Abstract
Glioblastomas are highly aggressive brain tumors for which therapeutic options are very limited. In a quest for new anti-glioblastoma drugs, we focused on specific structural modifications to the benzoyl-phenoxy-acetamide (BPA) structure present in a common lipid-lowering drug, fenofibrate, and in our first prototype glioblastoma drug, PP1. Here, we propose extensive computational analyses to improve the selection of the most effective glioblastoma drug candidates. Initially, over 100 structural BPA variations were analyzed and their physicochemical properties, such as water solubility (− logS), calculated partition coefficient (ClogP), probability for BBB crossing (BBB_SCORE), probability for CNS penetration (CNS-MPO) and calculated cardiotoxicity (hERG), were evaluated. This integrated approach allowed us to select pyridine variants of BPA that show improved BBB penetration, water solubility, and low cardiotoxicity. Herein the top 24 compounds were synthesized and analyzed in cell culture. Six of them demonstrated glioblastoma toxicity with IC50 ranging from 0.59 to 3.24 µM. Importantly, one of the compounds, HR68, accumulated in the brain tumor tissue at 3.7 ± 0.5 µM, which exceeds its glioblastoma IC50 (1.17 µM) by over threefold.
Subject terms: Cancer, Drug discovery, Chemistry
Introduction
Glioblastomas are the most aggressive brain neoplasms with a dismally low 5 year patient survival rate of below 5%1. According to the World Health Organization (WHO), glial tumors are classified as grade I and grade II (low-grade gliomas), grade III (anaplastic), and grade IV (glioblastoma)2. Current standard-of-care therapies include maximal surgical resection, followed by radiotherapy plus concomitant and maintenance treatment with temozolomide (TMZ)3. In addition, a large variety of different genetic and epigenetic modifications have been found in glioblastomas, among which p53, EGFR, PTEN, and IDH mutations are the most common4–9. However, these validated molecular targets, as well as immunotherapies, including immune checkpoint inhibitors10, tumor vaccines11, and chimeric antigen receptor T cell (CAR T) therapies12, have all been extensively studied, but failed to significantly improve the therapeutic outcome in glioblastoma patients.
There are several reasons why it is difficult to develop more effective glioblastoma therapies: (1) Glioblastomas are characterized by many dysregulated pathways that cannot be blocked simultaneously via a single therapy13: (2) Glioblastomas are highly infiltrating and heterogenous tumors that are very difficult to remove by surgical resection without compromising the function of surrounding brain areas14; (3) It is difficult to diagnose glioblastomas in their early stages, therefore, large highly infiltrating and vascularized tumors are often present at diagnosis15; (4) Use of rodent syngeneic and patient-derived models are common to optimize clinical protocols. One major problem is that these experimental tumors are typically ~ 103–104 smaller than actual tumors in humans. Therefore, data from drug delivery, drug retention, and tissue penetration experiments obtained from these small animal models are difficult to extrapolate to glioblastoma patients16; and finally, (5) The blood brain barrier (BBB) prevents the majority of anticancer drugs from reaching tumor sites at clinically relevant concentrations, and current methods to enhance BBB penetration are not very effective for glioblastoma patients17.
One drug that readily crosses the BBB is temozolomide (TMZ). Upon oral administration, TMZ maximum plasma concentration can be reached in about one hour, and the elimination half-life is approximately 1.8 h. Importantly, penetration efficiency of TMZ into the central nervous system (CNS) is experimentally estimated to be about 20% of plasma levels. This is important because applying this estimate to calculate logBB (Brain-Blood Distribution)18 produces a value of -0.7, which indicates a high capability of the compound to cross the BBB. In spite of these positive features, TMZ-treated glioblastoma patients develop TMZ-resistance and recurrent tumors are practically incurable19.
In addition, TMZ has been used in combination with other drugs, which enhanced its therapeutic effects. One example is a combination of TMZ with lipid lowering drugs, including statins20 and fibrates, like for instance fenofibrate (FF), which has strong anti-glioblastoma activity in cell culture, and in glioblastoma intracranial mouse models21. However, we have also found, that the ability of FF to cross the BBB is low, and the compound is quickly processed by blood and tissue esterases to form the PPARα agonist, fenofibric acid (FFA), which is no longer effective in triggering tumor cell death22. We made numerous modifications to the FF chemical structure and selected our first drug candidate, PP123, which similar to FF, blocks mitochondrial respiration and triggers a severe ATP depletion. With the use of both substances, ATP depletion is followed by phosphorylation/activation of AMP-activated protein kinase (AMPK—intracellular energy sensor), blockade of p70S6K phosphorylation (marker of active protein synthesis), activation of autophagy (p62 degradation), and extensive glioblastoma cell death24. In spite of these promising results, and in spite of the fact that we detected PP1 in the brain at therapeutically relevant concentrations (Supplementary materials, page 154), PP1 anti-glioblastoma therapeutic effects were only marginal when the treatment was applied to large intracranial tumors in mice24. These data indicate that additional adjustments to the BPA scaffold, focused on improving compound cytotoxicity, BBB penetration and retention in the brain tumor tissue, are continuously needed.
Results and discussion
In early stages of drug design it is important to evaluate relevant physicochemical properties of prospective drug candidates. For over two decades, Lipinski’s rule of five was the gold standard in drug design25. For drugs associated with the CNS, their ability to cross the brain blood barrier (BBB) is the most important characteristic and is not directly addressed by the Lipinski’s rule of five. Therefore, two computational scoring systems have been introduced to evaluate the probability of new drug candidates penetrating the CNS. One is CNS Multiparameter Optimization (CNS-MPO) algorithm26, which uses 6 physicochemical properties [ClogP (calculated partition coefficient—lipophilicity), ClogD (calculated distribution coefficient at physiological pH (7.4)—lipophilicity), MW (molecular weight), TPSA (topological polar surface area), HBD (hydrogen bond donor at pH = 7), and pKa (-log acid dissociation constant)] for estimating the probability of entering the CNS. The CNS-MPO score range is between 0 and 6, and values ≥ 4.0 have been used as cut-offs for compounds with increased ability to penetrate the CNS26. Another scoring system is the blood–brain barrier score (BBB_SCORE)27. BBB_SCORE is based on five physicochemical parameters including number of aromatic rings, heavy atoms, MWHBN (a value comprising molecular weight, hydrogen bond donor, and hydrogen bond acceptor), pKa and topological polar surface area. Similar to CNS-MPO, BBB_SCORE also considers the value of 4.0 as the cut-off for acceptable BBB penetration27. Consequently, CNS-MPO and BBB_SCORE both serve as complementary algorithms in which CNS-MPO provides information regarding probability of the compound to be found in the CNS, and BBB_SCORE is oriented towards specific physiochemical properties that increase probability of the compound crossing the BBB.
In regard to toxicity, one of the most common hurdles in drug testing is cardiotoxicity triggered by inhibition of the cardiac potassium channel that is coded by the human ether-à-go-go-related gene (hERG). This toxicity test became a mandatory requirement for drug design and development, and can be calculated by the hERG algorithm28,29. Based on the compound structure, the hERG algorithm gives a score which is indictive of the compound’s inhibitory effect towards hERG. In other words, for the drug to be considered safe, it’s hERG IC50 should be significantly higher than its therapeutic IC5028.
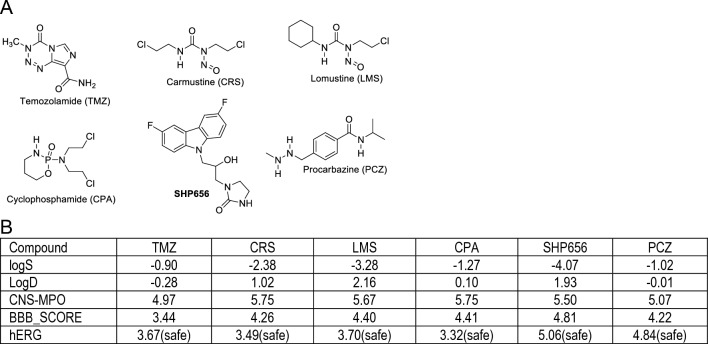
In order to determine glioblastoma-specific and therapeutically relevant values for all proposed algorithms/scores, including hERG, we have selected fifteen drugs (see Supplementary Materials, pages 57–79) that are either currently used as glioblastoma drugs, or are in clinical trials with glioblastoma patients30. Results in Fig. 1B show that six of these compounds (Fig. 1A) have relatively high water solubility (LogS < 0), low lipophilicity (logD < 3), acceptable values for CNS-MPO and BBB_SCORE (ranging between 3 and 5), and low cardiac toxicity (hERG ≤ 5.5). We used these critical values as a guideline for designing new anti-glioblastoma drug candidates, which are based on the BPA chemical structure present in a common lipid lowering drug, fenofibrate21,22,31,32, and in our first glioblastoma prototype drug, PP123,24.
