Abstract
Replicase gene expression by the human coronavirus 229E involves the synthesis of two large polyproteins, pp1a and pp1ab. Experimental evidence suggests that these precursor molecules are subject to extensive proteolytic processing. In this study, we show that a chymotrypsin-like enzyme, the virus-encoded 3C-like proteinase (3CLpro), cleaves within a common region of pp1a and pp1ab (amino acids 3490 to 4068) at four sites. trans-cleavage assays revealed that polypeptides of 5, 23, 12, and 16 kDa are processed from pp1a/pp1ab by proteolysis of the peptide bonds Q3546/S3547, Q3629/S3630, Q3824/N3825, and Q3933/A3934. Relative rate constants for the 3CLpro-mediated cleavages Q2965/A2966, Q3267/S3268, Q3824/N3825, and Q3933/A3934 were derived by competition experiments using synthetic peptides and recombinant 3CLpro. The results indicate that coronavirus cleavage sites differ significantly with regard to their susceptibilities to proteolysis by 3CLpro. Finally, immunoprecipitation with specific rabbit antisera was used to detect the pp1a/pp1ab processing end products in virus-infected cells, and immunofluorescence data that suggest an association of these polypeptides with intracellular membranes were obtained.
The human coronaviruses (HCV) are causative agents of upper respiratory tract infections in both adults and children (18, 30). Recent epidemiological data, however, also provide evidence for the involvement of both prototype strains, HCV 229E and HCV OC43, in lower respiratory tract and gastrointestinal diseases (9, 28, 32, 43).
The HCV 229E genome is a positive-sense RNA of 27,277 nucleotides (13). Gene 1, or the replicase gene, which is located towards the 5′ end of the genome, is composed of two large, overlapping open reading frames (ORFs), ORF 1a and ORF 1b. ORF 1a encodes a polyprotein, pp1a, with a calculated molecular mass of 454 kDa. The downstream ORF 1b is expressed as a fusion protein with pp1a by a mechanism involving a −1 ribosomal frameshift during translation (13, 14). The ORF 1a/1b gene product has a calculated molecular mass of 754 kDa and is referred to as polyprotein 1ab or pp1ab.
Analyses of the deduced amino acid sequences of pp1a and pp1ab revealed that the coronavirus replicase polyproteins contain motifs characteristic of both papain-like cysteine proteinases and a chymotrypsin-like enzyme, the 3C-like proteinase (3CLpro) (7, 10, 13, 20). A number of different experimental approaches have now established that these enzymatic activities are indeed responsible for the proteolytic processing of pp1a and pp1ab (1, 2, 11, 12, 16, 17, 22, 24, 25, 27, 31, 37, 44). The data from these studies also demonstrate that the virus-encoded 3CLpro is responsible for the majority of cleavages within the coronavirus replicase polyproteins. Consequently, this enzyme has been studied extensively to elucidate its biochemical properties. In this context, data suggesting that the coronavirus 3CLpro possesses a catalytic dyad consisting of His and Cys have been reported (23, 27, 38, 44, 45). An acidic amino acid residue, regarded as typical for chymotrypsin-like enzymes, has, however, not been identified for any coronavirus 3CLpro characterized to date (23, 26, 45). This finding clearly separates the coronavirus 3CLpro from other viral chymotrypsin-like enzymes.
Previously, Gorbalenya and coworkers (10) identified several putative 3CLpro cleavage sites in the ORF 1a-encoded regions of pp1a and pp1ab. These included sites that flank the 3CLpro domain itself and a number of downstream sites. Recent data obtained for both the mouse hepatitis virus (MHV) and the avian infectious bronchitis virus (IBV) replicase polyproteins suggest that several of these cleavage sites are functional (MHV pp1a/pp1ab 22-kDa polypeptide [25] and IBV pp1a/pp1ab 10- and 24-kDa polypeptides [24, 31]). In this article, we present an analysis of the proteolytic processing of a region corresponding to the carboxyl terminus of the HCV 229E polyprotein 1a and the central portion of the polyprotein 1ab (amino acids 3490 to 4068). Our in vitro studies show that, within this region, the HCV 229E 3CLpro cleaves the replicase polyproteins at four sites, giving rise to polypeptides of 5, 12, 16, and 23 kDa. Moreover, as evidenced by competition peptide cleavage assays, there appear to be kinetic differences in the conversion of these substrates by recombinant 3CLpro. Specifically, a noncanonical Q/N peptide bond was found to be cleaved far less efficiently than the canonical sites, Q/A,S. Finally, we have identified the processing products in virus-infected cells by using specific rabbit antisera.
MATERIALS AND METHODS
Virus and cells.
The methods for HCV 229E propagation in MRC-5 cells (ECACC 84101801) and concentration of virus with polyethylene glycol have been described previously (44).
Preparation of antigens and antisera.
The ORF 1a nucleotide sequences coding for amino acids 3547 to 3629, 3630 to 3824, and 3934 to 4068 were amplified by PCR from pBS-T16D8 plasmid DNA (13) by standard procedures. Each of the downstream PCR primers contained the complementary sequence of a translation stop codon followed by an EcoRI restriction site. The PCR products were treated with T4 DNA polymerase, phosphorylated with polynucleotide kinase, digested with EcoRI, and ligated with XmnI- and EcoRI-digested pMal-c2 DNA (New England Biolabs, Schwalbach, Germany). The resultant plasmids, pMal-p5, pMal-p23, and pMal-p16, encode the specified ORF 1a amino acids fused to the Escherichia coli maltose-binding protein (MBP) (Fig. 1). The plasmids were used to transform competent E. coli TB1 cells. The bacterial fusion proteins were expressed and purified as described previously (15, 44). Subsequently, the virus-specific polypeptides were released from MBP by cleavage with endoproteinase Xa (Amersham Pharmacia Biotech, Freiburg, Germany) and used to immunize rabbits as described previously (44). The resulting antisera were designated α-p5, α-p23, and α-p16, respectively.
FIG. 1.
