTCA-3′) and EL230 (5′-GGGCGGAGCCTATGGAAAA-3′), which are nested to 5′ and 3′ ITR sequences, respectively. In preliminary experiments, the sequence obtained from supercoiled plasmids by dideoxy sequencing was minimal with these primers. To this end, we employed an alternative strategy by which plasmids were digested with SmaI (which cuts within ITR sequences), and linear plasmids were gel isolated prior to sequencing. This maneuver resulted in longer sequence, which in some cases extended as far as the SmaI restriction site.
RESULTS
Construction of rAAV shuttle vector and isolation of circular intermediates.
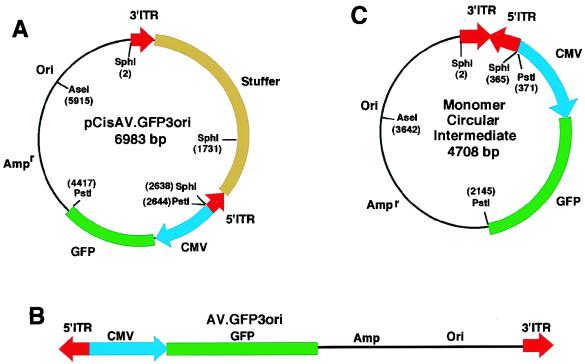
Circular intermediate forms of AAV have been previously proposed, based on the structure of integrated proviruses, as head-to-tail monomer and multimer genomes (3, 5, 18). Circular structures may play roles either as preintegration intermediates or as stable episomal forms resistant to nuclease digestion. However, verification of circular intermediate forms of AAV has remained elusive because of the difficulties in manipulating the wtAAV genome to allow for the retrieval of these structures. To circumvent this limitation, we developed an alternative strategy to “trap” circular intermediates by using an rAAV shuttle vector. Recombinant AV.GFP3ori virus (Fig. 1B) was generated from a cis-acting plasmid (pCisAV.GFP3ori [Fig. 1A] by cotransfection of 293 cells with trans-acting plasmids containing Rep and Cap virus genes. This virus vector (AV.GFP3ori) carried the GFP reporter gene, a bacterial origin of replication (ori), and the bacterial ampicillin resistance gene. Ori and ampicillin resistance sequences encoded in this virus allow for the rescue of circular AAV genomes formed during the transduction process. Despite the advantage of this “rescue” system, it is also important to recognize the inherent limitation associated with this selection scheme. Head-to-head or tail-to-tail multimer rAAV genomes, which contain replication origins in the opposite orientations, will likely not propagate in bacteria. Hence, we anticipate that head-to-tail circular genomes would have a preferential growth advantage according to this strategy.
FIG. 1.
Structure of the provirus shuttle vector and the predicted structure of rAAV circular intermediate monomers. With the aid of an rAAV cis-acting plasmid, pCisAV.GFP3ori (A), we produced the AV.GFP3ori recombinant virus (B). This vector encoded a GFP transgene cassette, an Ampr gene, and a bacterial replication origin (Ori). The predominant form of circular intermediates isolated following transduction of HeLa cells with AV.GFP3ori consisted of head-to-tail monomers (C).
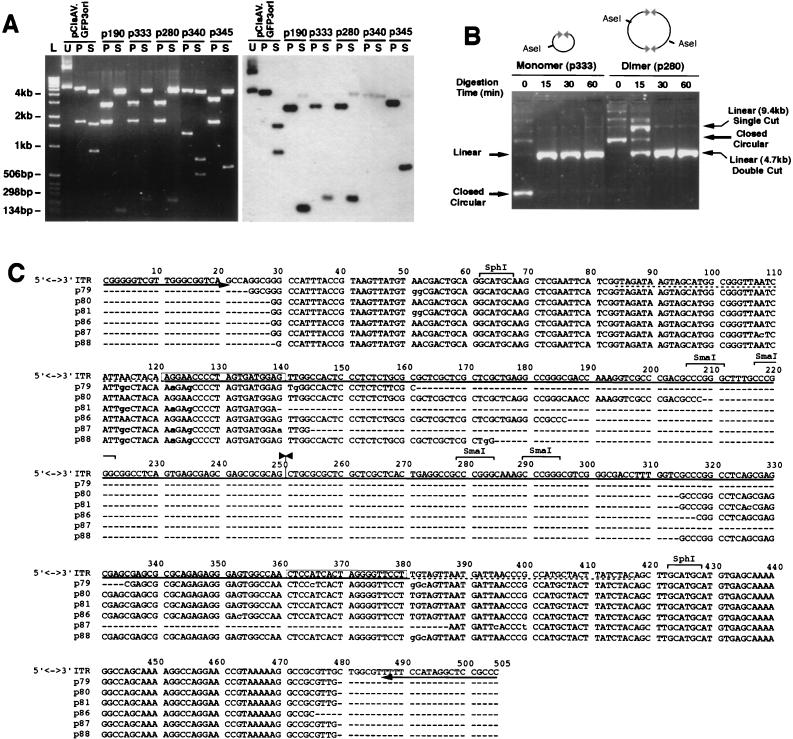
To test our hypothesis, HeLa cells were infected with AV.GFP3ori (MOI = 1,000 particles/cell), and the abundance of circular intermediates was evaluated following transformation of Escherichia coli SURE cells with low-molecular-weight cellular Hirt DNA. The formation of circular intermediates in infected HeLa cells was inferred by the presence of ampicillin-resistant bacterial colonies. Structural features of rescued circular form genomes were determined by restriction enzyme analysis and by Southern blotting with probes from various regions of the provirus, including GFP, stuffer, and ITR sequences. The predominant circular form isolated after transduction of HeLa cells with AV.GFP3ori consisted of 4.7-kb monomer-sized molecules (Fig. 1C). SphI digestions of these circular intermediates yielded characteristic 300-bp bands which hybridized to an ITR probe on Southern blots (Fig. 2A). PstI, SphI, and AseI single and double digests, together with Southern blot analysis using GFP, stuffer (data not shown), and ITR (Fig. 2A) probes, confirmed the structure of the circular intermediates as head-to-tail monomer genomes (Fig. 1C). In particular, PstI digests together with ITR Southern blots distinguished these head-to-tail circular monomers from head-to-head or tail-to-tail circular dimers. Similar results obtained from studies of AV.GFP3ori-infected 293 cells and primary fibroblasts have confirmed that monomer head-to-tail circular intermediates were also the most abundant form in these cell types (data not shown).