Figure 1.
(A) Chemical structures of the most common chemotherapy drugs used to treat brain and spinal cord and their computed psychopharmacological properties. (B) Physicochemical properties calculated for the compounds depicted in Panel A. LogS = water solubility; logD = distribution at pH 7.4; CNS-MPO = CNS multiparameter optimization algorithm; BBB_SCORE = blood–brain barrier penetration score; hERG = estimated pIC50 value for hERG (the human ether-a-go-go (hERG) potassium channel).
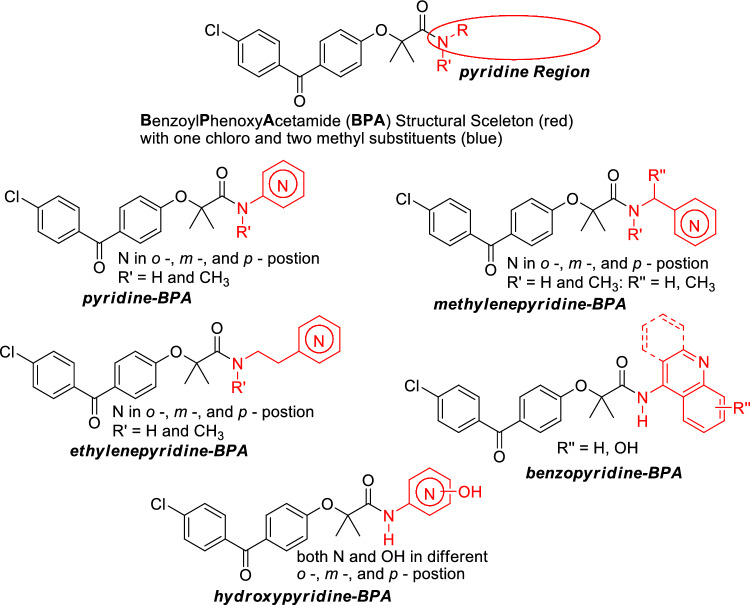
In the search for more effective benzoyl-phenoxy-acetamide (BPA) derivatives, we have decided to explore pyridine variants (Fig. 2). This is because the pyridine moieties are part of a diverse group of compounds with broad pharmacological applications33,34. In addition, it is well documented that pyridine-based compounds have a high probability of penetrating the CNS35. Herein, we focused on developing and testing five pyridine-base BPA variants, which are grouped according to their structural relationship with pyridine and amide moieties: (I) one with a directly attached unsubstituted pyridine moiety (pyridine-BPA), (II) and one with a separation unsubstituted pyridine moiety with one carbon (methylenepyridine-BPA), (III) with two carbons (ethylenepyridine), (IV) a benzo fused pyridine (benzopyridine-BPA, and (V) a hydroxy substituted pyridine (hydroxypyridine-BPA) (Fig. 2).
Figure 2.
The pyridine region of BPA skeleton was selected for modification (circle) in search of the optimal anti-glioblastoma drug.
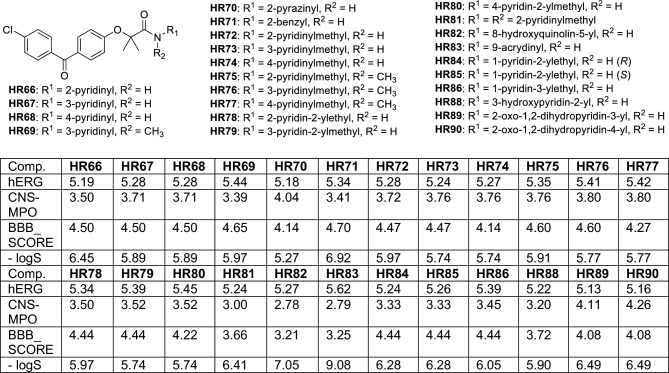
These structural variations of BPA-pyridines were designed and tested using calculated values from four algorithms: cardiotoxicity (hERG)36; brain penetration capability (CNS-MPO and BBB_SCORE), and water solubility (− logS). The values for the best 24 BPA-pyridines are presented in Fig. 3 (for complete list of calculated values see Supplementary Materials, pages 77–138). According to our initial calculations, compounds in Fig. 3 have low cardiotoxicity (all predicted hERG values are below 5.5), and acceptable water solubility (estimated -logS below 7.5). In addition, CNS-MPO scores for almost all BPA-pyrimidines are over 3, which indicates probability of brain penetration above 50%. Furthermore, these compounds have a BBB_SCORE between 4 and 5, which suggests high probability of crossing the BBB.
Figure 3.
Estimated cardio potassium channel toxicity (hERG), CNS penetrability (CNS-MPO), BBB penetrability (BBB_SCORE), and water solubility (-LogS) of the selected pyridine-based BPA variants.
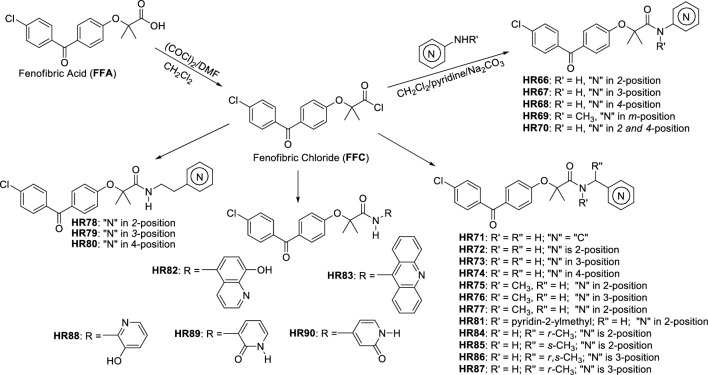
After determining that the proposed 24 BPA-pyridines have acceptable brain penetration ability, water solubility and low cardiotoxicity, we developed specific preparation methods for each compound. From our previous studies, we demonstrated that the carboxylic group of fenofibric acid (FFA) and its derivatives have exceptionally low reactivity toward nucleophilic acyl substitution. The obvious synthetic pathway for preparation of these compounds is by coupling FFA with corresponding amines37. There are two major problems in performing FFA coupling with aminopyridines: (a) FFA has exceptionally low reactivity due to steric constraints generated by the two methyl groups located alpha to the carbonyl group, and (b) amino groups of aminopyridines are exceptionally weak nucleophile due to amino group electron delocalization through the pyridine ring. The difficulty of performing this kind of coupling reaction was recently examined by others38. However, FFA can be converted into corresponding fenofibric chloride (Fig. 4), which is sufficiently reactive to aliphatic amines and activated anilines. But, coupling reactions with aminopyridines such as 4-aminopyridine is challenging at best. This is a major drawback to drug design because there are wide applications for pyridine scaffolds in medicinal chemistry34. For these reasons, we have decided to explore nucleophilicity of aminopyridines through their computational data.
Figure 4.
Procedure for preparation of pyridine derivatives of BPA (for more details see "Methods" and Supplementary Materials, pages 2–56).
In molecules of similar structural framework, it is possible to compare frontier molecular orbitals to determine order of nucleophilic and electrophilic reactivity39. We have used density functional theory to compute HOMO energies. Results provided in Supplementary Materials (page 153) indicate that aminopyridines should be less reactive (low HOMO) in comparison to aniline while aminophenol is more nucleophilic. Based on our calculations, the coupling of sterically hindered FFA through the reactive fenofibric chloride (FFC) with 4-aminophenol should give a product with a highly isolated yield. Our previous studies23,40 demonstrated that FFC is a viable, reactive FFA intermediate for amine coupling. For these reasons, we have developed a new synthetic methodology, which is shown in Fig. 4.
