SUMMARY
We describe a patient haploinsufficient for the neuronal RNA binding protein NOVA1 who developed a behavioral motor hyperactivity disorder, suggesting a role of NOVA1 in postnatal motor inhibition. To investigate Nova1’s action in adult Gad2+ inhibitory neurons, we generated a conditional Nova1-null mouse (Nova1-cKOGad2-cre). Strikingly, the phenotypes of these mice show many similarities to the NOVA1 haploinsufficient patient and identify a function of Nova1 in the hypothalamus. Molecularly, Nova1 loss in Gad2-positive neurons alters downstream expression of Impact mRNA, along with a subset of RNAs encoding electron transport chain-related factors and ribosomal proteins. NOVA1 stabilizes Impact mRNA by binding its 3′ UTR, antagonizing the actions of miR-138 and miR-124. Together, these studies demonstrate actions of NOVA1 in adult hypothalamic neurons, mechanisms by which it functions in translation and metabolism, including through direct binding to Impact mRNA, and illuminate its role in human neurologic disease.
Graphical abstract
In brief
Tajima et al. report that NOVA1 regulates motor inhibition and learning in humans. NOVA1 acts on Gad2+ neurons in the mouse hypothalamus, regulating physiology and behavior. Molecularly, NOVA1 stabilizes Impact mRNA through 3′ UTR binding, antagonizing miRNA binding, resulting in translational controls and effects on intracellular and systemic energy homeostasis.
INTRODUCTION
NOVA proteins were identified as the target antigens of paraneoplastic opsoclonus-myoclonus-ataxia (POMA), a spectrum of neurological disorders mediated by naturally occurring tumor immunity against cancer antigens that turns into an autoimmune attack against the nervous system.1–4 POMA patients have underlying gynecologic or lung tumors and are characterized neurologically as having a failure to properly inhibit brain stem neurons (opsoclonus) and spinal motor neurons (action-induced myoclonic tremors). The molecular function of NOVA proteins in adult neurons has been unclear, in part because Nova-null mice are postnatal lethal.
NOVA1 and its family member NOVA2 are RNA-binding proteins with three K Homology (KH) domains, and both are highly expressed in the central nervous system.4,5 They bind to YCAY repeats in RNA,6 and X-ray crystallography has uncovered structural details of the interaction between the KH domains of NOVA and YCAY sequences.7,8 NOVA1 and NOVA2 show distinct expression patterns in the mouse brain, where NOVA1 is highly expressed in the diencephalon/mesencephalon and hindbrain, and NOVA2 is highly expressed in the cortex and hippocampus.4 Cell-type-specific gene expression analysis revealed that Nova1 is enriched in Gad2+ inhibitory neurons, while Nova2 is enriched in Emx1+ excitatory neurons.9 These observations suggest that the actions of NOVA1 may be directly pertinent to the inhibitory phenotype of humans with POMA, although, prior to this study, genetic-clinical correlations of NOVA1 deficiency have not been described.
The biological roles of NOVA proteins in regulating the transcriptome have been analyzed through use of knockout mouse models10–12 coupled with high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) methods. Specifically, this approach enabled us to comprehensively identify NOVA-bound transcripts in the prenatal brain.13–15
Molecular biological functions of NOVA proteins include alternative splicing control, transcript localization, and regulation of transcript stability through binding to nonsense-mediated decay (NMD) exons and 3′ UTRs.10,16–18 In studies focused on regulation of alternative splicing, NOVA binds upstream or downstream of the splice sites of alternative exons on transcripts encoding neuronal synapse-associated molecules to regulate alternative exon exclusion or inclusion, respectively.15,19 The mechanisms and biological significance of the relationships between NOVA binding and splicing have been partially explored,11,20 and direct actions of NOVA have been described for many target transcripts.21 Interestingly, while nearly 80% of the binding sites for NOVA2 are located in the intron region of the transcript, NOVA1 binding sites lie predominantly at 3′ UTRs, and the splicing changes affected in Nova1-null mice are not as obvious as those observed in Nova2-null cells.9 These results suggest that NOVA1 and NOVA2 have non-redundant molecular functions related to their distinct expression patterns.9
Several human clinical studies have reported that abnormalities in chromosomal regions including NOVA1 cause severe psychiatric disorders and motor developmental abnormalities.22,23 Nova1 expression appears to be associated with autism19 and the progression and prognosis of cancers such as gastric cancer, hepatocellular carcinoma, and lymphoma, making studies of NOVA1 function interesting from multiple perspectives.24–31 In mice, Nova1 deficiency leads to a lethal phenotype within a few days after birth, with marked motor dysfunction (weakness and action-induced tremors) and neuronal apoptosis in the spinal cord and brain stem.32 Despite the implied importance of NOVA1 in maintaining neural activity and function in the brain, other than a role in Agrin splicing and development of the neuromuscular junction,11 the specific physiological functions of NOVA1 remain largely unknown.
Here we describe a human with NOVA1 haploinsufficiency with a series of neuro-behavioral deficits. We generated mouse model lacking Nova1 in Gad2+ inhibitory neurons to analyze the biological and molecular functions of NOVA1 in the adult brain. We report that NOVA1 stabilizes the Impact transcript by binding directly to the 3′ UTR in Gad2+ cells in the hypothalamus, supporting intracellular energy balance, including translation, resulting in maintainance of homeostasis in the animal.
RESULTS
A NOVA1 haploinsufficiency patient and generation of Nova1-deficient mice in Gad2+-inhibitory neurons
Our laboratory was contacted for clinical input regarding a 9-year-old boy with motor dysfunction and developmental delay. He began experiencing abnormal movements of the head, trunk, and extremities around the age of 3–4 years, which were more pronounced when he was emotionally agitated or anxious. He had been diagnosed with hypotonia during infancy and developmental delay, not babbling until age 2 and not speaking phrases until age 3. At age 2 he began speech, language therapy, and occupational/physical therapy. In evaluations between ages 4–7, his IQ was 87 (19th percentile), he had behaviors associated with anxiety, learning dysfunction (including difficulty with attention, attention shifting, behavioral inhibition, and abstract thinking), was transiently considered to have autistic features, and was evaluated as having atypical attention disorder. At age 8 he was evaluated for hypotonia, speech, and oculomotor apraxia. We precisely defined a haploid deletion of 2,299,695 bp on the long arm of chromosome 14, and NOVA1 was found to be the only protein-coding gene in this region (Figure 1A). Examination at Rockefeller University was remarkable for abnormal motor behavior, consisting of excess motor movements he could suppress volitionally, followed by rebound excess motor movement, suggestive of a motor-only Tourette’s-like phenomenon. Taken together, these findings were consistent with the hypothesis that NOVA1 is involved in inhibitory control of higher motor and cognitive functions. Follow-up histories revealed generally resolving phenotypes, including attenuation of the movement disorder, with attendance at an age-appropriate elementary school.