FIG. 2.
Structural analysis of rAAV circular intermediates in HeLa cells. Circular rAAV intermediate clones isolated from AV.GFP3ori-infected HeLa cells were analyzed by diagnostic restriction digestion with AseI, SphI, and PstI, together with Southern blotting against ITR, GFP, and stuffer 32P-labeled probes (only data for ITR probes are shown). In panel A, four clones representing the diversity of intermediates found (p190, p333, p280, and p345) gave a diagnostic PstI (P) restriction pattern (3- and 1.7-kb bands) consistent with a circular monomer or multimer intact genome (agarose gel [left] and Southern blot [right]). SphI (S) digestion demonstrated the existence of a single ITR (p190), two ITRs in a head-to-tail orientation (p333 and p280), or three ITRs (p345) in isolated circular intermediates. The restriction patterns of pCisAV.GFP3ori (U, uncut; P, PstI cut; S, SphI cut) and a 1-kb DNA ladder (L) are also given for comparison. One additional circular form (p340) that was repetitively seen had an unidentifiable structure which lacked intact ITR sequences. Circular concatemers were identified by partial digestion with AseI for clones p280 (dimer) and p333 (monomer), as is shown in panel B. Sequence analysis (C) of six clones with restriction patterns identical to that of p333 (A) was performed with primers (indicated by arrows) juxtaposed with the partial AAV 3′-polyadenylation sequences (dotted line) which flank the pSub201-derived ITRs (solid line). The top sequence represents the proposed head-to-tail structure of intact ITR arrays with sequence alignment derived from individual clones. Only partial sequence was achievable, due to the high secondary structure of ITRs (unknown sequence is marked by dashes in the sequence alignment). The junction of the inverted ITRs is marked by inverted arrowheads (at 251 bp). Several consistent base pair changes (shaded) were noted in the 5′ ITR D sequence (boxed) within four clones (p79, p81, p87, and p88). All base pair changes are indicated in lowercase letters.
Because the predicted size of an intact head-to-tail ITR SphI fragment would be approximately 360 bp, we hypothesize that the high secondary structure of inverted repeats within ITRs might lead to anomalous migration in agarose gels. The head-to-tail orientation of the ITRs, as predicted by Southern blot analysis, was also confirmed by sequencing. Because the high secondary structure prevented complete sequencing across these ITR arrays, several alternative sequencing strategies were developed. First, the SphI ITR-hybridizing fragment of circular intermediates was subcloned into a secondary plasmid vector and sequenced with primers outside the cloned ITR sequences. The results confirmed the head-to-tail orientation of ITRs (data not shown). Additionally, the sequence of ITR arrays was obtained directly from six monomer circular intermediate clones (Fig. 2C). In these studies, circular intermediates were digested with SmaI and the linear 4.6-kb plasmid was gel isolated prior to sequencing. SmaI digestion (which relaxed the secondary structure of ITRs) was necessary to obtain sequence information within the ITRs. These sequencing results, presented in Fig. 2C, confirmed the orientation of head-to-tail ITR arrays in these intermediates. However, given the inability to sequence across the ITR junction by this method, it is presently impossible to rule out potential deletions in the interior of ITRs. Interestingly, sequencing also revealed several consistent base pair changes in four of the six clones analyzed (Fig. 2C). These four clones (p79, p81, p87, and p88) had consistent 2-bp changes within the D sequence (G→A [122 bp] and A→G [125 bp]), which were always accompanied by base pair alterations just outside the D sequence (A→G [114 bp] and A→C [115 bp]). No other consistent base pair changes were noted; however, two clones (p79 and p88) demonstrated mutations just outside the 3′ ITR D sequence (T→G [381 bp] and T→C [383 bp]).