There are two distinctive groups of HR-compounds from the viewpoint of amine reactivity and their preparation (a) one with an aromatic heterocyclic ring directly attached to the nitrogen atom (HR66-HR70, HR82, HR83, HR88-HR90), and (b) one with separation of an aromatic heterocyclic ring by one or two aliphatic carbons (HR71-HR81, HR84-HR87). Preparation of the second group of compounds (with reactive amines) is straightforward by mixing FFC with corresponding amines in presence of a base. Because the corresponding amines are more nucleophilic than water and reactions are completed in several minutes at room temperature, these reactions can be performed with an environmentally friendly base such as sodium carbonate in water. Isolated yields are nearly quantitative and this method (Method A) is well suited for small (milligrams) and large (hundreds of grams) scale synthesis. Also, this synthetic procedure does not require any special precautions or preparations. Preparation of the first group, with the heterocyclic aromatic ring directly attached to the amino group, requires certain precautions. Because water is a better nucleophile than these amines’, reaction must be performed in water-free (dry) conditions. In addition, less nucleophilic amines are also prone to oxidation in basic conditions40. Because of this, such reactions should be carried out in an oxygen free atmosphere (Method B). Under dry conditions, a pyridine solution of a corresponding aminopyridine was mixed with anhydrous sodium carbonate and kept under nitrogen atmosphere overnight. A separate dichloromethane solution of FFC was also prepared in dry conditions under nitrogen atmosphere. The dichloromethane solution was slowly added to cold (~ 0–5 °C) pyridine suspension of a corresponding aminopyridine and sodium carbonate under a nitrogen atmosphere. Nearly quantitative yields were obtained with reactions performed at 0–5 °C for 3 h, then at room temperature overnight, and finally, at 60 °C for additional 3 h. The product was isolated after solvent evaporation and water addition by simple filtration and extensive water washing. This method gave products in high yield, more the 97% purity, and did not require additional purification by either extraction or chromatography. It is also applicable to milligram and multigram preparation scales for amines with wide ranges of reactivity (Fig. 4).
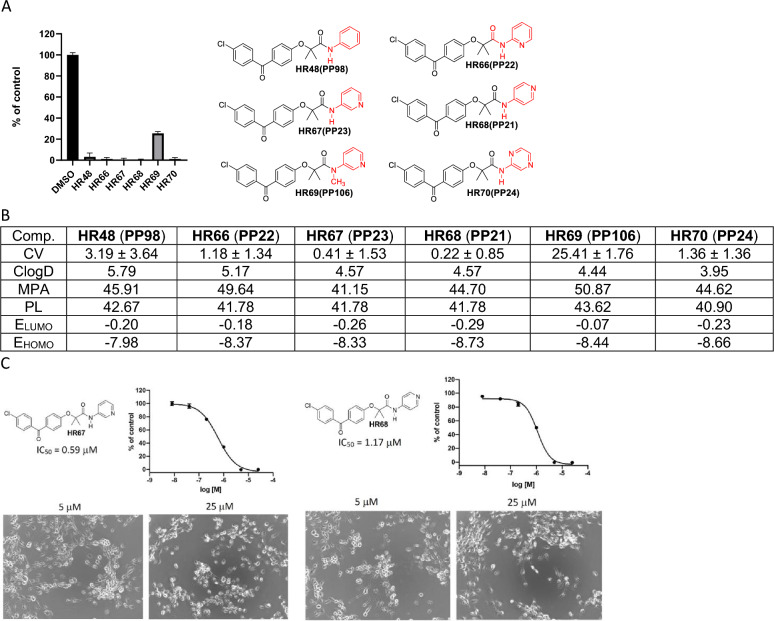
The first group of pyridine-BPA variants are presented in Fig. 5 in which glioblastoma cell viability (CV-based on MTT assay), estimated minimal projection area (MPA), lipophilicity (ClogD), molecular polarizability (PL), as well as, energy of Lowest Unoccupied Molecular Orbital (ELUMO) and energy of Highest Occupied Molecular Orbital (EHOMO) were determined. We have included MPA based on studies showing that if a compound does not interact with cell membranes and has a MPA lower than 60 Å2, it should be able to penetrate the CNS via passive diffusion41–43. Therefore, MPA is considered as a better parameter than molecular weight in discriminating the compound ability of entering the CNS44.
Figure 5.
BPA-based drug candidates with pyridine moiety. (A) Cell viability (MTT assay) evaluated following glioblastoma cell (LN229) exposure to 25 μM of the corresponding BPA-pyrimidines. Data indicate average values with standard deviation (n = 3). (B) Tabulated values for glioblastoma-relevant parameters. CV = Cell viability (% of control) mean ± SD at 25 μM; ClogD = calculated distribution coefficient at physiological pH (lipophilicity); MPA = Minimal Projection Area (Å2); PL = Molecular Polarizability (Å3); ELUMO = energy of LUMO (Lowest Unoccupied Molecular Orbital) (eV); EHOMO = Energy of HOMO (Highest Occupied Molecular Orbital) (eV). (C) IC50 graphs and pictures of the cells at 5 and 25 µM for HR67 and HR68, which are considered the most promising drug candidates in this group. Data represent average values with standard deviation (n = 3).
In the context of comparing compounds with high structural similarity, frontier orbital energies (HOMO and LUMO) can be used in determining which of the similar compounds may have a stronger protein binding ability, and therefore a higher chance of BBB penetration45,46.
In this regard, HR67 and HR68 (Fig. 5) both have low HOMO and LUMO energies, along with acceptable lipophilicity (ClogD), and very promising glioblastoma IC50 values (0.59 and 1.17 µM, respectively), indicating that these two compound could become leading candidates for the brain tumor drug development.
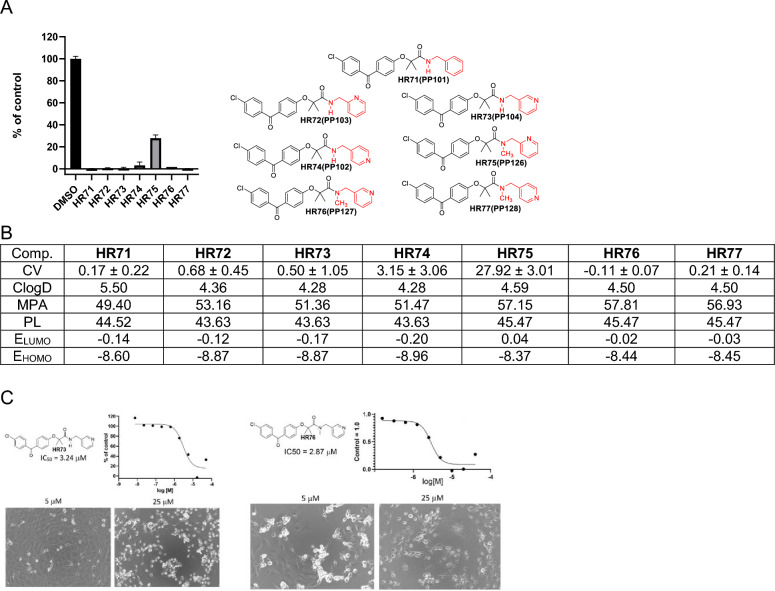
The next group of compounds belong to methylenepyridine-BPA (Fig. 6). All of these compounds show promising anti-glioblastoma activity at 25 µM, and their estimated distributions (ClogD) are relatively high, indicating high lipophilicity. MPA values are below 60, suggesting that there is no obstacle to CNS penetration regarding molecular size. Polarizability (PL) is between 40 and 50 indicating that these molecules can adapt to the binding area of a biomolecule through complementary polarization47. In the consideration of computed frontier orbital energies (LUMO and HOMO), HR74 should have the best binding ability. In addition, all HR compounds in Fig. 6 are highly cytotoxic at 25 µM except HR75, and corresponding IC50 values for the most promising compounds in this group, HR73 and HR76, are 3.24 and 2.87 µM, respectively.
Figure 6.
Drug candidates with methylenepyridine moiety. (A) Cell viability (MTT assay) evaluated following glioblastoma cell (LN229) exposure to 25 μM of the corresponding BPA-pyrimidines. Data indicate average values with standard deviation (n = 3). (B) Tabulated values for glioblastoma-relevant parameters. CV = Cell viability (% of control) mean ± SD at 25 μM; ClogD = calculated distribution coefficient at physiological pH (lipophilicity); MPA = Minimal Projection Area (Å2); PL = Molecular Polarizability (Å3); ELUMO = energy of LUMO (Lowest Unoccupied Molecular Orbital) (eV); EHOMO = Energy of HOMO (Highest Occupied Molecular Orbital) (eV). (C) IC50 graphs and pictures of the cells at 5 and 25 µM for HR73 and HR76, which are considered the most promising drug candidates in this group. Data represent average values with standard deviation (n = 3).