Figure 1. NOVA1 haploinsufficiency patient and generation of Nova1-deficient mice in Gad2+ inhibitory neurons.
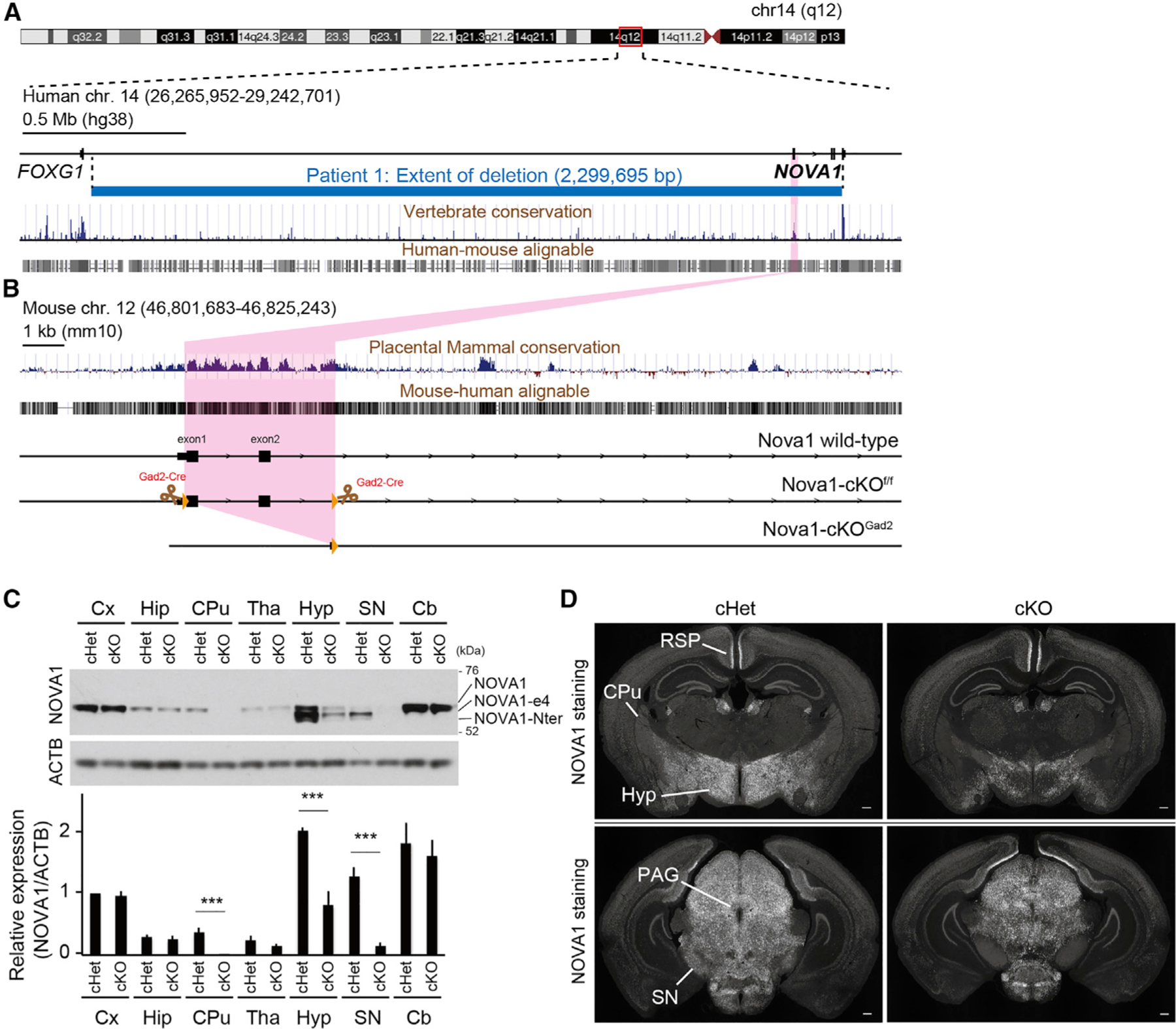
(A and B) Overview of the human (A) and mouse (B) loci. Physical mapping of the deletion at 14q12 (patient) is shown in (A). The detailed view of the region is derived from the University of California, Santa Cruz (UCSC) Genome Browser (GRCh38/hg38 December 2013 assembly). 100-vertebrate PhyloP scores measure vertebrate conservation. Genes are represented in black. Deletion in the patient is depicted by a horizontal blue line. The deletion region for the patient is approximately 2.3 Mb. Nova1-cKOf/f mice were designed with exons 1 and 2 of Nova1 flanked by LoxP sites.
(C) Western blot of NOVA1 in lysates of each dissected brain region of adult mice. NOVA1-e4 is NOVA1 without exon 4, and NOVA1-Nter is an isoform lacking the canonical N terminus. Bottom: quantification of NOVA1 western blot analysis (mean ± SEM, n = 3 biological replicates, unpaired t test, ***p < 0.005).
(D) Immunostaining of P21 cHet (Gad2cre-Nova1f/w) and cKO (Gad2cre-Nova1f/f) mouse brain with NOVA1 antibody. Hyp, hypothalamus; SN, substantia nigra; PAG, periaqueductal gray; CPu, caudate putamen; RSP, retrosplenial region. 40× magnification. Scale bar, 300 μm.