Although head-to-tail circular intermediates were the most abundant forms present in Hirt DNA from rAAV-infected HeLa cells, several less frequent structures were also detected. These included monomer circularized AAV genomes with one (p190) and three (p345) ITRs arranged in a head-to-tail fashion, as well as several clones with an unknown structure lacking complete ITRs (p340) (Fig. 2A). Such diversity within the ITR array may represent homologous recombination in vivo or in bacteria during amplification. However, previous studies demonstrating similar variations in ITR sequences of head-to-tail integrated genomes suggest that such changes in the length of the ITR array may occur in vivo (5). Additionally, less frequent head-to-tail circularized multimer forms were predicted by variations in the migration patterns of uncut plasmids exhibiting identical restriction patterns. Results in Fig. 2B confirmed the existence of monomer and dimer head-to-tail circular intermediates by using partial digestion with an enzyme which cuts once in the AAV genome (AseI). Cumulative analysis of more than 200 independently isolated circular intermediates from HeLa cells demonstrated that head-to-tail circular AAV genomes occurred in the greatest abundance as monomers (92%) and less frequently as multimers of more than one genome (8%). In addition, we have estimated the overall abundance of the head-to-tail circular intermediates in HeLa cells. Based on reconstitution experiments calculating the abundance of circular molecules (Table 1), we would expect that approximately 1 in 300 rAAV genomes are circularized in vivo in HeLa cells.
TABLE 1.
Yield of circular intermediates isolated from HeLa cells
| Bacterial transformation component(s) | Starting no. of plasmid or AAV genome molecules | Actual no. of Ampr CFU | Adjusted yield (CFU)e |
|---|---|---|---|
| Hirt DNA from rAAV-infected HeLa cellsa | 109 | 2 × 103 | 4 × 105f |
| Hirt DNA + 7.6 ng of LacZ plasmidbc | 109 | 6 × 105d | 1.2 × 108 |
| 7.6 ng of LacZ plasmidc | 109 | 5 × 106 |
The actual amount of Hirt DNA used for transformation was 1/10 of the entire Hirt DNA. The numbers have been adjusted to reflect the viral inoculum and yields for the entire 35-mm-diameter plate of infected cells.
Plasmid DNA was spiked into mock-infected cell homogenates prior to isolation of Hirt DNA. This reconstituted Hirt DNA was then used for transformation of bacteria.
The control LacZ plasmid was approximately 7,000 bp, with a molecular weight of 4.6 × 106 g/mol.
The average of several experiments indicates an approximate 10-fold reduction in the number of CFU recovered from bacterial transformations with DNA isolated from Hirt DNA extract spiked with plasmids, compared to transformation with an equivalent amount of plasmid DNA alone.
Adjusted yields take into account the relative efficiency of transformation with purified DNA (1 in 200 plasmid molecules leads to an Ampr CFU).
This adjusted yield indicates that approximately 1 in 300 AAV genomes circularize in vivo. This number takes into account both the transformation efficiency of pure plasmid DNA (1 in 200 molecules leads to a CFU) and the loss of plasmid DNA during Hirt DNA preparation (10-fold).
To establish that head-to-tail circular intermediates were formed in vivo and not by nonspecific bacterial recombination of linear AAV genomes present in the Hirt DNA, we performed a set of reconstitution experiments, including bacterial transformation with multiple forms of ssDNA and dsDNA AAV genomes, which were isolated either from pCisAV.GFP3ori plasmids or from purified virus DNA. In all cases, these control transformations yielded few replication-competent plasmids, none of which had head-to-tail orientations similar to those of the in vivo-isolated circular intermediates. Furthermore, when exogenous ligase was added to linear dsDNA forms of the AAV genome, circular head-to-tail intermediates with structures identical to those of in vivo forms could be isolated (6). These findings support the notion that circular intermediates do not arise from nonspecific recombination or ligation events with either ssDNA or dsDNA linear AAV genomes in bacteria. Additional control experiments, demonstrating the lack of stuffer hybridizing sequences in AAV circular intermediates by Southern blotting, also confirm that these structures do not arise from contamination of viral stocks with pCisAV.GFP3ori plasmid (data not shown).
The formation of head-to-tail circular AAV intermediates is augmented by superinfection with E1-deleted adenovirus.
Many aspects of the wtAAV growth cycle are affected by helper adenovirus, including AAV DNA replication, transcription, splicing, translation, and virion assembly. Such studies have provided concrete evidence that a subset of adenovirus early gene products provide helper functions for the wtAAV lytic cycle, including E1a, E1b, E2a, E4 ORF6, and VA1 RNA (18). In this regard, one of the most critical factors required for AAV replication is the 34-kDa E4 (ORF6) protein. Recent observations about the helper function of adenovirus protein in rAAV transduction have also demonstrated that adenovirus E4 ORF6 protein is essential for the augmentation of rAAV transgene expression seen with adenovirus coinfection (7, 8). According to these reports, the rate-limiting step enhanced by these adenovirus proteins is the conversion of single-stranded AAV genomes to double-stranded forms.
