Abstract
Background
The composition of the gut fungal microbiome, mycobiome, is likely associated with human health. Yet, the development of gut mycobiome is poorly understood in infants and children. Here we investigate how perinatal events influence the development of gut mycobiome.
Methods
In this prospective cohort study of 140 infants, we used ITS gene sequencing of fecal samples from birth to the age of 18 months. We compared gut mycobiome composition according to delivery mode and exposure to intrapartum antibiotics during vaginal delivery.
Results
At birth, gut mycobiome were dominated by the genus Candida, at 6-month stool samples by Malassezia and Cystofilobasidium, and the 18-month stool samples by Trichosporon and unidentified fungi. Perinatal factors altered mycobiome. At 18 months, gut mycobiome of infants born vaginally consisted mostly of Trichosporon (32%) and unidentified fungi (31%), while those born via Cesarean section delivery samples had mycobiome dominated by Saccharomyces (50%). At the age of 18 months, those exposed to intrapartum antibiotics had mycobiome dominated by Trichosporon (66%) not seen in those unexposed to antibiotics.
Conclusions
Delivery mode and exposure to intrapartum antibiotic prophylaxis were markedly associated with gut mycobiome composition from birth to 18 months of age.
Impact
The composition of the gut mycobiome is likely associated with human health. Yet, the development of gut mycobiome is poorly understood in infants and children.
In this prospective cohort study, delivery mode and exposure to intrapartum antibiotic prophylaxis were markedly associated with gut mycobiome composition from birth to 18 months of age.
The impact of intrapartum antibiotic prophylaxis on fungal microbiome in vaginally born infants, previously shown to influence gut bacteriome composition, may be explained by the interaction between bacteria and fungi.
Gut mycobiome composition likely deserves further investigation in relation to gut microbiome and health in children.
Introduction
The gut microbiome consists of bacteria, viruses, fungi, archaea, and protozoa, but while the human gut microbiome has been studied extensively, there are limited data available that extend beyond the bacterial microbiome, also known as the bacteriome. It is only recently that the number of studies of the gut mycobiome, i.e. fungi in the gut, has started to increase regarding both animals1–4 and humans.1,4–7
The composition of the gut mycobiome has been shown to be associated with human health. Decreased biodiversity and a higher prevalence of the genera Candida and Malassezia, as well as strong bacteria-fungi associations, have been linked with inflammatory bowel disease (IBD)8,9 and irritable bowel syndrome (IBS),8,10 and the gut mycobiome has been associated with obesity, particularly with fungi related to lipid and glucose metabolism.11 The presence of Candida in the gut microbiome has been thought to contribute to glucose metabolism disorders,12 and shifts in the gut mycobiome have also been associated with diseases, such as colorectal cancer,13 chronic liver diseases,14 atopic dermatitis,15 and multiple sclerosis.16 Fungal colonization of the gut likely starts at birth, at least partially through vertical transmission from mother to child.17,18 Willis et al. studied the presence of fungi in the feces of newborn infants and found a low biomass mycobiome.19 In preterm infants, mycobiome dominated by the genus Candida has been observed.20 There are limited data on early-life factors affecting gut mycobiome composition. We have shown previously that exposure to antibiotics at birth markedly changes the composition of the gut bacteriome21 and its main source shift from mother to other sources.22
We set out here to characterize the development of the gut mycobiome in a prospective cohort study of 140 newborn infants from birth to 18 months of age.
Materials and methods
Study design and population
This was a prospective cohort study of 140 newborn infants investigating the impact of early-life factors on the development of the gut mycobiome from birth to 18 months of age. The protocol was reviewed and found acceptable by Ethical Committee of Northern Ostrobothnia Hospital District at Oulu University Hospital, Finland, decision number 3/2016. The families gave their written informed consent in advance, and the study was conducted according to the relevant guidelines and restrictions for clinical research. Altogether 434 fecal samples were collected (Table 1), and 50 negative control samples (HyClone™ HyPure, Thermo Fisher Scientific, Waltham, MA) were prepared together with the fecal samples for contamination removal.
Table 1.