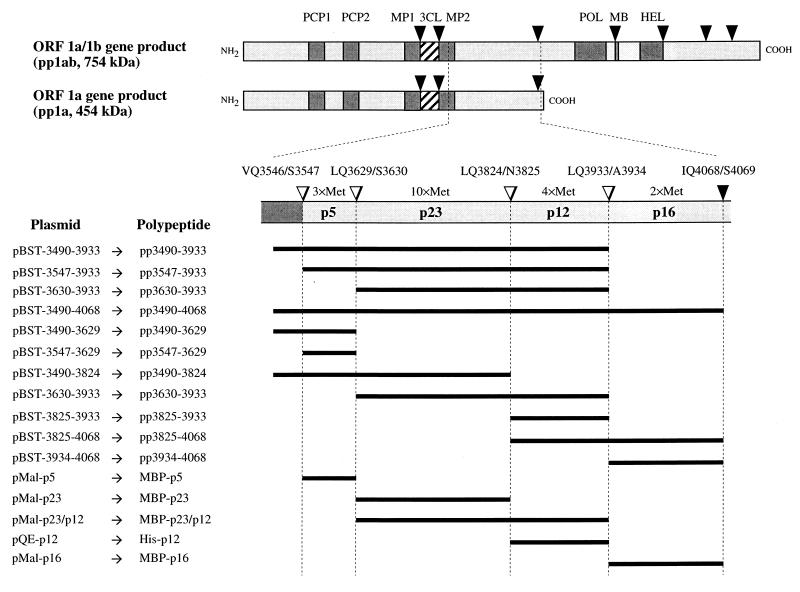
3CLpro-mediated processing of the replicase polyproteins pp1a and pp1ab. The triangles represent HCV 229E 3CLpro cleavage sites that have either been described previously (black) or identified during this study (white). The processing products described in this study are indicated, and their methionine residue numbers are given. The plasmids and polypeptides used are depicted on the left. The black lines designate these polypeptides in relation to their localization within pp1a and pp1ab. PCP1, papain-like cysteine proteinase 1; PCP2, putative papain-like cysteine proteinase 2; MP1, putative membrane protein 1; 3CL, 3C-like proteinase; MP2, putative membrane protein 2; POL, putative RNA polymerase domain; MB, putative metal ion-binding domain; HEL, putative helicase domain.
In order to express the ORF 1a-encoded amino acids 3825 to 3933, nucleotides 11765 to 12091 of the HCV 229E genomic sequence were amplified by PCR from pBS-T16D8 DNA. The upstream PCR primer contained a BamHI restriction site, and the downstream primer contained a translation stop codon and a HindIII restriction site. The PCR product was digested with BamHI and HindIII and ligated with BamHI- and HindIII-digested pQE10 DNA (Qiagen, Hilden, Germany). The resultant plasmid, pQE-p12, was used to express a histidine-tagged fusion protein, His-p12 (Fig. 1). Purification of the recombinant protein under denaturing conditions from E. coli M15 (pREP4) cells and immunization of rabbits have been described previously (44). The resulting antiserum was designated α-p12.
Metabolic labeling, cell lysis, and immunoprecipitation.
Infection or mock infection of MRC-5 cells was done essentially as described previously (44). Briefly, 6 × 105 MRC-5 cells were mock infected or infected with HCV 229E at a multiplicity of 15 PFU per cell. Radioactive labeling of newly synthesized proteins was done for 7 h at 33°C, between 5 and 12 h postinfection (p.i.). Before labeling, the cells were washed twice with methionine- and cysteine-free Dulbecco’s modified Eagle’s medium (Life Technologies, Eggenstein, Germany) supplemented with 2% dialyzed fetal bovine serum. Pro-Mix L-35S in vitro cell-labeling mixture (SJQ 0079; Amersham Pharmacia Biotech) was added to the labeling medium at concentrations of 100 μCi of l-[35S]methionine and 42 μCi of l-[35S]cysteine per ml. After labeling, the cells were lysed and immunoprecipitation was done essentially as described previously (44). To improve the specificity of immunoprecipitation with the α-p16 serum, the protocol was modified as follows. One hundred microliters of cell lysate was mixed with 400 μl of immunoprecipitation buffer (44) and incubated with 25 μl of protein A–Sepharose CL-4B (150 mg/ml) (P3391; Sigma, Deisenhofen, Germany). After 60 min at 4°C, the protein A-Sepharose was removed by centrifugation, and 5 μl of preimmune serum or 5 μl of α-p16 serum was added to the supernatant. This mixture was incubated for 105 min at 4°C. Thereafter, protein A-Sepharose (25 μl, 150 mg/ml) was used to isolate the immune complexes, which were washed and eluted as previously described (44). The immunoprecipitated proteins were analyzed by sodium dodecyl sulfate (SDS)–17.4% polyacrylamide gel electrophoresis and autoradiography.
Immunofluorescence assay.
MRC-5 cells were grown on coverslips, infected with HCV 229E at a multiplicity of 10 PFU per cell, and incubated at 33°C. At 11 h p.i., the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) and washed with 1% Nonidet P-40 in PBS. Following permeabilization with 0.2% Triton X-100 in PBS, indirect immunofluorescence assays were done with α-p5, α-p23, α-p12, α-p16, and the appropriate preimmune sera at a 1:100 dilution in PBS containing 1% Nonidet P-40 and 5% normal goat serum. A fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin (1:200 dilution; Dianova, Hamburg, Germany) was used as the secondary antibody.
Construction of expression plasmids, in vitro translation, and 3CLpro cleavage assay.