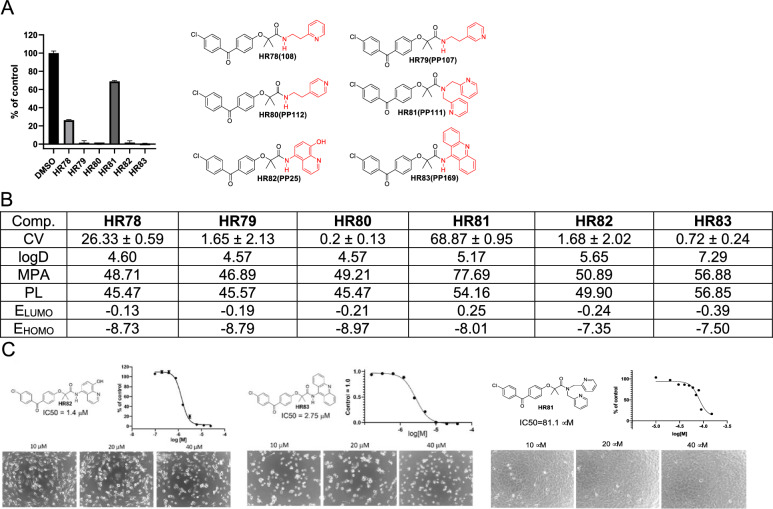
Next, we asked if the compound activity would change by adding ethylene linker to ethylenepyridine-BPA or by increasing the pyridine’s molecular delocalization in benzopyridine-BPA (Fig. 7). As expected, by increasing the number of carbon atoms, either by adding methylene group or additional aromatic ring, lipophilicity increased noticeably (for instance, ClogD for HR83 is 7.29). As the size of the molecule increases, MPA values follow, suggesting that HR81, HR82, and HR83 may have a low probabilities of penetrating the CNS. Considering the energies of both frontier molecular orbitals from three similar compounds (HR78, HR79, and HR80) HR80 should be the most active. Indeed, obtained cell viability (CV) data for these compounds correlated with computational data and suggest that best drug candidate from this group is HR80. Although both benzopyridine-BPAs (HR82 and HR83) have encouraging IC50 values, 1,4 and 2.75 µM, respectively, their computed physical properties such as ClogD and MPA suggest that there is very low probability for these compounds to penetrate the CNS, further supporting HR80 as the best glioblastoma drug candidate in this group. In addition, HR81, which has low glioblastoma-specific cytotoxicity (Fig. 7A) was used here as a negative control for IC50 calculations (Fig. 7C).
Figure 7.
Drug candidates with ethylenepyridine and benzopyridine moieties. (A) Cell viability (MTT assay) evaluated following glioblastoma cell (LN229) exposure to 25 μM of the corresponding BPA-pyrimidines. Data indicate average values with standard deviation (n = 3). (B) Tabulated values for glioblastoma-relevant parameters. CV = Cell viability (% of control) mean ± SD at 25 μM; ClogD = calculated distribution coefficient at physiological pH (lipophilicity); MPA = Minimal Projection Area (Å2); PL = Molecular Polarizability (Å3); ELUMO = energy of LUMO (Lowest Unoccupied Molecular Orbital) (eV); EHOMO = Energy of HOMO (Highest Occupied Molecular Orbital) (eV). (C) IC50 graphs and pictures of the cells at 10, 20, and 40 µM for HR82, HR83 and HR81 (negative control for glioblastoma-specific toxicity). Data represent average values with standard deviation (n = 3).
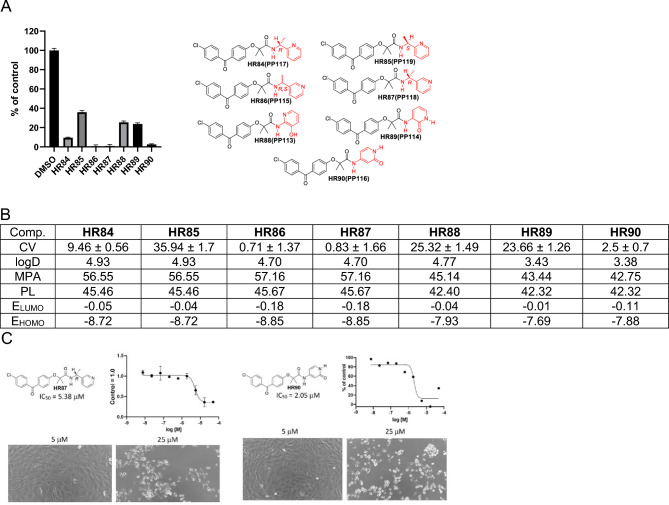
We have also explored the importance that chirality may have on the activity of these compounds (Figure 8). For instance, HR84 and HR85 are structural isomers of HR78 that show moderate cytotoxicity at 25 µM (CV = 26.33). There are noticeable differences between the two stereoisomers: R isomer HR84 being three times more cytotoxic than S isomer HR85 (Figure 8). On the other hand, there is no difference between racemic HR86 (both R&S) and optically pure R isomer HR87. This finding is reasonable because of the pyridine nitrogen proximity to the chiral center. One could argue that pyridine nitrogen binding is sterically diminished in HR85 in comparison with HR84. Because of the position of the pyridine nitrogen regarding chiral center, steric difference is diminished in HR87, which is associated with improved cytotoxicity. The hydoxypyridine-BPAs can exist in both the hydroxypyridine and the amide form with the amide form being preferable by 8.97 kcal/mol according to the DFT ωB97X-D/6-31G* computational method. Computational studies as well as NMR spectroscopy indicate that HR88 is in the hydroxypyridine form while HR89 and HR90 are in their amide form.
Figure 8.
Drug candidates with chiral methylenepyridine and hydroxypyridine moietis. (A) Cell viability (MTT assay) evaluated following glioblastoma cell (LN229) exposure to 25 μM of the corresponding BPA-pyrimidines. Data indicate average values with standard deviation (n = 3). (B) Tabulated values for glioblastoma-relevant parameters. CV = Cell viability (% of control) mean ± SD at 25 μM; ClogD = calculated distribution coefficient at physiological pH (lipophilicity); MPA = Minimal Projection Area (Å2); PL = Molecular Polarizability (Å3); ELUMO = energy of LUMO (Lowest Unoccupied Molecular Orbital) (eV); EHOMO = Energy of HOMO (Highest Occupied Molecular Orbital) (eV). (C) IC50 graphs and pictures of the cells at 5 and 25 µM for HR87 and HR90. Data represent average values with standard deviation (n = 3).
Of the three hydroxypyridine-BPAs, only computed data for HR89 and HR90 can be compared because they are in the amide form while HR88 is in the hydroxypyridine form. All computed parameters suggest that both HR89 and HR90 should easily penetrate the CNS. However, computed frontier orbital energies for HR90 are lower, indicating it should be more effective in penetrating the BBB. In addition, HR90-induced glioblastoma cytotoxicity is nearly 10-fold greater in comparison to HR89, making HR90 the better drug candidate. In conclusion, HR87 and HR90 are the best drug candidates in this group with corresponding, glioblastoma-specific IC50 values of 5.38 and 2.05 µM, respectively (Fig. 8C).
When pursuing new drug candidates, it is very important to envision the possible metabolites and estimate their physicochemical properties, including toxicity48. We used computational methods to generate metabolites of all the studied compounds then evaluated their toxicity, solubility, lipophilicity and CNS penetration using the BioTransformer method49. Only results for our best drug candidates, HR67 and HR68, are reported here (Supplementary Materials, page 155). Reaction types involved in Phase I metabolism are hydrolysis, oxidation, and reduction50. All predicted metabolites of HR67 and HR68 have better or comparable computed water solubility, as well as computed abilities to penetrate the CNS. Importantly, all these metabolites are predicted to be relatively safe (hERG values range 5.76 to 4.95) with the exception of amide hydrolysis, which may trigger some cardiac toxicity and should be further analyzed (hERG=3.91) (Supplementary Materials, page 155).