In mice, NOVA1 is mainly expressed in Gad2+ neurons,9 and the fact that POMA patients, the NOVA1 haploinsufficient patient, and Nova1-null mice show abnormal motor inhibition1,32 prompted us to develop a means to study NOVA1 function specifically in inhibitory neurons. For this purpose, we generated Gad2cre-Nova1f/f conditional knockout (cKO) mice. Gad2 encodes the enzyme glutamate decarboxylase, which converts glutamate into the inhibitory neurotransmitter gamma-aminobutyric acid (GABA), and Gad2-cre is often used as a marker of inhibitory neurons33 (we recognize that there may be some neurons where this distinction may be blurred,34 but for simplicity, here we refer to Gad2-Cre neurons as inhibitory neurons). In these mice, exons 1 and 2, spanning the start codon of the Nova1 locus, were deleted in a Cre-dependent manner in Gad2-expressing cells (Figure 1B). In contrast to Nova1-null mice, which die shortly after birth,32 77% of cKO mice survive at 12 weeks of age. Heterozygous littermates of Gad2cre-Nova1f/w (cHet) were used as controls in this study because the adults showed no clear abnormalities in survival, growth, or reproduction. While cHet mice may nonetheless have some subtle phenotypes and molecularly may have some splicing defects (as seen with Nova1-null mice32), our focus on mice in which NOVA1 is deleted only in inhibitory neurons allowed a focus on neurons of great clinical interest.
To investigate the regions within the cKO brain with the most prominent loss of NOVA1, western blotting and immunostaining were used (Figures 1C and 1D). The loss of NOVA1 expression was particularly pronounced in the hypothalamus and also observed in the periaqueductal gray and substantia nigra (Figures 1D and S1). Expression of NOVA1 in the caudate putamen was also almostentirely lost, although the frequency of NOVA1-expressing cells in control animals was low (Figures 1C and S1), and some neurons express NOVA1 in additional neurons, such as in the hypothalamus, retrosplenial region (Figure 1D) and elsewhere, that presumably do not express Gad2-Cre.
Nova1-deficient mice in inhibitory neurons have abnormalities related to the hypothalamus
cKO mice were observed to have marked developmental delay. cKO mice were born at the same weight as their control cHet littermates but showeda 25%–45% reduction in body weightas the animals grew, andthisdifferencepersistedthroughoutlife(Figure2A). RNA sequencing (RNA-seq) analysis revealed decreased expression of growth hormone (Gh) in the pituitary gland of cKO mice, likely contributing to the poor growth of these mice (Figure S2A). In addition, the expression of various other hormones, such as gonadotropins (follicle-stimulating hormone beta subunit [Fshb], luteinizing hormone beta subunit [Lhb], and chorionic gonadotropin alpha subunit [Cga]), was decreased in cKO mice (Figures S2A and S2B). Paradoxically, when the food intake of the cKO mice was measured, it was increased compared with cHet mice (Figure 2B). Notably, there was no evidence of NOVA1 expression in the pituitary, consistent with metabolic and hormonal changes having their origin in hypothalamic inhibitory neurons. Taken together, these observations suggest a functional alteration in the cKO brain, particularly in the hypothalamus, the CNS region controlling the endocrine system and appetite. This further suggested the possibility of a role of Nova1 in metabolic homeostasis.
Figure 2. Nova1-deficient mice in Gad2+ inhibitory neurons show abnormalities related to the hypothalamus.
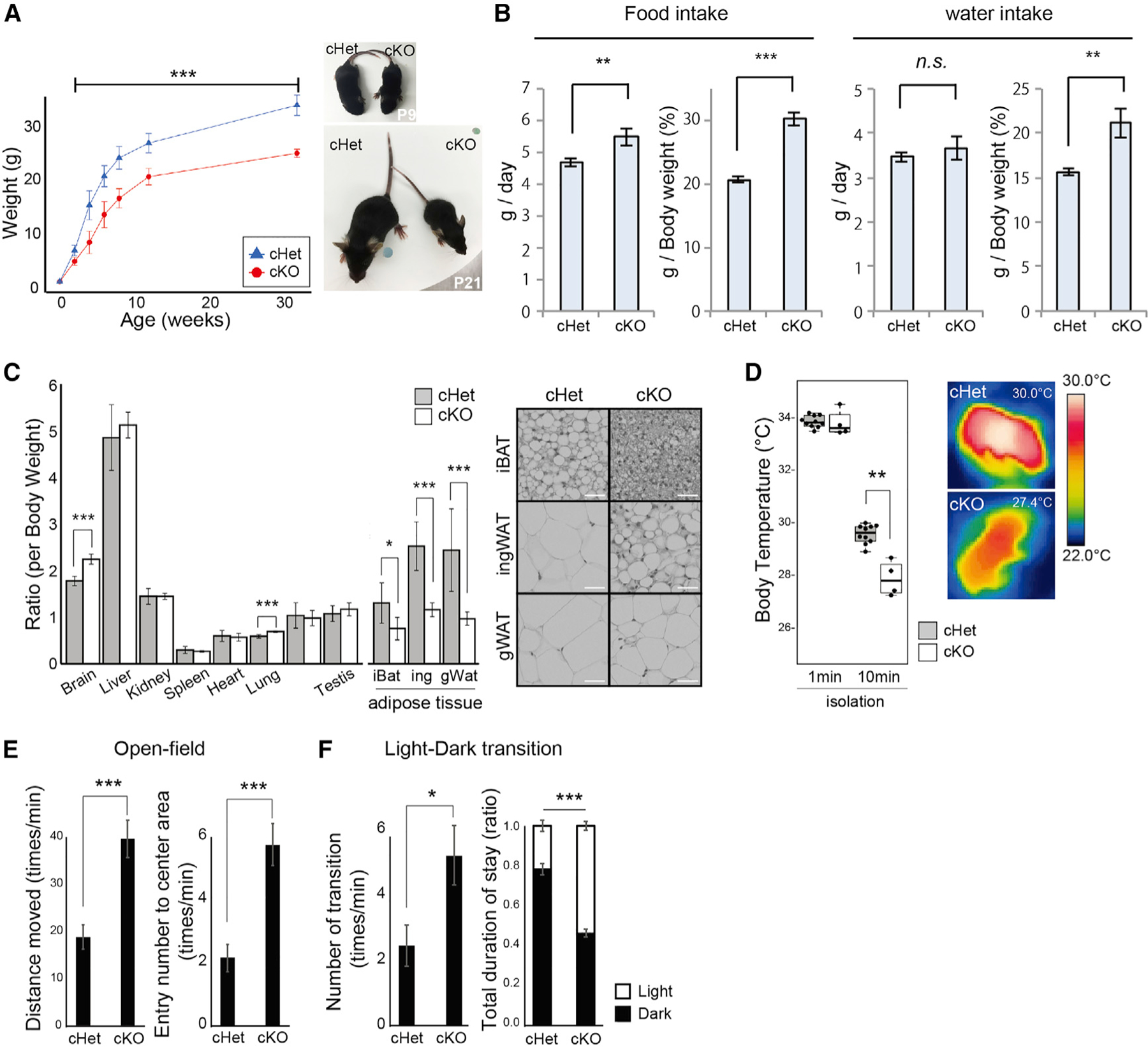
(A) Body weight transition of Gad2creNova1cKOff mice and littermate control Gad2creNova1cKOfw mice (mean ± SEM; cHet, n = 18; cKO, n = 23; unpaired t test, ***p < 0.005).