The expression plasmid pBS-T (11) facilitates the generation of synthetic RNAs with T7 RNA polymerase, and it provides each RNA with a translation start codon in an optimal context. Cloning of insert DNA into the unique BamHI restriction site of pBS-T leads to the expression of polypeptides with a vector-encoded amino-terminal sequence, Met-Asp-Pro. A set of PCR products representing different coding sequences of ORF 1a were ligated with BamHI- and EcoRI-digested pBS-T DNA (Fig. 1). Each of these PCR products was generated from pBS-T16D8 template DNA by using primers containing either a BamHI restriction site (upstream primer) or a translation stop codon followed by an EcoRI restriction site (downstream primer). The resultant 11 plasmids were designated pBST-3490-3933 through pBST-3934-4068 (Fig. 1). Capped RNAs derived from these plasmids were translated in a reticulocyte lysate (Promega, Heidelberg, Germany) in the presence of [35S]methionine. After 40 min, the translation reactions (15-μl mixtures) were stopped by the addition of 1.7 μl of 10× translation stop mix (0.1 mg of RNase A per ml, 10 mg of cycloheximide per ml, 5 mM [32S]methionine), and the mixtures were divided into two aliquots. Next, 1 μl of recombinant 3CLpro (10 mg/ml in 10 mM Tris-HCl [pH 7.35]–200 mM NaCl–0.1 mM EDTA–1 mM dithiothreitol) (45) was added to one of the aliquots. As a control, 1 μl of the identical buffer was added to the second aliquot. After 45- or 90-min incubation periods at 30°C, 0.2 μl of each reaction mixture was analyzed by SDS–17.4% polyacrylamide gel electrophoresis and autoradiography.
Cleavage of MBP-p23/p12 by recombinant 3CLpro and amino-terminal sequence analysis.
The nucleotide sequence coding for the pp1a/1ab amino acids 3630 to 3933 was amplified by PCR from pBS-T16D8 template DNA. The downstream primer contained a translation stop codon followed by an EcoRI restriction site. After T4 DNA polymerase and polynucleotide kinase treatment, the DNA was digested with EcoRI and inserted into XmnI- and EcoRI-digested pMal-c2 DNA. Expression and purification of the MBP fusion protein were done as described above. Approximately 30 μg of the affinity-purified fusion protein, MBP-p23/p12 (Fig. 1), was incubated with 20 μg of recombinant 3CLpro in a buffer containing 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol for 16 h at 20°C. Thereafter, the reaction mixture was separated on an SDS–17.4% polyacrylamide gel and transferred electrophoretically to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Munich, Germany). Protein staining and amino-terminal sequence analysis of the membrane-bound protein were done as described previously (11).
Peptide synthesis.
Four synthetic 15-mer peptides, SP1, SP4, SP5, and SP6, were used in this study. They contain the following pp1a/pp1ab-derived amino acids: SP1, VSYGSTLQAGLRKMA; SP4, QMFGVNLQSGKTTSM; SP5, CERVVKLQNNEIMPG; and SP6, IGATVRLQAGKQTEF. The peptides were prepared by solid-phase chemistry (29) and purified by high-performance liquid chromatography (HPLC) on a reversed-phase C18 silica column (Jerini Bio-Tools, Berlin, Germany). The identity and homogeneity of the peptides were confirmed by mass spectrometry and analytical reversed-phase chromatography.
Competition peptide cleavage assays for (Vmax/Km)rel determination.
The peptide cleavage reaction mixtures were incubated at 20°C in buffer consisting of 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, 0.5 mM EDTA, and 0.5 mM dithiothreitol. Recombinant HCV 229E 3CLpro (final concentration, 0.77 μM) was added to a mixture of two substrate peptides, both at 370 μM. The peptide SP1, which has been shown to be cleaved rapidly by recombinant 3CLpro (45), was used as the standard peptide in each assay. Reaction aliquots were separated by reversed-phase chromatography on a Delta Pak C18 column as described previously (45), and the elution profile was monitored at 215 nm. Quantitation of the peak areas was used to determine the extent of substrate conversion. The data obtained were analyzed as described by Pallai et al. (34) to give the (Vmax/Km)rel value. This was calculated for peptides X and SP1 by using the equation (Vmax/Km)x/ (Vmax/Km)SP1 = log(1 − Fx)/log(1 − FSP1), where F is the fraction of substrate that is converted to product (33). Each value reported is an average from at least four experiments and is reproducible to ±20%. In all competition experiments, the substrate and product peptides were separable from each other, and the area under the combined peaks was independent of the extent of conversion of the substrate.
RESULTS
Identification of 3CLpro cleavage sites within amino acids 3490 to 4068 of pp1a/pp1ab.
In our previous studies, we have used recombinant HCV 229E 3CLpro, in combination with trans-cleavage assays and microsequencing, to identify a total of seven 3CLpro cleavage sites in the HCV replicase polyproteins. These include three sites common to pp1a and pp1ab (including the two sites that flank the 3CLpro domain itself) and four sites unique to pp1ab (11, 16, 17, 44, 45). In this study, we extended this approach to the analysis of 3CLpro cleavage sites in the carboxyl-terminal portion of pp1a, which also corresponds to the central portion of pp1ab.
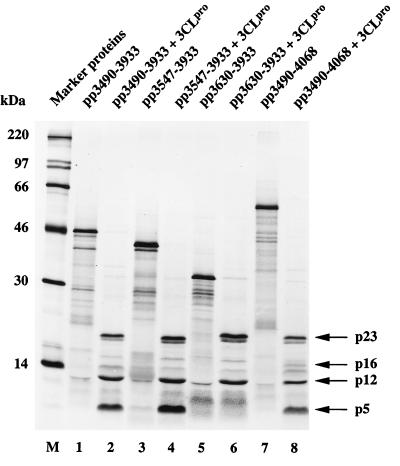
In an initial series of experiments, we constructed four plasmids, pBST-3490-3933, pBST-3547-3933, pBST-3630-3933, and pBST-3490-4068, that facilitated the in vitro synthesis of putative 3CLpro substrates in a reticulocyte lysate system (Fig. 1 and 2). The in vitro-translated substrate polypeptides were then incubated with highly purified, bacterially expressed HCV 229E 3CLpro. The results of this experiment (Fig. 2) show that all substrates (lanes 1, 3, 5, and 7) were proteolytically processed upon addition of recombinant 3CLpro (lanes 2, 4, 6, and 8). With substrates pp3490-3933 and pp3547-3933 (Fig. 2, lanes 2 and 4), three major cleavage products (p5, p12, and p23) could be identified. With substrate pp3630-3933 (Fig. 2, lane 6), two major cleavage products (p12 and p23) could be identified, and with substrate pp3490-4068 (lane 8), three major cleavage products (p5, p12, and p23) and one minor cleavage product (p16) could be identified. These initial data are consistent with the location of one cleavage site in pp3630-3933, two cleavage sites in pp3547-3933, at least two cleavage sites in pp3490-3933, and at least three cleavage sites in pp3490-4068. The data also suggest that the cleavage product p5 is amino terminal and the cleavage product p16 is carboxyl terminal. This interpretation is consistent with the predictions of 3CLpro cleavage sites made originally by Gorbalenya et al. (10) for IBV and with the recent results of Liu et al. (24), Lu et al. (25), and Ng and Liu (31).