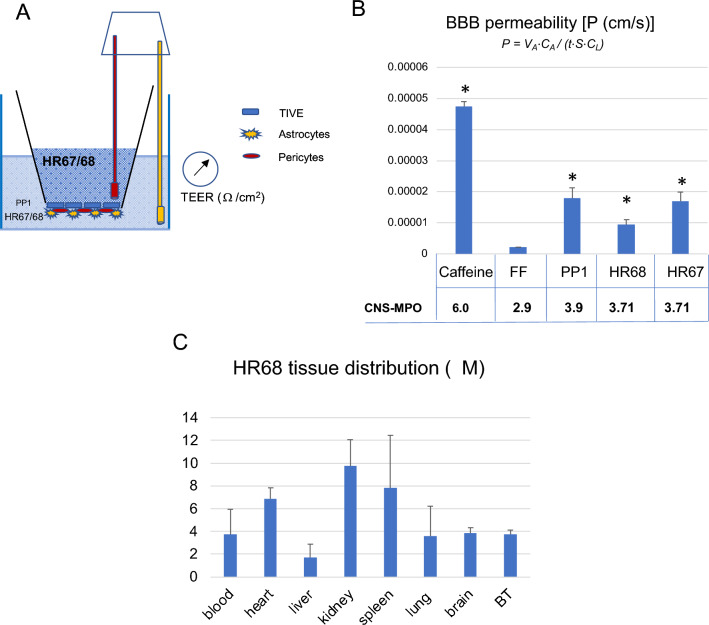
Our data presented in Figs. 1, 2, 3, 4, 5, 6, 7 and 8 allowed us to perform high throughput and unbiassed selection of BPA-based compounds, which have a good chance of becoming glioblastoma drug candidates. Two pyrimidine variants of BPA, HR67 (PP23) and HR68 (PP21), were subsequently tested for their ability to penetrate artificial BBB model membranes (Fig. 9A,B). In this experiment, BBB permeability values (P) were compared between the two experimental drugs, HR67 and HR68, our prototype drug candidate, PP124, and a positive control (caffeine). We also compared the negative control (FF), for which we have previously demonstrated an inability to accumulate in the brain tumor tissue22. Although CNS-MPO scores for HR67 (3.71) and HR68 (3.71) are slightly lower, compared to the CNS-MPO of our prototype drug, PP1 (CNS-MPO = 3.9)23,24, these two compounds can cross the BBB model membrane (Fig. 9B). Importantly, we have detected HR68 in the brain tumor tissue at concentrations over 3-fold higher than glioblastoma-specific IC50 for this compound (1.17 µM) (Fig. 9C), further indicating its potential as new glioblastoma drug candidate.
Figure 9.
Penetration of selected PP compounds across in vitro BBB model membrane: (A) Schematic representation of a triple-coculture model of the BBB, which consists of astrocytes, pericytes and epithelial cells cultured on 24-well transwell membranes with 3 μm pores. Trans-endothelial electric resistance (TEER) was measured using a EVOM2 meter with a STX3 electrode (World Precision Instruments). (B) BBB permeability (P) for the selected compounds was calculated using P = VA⋅CA/(t⋅S⋅CL) equation51 and normalized by TEER coefficient. Data represent average values from 2 independent experiments in triplicates (n = 6) with standard deviation SD. * indicates values significantly different from fenofibrate (negative control), and caffeine was used as positive control. Panel C: HR68 (PP21) tissue concentration evaluated in Foxn1 nude mice bearing intracranial glioblastoma (GBM12). Mice were treated intraperitoneally (ip) with HR68 diluted in 20% cyclodextrin at 15 mg/kg/day and the levels of HR68 in the blood, heart, liver, kidney, spleen, lung, brain and in brain tumor (BT), were evaluated by HPLC, as we previously reported21,24. Data represent average values with standard deviation (n = 3). Please note that average glioblastoma IC50 for HR68 is 1.17 µM, and we detected 3.7 ± 0.5 µM of HR68 in the brain tumor tissue.
In conclusion, introducing pyridine moieties to the BPA scaffold improves chemo- pharmacological properties of new drug candidates. In particular, water solubility and predicted CNS penetration of these pyridine-derivatives of BPA are higher than in previously studied alkyl and phenolic derivatives of BPA23,52. It was also demonstrated here that a properly positioned pyridine moiety in respect to the BPA scaffold increased anti-glioblastoma potency of these compounds with glioblastoma-specific IC50 values being close to 1 µM. Importantly, these specific modifications, which increased molecular flexibility and improved water solubility of the compounds, was achieved without compromising glioblastoma specific cytotoxicity. In addition, stereochemistry of the chiral center close to the pyridine moiety is important because of a difference in three- dimensional orientation of pyridine nitrogen, which can change compound interaction with targeted biomolecules.
Methods
Ethics and inclusion statement
All intellectual, experimental and collaborative work included in this manuscript have been approved by the Louisiana Board of Ethics, and approved in accordance with the relevant guidelines and regulations by the Louisiana State University (LSU) Institutional Biosafety Committee (IBC, protocol #4351), and LSU Institutional Animal Care and Use Committee (IACUC, protocol #4966). In addition, our IACUC approved animal experiments follow 10 essential recommendations included in the ARRIVE guideline.
Materials
All starting materials were reagent grade and purchased from AmBeed (https://www.ambeed.com), Millipore Sigma (https://www.sigmaaldrich.com), and TCI America (https://www.tcichemicals.com). 1H-NMR spectra were recorded on Varian Mercury 300 and Varian Mercury 400 Plus instruments in CDCl3 and DMSO-d6, using the solvent chemical shifts as an internal standard. NMR solvents were purchased from Cambridge Isotope (https://www.isotope.com). In DMSO-d6 solvent our final compounds such as HR68 show two sets of signals due to restricted rotation of C-N amide bond53. This restriction is not present in CDCl3 as NMR solvent [See supplementary data; pp 9–13]. All studied compounds have 96% or higher purity as determined with H-NMR and HPLC. All computed molecular descriptors were generated by ChemAxon MarvinSketch version 22.21 (https://chemaxon.com/products/marvin). All calculated values for each and every compound were performed with MarvinSketch and are included in the Supplementary materials. Frontier orbital energies, conformational studies, energy differences between various isomers and their electrostatic potential maps were calculated with ωB97X-D/6-31G* Density Functional Theory (DFT) method as implemented in Spartan ’18 v 1.1.0 (https://www.wavefun.com) and are included in the Supplementary Materials (pages 150–152). 1H-NMR and 13C-NMR spectra for all HR compounds generated in this study are included in Supplementary Materials. Phase I metabolites prediction was performed with BioTransformer 3.0 (https://biotransformer.ca/new) [Wishart DS, Tian S, Allen D, Oler E, Peters H, Lui VW, Gautam V, Djoumbou-Feunang Y, Greiner R, Metz TO. BioTransformer 3.0-a web server for accurately predicting metabolic transformation products54.
Method A
(General method applicable for preparation of all HR66-HR90 compounds in large scale without extraction or crystallization). Preparation of 2-[4-(4-chlorobenzoyl)phenoxy]-2-methyl-N-(pyridin-3-yl)propenamide (HR67). Dry pyridine suspension of 3-aminopyridine (14.2 g; 0.15 mol) and anhydrous sodium carbonate (42.4 g; 0.4 mol) was sonicated for thirty minutes and left at room temperature in a closed system overnight under nitrogen atmosphere to ensure that the pyridine suspension remained dry. Separately, fenofibric chloride (FFC) was prepared as follows: Dichloromethane (500 ml) suspension of fenofibric acid (47.4 g; 0.15 mol), oxalyl chloride (25.7 ml; 38.1 g; 0.3 mol), and DMF (few drops) were stirred at room temperature overnight. After approximately 1.5 h, the reaction mixture became light brown. The majority of solvent was removed by distillation at atmospheric pressure and the remaining solvent was removed under Argon flow at room temperature. The resulting solid material was dissolved in dichloromethane (150 ml), under nitrogen atmosphere and with slow stirring, was added to the previously prepared pyridine suspension of 3-aminopyridine and sodium carbonate cooled with ice-water. The resulting suspension was stirred at 0–5 °C for 3 h, then at room temperature overnight, and was followed by stirring at 60 °C for additional 3 h. The resulting solvent was removed under reduced pressure to separate the solid residue. This residue was mixed with water (1 L) and stirred via sonication for 4 h. The insoluble white crystalline product was separated by filtration, extensively washed with water (20 × 100 ml), and dried at 60 °C, under vacuum. The isolated yield was 90% (53.3 g). 1H-NMR (DMSO-d6, 400 MHz) δ 10.31 (1H, s), 8.83 (1H, s), 8.26 (1H, d, J = 4.4 Hz), 8.06 (1H, d, J = 8.4 Hz), 7.72 (2H, d, J = 8.4 Hz), 7.67 (2H, d, J = Hz), 7.54 (2H, d, J = 8.4 Hz), 7.31 (1H, d of d, J1 = 8.4 Hz, J2 = 4.4 Hz), 7.04 (2H, d, J = 8.8 Hz), and 1.63 (6H, s) ppm. 13C-NMR (DMSO-d6, 100 MHZ) δ 193.7, 172.9, 159.4, 145.2, 142.6, 137.6, 136.6, 135.6, 132.3, 131.6, 130.6, 129.0, 128.0, 123.9, 118.8, 81.5, and 25.2 ppm.