(B) Experiment to measure food intake. The 3-day average for each mouse was calculated. Total food/water intake and food/water intake per body weight are shown (mean ± SEM; cHet, n = 10; cKO, n = 9; unpaired t test, **p < 0.01, ***p < 0.005).
(C) Comparison of the percentage of each organ per body weight and H&E staining of each tissue in 12-week-old mice. iBAT, interscapular brown adipose tissue; ingWAT, inguinal white adipose tissue; gWAT, gonadal white adipose tissue (mean ± SEM; cHet, n = 7; cKO, n = 6; unpaired t test, *p < 0.05, **p < 0.01, ***p < 0.005). 2003 magnification. Scale bar, 30 μm.
(D) Heat production in neonatal (P5) mice. The body surface temperature of newborn mice 0 and 10 min after separation from the nest/mother was measured (median values are indicated as horizonal lines with 25th–75th percentiles and whiskers [minimum and maximum values]; cHet, n = 10; cKO, n = 4; Wilcoxon test, **p < 0.01).
(E) Open field test. Free exploration of the mice was recorded for 5 min. The number of times the animals entered the center area of the box was also recorded (mean ± SEM; cHet, n = 8; cKO, n = 12; unpaired t test, ***p < 0.005).
(F) Light/dark transition test. The number of entries into the bright chamber and the duration of time spent there were measured (mean ± SEM; cHet, n = 5; cKO, n = 5; unpaired t test, *p < 0.05, ***p < 0.005).
We therefore further examined the metabolism of cKO mice and found a significant reduction in adipose tissue weight among other organs (Figure 2C). Histological analysis of individual adipose tissues indicated reduced lipid accumulation in cKO adipose tissues. These data suggested that systemic energy homeostasis was affected in cKO mice. Because lipid handling in adipose tissues is known to be critical for brown fat thermogenesis,35,36 we examined the thermoregulatory function in cKO mice in the neonatal period (5 days old) by temporally isolating them from their mother and nests. The cKO mice displayed a significantly lower body surface temperature 10 min after isolation compared with cHet mice, although they did not differ in their body surface temperature at the time of separation (Figure 2D). This indicated a decrease in heat-producing functions associated with a decrease in brown adipose tissue in cKO mice.
cKO mice showed an obvious paw-clasping phenotype (bat-like posture, clenching the limbs and curling the entire body) when lifted vertically by the tail (Figure S2C). This phenotype was observed from at least 9 days of age and remained throughout life. This phenotype is often observed in mice with lesions in the cerebellum, basal ganglia, and neocortex and is thought to involve the cerebellar-cortical-reticular pathway and the cortical-striatal-pallidal-reticular pathway.37 In cKO mice, these pathways may be damaged in inhibitory neurons contributing to these circuits.
We examined the changes in learning and memory of cKO mice by the Y maze test. It is known that mice usually prefer to visit new environments.38 cKO mice showed a significantly lower probability of selecting different arms in the Y maze consecutively compared with control cHet mice, suggesting that they have lower working memory (Figure S2D). cKO mice moved more frequently and were more active than cHet mice. We also conducted an open field test and a light/dark transition test.39,40 In both tests, cKO mice showed significantly more distance/number of movements than cHet mice, suggesting that cKO mice have a hyperactive phenotype (Figures 2E and 2F). Furthermore, in the open field test, cKO mice penetrated the center area of the field significantly more often, suggesting a decrease of anxiety in cKO mice compared with cHet mice. The phenotype of reduced anxiety in cKO mice was also supported by the longer time spent in the brighter box compared with cHet mice in the light/dark transition test. These phenotypes, such as abnormal movements during tension (paw clasping), learning dysfunction, and hyperactivity, are similar to those observed in autism, suggesting that the link observed previously between Nova1 and autism may be mediated in part through its action in inhibitory neurons.
Loss of Nova1 in inhibitory neurons indirectly affects gene expression of ribosomal proteins
To characterize the molecular changes in cKO mice, we examined gene expression changes in the cKO mouse brain with RNA-seq analysis. In particular, we dissected regions of the hypothalamus, substantia nigra, periaqueductal gray, and caudate putamen, where the loss of Nova1 expression was particularly prominent (Figures 1D, S1A).
We first recognized changes in the expression of neuropeptide molecules in the hypothalamus. The hypothalamus is important for homeostasis controlling the autonomic nervous system, endocrine system, feeding, drinking, and body temperature,41–44 and it is known that various neuropeptides act as neurotransmitters in the regulation of these processes. Interestingly, levels of RNAs encoding various neuropeptides and hormones were altered in cKO mice (Figure S3A). For example, expression of hormones such as Npy and Agrp, which promote feeding behavior,45,46 and Sst, which inhibits hormone secretion in the pituitary and thyroid gland,47,48 was upregulated in cKO mice. It should be noted that, although these neuropeptides are produced in the hypothalamus and peripheral sympathetic nervous system, Nova1 is not expressed in the latter3; hence, their actions in cKO mice are likely to be central.