FIG. 2.
Identification of 3CLpro cleavage sites by trans-cleavage assay. In vitro-translated substrates encoded by HCV 229E ORF 1a were incubated with buffer (lanes 1, 3, 5, and 7) or buffer containing recombinant 3CLpro (lanes 2, 4, 6, and 8) for 90 min at 30°C as described in Materials and Methods. The reaction mixtures were separated by SDS–17.4% polyacrylamide gel electrophoresis and analyzed by autoradiography. Sizes of molecular mass markers (lane M) (CFA 626; Amersham Pharmacia Biotech) are given on the left, and the major cleavage products are indicated by arrows on the right.
Mapping of the cleavage sites by trans-cleavage assay with recombinant 3CLpro.
To identify each of the cleavage products observed in the experiment described above and to localize the cleavage sites more precisely, we have produced an additional series of four plasmids (pBST-3490-3629, pBST-3490-3824, pBST-3630-3933, and pBST-3825-4068) that can be used to generate synthetic substrates by in vitro translation (Fig. 1). With one exception (pp3490-3824), each of these substrates contained only one predicted cleavage site, and their amino and carboxyl termini were also chosen according to the positions of predicted cleavage sites. In parallel, the peptide sequences of putative cleavage end products were translated from the plasmids pBST-3547-3629, pBST-3825-3933, and pBST-3934-4068 (Fig. 1). These translation products were used to show that the polypeptides were indeed processing end products (i.e., that they were not susceptible to further 3CLpro cleavage), and they also served as size markers in SDS-polyacrylamide gel electrophoresis.
The results shown in Fig. 3A (lanes 1 and 2) indicate that the substrate pp3490-3629 was cleaved specifically by 3CLpro to yield a p5 cleavage product. This polypeptide migrated with the ORF 1a-encoded peptide sequence from amino acid 3547 to 3629, translated from pBST-3547-3629 (Fig. 3A, lanes 3 and 4). This result suggests that p5 encompasses the pp1a/pp1ab amino acids 3547 to 3629, after cleavages at the predicted Q3546/S3547 and Q3629/S3630 residues. Also, p5 represents a processing end product, as no further processing could be seen after incubation with 3CLpro (Fig. 3A, lanes 3 and 4). It should be noted that the apparent molecular mass of p5 differs significantly from the calculated size of 9.3 kDa. Also, in this experiment, no second cleavage product was identified. We surmise that the amino-terminal cleavage product, presumably encompassing pp1a/pp1ab amino acids 3490 to 3546 and thus representing the carboxyl terminus of the so-called membrane protein 2 (20), either comigrates with p5 or is not resolved in our gel system.
FIG. 3.
Mapping of 3CLpro cleavage sites. (A) trans-cleavage assays of pp1a/pp1ab amino acids 3490 to 3824. (B) trans-cleavage assays of pp1a/pp1ab amino acids 3630 to 4068. In vitro-translated substrate polypeptides were incubated with buffer (lanes 1, 3, and 5 in panel A and lanes 1, 3, 5, and 7 in panel B) or buffer containing recombinant 3CLpro (lanes 2, 4, and 6 in panel A and lanes 2, 4, 6, and 8 in panel B) for 45 min at 30°C, separated by SDS–17.4% polyacrylamide gel electrophoresis, and analyzed by autoradiography. Sizes of molecular mass markers (lane M) (CFA 626 and 645; Amersham Pharmacia Biotech) are given, and the major cleavage products are indicated.
The second substrate analyzed was pp3490-3824. In this case, 3CLpro cleavage resulted in two products, p5 and p23 (Fig. 3A, lanes 5 and 6). From these data, we concluded that p23 represents the adjacent cleavage product that is released from p5 at the predicted Q3629/S3630 peptide bond. Additionally, in this experiment, a 14-kDa intermediate was observed, most probably because the amino-proximal cleavage site remained uncleaved, thus giving rise to a polypeptide encompassing amino acids 3490 to 3629. This interpretation is supported by the comigration of the 14-kDa polypeptide with pp3490-3629 (Fig. 3A, lane 1).
The third substrate analyzed was pp3630-3933, a polypeptide delimited by the predicted cleavage sites Q3629/S3630 and Q3933/A3934. After incubation with 3CLpro (Fig. 3B, lanes 1 and 2) two polypeptides, p23 and p12, could be readily identified, despite a high background resulting from (presumably) premature termination products of substrate translation. From its electrophoretic mobility (compared to the 23-kDa cleavage product of pp3490-3824), p23 was identified as the amino-terminal cleavage product. Thus, it appeared that, as has been shown for the IBV replicase polyproteins by Liu et al. (24), the HCV 229E replicase polyproteins are also processed at this noncanonical Q3824/N3825 cleavage site. This conclusion was confirmed by the comigration of the pp1a/pp1ab amino acids 3825 to 3933 with p12 (Fig. 3B, lanes 2, 3, and 4). Again, p12 represents an end product, since no further processing was observed upon incubation with 3CLpro (Fig. 3B, lanes 3 and 4).