Fecal samples collected for the study by sampling time, delivery mode, and exposure to intrapartum antibiotics at birth (first-pass meconium, 6-month stool, 18-month stool) and by delivery mode and antibiotic usage (vaginal delivery, vaginal delivery with intrapartum antibiotics, C-section delivery with intrapartum antibiotics).
| At birth | At 6 months | At 18 months | |
|---|---|---|---|
| Vaginal delivery | 56 | 59 | 59 |
| Vaginal delivery with intrapartum antibiotics | 24 | 33 | 23 |
| C-section delivery with intrapartum antibiotics | 60 | 60 | 60 |
Sample collection
Meconium samples were obtained from a diaper by a midwife in the delivery room or by the nurse responsible for the child in the labor ward. The infant stool samples at 6 and 18 months were collected by the families at home. All the samples were stored at −80 °C until further processing.
DNA extraction
DNA was extracted from the meconium and stool samples using the DNeasy PowerSoil Pro kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. In all, 200 mg of sample was weighed out, and 1 ml of phosphate-buffered saline was added to each sample. The samples were homogenized by bead beating with Tissuelyzer (Qiagen) for 2 min at 25 Hz and incubated in ice for 1 min. This homogenization was repeated 1–3 times. The negative control samples and meconium samples with little material were not put through the Tissuelyzer homogenization, but instead the samples followed the protocol for vortex adapter homogenization. After homogenization, extraction was performed on a QIAcube Connect extraction machine (Qiagen) with the final elution set to 100 µl. The quantity and quality of the DNA were measured using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific).
PCR, sequencing, and analysis
Sequencing of the internal transcribed spacer 2 (ITS2) gene was performed using the primer fITS7b (5’-GTGARTCATCGAATCTTTG-3’) and the primers ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) with unique barcodes. PCR was conducted using the Phusion Flash High-Fidelity PCR master mix (Thermo Fisher Scientific) according to the manufacturer’s protocol. An additional negative control (sterile water, HyClone™ HyPure, Thermo Fisher Scientific) and a positive control, Mycobiome Genomic DNA Mix (msa-1010, ATCC, VA) were added to each PCR plate. The reactions were performed with an Applied Biosystems™ Veriti 96-Well Thermal Cycler machine (Thermo Fisher Scientific). The PCR program started with 2 min of initialization, followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 54 °C for 20 s and elongation at 72 °C for 30 s. The final elongation at 72 °C lasted for 7 min. The PCR products were imaged by agarose gel electrophoresis in 1.5% TAE agarose gel with ethidium bromide dye using a VersaDoc (Bio-Rad, Hercules, CA) imaging machine.
Sequencing was performed on an IonTorrent PGM platform using a method described previously,23 and the sequence data were analyzed using the QIIME2 platform (version 2021.2).24 Sequences less than 200 bp long were omitted from the analysis. QIIME2-implemented DADA2 was used to demultiplex and denoise the sequence data.25 Reads were trimmed at 15 and truncated at 160, and chimeric reads were filtered out. As microbiome studies are sensitive to contamination,26 we removed instances of environmental contamination from the samples with the R package decontam (version 1.14.0) using 50 negative controls (sterile water), using a prevalence-based method with a threshold of 0.5.27
To calculate the within-sample diversity or alpha diversity, we rarefied the samples at 1004 and used the Shannon Index and observed features as metrics, with Kruskal–Wallis H as the statistical test. Between-sample diversity, or beta diversity, was calculated using Principal Coordinate Analysis with Bray–Curtis Dissimilarity. PERMANOVA was used to measure the statistical significance of these results. All the statistical tests were further evaluated with a p value, for which 0.05 was considered statistically significant. For alpha diversity tests, we used a p value with a Benjamini–Hochberg false discovery rate. We used the UNITE database (version 8.3) for the taxonomic analysis,28 and analysis of composition of microbiomes (ANCOM)29 together with the Mann–Whitney U-test to analyze the statistical significance of the taxonomic differences between the sample groups. The images were produced using Rstudio (R version 4.2.2) with the package ggplot2 (version 3.4.0), and the panel images were finalized with Inkscape (version 1.1). The raw sequences were uploaded in BioProject with the accession number PRJNA831656.
Results
We enrolled 56 infants born vaginally, 24 born vaginally and exposed to intrapartum antibiotics, and 60 born via Cesarean section. The characteristics of the cohort are presented in Table 2.
Table 2.