We examined the most highly affected pathways in the cKO brain for their biological relevance (Figures 3A and S3B). Metabolic pathways such as cholesterol were significantly downregulated, whereas translation-related genes, including many ribosomal proteins, were significantly upregulated. Strikingly, upregulation of translation-related genes was common to regions where NOVA1 expression was largely lost (Figures 3A, 3B, and 1D), including the hypothalamus, periaqueductal gray, and substantia nigra, and less prominent in the caudate putamen. Notably, mRNAs encoding ribosomal proteins and translation initiation factors were upregulated in cKO mice, while factors involved in translation elongation and termination were unchanged (Figure 3B).
Figure 3. Loss of Nova1 in inhibitory neurons affects gene expression of ribosomal proteins.
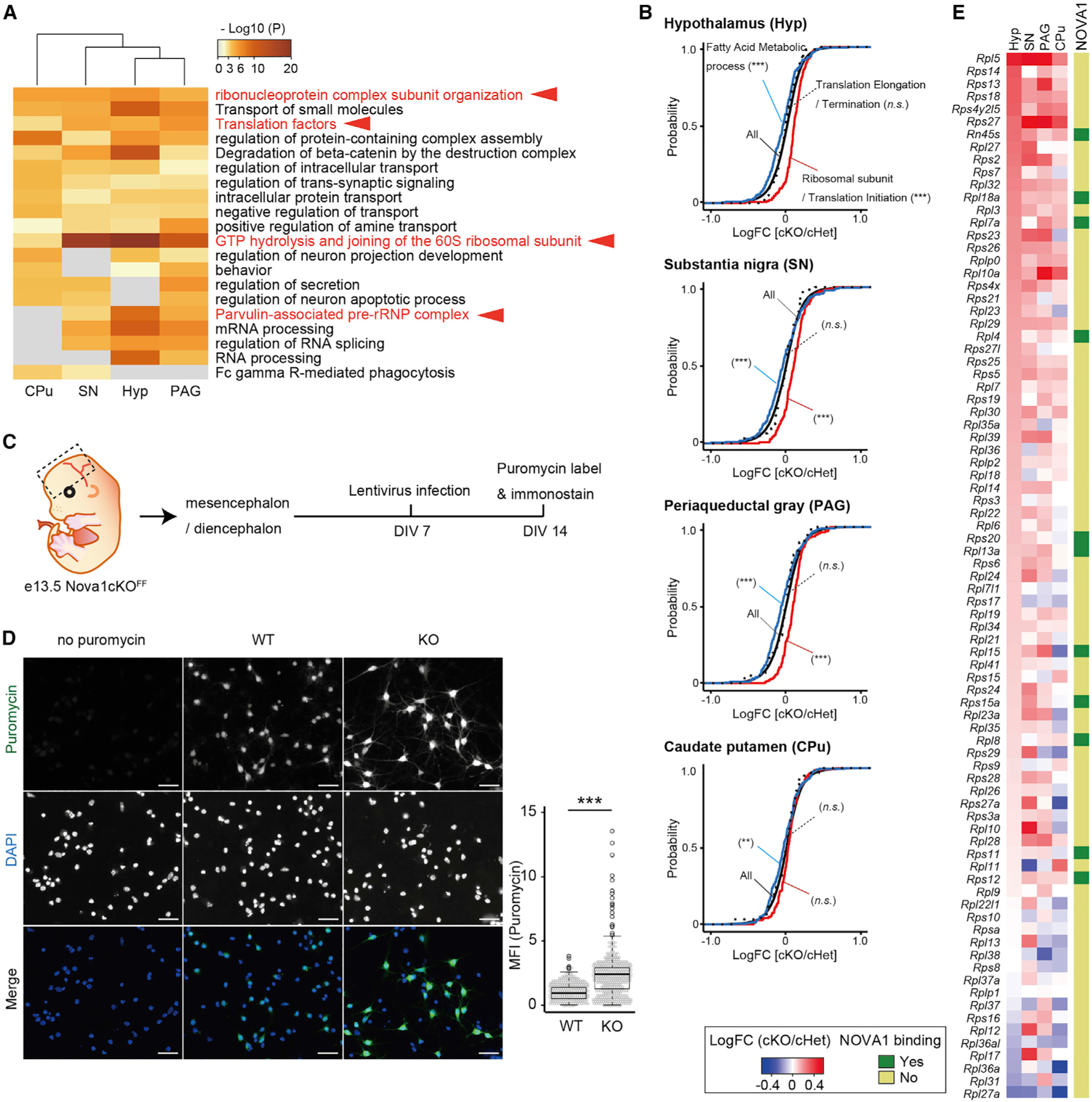
(A) GO analysis of genes significantly upregulated in cKO relative to control (cHet) in P21 Hyp, SN, PAG, and CPu RNA-seq data. Terms involved in translation are shown in red.
(B) mRNA expression changes upon Nova1 deficiency in the Gad2 lineage determined by RNA-seq. Shown is the empirical cumulative distribution function (CDF) binned by genes of fatty acid metabolic process (blue), ribosomal subunit and translation initiation (red), and translation elongation and termination (dotted line) compared with all expressed genes (black). The p values were determined by Mann-Whitney U test (***p < 0.005).
(C) Diagram of experiments using puromycin to label nascent protein synthesis in primary neuronal cultures of the mouse diencephalon/mesencephalon.
(D) Left: immunostaining image of anti-puromycin antibody-stained primary neurons. Cells lacking Nova1 (via the Cre expression lentivirus [KO]) were compared with control neurons infected with empty vector (WT). Cells without puromycin treatment were used as a negative control for staining. WT, empty-lentivirus-infected cells; KO, Cre-expressing lentivirus-infected cells. 200× magnification. Scale bar, 30 μm. Right: relative protein synthesis was quantitated by measuring the mean fluorescence intensity (MFI; arbitrary units) obtained from puromycin immunostaining of individual neurons. The fluorescence of puromycin in cells (demarcated by DAPI positivity) was quantified. The mean value of the empty-vector-infected cell group was set to 1. Each dot represents one cell. Median values are indicated as horizonal lines with 25th–75th percentiles and whiskers (minimum and maximum values). The p values were determined by Wilcoxon test (***p < 0.005).
(E) Heatmap showing the expression changes of each ribosomal protein gene in cKO mice and the presence or absence of Nova1 binding peaks (indicated by dark green in the side column) for each transcript.