Method B
General preparation of HR71-HR81 and HR84-HR87 in large scale with sodium carbonate in water as base. Preparation of 2-(4-(4-chlorobenzoyl)phenoxy)-2-methyl-N-(2-(pyridin-4-yl)ethyl)propenamide (HR80). Fenofibric chloride (FFC) (0.3 mol) was prepared by the following procedure described in Method A from fenofibric acid (95.6 g; 0.3 mol) and oxalyl chloride (63.5 g; 43 ml) in dichloromethane (1 L). Prepared FFC (0.3 mmol) was slowly added dropwise over a period of 45 min at ice-water bath temperature into a magnetically stirred mixture of sodium carbonate (106 g; 1 mol) in water (500 ml) and 1-(pyridine-4-yl) ethanamine (24.4 g; 0.2 mmol) in tetrahydrofuran (500 ml). After addition was complete, the resulting reaction mixture was stirred at room temperature overnight. The reaction mixture volume was reduced by 75% via solvent evaporation under air flow (produced by air pump). The resulting white suspension was mixed with water (500 ml) and the insoluble product was separated by filtration, washed with water (10 × 50 ml), and dried at 50 °C overnight. The isolated yield was 97% (82 g) of pure product. The filtrate was acidified with hydrochloric acid to pH = 2. The resulting white solid precipitate was separated by filtration, washed with water (10 × 20 ml) and dried at 50 °C overnight to give 34.2 g (95% recovery) of fenofibric acid. 1H-NMR (DMSO-d6, 400 MHz) δ 8.36 (2H, d, J = 4.8 Hz), 8.21 (1H, t, J = 5.2 Hz), 7.66 (6H, m), 7.13 (2H, d, J = 5.2 Hz), 6.86 (2H, d, J = 8.0 Hz), 3.85 (2H, m), 2.73 (2H, t, J = 7.2 Hz), and 1.45 (6H, s) ppm. 13C-NMR (DMSO-d6, 400 MHz) δ 193.7, 173.1, 159.6, 149.7, 148.7, 137.5, 136.7, 132.1, 131.6, 130.2, 129.1, 124.6, 118.7, 81.1, 39.4, 34.3, and 25.5 ppm.
Method C
Small scale preparation applicable to all HR66-HR90. Preparation of 2-(4-(4-chlorobenzoyl)phenoxy)-2-methyl-N-(2-oxo-1,2-dihydropyridin-3-yl)propanamide (HR89). Freshly prepared dichloromethane (5 ml) solution of fenofibric acid chloride was made from fenofibric acid (80 mg; 0.25 mmol) and oxalyl chloride (1 mmol) as described above. These were added, under a nitrogen atmosphere while stirring, to a pyridine (10 ml)—tetrahydrofuran (10 ml)-sodium carbonate (212 mg; 2 mmol) of 3-aminopyridin-2(1H)-one (27.5 mg; 0.25 mmol) solution. The resulting mixture was stirred at room temperature in the nitrogen atmosphere for 3 h, followed by stirring under the nitrogen atmosphere at 60 °C for an additional 3 h. After cooling to room temperature, the solvent was evaporated under air flow (produced by air pump) at room temperature yielding a solid residue. This solid residue was mixed with dichloromethane (30 ml) and? water (100 ml). The water layer was discarded, and the organic layer was washed with water (3 × 100 ml), 5% sodium carbonate (3 × 100 ml), and dried over anhydrous sodium carbonate. The drying material was separated by filtration. The volume of the filtrate was reduced to ~ 2 ml, then hexanes (~ 10 ml) were added. The resulting solution was left uncovered at room temperature, allowing the solvent to slowly evaporate. The resulting white crystalline product was separated by filtration, washed with hexane (3 × 3 ml) and air-dried overnight. Isolated yield = 93% (95 mg). 1H-NMR (DMSO-d6, 400 MHz) δ 12.08 (1H, s), 9.29 (1H, s), 8.24 (1H, d of d, J1 = 7.2 Hz, J2 = 1.6 Hz), 7.73 (2H, d, J = 8.8 Hz), 7.71 (2H, d, J = 8.8 Hz), 7.59 (2H, d, J = 8.4 Hz), 7.12 (1H, m), 7.10 (2H, d, J = 8.8 Hz), 6.26 (1H, t, J = 7.2 Hz), and 1.58 (6H, s) ppm. 13C-NMR (DMSO-d6, 100 MHz) δ 193.8, 172.4, 158.5, 157.6, 137.7, 136.4, 132.3, 131.7, 131.6, 129.1, 128.7, 128.6, 123.1, 120.3, 105.9, 82.3, and 25.2 ppm.
Cell culture and viability assays
Human glioblastoma LN-229 cells (ATCC CRL-2611) were maintained as a semi-confluent monolayer culture in DMEM with 1 g/L glucose, sodium pyruvate and L-glutamine (Corning), supplemented with 10% heat-inactivated FBS (Gibco) and P/S (50 units/mL of penicillin and 50 µg/mL of streptomycin) at 37 °C in a 5% CO2 atmosphere. Prior to treatment with HR compounds, cells were plated in 96-well plates (BD Falcon) at an initial density of 2 × 104 cells/cm2. Twenty-four hours after plating, stock solutions of HR compounds were prepared in DMSO, diluted in cell culture medium and added to previously plated cells in triplicate for every experimental condition (final concentration 25 µM). DMSO (0.5%) was used as vehicle control. After 72h incubation, an MTT assay was performed to measure cell metabolic activity (surrogate for cell viability). Following a 1.5 h incubation with 0.5 mg/ml MTT in serum free low glucose DMEM, the resulting formazan crystals were dissolved in 5mM HCl in isopropanol and the absorbance read at 540 nm. Data represent mean values expressed as the percentage of vehicle control ± SD. Phase contrast images of treated cells were taken 72 h after treatment with HR compounds using a BZ-X800 fluorescence microscope (Keyence) equipped with a 20 × objective. The drug dose resulting in 50% inhibition of cell metabolic activity (surrogate for cell viability) was measured using MTT assay, at 72-h time point, and half maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 8.
In vitro model of the blood brain barrier (BBB)
The BBB was re-created in vitro using a modified protocol provided by Stone et al.55. Briefly, 24-well transwell inserts (Falcon, catalog number 353096) were coated with 10 μg/cm2 of Collagen Type IV (Sigma) for 24 h at 4 °C. Inserts were washed with sterile water and air-dried for 2 h. Next, the inserts were coated with 2 μg/cm2 poly-l-lysine (ScienCell) for 1 h at 37 °C, then washed twice with sterile H2O and air-dried for 2 h. Primary human astrocytes (1.5 × 105) and 3 × 104 primary human pericytes (both ScienCell) were resuspended in 25 μl of astrocyte medium and pericyte medium (ScienCell), respectively, then combined in a 1:1 ratio for 50 μl total volume. Dried, coated inserts were turned upside down such that the basolateral surface was exposed at the top, and 50 μl of the cell mixture was added to the membrane, covered with the plate lid, and incubated for 2 h at 37 °C to allow cell adhesion. Any medium remaining on top of the membrane was carefully removed before returning inserts to their upright position with the apical surface facing upward, as they were placed in a 24-well plate containing 500 μl per well of astrocyte/pericyte medium (1:1). An additional 300 μL of medium was added to the apical compartment. Four days after plating, the apical compartment medium was removed, and 3.75 × 104 of telomerase-immortalized vain endothelial cells (TIVE; provided by Dr. Rolf Renne) in 50 μl of TIVE medium56 were added and incubated for 5 h at 37 °C to allow cell attachment, followed by the addition of an extra 250 μl of TIVE medium. Half the volume of the corresponding media in the lower and upper compartment was replaced with fresh media every third day. Ten days after initial plating, trans-endothelial electric resistance (TEER) was measured using a EVOM2 meter with a STX3 electrode (World Precision Instruments). The ability of selected HR compounds to pass through the in vitro BBB was tested using inserts with effectively reconstructed BBB as confirmed by TEER values55,57.
High performance liquid chromatography (HPLC)-based detection of selected HR compounds
Following TEER measurement, the medium from the apical compartment (insert) of the in vitro BBB model (Fig. 9A) was replaced with 350 µL of fresh TIVE medium containing corresponding compounds [HR67 (PP23), HR68 (PP21), both used at 25 µM. In addition, 25 µM fenofibrate (FF), which does not cross the BBB22, was used as a negative control, and 50 µM caffeine was used as a positive control58. Plates containing the inserts were returned to the incubator (37 °C, 5% CO2), and after 3 h of incubation, conditioned media from the well and insert (Fig. 9A) were collected. The aliquots (100 μl) of the collected samples were subsequently mixed with 100 μl of 100% acetonitrile, centrifuged (16,000 rpm at 4 °C for 10 min) and supernatants collected for HPLC -based detection of HR67 and HR68.