Finally, the polypeptide substrate pp3825-4068, which contained the remaining pp1a/pp1ab sequence up to the cleavage site liberating the amino terminus of the putative RNA polymerase (11), was incubated with 3CLpro. In this case, the processing products p12 and p16 (Fig. 3B, lanes 5 and 6) were generated. Comigration of p12 with pp3825-3933 (Fig. 3B, lanes 3 and 4) indicates an amino-terminal location in pp3825-4068. The comigration of p16 with pp3934-4068 (Fig. 3B, lanes 7 and 8) verifies its identity and confirms that it is a processing end product. We concluded that p16, which contains a putative growth factor-like domain (10), is released from pp1a/pp1ab by cleavage at Q3933/A3934. Previously, the carboxyl terminus of p16 had been determined indirectly by amino-terminal sequence analysis of the downstream-encoded, 105-kDa polymerase polypeptide (11).
In summary, the data show that four polypeptides, p5, p23, p12, and p16, are processed from pp1a/pp1ab by 3CLpro-mediated proteolysis of the peptide bonds Q3546/S3547, Q3629/S3630, Q3824/N3825, and Q3933/A3934.
Identification of a noncanonical HCV 229E 3CLpro cleavage site by amino-terminal sequence analysis.
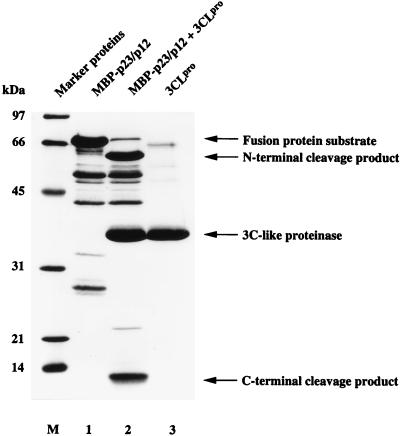
This study and the results of Liu et al. (24) and Lu et al. (25) provide strong, but nevertheless indirect, evidence that a noncanonical Q/N peptide bond represents a functional substrate of coronavirus 3CLpro. To substantiate this conclusion, we have expressed the pp1a/pp1ab amino acids 3629 to 3933 fused to E. coli MBP (MBP-p23/p12) in bacteria. After amylose affinity purification (Fig. 4, lane 1), the recombinant fusion protein, containing the putative Q3824/N3825 cleavage site, was incubated with recombinant 3CLpro for 16 h at 20°C. As Fig. 4 (lane 2) shows, the substrate was converted into two major products. The smaller cleavage product had, as expected, an apparent molecular mass of 12 kDa (compare Fig. 2, 3B, and 4). After transfer of the 12-kDa polypeptide to a polyvinylidene difluoride membrane, the amino-terminal sequence was determined by automated Edman degradation and HPLC analysis. The chromatographic analysis of the first five sequencing cycles is shown in Fig. 5. It shows conclusively that the amino-terminal residues of p12 are Asn-Asn-Glu-Ile-Met. This result is consistent with a 3CLpro-mediated cleavage at the pp1a/pp1ab Q3824/N3825 residues, and it confirms that the HCV 229E 3CLpro has a broader specificity than previously assumed (45).
FIG. 4.
Proteolytic cleavage of bacterially synthesized MBP-p23/p12. The substrate fusion protein containing the HCV 229E ORF 1a-encoded amino acids 3630 to 3933 was incubated with buffer (lane 1) or buffer containing recombinant 3CLpro (lane 2) for 16 h at 20°C and analyzed by SDS–17.4% polyacrylamide gel electrophoresis. Recombinant 3CLpro was electrophoresed in lane 3. Sizes of molecular mass markers (lane M) (Bio-Rad Laboratories) are given on the left. The positions of the fusion protein substrate, the major cleavage products, and 3CLpro are indicated by arrows on the right.
FIG. 5.
Amino-terminal sequence analysis of the 12-kDa, carboxyl-terminal MBP-p23/p12 cleavage product. After cleavage of MBP-p13/p12 with recombinant 3CLpro, separation by SDS-polyacrylamide gel electrophoresis, and transfer to a polyvinylidene difluoride membrane, the 12-kDa cleavage product was subjected to Edman degradation. Phenylthiohydantoin-amino acids generated during each reaction cycle were detected by their absorbance at 269 nm (expressed as milli-absorbance units [mAU]) and identified by their characteristic retention times on a reversed-phase HPLC support. (a) Chromatogram of phenylthiohydantoin-amino acid standards; (b to f) chromatograms of phenylthiohydantoin-amino acids from reaction cycles 1 to 5, respectively. Specific peaks of phenylthiohydantoin-amino acids are indicated in the single-letter code.
Relative rates of cleavage of pp1a/pp1ab-derived 3CLpro substrates.
Having identified a large number of cleavage sites of the HCV 229E 3CLpro (11, 16, 17, 44, 45), we were interested to see if different sites are equally susceptible to proteolytic cleavage. Clearly, differences in the susceptibility of cleavage sites may translate into differences in the relative accumulation of intermediary and end products of replicase polyprotein processing, and this, in turn, could represent an important regulatory mechanism during virus replication. Our approach was again based upon the trans-cleavage activity of recombinant 3CLpro, in combination with synthetic peptide substrates. Thus, we have chosen to synthesize four different 15-mer peptides, each containing one pp1a/pp1ab cleavage site. Two of these peptides (SP1 and SP4) contained the cleavage sites flanking the 3CLpro domain itself, the third (SP5) represented the noncanonical Q3824/N3825 cleavage site, and the fourth (SP6) represented the Q3933/A3934 cleavage site separating p12 from p16.
Relative Vmax/Km values for all four substrates were obtained in experiments where two peptides, SP1 and another, were incubated simultaneously with 3CLpro. The substrates thus competed for the active site. This approach is economical and is not affected by variations in enzyme activity in different experiments. Competition of peptide SP1 with each of the three other peptides resulted in the relative Vmax/Km values that are shown in Table 1. The data show that SP1 and SP4 are converted significantly faster than SP5 and SP6. Thus, we conclude, for example, that the 3CLpro-flanking cleavage sites are processed approximately 10-fold more actively than the cleavage site represented by the peptide SP5. It is important to note that, besides the primary structure of the cleavage site, the polyprotein conformation, as well as intramolecular versus intermolecular cleavage, may also contribute significantly to the accessibility of specific cleavage sites.