Population characteristics of the participating infants (n = 140).
| Vaginal delivery n = 56 | Vaginal delivery with antibiotics n = 24 | C-section with antibiotics n = 60 | |
|---|---|---|---|
| Maternal characteristics | |||
| Maternal age (years), mean (SD) | 29.7 (5.4) | 28.5 (5.2) | 31.6 (6.5) |
| Number of siblings, mean (SD) | 1.5 (2.6) | 1.0 (1.7) | 1.3 (1.4) |
| Maternal asthma, N (%) | 5 (8.9) | 1 (4.2) | 11 (18.3) |
| Maternal allergy, N (%) | 14 (25.0) | 5 (20.8) | 20 (33.3) |
| GDM | 5 (8.9) | 6 (25.0) | 16 (26.7) |
| Smoking during pregnancy | 3 (5.4) | 2 (8.3) | 12 (20.0) |
| Str. agalactiae positivea | 1 (1.8) | 21 (87.5) | 9 (15.0) |
| Antibiotics during pregnancyb | 8 (14.3) | 12 (50) | 16 (26.7) |
| Newborn characteristics | |||
| Female (%) | 24 (42.9) | 10 (41.7) | 29 (48.3) |
| Gestational age (weeks), mean (SD) | 39.9 (1.1) | 39.9 (1.3) | 38.9 (1.3) |
| Birth weight (g), mean (SD) | 3480 (360) | 3540 (550) | 3490 (610) |
| Apgar 1 min, mean (SD) | 8.8 (1.0) | 8.3 (1.6) | 8.7 (1.0) |
| Apgar 5 min, mean (SD) | 9.2 (0.8) | 9.0 (0.9) | 9.1 (0.6) |
| Apgar 15 min, mean (SD) | 9.5 (0.6) | 9.2 (0.9) | 9.4 (0.5) |
| Perinatal antibiotics, N (%)c | 0 | 1 (4.2) | 1 (1.7) |
GDM gestational diabetes mellitus.
aMaternal S. agalactiae screening was not performed for 24 mothers in the C-section group.
bOne mother in the vaginal delivery group had received cephalexin, 1 amoxicillin, 1 nitrofurantoin, 1 pivmecillinam, and 1 topical metronidatzole. For 4 mothers, the antibiotic used was not recorded. Two mothers in the vaginal delivery with antibiotics group had received pivmecillinam and 1 had received amoxicillin, but the antibiotic used was not recorded for 9 mothers. Two mothers in the C-section group had received cephalexin, 2 amoxicillin, 2 topical metronidatzole, 1 nitrofurantoin, and 1 clindamycin. For 8 mothers, the antibiotic had not been recorded.
cBoth children received a combination of benzyl penicillin and tobramycin.
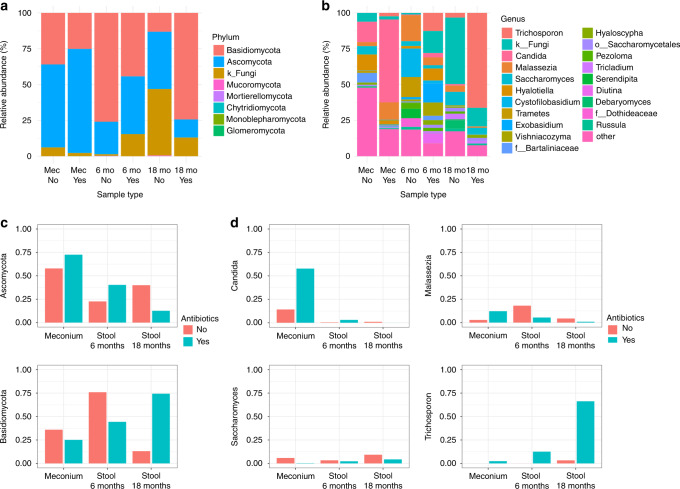
We compared the fungal taxonomies at three time points: at birth (first-pass meconium), at 6 months of age, and at 18 months of age. The most abundant phyla in the meconium samples were Ascomycota (60%) and Basidiomycota (35%) (Fig. 1a and Supplementary Information 1), while the most common genera were Candida (24%), Hyalotiella (8.9%), and Malassezia (7.7%) (Fig. 1b and Supplementary Information 1). At the age of 6 months, Basidiomycota was the most common phylum (61%), followed by Ascomycota (32%) (Fig. 1a and Supplementary Information 1), and Candida did not appear as the most abundant genus any longer, being replaced by Malassezia (17%), Cystofilobasidium (11%), Trametes (7.7%), and others (Fig. 1b and Supplementary Information 1). At the age of 18 months, Ascomycota and Basidiomycota were still the most abundant phyla (39% and 35%, respectively), but unidentified fungi had risen in prevalence (26%) (Fig. 1a and Supplementary Information 1). Trichosporon became the most abundant taxon (26%), followed by unidentified fungi (26%) and Saccharomyces (17%) (Fig. 1b and Supplementary Information 1). ANCOM analysis showed significant temporal changes in Ascomycota and Basidiomycota abundances from birth to the age of 18 months (Supplementary Information 2), and a total of 29 fungal genera showed differential abundances between the meconium, the 6-month stool and the 18-month stool (Supplementary Information 2).