HPLC analyses were performed using an UltiMate 3000 system (Thermo Scientific) equipped with an analytical YMCbasic, 3µm, 150 × 4.6 mm column (octyl silane C8; YMC America, Inc.). Isocratic elution of the compounds was performed using a mobile phase composed of solvent A (50 mM acetic acid in dH2O) and solvent B (acetonitrile) mixed at predetermined ratios for each compound (Table 1). All separations were carried out with 5μl sample volume at a flow rate of 1 ml/min, at 25°C. The concentration of each compound was calculated using serial dilutions of the known concentration of the compound separated at the same run with experimental and control samples. After separation, integrated areas under the peak were used to prepare calibration curves and to determine concentration of the compounds.
Table1.
Details of the HPLC method for selected HR compounds.
| Compound | Method length (min) | Concentration solvent B (%) | Detection wavelength (nm) | Retention time (min) |
|---|---|---|---|---|
| Caffeine | 5 | 25 | 272 | 2.54 |
| Fenofibrate | 10 | 70 | 288 | 5.84 |
| HR67 | 10 | 60 | 268 | 4.45 |
| HR68 | 10 | 60 | 266 | 4.45 |
Intracranial glioblastoma and tissue extraction
Foxn1nu female immunodeficient mice at 6 to 8 weeks of age were used in this study (both male and female are similarly affected by glioblastoma). The mice were inoculated with patient-derived glioblastoma, GBM12-TMZ-resistant, which stably express luciferase reporter24,59, were kindly provided by Dr. Sarkaria (Cleveland Clinic, Brain Tumor National Resource) and were cultured and propagated according to the recommended protocols59. The cells were injected into the striatum region using 5 μl of PBS containing 1 × 105 of the tumor cells guided by the stereotactic approach [1.5 mm posterior to Bregma; 1.5 mm lateral to Sagital suture; 3 mm down from surface] as reported in our previous study24. The treatment started when the intracranial tumors were well-established (evaluated by the Optical Image System for small animals (Xenogen IVIS CT). HR68 injection solution was prepared from the 50 mM DMSO stock solution diluted in 20% cyclodextrin (2-Hydroxypropyl-β-cyclodextrin) in sterile PBS and delivered intraperitoneally (ip) at 15 mg/kg. Blood, liver, kidneys, spleen, heart, brain and brain tumor were subsequently collected, solid tissues were washed from blood in PBS, and placed on ice for an immediate sample preparation for HPLC analysis (see above). Tissues were prepared for HPLC by mixing 150 μl of sample tissue (~ 120 mg) that had been mixed with 3 volumes of Methanol : H2O mix (4:1), well-blended using TissueRuptor II (Qiagen), and centrifuged at 15,000g for 10 min at 4 °C. Supernatants were collected in 1.5 ml Eppendorf tubes and incubated at 95 °C for 3 min. Following flash cool on ice, samples were centrifuged again at 15,000g for 10 min at 4 °C and supernatants were used for HPLC-based measurement.
Statistical analysis
The data were analyzed with a homoscedastic Student t test. Differences between control and experimental groups were considered significant at P values of ≤ 0.05.
Supplementary Information
Acknowledgements
This work was supported by P20-GM121288-01 (KR), 1R41CA275433 (KR), and LSU-2022-CCRI-8 (KR/GK). All chemistry and computational studies were supported by STEPFARM, LLC. (BSJ).
Author contributions
C.I.: participated in performing specific chemical reactions designed by BSJ. Performed and helped in design of cell culture analyses, cell toxicity tests, microscopy, imaging, data analyses, interpretation of in vitro studies, and preparation of figures, as well as, editing and conceptual efforts related to final design of the manuscript; J.S.: performed and help in design of cell culture analyses, cell toxicity tests, cell imaging, data analyses and preparation of figures for cell culture experiments; M.R.: performed multiple cell culture experiments, cell toxicity tests and cell imaging following departure of J.S. S.C.C.: participated in performing specific chemical reactions designed by B.S.J.; A.L.: HPLC-based evaluation of HR68 concentration in cell culture and in mouse tissues; S.B.C.: editing and conceptual effort related to the final design of the manuscript. F.P.: Both conceptual and experimental contribution during revision. All Authors reviewed the manuscript; K.R.: helped designing cell culture experiments, authored manuscript sections related to glioblastoma, cell culture, cell toxicity, and interpretation of cell culture studies; B.S.J.: designed chemical strategies for the development of new HR compounds, performed corresponding chemical reactions, computational analyses of the new compounds, authored the section of the manuscript related to development of chemical modifications and computational analyses.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
Dr. Branko Jursic is associated with Stepharm LLC, P.O. Box 24220, New Orleans, LA; Dr. Reiss is associated with WayPath Pharma LLC. 217 Sena Dr. Metairie LA 70005. Dr. Krzysztof Reiss and Dr. Branko Jursic have an LSU provisional patent for HR compounds presented in this manuscript (“Anticancer Composition and methods of use” 2932719-056-us2). Other authors do not have any competing interest in relation to this submission.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Krzysztof Reiss, Email: kreiss@lsuhsc.edu.
Branko S. Jursic, Email: bjursic1@uno.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-39236-w.
References
- 1.Gould J. Breaking down the epidemiology of brain cancer. Nature. 2018;561:S40–S41. doi: 10.1038/d41586-018-06704-7. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2021 WHO classification of tumors of the central nervous system: A summary. Neuro Oncol. 2021;23:1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandes, C. et al. In Glioblastoma (ed S. De Vleeschouwer) (2017).
- 4.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network TC. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2013;494:506. doi: 10.1038/nature11903. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romanidou O, Kotoula V, Fountzilas G. Bridging cancer biology with the clinic: Comprehending and exploiting IDH gene mutations in gliomas. Cancer Genom. Proteom. 2018;15:421–436. doi: 10.21873/cgp.20101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F, et al. Molecular classification of IDH-mutant glioblastomas based on gene expression profiles. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L, et al. Integrated metabolomics and lipidomics analyses reveal metabolic reprogramming in human glioma with IDH1 mutation. J. Proteome Res. 2019;18:960–969. doi: 10.1021/acs.jproteome.8b00663. [DOI] [PubMed] [Google Scholar]
- 10.Romani M, Pistillo MP, Carosio R, Morabito A, Banelli B. Immune checkpoints and innovative therapies in glioblastoma. Front. Oncol. 2018;8:464. doi: 10.3389/fonc.2018.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuoco JA, et al. Vaccine-based immunotherapeutics for the treatment of glioblastoma: Advances, challenges, and future perspectives. World Neurosurg. 2018;120:302–315. doi: 10.1016/j.wneu.2018.08.202. [DOI] [PubMed] [Google Scholar]
- 12.Bagley SJ, O'Rourke DM. Clinical investigation of CAR T cells for solid tumors: Lessons learned and future directions. Pharmacol. Ther. 2019 doi: 10.1016/j.pharmthera.2019.107419. [DOI] [PubMed] [Google Scholar]
- 13.Drakulic D, et al. Current opportunities for targeting dysregulated neurodevelopmental signaling pathways in glioblastoma. Cells. 2022 doi: 10.3390/cells11162530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seker-Polat F, Pinarbasi Degirmenci N, Solaroglu I, Bagci-Onder T. Tumor cell infiltration into the brain in glioblastoma: From mechanisms to clinical perspectives. Cancers. 2022 doi: 10.3390/cancers14020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly PJ. Gliomas: Survival, origin and early detection. Surg. Neurol. Int. 2010;1:96. doi: 10.4103/2152-7806.74243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahu U, Barth RF, Otani Y, McCormack R, Kaur B. Rat and mouse brain tumor models for experimental neuro-oncology research. J. Neuropathol. Exp. Neurol. 2022;81:312–329. doi: 10.1093/jnen/nlac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arvanitis CD, Ferraro GB, Jain RK. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat. Rev. Cancer. 2020;20:26–41. doi: 10.1038/s41568-019-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naef R. A generally applicable computer algorithm based on the group additivity method for the calculation of seven molecular descriptors: heat of combustion, LogPO/W, LogS, refractivity, polarizability, toxicity and LogBB of organic compounds; scope and limits of applicability. Molecules. 2015;20:18279–18351. doi: 10.3390/molecules201018279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh N, Miner A, Hennis L, Mittal S. Mechanisms of temozolomide resistance in glioblastoma—A comprehensive review. Cancer Drug Resist. 2021;4:17–43. doi: 10.20517/cdr.2020.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shojaei S, et al. Simvastatin increases temozolomide-induced cell death by targeting the fusion of autophagosomes and lysosomes. FEBS J. 2020;287:1005–1034. doi: 10.1111/febs.15069. [DOI] [PubMed] [Google Scholar]