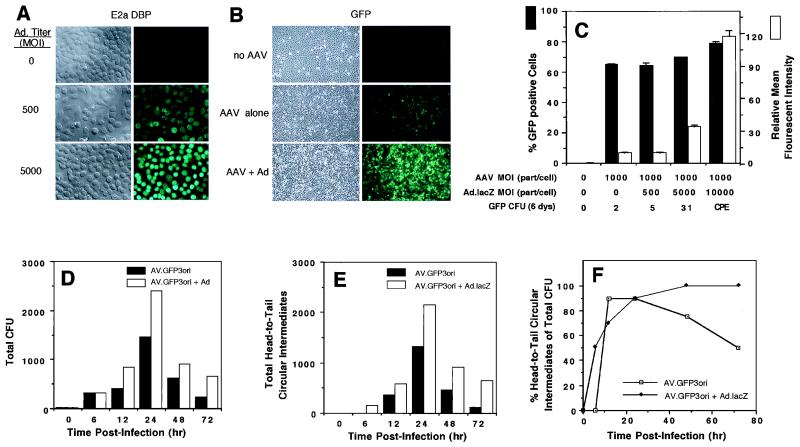
Studies evaluating the kinetics of rAAV circular intermediate formation demonstrated a time-dependent increase in abundance, which peaked at 24 h postinfection in HeLa cells and coincided with the onset of GFP transgene expression (Fig. 3). To better understand the cellular mechanisms associated with AAV circular intermediate formation, we sought to evaluate the effects of adenovirus coinfection on this process. To this end, we compared the extents of transgene expression and circular intermediate formation in AV.GFP3ori-infected HeLa cells with or without coinfection with E1-deleted recombinant adenovirus. Although E1-deleted adenoviruses are severely handicapped in their ability to synthesize virus gene products, at high MOIs of ≥5,000, significant E2a protein expression was noted (Fig. 3A). As an indicator of transgene expression, the abundance and average relative intensity of GFP-positive cells were determined relative to those of mock-infected controls by fluorescent microscopy (Fig. 3B) and FACS analysis (Fig. 3C) at 72 h postinfection. As anticipated from previous reports demonstrating augmentation in rAAV transgene expression by adenovirus (7, 8), the extent of GFP transgene expression was dramatically increased at doses of adenovirus which led to virus gene expression (MOI, ≥5,000 [Fig. 3A to C). Additionally, persistence of rAAV transgene expression was also augmented by coinfection with E1-deleted adenovirus, as determined by GFP-expressing colony formation following serial passages (Fig. 3C).
FIG. 3.
Adenovirus augments AAV circular intermediate formation in HeLa cells. Infection of HeLa cells with increasing doses (0, 500, and 5,000 particles/cell) of recombinant E1-deleted adenovirus (Ad.CMVlacZ) led to substantial expression of E2a 72-kDa DBP, as demonstrated by indirect immunofluorescent staining for DBP at 72 h postinfection (A). Coinfection of HeLa cells with Ad.CMVlacZ (5,000 particles/cell) and AV.GFP3ori (1,000 DNA particles/cell) led to substantial augmentation of rAAV GFP transgene expression (B). Augmentation of rAAV GFP transgene expression in the presence of increasing amounts (0, 500, 5,000, and 10,000 particles/cell) of recombinant Ad.CMVlacZ was quantified by FACS analysis at 72 h postinfection (C). Results demonstrate the mean (± standard error) for two experiments performed in duplicate. In addition, an aliquot of cells was split (1:10) at the time of FACS analysis, and GFP CFU per ×10 field were quantified at 6 days. (CPE denotes significant CPE at an adenovirus MOI of 10,000 particles/cell and was not quantified for GFP colonies.) Hirt DNAs from AV.GFP3ori (1,000 DNA particles/cell)-infected HeLa cells with or without coinfection with Ad.CMVlacZ (5,000 particles/cell) were used to transform E. coli. The total number of ampicillin-resistant bacterial CFU (D) and total number of head-to-tail circular intermediate CFU (E) are given for a representative experiment. More than 20 clones for each time point were evaluated by Southern blotting (see Fig. 2 for details). Zero hour control experiments were performed by mixing an amount of AV.GFP3ori virus equivalent to that used in experiments with mock-infected cellular lysates prior to Hirt DNA purification. (F) Abundance of head-to-tail circular intermediates as a percentage of total ampicillin-resistant bacterial CFU isolated from Hirt DNA.
We hypothesized that if circular intermediates represent a molecular form of rAAV important for efficient and/or persistent transgene expression, augmentation of rAAV transgene expression by adenovirus might also modulate circular intermediate formation. In these studies, the abundance and time course of AAV circular intermediate formation were evaluated following superinfection with Ad.CMVlacZ. Results from these experiments are shown in Fig. 3D, which represents the total number of bacterial colonies (per 35-mm-diameter plate) obtained following transformation of E. coli with Hirt DNA isolated from HeLa cells infected with AV.GFP3ori (1,000 DNA particles/cell), with or without coinfection with Ad.CMVlacZ (5,000 particles/cell). An MOI of 5,000 adenovirus particles/cell was chosen for these experiments, since this level of adenovirus led to minimal cytopathic effect (CPE) but high levels of E2a expression. These studies demonstrated a nearly twofold augmentation by Ad.CMVlacZ in the total abundance of AAV-rescued plasmid intermediates in E. coli (Fig. 3D). Southern blot restriction enzyme analysis indicated that the predominant forms, in both the presence and absence of adenovirus, were head-to-tail monomer circular intermediates containing the diagnostic 300-bp ITR fragment following SphI digestion (Fig. 3E). Additionally, the results established that adenovirus coinfection led to an earlier time of onset and increased stability of AAV head-to-tail monomer circular intermediates (Fig. 3E and F). For example, at 6 h postinfection, head-to-tail circular intermediates were only present in HeLa cells coinfected with adenovirus. Furthermore, a decline in the percentage of head-to-tail circular intermediate clones was seen at 48 to 72 h post-AAV infection in the absence of adenovirus. In contrast, this decline was significantly blunted by the presence of helper adenovirus (Fig. 3F). Based on these findings, we concluded that certain adenovirus proteins produced by superinfection with E1-deleted adenovirus were capable of modulating circular intermediate formation and stability during rAAV transduction.