Fig. 1. Taxonomic figures, alpha diversity, and beta diversity of all the stool samples by age at sampling.
a All the phyla present in the samples for each time point. b The 20 most common genera in the samples for each time point. The rest of the genera are collapsed in the “other” group. c Shannon Index, using Kruskal–Wallis H as the statistical test. d Observed features, using Kruskal–Wallis H as the statistical test. e Bray–Curtis Dissimilarity between samples based on the sample age group. PERMANOVA was used as the statistical test for beta diversity.
The alpha diversity also showed significant differences between the three time points. The meconium samples showed the most diversity in terms of both the Shannon Index and observed features, while there was a drop in diversity in the 6-month samples followed by a rise in the 18-month samples (Fig. 1c, d). In the case of beta diversity, however, the samples obtained at birth and at the age of 6 months were clustered together, whereas the 18-month stool samples showed more diversity (Fig. 1e).
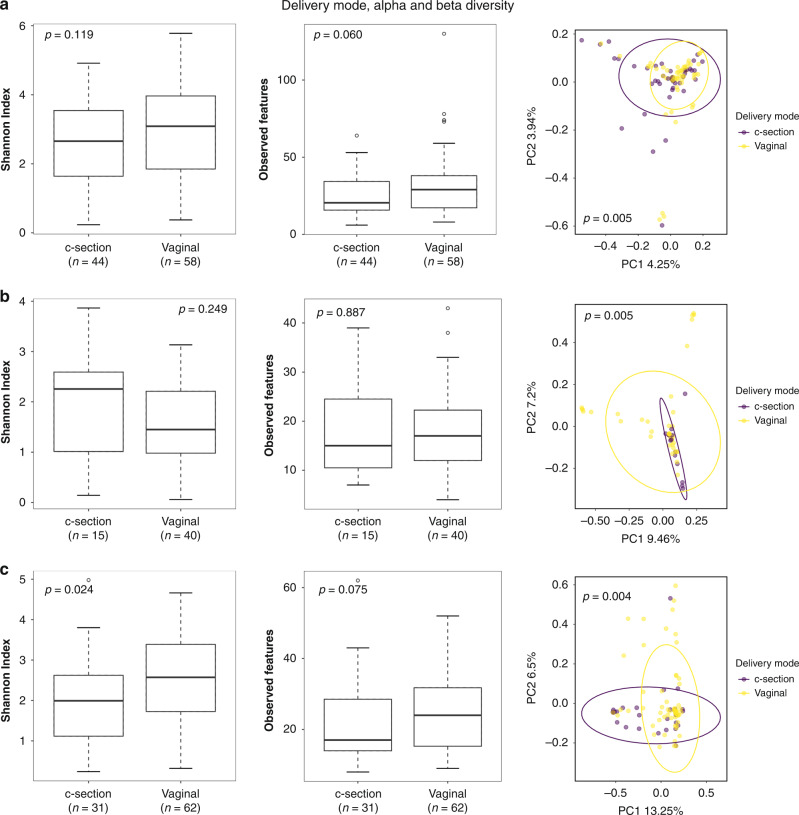
Delivery mode affects the gut mycobiome of newborns and infants
The stool samples differed at all three time points based on the delivery mode. In meconium samples, Candida was the most abundant genus in the vaginal samples (27%) and differed most notably between the vaginal and Cesarean delivery samples, whereas Malassezia (17%) was the most abundant genus in the Cesarean delivery samples (Fig. 2 and Supplementary Information 3). In 6-month stool samples, the most abundant genera were Cystofilobasidium (14%) and Malassezia (14%), while Malassezia remained the most abundant genus in the Cesarean delivery samples (29%) (Fig. 2 and Supplementary Information 3). In 18-month stool samples, the differences had grown, and the vaginal delivery samples mostly consisted of Trichosporon (32%) and unidentified fungi (31%), while the Cesarean delivery samples were dominated by Saccharomyces (50%) (Fig. 2 and Supplementary Information 3). The ANCOM and Mann–Whitney U-tests showed that differential abundances were found in 5 genera in the meconium, in 6 phyla and 3 genera in the 6-month stool samples, and 6 phyla and 4 genera in the 18-month stool samples (Supplementary Information 4).