To test whether the increased expression of transcripts encoding ribosomal proteins indeed affects translational activity, we assessed the incorporation of puromycin into nascent peptides to examine the changes in protein synthesis (using surface sensing of translation: SUnSET method)49 in primary neurons isolated from embryonic day 13.5 (E13.5) diencephalon/mesencephalon (including the hypothalamus) (Figure 3C). Primary neurons from Nova1-floxed mice were infected with a Cre-expressing lentivirus to knock out Nova1, and puromycin labeling and immunostaining were performed to assess protein synthesis. In cells lacking Nova1, the intensity of puromycin staining was increased compared withthose infectedwith a control vector virus, indicating that there was more denovo protein synthesis in the steady state in the absence of Nova1 (Figure 3D). Taken together, these results demonstrate that the loss of NOVA1 in inhibitory neurons causes an increase in expression of ribosomal genes to induce functional changes in cellular translational activity.
To identify direct binding targets of NOVA1, we performed CLIP analysis in the diencephalon/mesencephalon of post-natal day 21 (P21) mice (Figure S3C). NOVA1 was mainly distributed in the 3′ UTR, coding sequence (CDS), and introns, with YCAY being the most strongly enriched binding motif, as reported previously (Figures S3D and S3E).6,12 NOVA1 target transcripts were highly enriched in sets of genes involved in synapse formation and transmission and in development of neuronal projections (Figure S3F). Overlaying the results of the NOVA1 CLIP analysis with the list of expression changes in cKO mice revealed that approximately 45%–47% of the variable genes had NOVA1 binding peaks on their transcripts (Figure S3G). Interestingly, few of the ribosomal protein genes that were upregulated in cKO mice had NOVA1 binding peaks (Figures 3E and S3H). This suggested that the elevated expression of ribosomal proteins detected in the cKO brain was an indirect but strong consequence of loss of NOVA1.
Nova1 is required for Impact expression in inhibitory neurons
To quantify transcripts directly affected by loss of NOVA1 specifically in inhibitory neurons, CLIP data were combined with a quantitative measure of translated mRNAs. Targeted purification of polysomal mRNA (TRAP-seq) was performed in whole brains of cKO mice together with samples from control (cHet) littermates. TRAP-seq analysis of Gad2 cell-specific translated RNA revealed that 272 transcripts were significantly altered in the absence of Nova1 (184 decreased and 88 increased) (Figure 4A; Table S1). Among these, 80 RNAs had NOVA1 binding peaks on their transcripts (63 down-regulated, 17 up-regulated), indicating that NOVA1 can directly regulate their expression and translation. Notably, similar to the results of bulk RNA-seq analysis, the expression of translation-related factors, especially transcripts encoding ribosomal protein subunits, was up-regulated in the absence of Nova1 (Figure 4B).
Figure 4. Nova1 is required for Impact expression in inhibitory neurons.
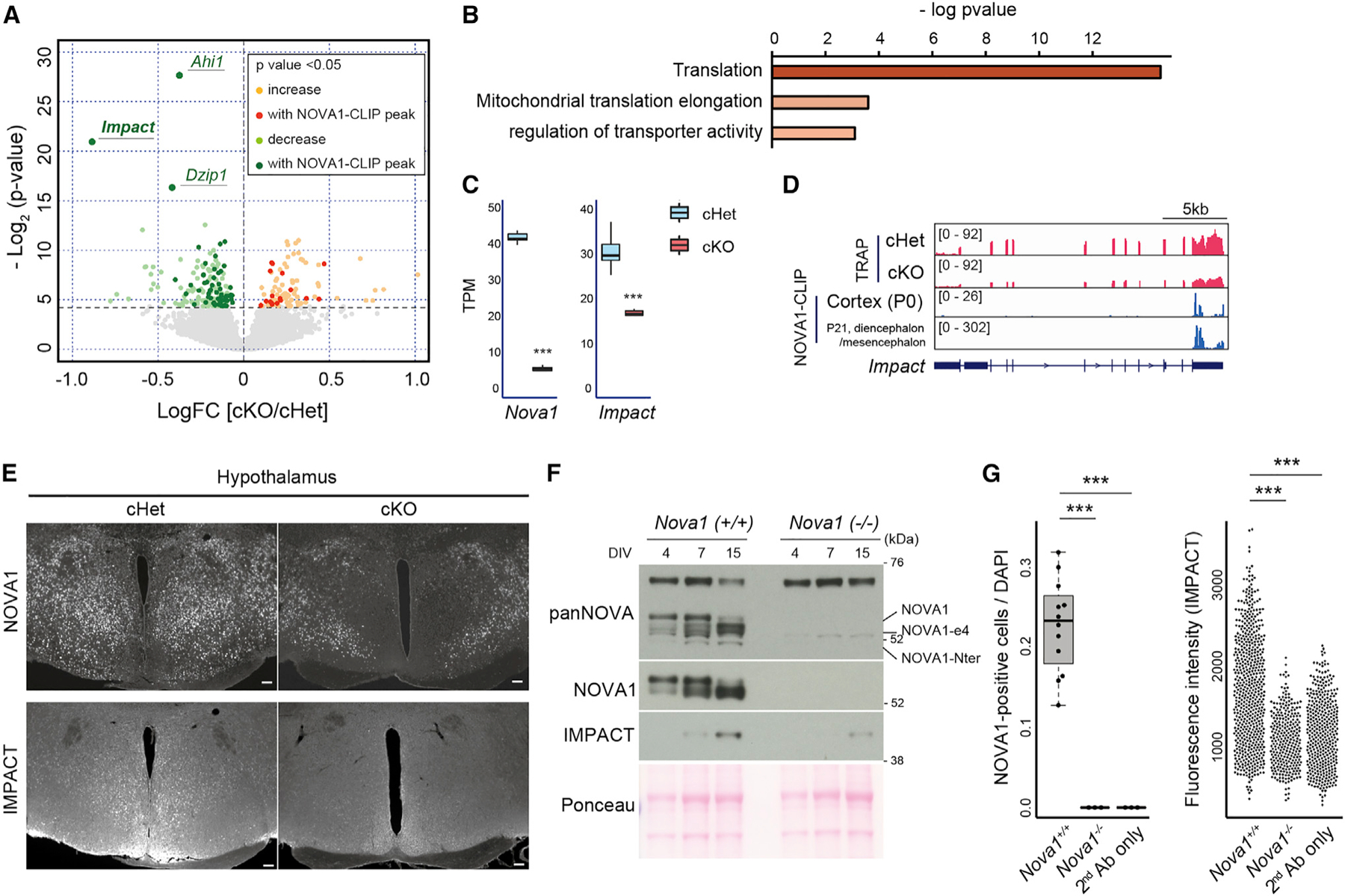
(A) Volcano plots of TRAP data from P0 whole brain for cHet and cKO. Yellow and light green indicate genes thatare significantly up- or down-regulated in cKO, and red and green indicate those with NOVA1 CLIP peaks on the genes, respectively. NOVA1 CLIP data from the cerebral cortex of P0 mice (Saito et al.12) were used.