The E2a gene product is responsible for augmentation of AAV circular intermediate formation by adenovirus.
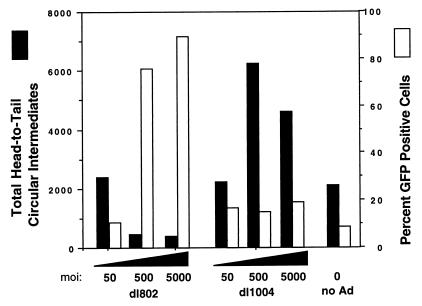
Previous studies have demonstrated that the E4 ORF6 protein (or the cellular factors induced by this protein) is important for the conversion of single-stranded AAV genomes to double-stranded replication form monomers (Rfm) and dimers (Rfd) (7, 8). This finding provides the mechanistic foundation for augmentation of rAAV transgene expression in the presence of adenovirus. Based on these studies, we hypothesized that E4 ORF6 might also enhance the abundance of circular intermediates. To further investigate adenovirus proteins responsible for the augmentation of rAAV circular intermediate formation, two adenovirus mutants, dl1004 (E4 deleted) and dl802 (E2a deleted), were used to coinfect HeLa cells with AV.GFP3ori. Contrary to our initial hypothesis, superinfection with E4-deleted adenovirus led to a two- to threefold increase in the abundance of head-to-tail circular intermediates at 24 h postinfection, compared to that in controls (Fig. 4). In contrast, coinfection with dl802 mutant virus (E2a deleted) led to a dose-dependent fivefold decrease in the total number of head-to-tail circular intermediates (Fig. 4). At MOIs of 500 to 5,000 particles/cell, there was a greater than 10-fold difference in the relative abundance of head-to-tail AAV circular intermediates resulting from coinfections with the dl802 or dl1004 mutant. These findings suggest that the E2a DBP is at least in part responsible for the augmentation of rAAV circular intermediate formation produced by coinfection with adenovirus. Furthermore, our results also indicate that the adenovirus E4 ORF6 protein has an inhibitory effect on the formation of circular intermediates.
FIG. 4.
Identification of adenovirus genes responsible for augmentation of AAV circular intermediate formation. HeLa cells were infected with AV.GFP3ori (1,000 DNA particles/cell) in the presence of dl802 (E2a deleted) and dl1004 (E4 deleted) adenovirus (at the indicated MOIs). The total number of head-to-tail circular intermediates from Hirt DNA and the level of augmentation of GFP transgene expression (as determined by FACS) were quantified at 24 h postinfection. Results are the average of duplicate experiments.
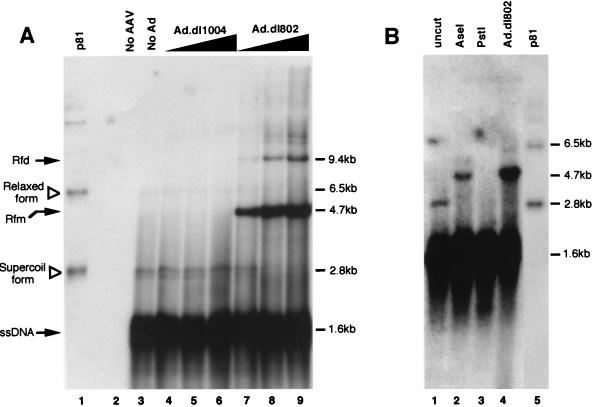
To gain molecular evidence for the functional experiments described above, which evaluated the abundance of circular form rAAV genomes by using bacterial transformation as an end point, Southern blot analyses of Hirt DNA samples were performed. In these studies, one-half of the Hirt DNA samples isolated from a 35-mm-diameter plate under different infection conditions were resolved in a 1% agarose gel and Southern blots hybridized with GFP probe. Consistent with previous observations by other groups (6, 7), our findings also demonstrated that E2-deleted adenovirus (dl802) led to significant augmentation in the conversion of ssDNA AAV genomes to double-stranded Rfm and Rfd forms, while coinfection with E4-deleted virus (dl1004) gave undetectable levels of Rf genomes, which was similar to what was found with uninfected controls (Fig. 5A). Interestingly, in rAAV and rAAV-Ad.dl1004-coinfected cultures, we also detected a set of bands which comigrated with a 2.8-kb form of supercoiled monomer head-to-tail circular AAV plasmid (p81) rescued in bacteria (Fig. 5A). The intensity of these candidate AAV circular intermediate bands appeared to mirror the abundance of rescued bacterial CFU from Hirt DNA following adenovirus coinfection (Fig. 5A). Specifically, a dose-dependent decrease in the abundance of this candidate circular form genome was seen in Ad.dl802-coinfected HeLa cells which coincided with a fivefold reduction in rescued head-to-tail circular plasmids. In contrast, coinfection with Ad.dl1004 led to an increased intensity of these bands at the highest MOIs used, which coincided with a threefold increase in rescued head-to-tail circular plasmids. Although the intensity of candidate AAV circular genome bands on Hirt Southern blots agreed with the abundance of rescued head-to-tail CFU following modulation by adenovirus infection, we sought to confirm the structure of this band by using restriction enzyme analysis of Hirt DNA. As was shown in Fig. 5B, AseI digestion (which cuts once in the viral genome) of Hirt DNA altered the migration of the 2.8-kb candidate circular form, giving rise to a 4.7-kb fragment consistent with the length of the linearized circular intermediate monomer. As expected, this linearized circular form also comigrated with the Rfm band seen in Ad.dl802-infected cells (Fig. 5B). In contrast, when Hirt DNA was predigested with PstI prior to Southern blotting, the candidate circular molecule would be expected to be cleaved into 1.7- and 3-kb fragments. Although GFP hybridization to this 1.7-kb fragment was masked by signal from single-stranded AAV genomes (Fig. 5B), additional Southern hybridization with an Ampr gene probe revealed the existence of a 3-kb band (data not shown). Furthermore, bacterial transformation of either AseI- or PstI-digested Hirt DNA produced no CFU, while transformation of mock-digested Hirt DNA samples in the absence of enzyme did not significantly alter the abundance of rescued CFU (data not shown). Taken together, these experiments strongly suggested that circular form rAAV genomes in this study exist as supercoiled molecules in vivo and migrate at approximately 2.8 kb on Hirt DNA Southern blots. In summary, our findings demonstrated an indirect correlation between the augmentation of Rf genomes by second-strand synthesis and circular intermediate formation. Furthermore, these results suggest that processes which are responsible for adenovirus augmentation of rAAV transgene expression (i.e., E4-enhanced second strand synthesis) may be distinct from those involving E2a-enhanced circular intermediate formation. Additionally, although circular forms represent a minor proportion of AAV genomes in the presence of E4 ORF6, their abundance in the presence of E2a DBP was greater than that of replication form intermediates.