Fig. 2. Taxonomic figures for each sample type based on the mode of delivery.
a Relative abundances of the taxa at the phylum level. b The top 20 most abundant genera with the rest collapsed into the “other” group. c Changes in the abundance of Ascomycota (top) and Basidiomycota (bottom) over time in the vaginal delivery and Cesarean delivery groups. d Changes in the abundance of Candida (top left), Malassezia (top right), Saccharomyces (bottom left) and Trichosporon (bottom right) over time in the vaginal delivery and Cesarean delivery groups.
No significant differences in alpha diversity were seen between the delivery modes until the age of 18 months (Fig. 3), but the beta diversity showed significant differences between the delivery modes at all three time points (Fig. 3). The Cesarean delivery samples showed higher diversity in meconium, but by 6 and 18 months the vaginal delivery samples had diversified more (Fig. 3).
Fig. 3. Alpha and beta diversity of all the samples by the mode of delivery.
a Meconium, b 6-month stool, c 18-month stool. Alpha diversity metrics used: Shannon Index, observed features, with the Kruskal–Wallis H used as a statistical test. Beta diversity metric used: Bray–Curtis Dissimilarity, with PERMANOVA used as a statistical test. p values are shown in the respective figures.
Intrapartum antibiotics affect the gut mycobiome in newborns and infants
Differences based on intrapartum antibiotic usage were also found between the vaginal delivery samples. The meconium samples from the vaginal births with no intrapartum antibiotic exposure consisted of Candida (14%) and Hyalotiella (11%) as well as many other low-abundance taxa (44%), while in the vaginal samples exposed to intrapartum antibiotic treatment over half of the fungi consisted of Candida (58%), followed by Malassezia (12%) (Fig. 4 and Supplementary Information 5).
Fig. 4. Taxonomic figures for each time point showing the effect of intrapartum antibiotic exposure in vaginal delivery samples.
a Relative abundances of the taxa at the phylum level. b The top 20 most abundant genera with the rest collapsed into the “other” group. c Changes in the abundance of Ascomycota (top) and Basidiomycota (bottom) over time in the groups with and without intrapartum antibiotics. d Changes in the abundance of Candida (top left), Malassezia (top right), Saccharomyces (bottom left), and Trichosporon (bottom right) over time in the groups with and without intrapartum antibiotic exposure.
At the age of 6 months, the vaginal samples with no intrapartum antibiotic exposure had an even distribution of Cystofilobasidium (20%), Malassezia (18%), and Trametes (14%), and those with intrapartum antibiotic exposure had an even distribution of unidentified fungi (15%), Exobasidium (13%), and Trichosporon (13%) (Fig. 4 and Supplementary Information 5).
At the age of 18 months, the vaginal delivery samples with no intrapartum antibiotic treatment were dominated by unidentified fungi (46%), while those exposed to intrapartum antibiotics were dominated by Trichosporon (66%), followed by unidentified fungi (13%) (Fig. 4 and Supplementary Information 5). The ANCOM and Mann–Whitney U-tests showed differences in abundance in 3 genera in the meconium, 3 phyla and 5 genera in the 6-month stool samples, and 5 phyla and 1 genus in the 18-month stool samples (Supplementary Information 4).
When measuring alpha diversity in the vaginal samples based on the usage of intrapartum antibiotics, significant differences were found in the 6-month samples (Shannon Index p = 0.025) and 18-month samples (Shannon Index p = 0.046, observed features p = 0.013) (Fig. 5). In the case of beta diversity, on the other hand, intrapartum antibiotic exposure showed significantly greater diversity between samples in the meconium (p = 0.011), but at the age of 6 months the samples with no exposure to intrapartum antibiotics were more diverse (p = 0.009; Fig. 5). By 18 months, the effect of exposure to intrapartum antibiotics on beta diversity was no longer significant (p = 0.061; Fig. 5).
Fig. 5. Alpha and beta diversity of vaginal delivery samples showing the effect of the intrapartum antibiotic exposure.
a Meconium samples. b 6-month stool samples. c 18-month stool samples. Alpha diversity metrics used: Shannon Index, observed features, with Kruskal–Wallis H used as a statistical test. Beta diversity metric used: Bray–Curtis Dissimilarity, with PERMANOVA used as a statistical test. p values are shown in the respective figures.