TABLE 1.
Relative rates of cleavage of HCV 229E 3CLpro peptide substrates
| Cleavage sitea | Peptide | Sequenceb
|
(Vmax/Km)relc | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P8 | P7 | P6 | P5 | P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | P5′ | P6′ | P7′ | ||||
| MP1/3CLpro | SP1 | V | S | Y | G | S | T | L | Q | ↓ | A | G | L | R | K | M | A | 1.00 |
| 3CLpro/MP2 | SP4 | Q | M | F | G | V | N | L | Q | ↓ | S | G | K | T | T | S | M | 1.17 |
| p23/p12 | SP5 | C | E | R | V | V | K | L | Q | ↓ | N | N | E | I | M | P | G | 0.09 |
| p12/p16 | SP6 | I | G | A | T | V | R | L | Q | ↓ | A | G | K | Q | T | E | F | 0.51 |
MP1, putative membrane protein 1 (10, 13, 20); 3CLpro, 3C-like proteinase (44, 45); MP2, putative membrane protein 2 (10, 13, 20); p23, ORF 1a-encoded, 23-kDa processing product (this study); p12, ORF 1a-encoded, 12-kDa processing product (this study); p16, ORF 1a-encoded, 16-kDa processing product (this study).
Amino acids flanking the proteinase cleavage sites are designated according to the scheme introduced by Schechter and Berger (36). The residues are numbered toward the carboxyl terminus as follows: P3-P2-P1↓P1′-P2′-P3′.
Relative Vmax/Km values were determined by using competition experiments as described in Materials and Methods with SP1 as the reference peptide.
Detection of ORF 1a-encoded polypeptides in virus-infected cells.
Our in vitro studies with recombinant HCV 229E 3CLpro have allowed us to establish a tentative processing scheme for the regions of pp1a and pp1ab encompassing amino acids 3490 to 4068. To support this scheme, we sought to confirm our in vitro data by the identification of the corresponding processing products in vivo, i.e., in HCV-infected MRC-5 cells. To do this, we first expressed four polypeptides, MBP-p5, MBP-p23, MBP-p16, and His-p12 (Fig. 1), in E. coli. These bacterial fusion proteins contained the exact peptide sequences of the p5, p23, p12, and p16 processing products, as defined by the in vitro trans-cleavage assay described above. The proteins were affinity purified on either amylose columns (MBP-p5, MBP-p23, and MBP-p16) or nickel-nitrilotriacetic acid-agarose columns (His-p12) and used to produce specific antisera in rabbits. These antisera were initially used to immunoprecipitate ORF 1a-encoded polypeptides from HCV 229E-infected MRC-5 cells.
The results of this experiment are shown in Fig. 6A. The p5-specific antiserum, α-p5, precipitated a protein of 5 kDa (Fig. 6A, lane 4). Antiserum α-p23 precipitated a protein of 23 kDa (Fig. 6A, lane 8), and antiserum α-p12 precipitated a protein of 12 kDa (lane 12). A number of controls were included to demonstrate the specificity of the immunoprecipitations. As expected, the proteins were not detected with the appropriate preimmune sera or in mock-infected cells (Fig. 6A, lanes 1 to 3, 5 to 7, and 9 to 11).
FIG. 6.
Detection of ORF 1a-encoded 3CLpro cleavage products in HCV 229E-infected cells. (A) Metabolically labeled lysates from mock-infected (M) (lanes 1, 3, 5, 7, 9, 11, 13, and 15) or HCV 229E-infected (I) (lanes 2, 4, 6, 8, 10, 12, 14, and 16) MRC-5 cells were analyzed by SDS–17.4% polyacrylamide gel electrophoresis after immunoprecipitation with the pp1a-specific rabbit antisera α-p5, α-p23, α-p12, and α-p16 or the corresponding preimmune sera. The cells were labeled from 5 to 12 h p.i. Either 180 μl (lanes 1 to 4) or 70 μl (lanes 5 to 16) was analyzed after immunoprecipitation with preimmune serum (lanes 1, 2, 5, 6, 9, 10, 13, and 14) or with the appropriate antiserum as indicated (lanes 3, 4, 7, 8, 11, 12, 15, and 16). (B) Metabolically labeled lysates (100 μl) from mock-infected (lanes 1 and 2) or HCV 229E-infected (lanes 3 and 4) MRC-5 cells were analyzed by SDS–17.4% polyacrylamide gel electrophoresis after immunoprecipitation with antiserum α-p16 (lanes 2 and 4) or the corresponding preimmune serum (lanes 1 and 3). The cells were labeled from 5 to 12 h p.i. Sizes of protein molecular mass markers (lanes PM) (CFA626 and CFA645; Amersham Pharmacia Biotech) and the processing products p5, p12, p23, and p16 are indicated.
In the experiment shown in Fig. 6A, the p16-specific antiserum, α-p16, failed to produce clear results. Therefore, we used a modified immunoprecipitation protocol. This modification substantially improved the specificity of the assay and allowed the detection of the 16-kDa polypeptide in virus-infected cells, as shown in Fig. 6B. Taken together, these data confirm the pp1a/pp1ab processing scheme established on the basis of our in vitro experiments.