FIG. 5.
Southern blot identification of circular and Rf rAAV genomes in Hirt DNA. To identify circular intermediates, one-half of the total Hirt DNA yield from a 35-mm-diameter plate of HeLa cells was loaded onto a 1% agarose gel and electrophoresed at 35 V for 14 h prior to Southern transfer. HeLa cell infections were carried out identically to that described in the legend to Fig. 4. GFP 32P-labeled probes were used for hybridization of Southern blots. The rAAV circular intermediate plasmid (p81) isolated from bacterial transformation was used as a marker for the migration of the supercoiled (closed circular) and relaxed (open circular or nicked) form molecules (indicated by open triangles in lane 1, panel A, and lane 5, panel B). A negative control of Hirt DNA isolated from uninfected parental HeLa cells was loaded in lane 2 of panel A. Also included in panel A are samples of HeLa cell Hirt DNA following rAAV infection alone (lane 3); coinfection with rAAV and Ad.dl1004 at MOIs of 50, 500, and 5,000 (lanes 4, 5, and 6, respectively); and coinfection with rAAV and Ad.dl801 at MOIs of 50, 500, and 5,000 (lanes 7, 8, and 9, respectively). The size of circular intermediates based on the migration of rescued bacterial plasmids is approximately 2.8 kb. Solid arrows mark replication form dimers (Rfd), replication form monomers (Rfm), and single-stranded AAV genomes (ssDNA). (B) Hirt DNA Southern blots from HeLa cells infected with rAAV alone prior to (lane 1) and following restriction digestion with AseI (lane 2) and PstI (lane 3). In addition to p81 as a marker for migration of rescued circular form genomes (lane 5), undigested Hirt DNA from cells coinfected with rAAV and Ad.dl802 (MOI, 50) is also shown as a marker for replication form monomer (4.7 kb) linear-length dsDNA genomes.
DISCUSSION
In the present study, we have begun to delineate mechanisms of AAV circular intermediate formation through structural and functional characterization in the presence of adenovirus proteins. As recently reported in muscle (6), circularization of rAAV genomes in HeLa cells appears to predominately occur as head-to-tail monomer genomes. However, the existence of less abundant circular multimer forms suggests that recombinational events subsequent to the initial infection may drive concatemerization of circular genomes. The diversity in the length of ITR arrays found within circular intermediates (i.e., one to three ITRs) also supports the notion that these forms may be highly recombinagenic. Of mechanistic interest in the formation of circular intermediates are the uniformity of mutations observed in the D sequences and the confinement of these mutations to the 5′ ITRs. Although the etiology of these base pair changes is unknown, their uniformity suggests that they may have a direct role in the formation of circular intermediates. Recent findings suggesting that an endogenous host single-strand D sequence binding protein is important in rAAV transduction lend support to the potential involvement of this sequence in circular intermediate formation (22, 28).
Molecular characterization of rAAV circular intermediates in Hirt DNA has suggested that these molecules contain a high level of supercoiling similar to that found in bacterial amplified plasmids. These circular form genomes have a distinctly different mobility in agarose gels from that of duplex replication form monomers, which are characteristic of lytic-phase AAV replication. Furthermore, the apparent size of circular monomer genomes in HeLa cells (2.8 kb) is similar to that which was observed in size-fractionated Hirt DNA isolated from muscle at 22 days postinfection with the same vector (6). However, concatemerization into large multimer circular genomes in muscle reached levels as high as 50% of the total circular genomes by 80 days. Since the extent of concatemerization seen in HeLa cells was much less (8% of total circular forms), one might conclude either cell-specific factors or extended lengths of time are needed to drive the concatemerization process.
By analogy, retrovirus transduction intermediates have striking similarities to the current findings with AAV. Three DNA forms have been isolated following retrovirus infection, including linear DNA with long terminal repeats (LTRs) at both ends, circular DNA with one LTR, and circular DNA with multiple LTRs (19). Although which of these forms is the direct precursor to integration is disputed, the existence of circular retrovirus genomes with repeat regions similar to those of AAV suggests the potential for common mechanisms guiding the formation of both retrovirus and AAV circular intermediates. These AAV circular intermediates could act as integration precursors and/or stable episomal genomes.