Discussion
This prospective cohort study characterized the development of the gut fungal microbiome, or the gut mycobiome, from birth to the age of 18 months. Delivery mode and exposure to intrapartum antibiotic prophylaxis were markedly associated with gut mycobiome composition from birth to 18 months of age.
Previous studies have shown that perinatal antibiotic exposure changes the gut bacterial microbiome, or bacteriome, in children.21,22,30 Fungi and bacteria likely interact in complex ways in the gut microbiome, either through mutualism or competition.31 Interestingly, antibacterial agents administered at birth, without having any direct antimicrobial effect on fungi, appeared to influence the development of the gut mycobiome in vaginally delivered infants as well, possibly through interaction with the gut bacteriome.
We found here that the meconium was dominated by the genus Candida, which has been found earlier to colonize the gut,32 and has been shown to be the most common fungal colonizer in the infant gut mycobiome according to a Swedish longitudinal study of 133 infants.33 The abundance of Candida had dropped by the age of 6 months, however, and the abundance of Malassezia and Cystofilobasidium had risen. Malassezia has been found to be a common gut colonizer, shifts in the abundance of which have previously been associated with IBD and IBS,10,34 and Cystofilobasidium has also been identified in the gut mycobiome of young children.18
At the age of 18 months, the stool samples were dominated by the genus Trichosporon, unidentified fungi and Saccharomyces. Trichosporon has been found to be associated with various infections in humans, and specifically T. asahii has been recognized as a common gut colonizer.35 Saccharomyces is a genus that includes many yeasts, and the species S. boulardii is known to act as a probiotic in the gut mycobiome.36 We found high abundances of the phyla Ascomycota and Basidiomycota from birth to 18 months of age. Phyla Ascomycota and Basidiomycota have previously been reported as dominant phyla colonizing the human gut, the ratio between which has been reported to affect the prevalence of IBD.34
The mode of delivery has been found previously to affect the bacterial composition of the human gut,37–41 and our results show in addition that it can markedly affect the newborn and infant gut mycobiome. Infants born by the vaginal route had the highest amounts of the genus Candida in their gut mycobiome at birth, whereas those born via Cesarean delivery had a much lower abundance of Candida in their meconium samples. Candida is a common yeast pathogen and colonizer of the vagina,42 and the present results suggest that vaginal Candida is likely to be one of the main sources of initial gut mycobiome development in vaginally delivered newborns. Interestingly, however, the effect of delivery mode on the gut mycobiome seemed to remain detectable until the age of 18 months of life, suggesting possible long-term effects.
Given that exposure to antibiotics at birth has previously been shown to influence the development of the gut bacteriome,21,22,30,43 we found that intrapartum antibacterial antibiotics, without having any direct effect on fungi, significantly affected the gut mycobiome of vaginally born newborns for at least 18 months after birth. Exposure to antibacterial agents has previously been shown to have a long-term influence on the human gut mycobiome in a cohort of 14 healthy adults, possibly by influencing bacterial-fungal interactions, including disruption and resilience of fungal community compositions.31 Our analogous finding was that antibiotic exposure at birth altered the development of the gut mycobiome at least up to 18 months of age, most likely by influencing bacterial–fungal interactions.
There are still limited data available on the role of the mycobiome in the human gut microbiome because most existing observations have been focused on the bacteriome. This is understandable, since the human gut microbiome has been estimated to consist of 99% bacteria.44 The role of other microbes may also be important, however, either through direct host–microbe interactions, or alternatively, through microbe–microbe interactions. This has been demonstrated by specifically a gut commensal Candida albicans, which has been shown to interact with gut bacteria and cause both bacterial and fungal dysbiosis.45 Environmental effects such as diet and the use of antibiotics have already been observed to affect the mycobiome and bacteriome alike, which may be partly caused by the mutualistic relationship between fungi and bacteria.46–48
The strength of our work is that it is one of the first prospective cohort studies comparing the effect of delivery mode and intrapartum antibiotic exposure on the development of the gut mycobiome in infants and young children. Furthermore, we had a sizeable cohort which enabled us to make reliable and meaningful comparisons. We also included a large negative control group in order to identify and filter out possible contaminant fungi present in the reagents, since the effect of contaminant DNA, especially on low biomass samples, has been recognized as an important factor to consider during the analysis of bacteriome findings.26 One limitation of the present study is that we cannot present exact taxonomic data on the fungi. Although UNITE is one of the most comprehensive fungal databases, it still lacks data on the lower taxonomic levels, especially the genus and species levels, when it comes to characterizing the gut mycobiome. Furthermore, like bacteriome studies,49 mycobiome studies suffer from primer bias, as the ITS and 18S genes that are used as fungal identifiers yield varying results.50 As there is no clear consensus on which gene to target for mycobiome analysis, a combination of both might yield the most accurate results.