To conclude our study, we took advantage of the α-p5, α-p23, α-p12, and α-p16 antisera to examine the intracellular localization of these pp1a/pp1ab-derived cleavage products. To this end, MRC-5 cells were infected with HCV 229E at a multiplicity of 10 PFU per cell, fixed, permeabilized, and analyzed by indirect immunofluorescence. As early as 4 h p.i. (data not shown), pp1a/pp1ab-derived polypeptides accumulated in the infected cell and produced a typical punctate staining pattern (16). Over the course of infection the punctate staining became increasingly apparent. At 11 h p.i., this staining pattern was readily obtained with all four pp1a/pp1ab-specific antisera. As an example, a specific immunostaining of the p23 protein at this time point is shown in Fig. 7. The staining pattern confirms our previous results on the intracellular localization of HCV 229E pp1a/pp1ab proteins (16) and is consistent with the results reported for replicase gene-encoded polypeptides of other nidoviruses (37, 41). An association of nidovirus replicase polypeptides with intracellular membranes, probably the endoplasmic reticulum or the intermediate compartment, has been suggested (41).
FIG. 7.
Indirect immunofluorescence analysis of mock-infected (A) or HCV 229E-infected (B) MRC-5 cells. The cells were immunostained at 11 h p.i. with α-p23 rabbit antiserum and fluorescein isothiocyanate-conjugated goat antirabbit antibodies.
DISCUSSION
The expression of precursor polypeptides (polyproteins) that are proteolytically processed to yield individual functional polypeptides is a strategy employed by many positive-sense RNA viruses, several double-stranded RNA viruses, and all retroviruses (6, 19). In this process, multiple, mostly replicative functions are activated from a single precursor molecule. Both the Coronaviridae and the Arteriviridae, which have been recently united in the order Nidovirales (3), express complex replicase polyproteins that are extensively processed by a number of different virus-encoded proteinases (reviewed in reference 5).
In the case of HCV 229E, it has been previously shown that a virus proteinase, 3CLpro, cleaves the ORF 1b-encoded region of pp1ab at four sites, yielding polypeptides of 105, 71, 58, 41, and 34 kDa. Three of the five pp1ab cleavage end products have been identified in virus-infected cells (11, 16, 17). It is also known that three sites common to pp1a and pp1ab are cleaved by 3CLpro. These are the two sites flanking the 3CLpro domain itself and the cleavage site that gives rise to the amino terminus of p105 (11, 44, 45).
In the experiments reported here, we now extend the HCV replicase polyprotein processing scheme by showing that 3CLpro cleaves at four additional sites in the region between 3CLpro and the carboxyl terminus of pp1a. We were also able to show that the predicted cleavage end products, p5, p23, p12, and p16, are synthesized in virus-infected cells and that they (or their precursors) are membrane associated. A membrane association, most probably in the endoplasmic reticulum or intermediate compartment, has also been proposed for the MHV (37) and arterivirus (8, 41) replication complexes.
We feel it is important to state that, as reported earlier, the HCV 229E replicase cleavage products were present in very small amounts in virus-infected cells. Consequently, exceedingly long labeling periods had to be used in order to identify them. With the cell culture system available for HCV 229E, it is, in our opinion, unrealistic to attempt to perform pulse-chase experiments in order to investigate precursor-product relationships, as has been done, for example, for the arterivirus (39, 41, 42) and MHV replicase (4, 25, 37) polyproteins. To overcome these problems, we are currently developing a system to overexpress the HCV 229E replicase polyproteins by using vaccinia virus recombinants.
With regard to the localization of the coronavirus replicase complex, two hydrophobic domains have been identified in the ORF 1a-encoded region of pp1a/pp1ab (10, 13), and it has been shown that these domains are required for full 3CLpro activity in reticulocyte lysate systems (35, 40). One of these hydrophobic domains directly precedes the pp1a/pp1ab polypeptides analyzed in this study. It is reasonable to speculate that these membrane domains may determine the localization of the viral replication complex in the infected cell.
This study also contributes to the characterization of the substrate specificity of the HCV 229E 3CLpro. The data indicate, for example, that valine is tolerated at the P2 position. So far, only leucine or isoleucine has been identified as a functional residue at this position. However, our data also suggest that this substitution is likely to reduce the conversion rate of the substrate. Thus, our in vitro cleavage reactions with polypeptide substrates have revealed intermediate products containing a noncleaved V-Q3546/S3547 peptide bond (Fig. 2 and 3). In contrast, the canonical L-Q3629/S3630 and L-Q3933/A3934 cleavage sites are converted completely under the same experimental conditions. Another example of a reduced cleavage activity is the noncanonical L-Q3824/N3825 cleavage site. Again, under the conditions we have used, a substantial fraction of the polypeptide substrate remained uncleaved. In this particular case, peptide cleavage data (Table 1) support the idea that the observed reduction of conversion is due to the properties of the cleavage site itself rather than to the overall conformation of the polypeptide and the accessibility of the cleavage site.
The peptide cleavage data shown in Table 1 provide, for the first time, kinetic parameters of coronavirus 3CLpro-mediated cleavages. The differences we have observed might have important functional implications. Thus, they might suggest that a sequential, 3CLpro-dictated release of specific polypeptides leads to a timely, coordinated generation of replicative functions. Such proteinase-mediated replication regulation has also been reported for other positive-stranded RNA viruses. For example, the shutoff of the alphavirus minus-strand synthesis is believed to be controlled in this way (21). Recent data from Wassenaar et al. (42) on the alternative processing pathways of the arterivirus ORF1a polyprotein highlight the possible complexity of such processes during nidovirus replication.
In summary, this study confirms that the coronavirus 3CLpro plays a central role in the formation of a functional replication complex and the virus life cycle. Thus, this enzyme represents, in our opinion, the ideal target for the design of synthetic inhibitors to control coronavirus infections in both humans and animals.
ACKNOWLEDGMENTS
We thank V. Hoppe for protein sequence data.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 165/B1 and SI 357/2-1).