The head-to-tail ITR structures found in AAV circular intermediates are most characteristic of latent integrated AAV genomes. In contrast, lytic phases of AAV growth are typically associated with head-to-head and tail-to-tail replication form genomes. Hence, it is likely that circular intermediates represent a latent aspect of the AAV life cycle. The finding that coinfection with adenovirus leads to increased abundance and stability of AAV circular intermediates suggests a novel link between adenovirus helper functions and latent infection of AAV. By using mutant forms of adenovirus to dissect the etiology of this augmentation, we have demonstrated that pathways which enhance AAV replication form head-to-head and tail-to-tail intermediate formation by E4 ORF6 are distinct from those mediating E2a augmentation of AAV circular forms (Fig. 6). The fact that E4 ORF6 expression leads to a decreased abundance of circular forms, as detected by both Southern blots of Hirt DNA and CFU rescue assays, suggests that pathways which drive formation of Rf genomes may complete with those involved in circular intermediate formation. Furthermore, Southern blot identification of circular AAV genomes in Hirt DNA suggests that in the absence of adenovirus helper functions, circular genome formation may be the predominant pathway of rAAV transduction, as opposed to replication form second-strand synthesis.
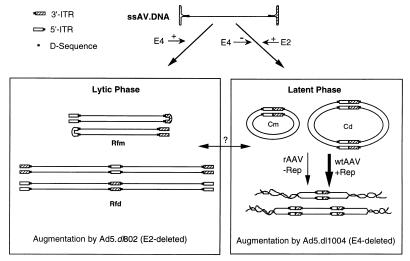
FIG. 6.
Model for independent mechanistic interactions of adenovirus with lytic- and latent-phase aspects of the AAV life cycle. The adenovirus E4 gene has been shown to augment the level of rAAV second-strand synthesis, giving rise to replication form dimers (Rfd) and monomers (Rfm). This augmentation leads to substantial increases in transgene expression from rAAV vectors and most closely mirrors lytic-phase replication of wtAAV as head-to-head and tail-to-tail concatemers. In contrast, E4 expression inhibits the formation of head-to-tail circular intermediates of AAV. Hence, it appears that increases in the amount of Rfd and Rfm dsDNA genomes do not increase the extent of circular intermediate formation. Such findings suggest that conversion of Rfm and Rfd to circular intermediates does not likely occur and implicate two mechanistically distinct pathways for their formation. In support of this hypothesis, adenovirus E2a gene expression does not enhance the formation of Rfm and Rfd genomes, but rather increases the abundance and/or stability of head-to-tail circular intermediates. Cm, monomer circular intermediate; Cd, dimer circular intermediate. Furthermore, in the absence of E4, E2a gene expression does not lead to augmentation of rAAV transgene expression. Given the similarities in genome structure of AAV circular intermediates to those of integrated proviruses, we hypothesize that circular form AAV genomes may represent preintegration complexes important in latent-phase persistence. In the presence of Rep, these circular intermediates may have a higher predisposition for integration into the host genome.
The E2 genes of adenovirus encode the 140-kDa virus DNA polymerase (E2b), the 80-kDa preterminal protein (E2b), and the 72-kDa single-strand DBP (E2a). The 72-kDa DBP may elicit functional augmentation of circular intermediate formation through direct interactions with rAAV genomes or by indirectly affecting the expression of cellular genes. Interestingly, previous reports have demonstrated that AAV DNA colocalizes with the adenovirus E2a 72-kDa DBP in nuclear inclusions composed of cellular factors, virus nucleic acid, and virus proteins (20, 29). The functional significance of associated DBP and single-stranded AAV genomes is currently unknown, but may represent a direct link between this protein and circular intermediate formation.
Interestingly, several observations regarding rAAV transgene expression in liver lend support to independent mechanisms for short-term augmentation of transgene expression by E4 ORF6 and long-term persistence of transgene expression in vivo (26). These observations include the fact that short-term adenovirus augmentation of rAAV transgene expression (presumably through increases in Rfd and Rfm genomes) does not lead to long-term increases in transgene expression, compared to that in livers infected with rAAV alone. Furthermore, the percentage of virus genomes which persist long term in the liver represent a minority of input virus DNA (approximately 1 in 1,000 genomes persist), and they occur as large head-to-tail integrated concatemers (17). These findings are consistent with both the abundance and genomic organization of AAV circular intermediates described in this study.
Although the function of AAV circular intermediates is currently unknown, aspects of inverted head-to-tail ITRs, which include palindromic hairpins similar in structure to “Holliday-like” junctions, might impart recombinagenic activity, which aids in virus integration. Such Holliday junctions have been shown to play critical roles in directing homologous recombination in bacteria through the processing of recombination intermediates by RuvABC proteins (13, 30). Interestingly, a mammalian endonuclease that is analogous to bacterial RuvC resolvase has also been isolated from cell lines (11). Despite the theoretical considerations which might suggest that circular AAV genomes have characteristics of preintegration intermediates, a study with recombinant retrovirus has demonstrated that palindromic LTR-LTR junctions of Moloney murine leukemia virus are not efficient substrates for provirus integration (15). Nonetheless, circular AAV genomes have been previously proposed as integration intermediates based on provirus structure (14) and are consistent with recent in vivo findings in the liver (17). Future studies elucidating the function of circular AAV intermediates in vivo will lend insights into their importance in the AAV life cycle and gene targeting.