Conclusion
In conclusion, both delivery mode and antibacterial agents could be shown to alter the development of the gut mycobiome in this prospective study of infants and young children. As the gut mycobiome composition has previously been associated with human health,8–16 the present findings suggest that the fungal microbiome is likely to be relevant when investigating the role of perinatal factors and the gut microbiome in health.
Supplementary information
Acknowledgements
We would like to thank Kaisa Nurminen for the laboratory work, including DNA extraction from the samples and PCR testing; Leena Okkonen for handling the collection of the samples; and Malcolm Hicks for a language check for the manuscript.
Author contributions
M.V.T. and T.T. designed the study; N.P. provided the participant information and metadata; J.T. supervised the DNA extraction, performed PCR, analyzed the sequence data, and wrote the manuscript with insights from N.P., J.R., T.T., and M.V.T.
Funding
M.V.T. thankfully acknowledges a grant from the Päivikki and Sakari Sohlberg Foundation. T.T. would like to thank the Academy of Finland for a Clinical Research grant in 2018–2022 and a Pediatric Research Foundation grant in 2019–2022. Open Access funding provided by University of Oulu including Oulu University Hospital.
Data availability
The raw sequences were uploaded in BioProject with the accession number PRJNA831656.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study has been approved by the Ethical Committee of Northern Ostrobothnia Hospital District at Oulu University Hospital, Finland, with the decision number EETTMK:3/2016. The participants gave their informed, written consent in advance.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Terhi Tapiainen, Mysore V. Tejesvi.
Supplementary information
The online version contains supplementary material available at 10.1038/s41390-023-02471-y.
References
- 1.Limon JJ, Skalski JH, Underhill DM. Commensal fungi in health and disease. Cell Host Microbe. 2017;22:156–165. doi: 10.1016/j.chom.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mims TS, et al. The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun. Biol. 2021;4:281. doi: 10.1038/s42003-021-01820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun B, et al. Captivity is associated with gut mycobiome composition in Tibetan macaques (Macaca thibetana) Front. Microbiol. 2021;12:665853. doi: 10.3389/fmicb.2021.665853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutierrez MW, Arrieta M-C. The intestinal mycobiome as a determinant of host immune and metabolic health. Curr. Opin. Microbiol. 2021;62:8–13. doi: 10.1016/j.mib.2021.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huseyin CE, O’Toole PW, Cotter PD, Scanlan PD. Forgotten fungi—the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017;41:479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 7.Vemuri R, Shankar EM, Chieppa M, Eri R, Kavanagh K. Beyond just bacteria: functional biomes in the gut ecosystem including Virome, Mycobiome, Archaeome and Helminths. Microorganisms. 2020;8:483. doi: 10.3390/microorganisms8040483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matijašić M, et al. Gut microbiota beyond bacteria—Mycobiome, Virome, Archaeome, and eukaryotic parasites in IBD. Int. J. Mol. Sci. 2020;21:2668. doi: 10.3390/ijms21082668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beheshti‐Maal A, et al. Gut mycobiome: the probable determinative role of fungi in IBD patients. Mycoses. 2021;64:468–476. doi: 10.1111/myc.13238. [DOI] [PubMed] [Google Scholar]
- 10.Das A, O’Herlihy E, Shanahan F, O’Toole PW, Jeffery IB. The fecal mycobiome in patients with Irritable Bowel Syndrome. Sci. Rep. 2021;11:124. doi: 10.1038/s41598-020-79478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mar Rodríguez M, et al. Obesity changes the human gut mycobiome. Sci. Rep. 2015;5:14600. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanaki C, Peppa M, Mastorakos G, Chrousos GP. Examining the gut bacteriome, virome, and mycobiome in glucose metabolism disorders: are we on the right track? Metabolism. 2017;73:52–66. doi: 10.1016/j.metabol.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Qin X, et al. Gut mycobiome: a promising target for colorectal cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1875:188489. doi: 10.1016/j.bbcan.2020.188489. [DOI] [PubMed] [Google Scholar]
- 14.Jiang L, et al. The gut mycobiome: a novel player in chronic liver diseases. J. Gastroenterol. 2021;56:1–11. doi: 10.1007/s00535-020-01740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mok K, et al. ITS2 sequencing and targeted meta-proteomics of infant gut mycobiome reveal the functional role of Rhodotorula sp. during atopic dermatitis manifestation. J. Fungi. 2021;7:748. doi: 10.3390/jof7090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah S, et al. Alterations of the gut mycobiome in patients with MS. EBioMedicine. 2021;71:103557. doi: 10.1016/j.ebiom.2021.103557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr. Infect. Dis. J. 2008;27:231–235. doi: 10.1097/INF.0b013e31815bb69d. [DOI] [PubMed] [Google Scholar]