- 21.Wilk A, et al. Molecular mechanisms of fenofibrate-induced metabolic catastrophe and glioblastoma cell death. Mol. Cell. Biol. 2015;35:182–198. doi: 10.1128/MCB.00562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grabacka M, et al. Fenofibrate subcellular distribution as a rationale for the intracranial delivery through biodegradable carrier. J. Physiol. Pharmacol. 2015;66:233–247. [PMC free article] [PubMed] [Google Scholar]
- 23.Stalinska J, et al. Exploring anticancer activity of structurally modified benzylphenoxyacetamide (BPA); I: Synthesis strategies and computational analyses of substituted BPA variants with high anti-glioblastoma potential. Sci. Rep. 2019;9:17021. doi: 10.1038/s41598-019-53207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stalinska J, et al. Chemically modified variants of fenofibrate with antiglioblastoma potential. Transl. Oncol. 2019;12:895–907. doi: 10.1016/j.tranon.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullard A. Re-assessing the rule of 5, two decades on. Nat. Rev. Drug Discov. 2018;17:777. doi: 10.1038/nrd.2018.197. [DOI] [PubMed] [Google Scholar]
- 26.Wager TT, Hou XJ, Verhoest PR, Villalobos A. Central nervous system multiparameter optimization desirability: Application in drug discovery. ACS Chem. Neurosci. 2016;7:767–775. doi: 10.1021/acschemneuro.6b00029. [DOI] [PubMed] [Google Scholar]
- 27.Gupta M, Lee HJ, Barden CJ, Weaver DF. The blood-brain barrier (BBB) score. J. Med. Chem. 2019;62:9824–9836. doi: 10.1021/acs.jmedchem.9b01220. [DOI] [PubMed] [Google Scholar]
- 28.Lanevskij K, Didziapetris R, Sazonovas A. Physicochemical QSAR analysis of hERG inhibition revisited: Towards a quantitative potency prediction. J. Comput. Aided Mol. Des. 2022;36:837–849. doi: 10.1007/s10822-022-00483-0. [DOI] [PubMed] [Google Scholar]
- 29.Garrido A, Lepailleur A, Mignani SM, Dallemagne P, Rochais C. hERG toxicity assessment: Useful guidelines for drug design. Eur. J. Med. Chem. 2020;195:112290. doi: 10.1016/j.ejmech.2020.112290. [DOI] [PubMed] [Google Scholar]
- 30.Miller S, et al. CRY2 isoform selectivity of a circadian clock modulator with antiglioblastoma efficacy. Proc. Natl. Acad. Sci. U. S. A. 2022;119:e2203936119. doi: 10.1073/pnas.2203936119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabacka MM, et al. Fenofibrate induces ketone body production in melanoma and glioblastoma cells. Front. Endocrinol. 2016;7:5. doi: 10.3389/fendo.2016.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilk A, et al. Fenofibrate-induced nuclear translocation of FoxO3A triggers Bim-mediated apoptosis in glioblastoma cells in vitro. Cell Cycle. 2012;11:2660–2671. doi: 10.4161/cc.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De S, et al. Pyridine: The scaffolds with significant clinical diversity. RSC Adv. 2022;12:15385–15406. doi: 10.1039/d2ra01571d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahu R, et al. Pyridine moiety: An insight into recent advances in the treatment of cancer. Mini Rev. Med. Chem. 2022;22:248–272. doi: 10.2174/1389557521666210614162031. [DOI] [PubMed] [Google Scholar]
- 35.Bortolami M, et al. New pyrimidine and pyridine derivatives as multitarget cholinesterase inhibitors: Design, synthesis, and in vitro and in cellulo evaluation. ACS Chem. Neurosci. 2021;12:4090–4112. doi: 10.1021/acschemneuro.1c00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HM, et al. Computational determination of hERG-related cardiotoxicity of drug candidates. BMC Bioinform. 2019;20:250. doi: 10.1186/s12859-019-2814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahjour B, Shen Y, Liu W, Cernak T. A map of the amine-carboxylic acid coupling system. Nature. 2020;580:71–75. doi: 10.1038/s41586-020-2142-y. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh AK, Shahabi D. Synthesis of amide derivatives for electron deficient amines and functionalized carboxylic acids using EDC and DMAP and a catalytic amount of HOBt as the coupling reagents. Tetrahedron Lett. 2021 doi: 10.1016/j.tetlet.2020.152719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Su NQ, Yang W. Describing chemical reactivity with frontier molecular orbitalets. JACS Au. 2022;2:1383–1394. doi: 10.1021/jacsau.2c00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raczynska ED, Stepniewski TM, Kolczynska K. DFT studies on one-electron oxidation and one-electron reduction for 2- and 4-aminopyridines. J. Mol. Model. 2012;18:4367–4380. doi: 10.1007/s00894-012-1446-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gosselet F, Loiola RA, Roig A, Rosell A, Culot M. Central nervous system delivery of molecules across the blood-brain barrier. Neurochem. Int. 2021;144:104952. doi: 10.1016/j.neuint.2020.104952. [DOI] [PubMed] [Google Scholar]
- 42.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 43.Mikitsh JL, Chacko AM. Pathways for small molecule delivery to the central nervous system across the blood-brain barrier. Perspect. Med. Chem. 2014;6:11–24. doi: 10.4137/PMC.S13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsson P, Kihlberg J. How big is too big for cell permeability? J. Med. Chem. 2017;60:1662–1664. doi: 10.1021/acs.jmedchem.7b00237. [DOI] [PubMed] [Google Scholar]
- 45.Smith QR, Fisher C, Allen DD. The Role of Plasma Protein Binding in Drug Delivery to Brain. Springer; 2001. pp. 311–321. [Google Scholar]
- 46.Du X, et al. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016 doi: 10.3390/ijms17020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansch C, et al. On the role of polarizability in chemical-biological interactions. J. Chem. Inf. Comput. Sci. 2003;43:120–125. doi: 10.1021/ci020378b. [DOI] [PubMed] [Google Scholar]
- 48.Kirchmair J, et al. Predicting drug metabolism: Experiment and/or computation? Nat. Rev. Drug Discov. 2015;14:387–404. doi: 10.1038/nrd4581. [DOI] [PubMed] [Google Scholar]
- 49.Djoumbou-Feunang Y, et al. Biotransformer: A comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform. 2019;11:2. doi: 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodges RE, Minich DM. Modulation of metabolic detoxification pathways using foods and food-derived components: A scientific review with clinical application. J. Nutr. Metab. 2015;2015:760689. doi: 10.1155/2015/760689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, et al. An experimentally validated approach to calculate the blood-brain barrier permeability of small molecules. Sci. Rep. 2019;9:6117. doi: 10.1038/s41598-019-42272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stalinska J, et al. Anti-glioblastoma effects of phenolic variants of benzoylphenoxyacetamide (BPA) with high potential for blood brain barrier penetration. Sci. Rep. 2022;12:3384. doi: 10.1038/s41598-022-07247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quintanilla-Licea R, et al. NMR detection of isomers arising from restricted rotation of the C-N amide bond of N-formyl-o-toluidine and N,N'-bis-formyl-o-tolidine. Molecules. 2002;7:662–673. doi: 10.3390/70800662. [DOI] [Google Scholar]
- 54.Wishart DS, et al. Biotransformer 3.0-a web server for accurately predicting metabolic transformation products. Nucleic Acids Res. 2022;50:W115–123. doi: 10.1093/nar/gkac313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stone NL, England TJ, O'Sullivan SE. A novel transwell blood brain barrier model using primary human cells. Front. Cell Neurosci. 2019;13:230. doi: 10.3389/fncel.2019.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An FQ, et al. Long-term-infected telomerase-immortalized endothelial cells: A model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 2006;80:4833–4846. doi: 10.1128/JVI.80.10.4833-4846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasan B, et al. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015;20:107–126. doi: 10.1177/2211068214561025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garberg P, et al. In vitro models for the blood-brain barrier. Toxicol In Vitro. 2005;19:299–334. doi: 10.1016/j.tiv.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Carlson BL, Pokorny JL, Schroeder MA, Sarkaria JN. Establishment, maintenance and in vitro and in vivo applications of primary human glioblastoma multiforme (GBM) xenograft models for translational biology studies and drug discovery. Curr. Protoc. Pharmacol. 2011 doi: 10.1002/0471141755.ph1416s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.