ACKNOWLEDGMENTS
We gratefully acknowledge comments from Michael Welsh and Beverly Davidson in the preparation of the manuscript.
This work was supported by NIH R01 DK/HL58340 (J.F.E.).
REFERENCES
- 1.Afione S A, Conrad C K, Kearns W G, Chunduru S, Adams R, Reynolds T C, Guggino W B, Cutting G R, Carter B J, Flotte T R. In vivo model of adeno-associated virus vector persistence and rescue. J Virol. 1996;70:3235–3241. doi: 10.1128/jvi.70.5.3235-3241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett J, Duan D, Engelhardt J F, Maguire A M. Real-time, noninvasive in vivo assessment of adeno-associated virus mediated retinal transduction. Invest Ophthalmol Vis Sci. 1997;38:2857–2863. [PubMed] [Google Scholar]
- 3.Berns K I, Giraud C. Adeno-associated virus (AAV) vectors in gene therapy. Berlin, Germany: Springer; 1996. [Google Scholar]
- 4.Conrad C K, Allen S S, Afione S A, Reynolds T C, Beck S E, Fee-Maki M, Barrazza-Ortiz X, Adams R, Askin F B, Carter B J, Guggino W B, Flotte T R. Safety of single-dose administration of an adeno-associated virus (AAV)-CFTR vector in the primate lung. Gene Ther. 1996;3:658–668. [PubMed] [Google Scholar]
- 5.Duan D, Fisher K J, Burda J F, Engelhardt J F. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 1997;48:41–56. doi: 10.1016/s0168-1702(96)01425-6. [DOI] [PubMed] [Google Scholar]
- 6.Duan D, Sharma P, Yang J, Yue Y, Dudus L, Zhang Y, Fisher K J, Engelhardt J F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle. J Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari F K, Samulski T, Shenk T, Samulski R J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher K J, Gao G-P, Weitzman M D, DeMatteo R, Burda J F, Wilson J M. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol. 1996;70:520–532. doi: 10.1128/jvi.70.1.520-532.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flotte T R, Afione S A, Zeitlin P L. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Respir Cell Mol Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 10.Herzog R W, Hagstrom J N, Kung S H, Tai S J, Wilson J M, Fisher K J, High K A. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc Natl Acad Sci USA. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyde H, Davies A A, Benson F E, West S C. Resolution of recombination intermediates by a mammalian activity functionally analogous to Escherichia coli RuvC resolvase. J Biol Chem. 1994;269:5202–5209. [PubMed] [Google Scholar]
- 12.Kaplitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O’Malley K L, During M J. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 13.Lee J, Voziyanov Y, Pathania S, Jayaram M. Structural alterations and conformational dynamics in holliday junctions induced by binding of a site-specific recombinase. Mol Cell. 1998;1:483–493. doi: 10.1016/s1097-2765(00)80049-0. [DOI] [PubMed] [Google Scholar]
- 14.Linden R M, Winocour E, Berns K I. The recombination signals for adeno-associated virus site-specific integration. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobel L I, Murphy J E, Goff S P. The palindromic LTR-LTR junction of Moloney murine leukemia virus is not an efficient substrate for proviral integration. J Virol. 1989;63:2629–2637. doi: 10.1128/jvi.63.6.2629-2637.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao C H, Snyder R O, Schowalter D B, Patijn G A, Donahue B, Winther B, Kay M A. The kinetics of rAAV integration in the liver. Nat Genet. 1998;19:13–15. doi: 10.1038/ng0598-13. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 18.Muzyczka N. Viral expression vectors. Berlin, Germany: Springer-Verlag; 1992. [Google Scholar]
- 19.Panganiban A T. Retroviral DNA integration. Cell. 1985;42:5–6. doi: 10.1016/s0092-8674(85)80092-1. [DOI] [PubMed] [Google Scholar]
- 20.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 1994;13:5075–5085. doi: 10.1002/j.1460-2075.1994.tb06837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X S, Srivastava A. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 22.Qing K, Wang X S, Kube D M, Ponnazhagan S, Bajpai A, Srivastava A. Role of tyrosine phosphorylation of a cellular protein in adeno-associated virus 2-mediated transgene expression. Proc Natl Acad Sci USA. 1997;94:10879–10884. doi: 10.1073/pnas.94.20.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reich N C, Sarnow P, Duprey E, Levine A J. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology. 1983;128:480–484. doi: 10.1016/0042-6822(83)90274-x. [DOI] [PubMed] [Google Scholar]
- 24.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genet Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 25.Samulski R J, Chang L-S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder R O, Miao C H, Patijn G A, Spratt S K, Danos O, Nagy D, Gown A M, Winther B, Meuse L, Cohen L K, Thompson A R, Kay M A. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 27.Walsh C E, Nienhuis A W, Samulski R J, Brown M G, Miller J L, Young N S, Liu J M. Phenotypic correction of Fanconi anemia in human hematopoietic cells with a recombinant adeno-associated virus vector. J Clin Investig. 1994;94:1440–1448. doi: 10.1172/JCI117481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X-S, Qing K, Ponnazhagan S, Srivastava A. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]