- 18.Schei K, et al. Early gut mycobiota and mother-offspring transfer. Microbiome. 2017;5:107. doi: 10.1186/s40168-017-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis KA, et al. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J. 2019;33:12825–12837. doi: 10.1096/fj.201901436RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stephen AJ, et al. Preterm infants harbour a rapidly changing mycobiota that includes Candida pathobionts. J. Fungi. 2020;6:273. doi: 10.3390/jof6040273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tapiainen T, et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep. 2019;9:10635. doi: 10.1038/s41598-019-46964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, et al. Vertical transmission of gut microbiome and antimicrobial resistance genes in infants exposed to antibiotics at birth. J. Infect. Dis. 2021;224:1236–1246. doi: 10.1093/infdis/jiaa155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turunen J, et al. Presence of distinctive microbiome in the first-pass meconium of newborn infants. Sci. Rep. 2021;11:19449. doi: 10.1038/s41598-021-98951-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan BJ, et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kõljalg U, et al. UNITE: a database providing web‐based methods for the molecular identification of ectomycorrhizal fungi. N. Phytol. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- 29.Mandal S, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ainonen S, et al. Antibiotics at birth and later antibiotic courses: effects on gut microbiota. Pediatr. Res. 2022;91:154–162. doi: 10.1038/s41390-021-01494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seelbinder B, et al. Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome. 2020;8:1–133. doi: 10.1186/s40168-020-00899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romo JA, Kumamoto CA. On commensalism of Candida. J. Fungi. 2020;6:16. doi: 10.3390/jof6010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondori N, et al. Candida species as commensal gut colonizers: a study of 133 longitudinally followed Swedish infants. Med. Mycol. 2020;58:485–492. doi: 10.1093/mmy/myz091. [DOI] [PubMed] [Google Scholar]
- 34.Sokol H, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho O, Matsukura M, Sugita T. Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 2015;39:87–88. doi: 10.1016/j.ijid.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Pais P, Almeida V, Yılmaz M, Teixeira MC. Saccharomyces boulardii: what makes it tick as successful probiotic? J. Fungi. 2020;6:78. doi: 10.3390/jof6020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundgren SN, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y-C, et al. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci. Rep. 2018;8:3212–3255. doi: 10.1038/s41598-018-21657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyman M, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019;10:4997. doi: 10.1038/s41467-019-13014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao Y, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cauchie M, Desmet S, Lagrou K. Candida and its dual lifestyle as a commensal and a pathogen. Res. Microbiol. 2017;168:802–810. doi: 10.1016/j.resmic.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 43.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 44.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez JC. The interplay between gut bacteria and the yeast Candida albicans. Gut Microbes. 2021;13:1979877. doi: 10.1080/19490976.2021.1979877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sam Q, Chang M, Chai L. The fungal mycobiome and its interaction with gut bacteria in the host. Int. J. Mol. Sci. 2017;18:330. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santus W, Devlin JR, Behnsen J. Crossing kingdoms: how the mycobiota and fungal-bacterial interactions impact host health and disease. Infect. Immun. 2021;89:e00648–20. doi: 10.1128/IAI.00648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frau A, et al. Inter-kingdom relationships in Crohn’s disease explored using a multi-omics approach. Gut Microbes. 2021;13:1930871. doi: 10.1080/19490976.2021.1930871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuczynski J, et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 2011;13:47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frau A, et al. DNA extraction and amplicon production strategies deeply inf luence the outcome of gut mycobiome studies. Sci. Rep. 2019;9:9328. doi: 10.1038/s41598-019-44974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequences were uploaded in BioProject with the accession number PRJNA831656.