REFERENCES
- 1.Baker S C, Shieh C-K, Soe L H, Chang M-F, Vannier D M, Lai M M C. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonilla P J, Hughes S A, Weiss S R. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 4.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty W G, Semler B L. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eleouet J-F, Rasschaert D, Lambert P, Levy L, Vende P, Laude H. Complete sequence (20 kilobases) of the polyprotein-encoding gene 1 of transmissible gastroenteritis virus. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faaberg K S, Plagemann P G W. Membrane association of the C-terminal half of the open reading frame 1a protein of lactate dehydrogenase-elevating virus. Arch Virol. 1996;141:1337–1348. doi: 10.1007/BF01718835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falsey A R, McCann R M, Hall W J, Criddle M M, Formica M A, Wycoff D, Kolassa J E. The “common cold” in frail older persons: impact of rhinovirus and coronavirus in a senior daycare center. J Am Geriatr Soc. 1997;45:706–711. doi: 10.1111/j.1532-5415.1997.tb01474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grötzinger C, Heusipp G, Ziebuhr J, Harms U, Süss J, Siddell S G. Characterization of a 105-kDa polypeptide encoded in gene 1 of the human coronavirus HCV 229E. Virology. 1996;222:227–235. doi: 10.1006/viro.1996.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold J, Gorbalenya A E, Thiel V, Schelle B, Siddell S G. Proteolytic processing at the amino terminus of human coronavirus 229E gene 1-encoded polyproteins: identification of a papain-like proteinase and its substrate. J Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herold J, Raabe T, Schelle-Prinz B, Siddell S G. Nucleotide sequence of the human coronavirus 229E RNA polymerase locus. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herold J, Siddell S G. An ‘elaborated’ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold J, Siddell S G, Ziebuhr J. Characterization of coronavirus RNA polymerase gene products. Methods Enzymol. 1996;275:68–89. doi: 10.1016/S0076-6879(96)75007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heusipp G, Grötzinger C, Herold J, Siddell S G, Ziebuhr J. Identification and subcellular localization of a 41 kDa, polyprotein 1ab processing product in human coronavirus 229E-infected cells. J Gen Virol. 1997;78:2789–2794. doi: 10.1099/0022-1317-78-11-2789. [DOI] [PubMed] [Google Scholar]
- 17.Heusipp G, Harms U, Siddell S G, Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J Virol. 1997;71:5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston S, Holgate S. Epidemiology of viral respiratory tract infections. In: Myint S, Taylor-Robinson D, editors. Viral and other infections of the human respiratory tract. London, United Kingdom: Chapman and Hall; 1996. pp. 1–38. [Google Scholar]
- 19.Kräusslich H-G, Wimmer E. Viral proteinases. Annu Rev Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 20.Lee H J, Shieh C-K, Gorbalenya A E, Koonin E V, la Monica N, Tuler J, Bagdzhadzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D X, Brierley I, Tibbles K W, Brown T D K. A 100-kilodalton polypeptide encoded by open reading frame (ORF) 1b of the coronavirus infectious bronchitis virus is processed by ORF 1a products. J Virol. 1994;68:5772–5780. doi: 10.1128/jvi.68.9.5772-5780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu D X, Brown T D K. Characterization and mutational analysis of an ORF 1a-encoding proteinase domain responsible for proteolytic processing of the infectious bronchitis virus 1a/1b polyprotein. Virology. 1995;209:420–427. doi: 10.1006/viro.1995.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D X, Xu H Y, Brown T D K. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J Virol. 1997;71:1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X T, Sims A C, Denison M R. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J Virol. 1998;72:2265–2271. doi: 10.1128/jvi.72.3.2265-2271.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Y, Denison M R. Determinants of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makela M J, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimaki M, Blomqvist S, Hyypia T, Arstila P. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrifield R B. Automated synthesis of peptides. Science. 1965;150:178–185. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- 30.Myint S H. Human coronavirus infections. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 389–401. [Google Scholar]
- 31.Ng L F P, Liu D X. Identification of a 24-kDa polypeptide processed from the coronavirus infectious bronchitis virus 1a polyprotein by the 3C-like proteinase and determination of its cleavage sites. Virology. 1998;243:388–395. doi: 10.1006/viro.1998.9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson K G, Kent J, Hammersley V, Cancio E. Acute viral infections of upper respiratory tract in elderly people living in the community: comparative, prospective, population based study of disease burden. Br Med J. 1997;315:1060–1064. doi: 10.1136/bmj.315.7115.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Leary M H, Baughn R L. Acetoacetate decarboxylase. Identification of the rate-determining step in the primary amine catalyzed reaction and in the enzymic reaction. J Am Chem Soc. 1972;94:626–630. doi: 10.1021/ja00757a051. [DOI] [PubMed] [Google Scholar]
- 34.Pallai P V, Burkhardt F, Skoog M, Schreiner K, Bax P, Cohen K A, Hansen G, Palladino D E H, Harris K S, Nicklin M J, Wimmer E. Cleavage of synthetic peptides by purified poliovirus 3C proteinase. J Biol Chem. 1989;264:9738–9741. [PubMed] [Google Scholar]
- 35.Piñón J D, Mayreddy R R, Turner J D, Khan F S, Bonilla P J, Weiss S R. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology. 1997;230:309–322. doi: 10.1006/viro.1997.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schechter I, Berger A. On the size of the active site in proteases. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 37.Schiller J J, Kanjanahaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seybert A, Ziebuhr J, Siddell S G. Expression and characterization of a recombinant murine coronavirus 3C-like proteinase. J Gen Virol. 1997;78:71–75. doi: 10.1099/0022-1317-78-1-71. [DOI] [PubMed] [Google Scholar]
- 39.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tibbles K W, Brierley I, Cavanagh D, Brown T D K. Characterization in vitro of an autocatalytic processing activity associated with the predicted 3C-like proteinase domain of the coronavirus avian infectious bronchitis virus. J Virol. 1996;70:1923–1930. doi: 10.1128/jvi.70.3.1923-1930.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X M, Herbst W, Kousoulas K G, Storz J. Biological and genetic characterization of hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. 1994;44:152–161. doi: 10.1002/jmv.1890440207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziebuhr J, Herold J, Siddell S G. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J Virol. 1995;69:4331–4338. doi: 10.1128/jvi.69.7.4331-4338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziebuhr J, Heusipp G, Siddell S G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J Virol. 1997;71:3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]