(B) GO analysis of genes whose expression was upregulated in cKO mice in TRAP-seq.
(C) Box-and-whisker plots of Nova1, Nova2, and Impact RNA-seq reads from TRAP data. TPM, transcripts per million. Median values are indicated as horizonal lines with 25th–75th percentiles and whiskers (minimum and maximum values). The p values were determined by Wilcoxon test (***p < 0.005).
(D) Integrative Genomics Viewer (IGV)50 tracks of the Impact locus. P0 TRAP data and NOVA1 CLIP data (P0 cortex and P21 diencephalon/mesencephalon) are shown.
(E) Immunostaining for NOVA1 and IMPACT in the hypothalamus of 12-week-old mice. Serial sections were used in each genotype. 40× magnification. Scale bar, 100 μm.
(F) Western blot analysis of NOVA1 and IMPACT against lysates from primary neuron culture of control (Nova1+/+) and Nova1 KO (Nova1−/−) mice.
(G) Comparison of the ratio of NOVA1+ cells in culture (left) (median values are indicated as horizonal lines with 25th–75th percentiles and whiskers [minimum and maximum values]) and IMPACT staining intensity (right) in primary neurons obtained from Nova1+/− and Nova1−/− mice on day 15 of culture. The p values were determined by Wilcoxon test (***p < 0.005).
The top three genes most significantly down-regulated in inhibitory neurons in cKO mice, Impact, Ahi1, and Dzip1, contained NOVA1 binding peaks (Figure 4A). We focused on the gene with the largest fold change, Impact (Figures 4C and 4D). Impact is expressed in the central nervous system and is thought to be a translational regulator that inhibits GCN2, a kinase of eIF2a.51,52 Impact KO mice show a phenotype similar to that of Gad2 cell-specific Nova1 KO mice: lean with reduced adipose tissue and impaired thermoregulation.53 These observations suggested the possibility that Impact could contribute to the phenotype of cKO mice. Analysis of IMPACT protein expression showed that IMPACT is strongly expressed in the hypothalamus, and its expression was decreased in Nova1 cKO mice (Figures 4E and S4A). We also observed reduced expression of IMPACT in regions such as the globus pallidus and subthalamic nucleus, both of which coincided with areas presenting clear NOVA1 loss (Figure S4B).
To further validate the relationship between NOVA1 and IMPACT, we examined the expression of each using primary cultured diencephalon/mesencephalon neurons. In primary neurons, we observed an upregulation of IMPACT protein levels in the late phase of culture, mirroring the expression pattern of NOVA1. Interestingly, NOVA1 was also observed to switch to an alternative isoform lacking exon 4 (NOVA1-ex4; Figures S4C and 4F), a NOVA1 autoregulated exon that encodes a serine-threonine-rich phosphorylated domain of unknown function.16 At the cellular level, IMPACT was distributed in the nucleus and cell body of NOVA1-expressing cells (Figure S4D). IMPACT expression was also suppressed in primary neurons prepared from Nova1 KO mice (Figures 4F, 4G, and S4C). Taken together, these results indicate that loss of Nova1 leads to downregulation of IMPACT protein expression in vivo and in vitro.
Ectopic Impact expression restores the expression of ribosomal and electron transport genes in Nova1 knockout neurons
To examine the effect of Nova1 loss on Impact expression in neurons, we infected diencephalon/mesencephalon primary neurons from Nova1-floxed mice with a Cre-expressing lentivirus (to remove Nova1) and Cre-T2A-Impact-expressing lentivirus (to induce Impact expression in Nova1-deficient cells) (Figure 5A). Similar to our previous results, Cre-mediated removal of Nova1 significantly reduced the expression of Impact, while infection with the Cre-T2A-Impact-expressing lentivirus induced Impact expression in the same Nova1-null cells and led to a 7-fold induction of Impact levels (Figure 5B).
Figure 5. Ectopic Impact expression restores the expression of ribosomal and electron transport genes in Nova1 KO neurons.
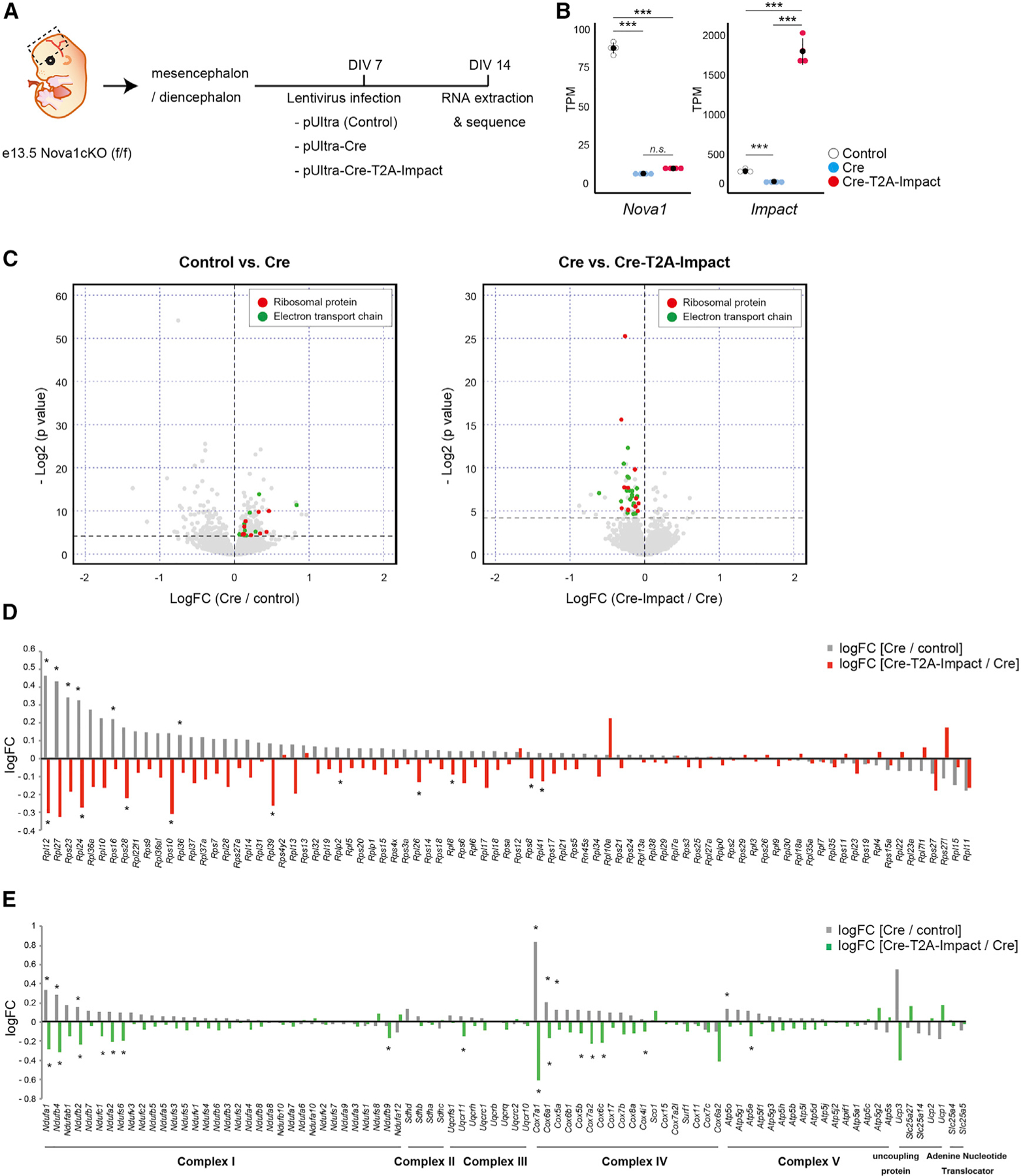
(A) Diagram of the experiment.
(B) Comparison of Nova1 and Impact mRNA expression in cells infected with each lentivirus (mean ± SEM, n = 4, Wilcoxon test, ***p < 0.005).
(C) Volcano plots comparing control versus Nova1-null cells (cells infected with a control or Cre-expressing lentivirus, left) and Nova1-null versus Nova1-null + Impact (cells infected with a Cre-expressing lentivirus or Cre-T2A-Impact-expressing lentivirus, right). Genes encoding ribosomal proteins are shown in red, and genes encoding electron transport chains are shown in green.
(D) Bar graph showing changes in expression of ribosomal protein genes. The gray bars show the log fold change (logFC) of Cre-lentivirus-infected cells relative to controls, and the red bars show the logFC of Cre-T2A-Impact lentivirus-infected cells relative to Cre-lentivirus-infected cells. Asterisks indicate significant changes (Wilcoxon test, *p < 0.05).
(E) Bar graph showing changes in expression of genes belonging to the electron transport chain. The gray bars show the logFC of Cre-lentivirus-infected cells relative to controls. The green bars show the logFC of Cre-T2A-Impact lentivirus-infected cells relative to Cre-lentivirus-infected cells. Asterisks indicate significant changes (Wilcoxon test, *p < 0.05).
We first examined the RNA changes in Nova1-null cells, in which Impact levels were reduced relative to control, and identified 288 genes that were altered (178 up-regulated and 110 down-regulated; Figure 5C, left; Table S2), and Gene Ontology (GO) analysis identified them as encoding pathways involved in a number of diverse functions (Figure S5A). Importantly, similar to the in vivo cKO data (Figures 3A, 3B, and S3B), transcripts involved in fatty acid metabolism pathways were significantly down-regulated, while those of ribosome and translation initiation factors were significantly upregulated (Figure S5C). These findings indicated that the transcriptomic features observed in culture recapitulate those observed in vivo. In addition to these changes, the electron transport pathway was significantly upregulated in Nova1-null cells in culture.
In Nova1-null cells where Impact was rescued, 68 gene expression changes (15 up-regulated and 53 down-regulated) compared with Nova1-null cells were observed (Figure 5C, right) and these genes were specifically highly enriched in translation-related and electron transport chain-related factors (Figures 5C–E and S5D; Table S3). This suggests that the NOVA1-IMPACT pathway regulates the expression of genes encoding ribosomal translation initiation factors and the electron transfer pathway.
NOVA1 regulates Impact expression through the 3′ UTR
We next explored the mechanism by which NOVA1 regulates Impact expression. NOVA1 CLIP analysis showed that NOVA1 has binding peaks in the 3′ UTR of the Impact transcript, and this binding was detected in the cortex at P0 and in the diencephalon/mesencephalon at P21 (Figure 6A). This suggested that NOVA1 may directly regulate the expression of Impact through 3′ UTR binding. We cloned the Impact 3′ UTR downstream of the luciferase gene and compared its expression in the presence and absence of NOVA1. First, knockdown of Nova1 by small interfering RNA (siRNA) in primary neurons from the diencephalon/mesencephalon of E13.5 wild-type mice significantly reduced the bioluminescence intensity of luciferase (Figure 6B). We then prepared primary neurons from Nova1-deficient embryos and overexpressed Nova1 using a lentivirus, which effectively rescued luciferase reporter activity (Figure 6C). Interestingly, expression of the NOVA1-ex4 isoform, which is particularly highly expressed in the diencephalon/mesencephalon, had a greater effect on increasing the reporter activity.
Figure 6. NOVA1 regulate Impact expression through the 3′ UTR.
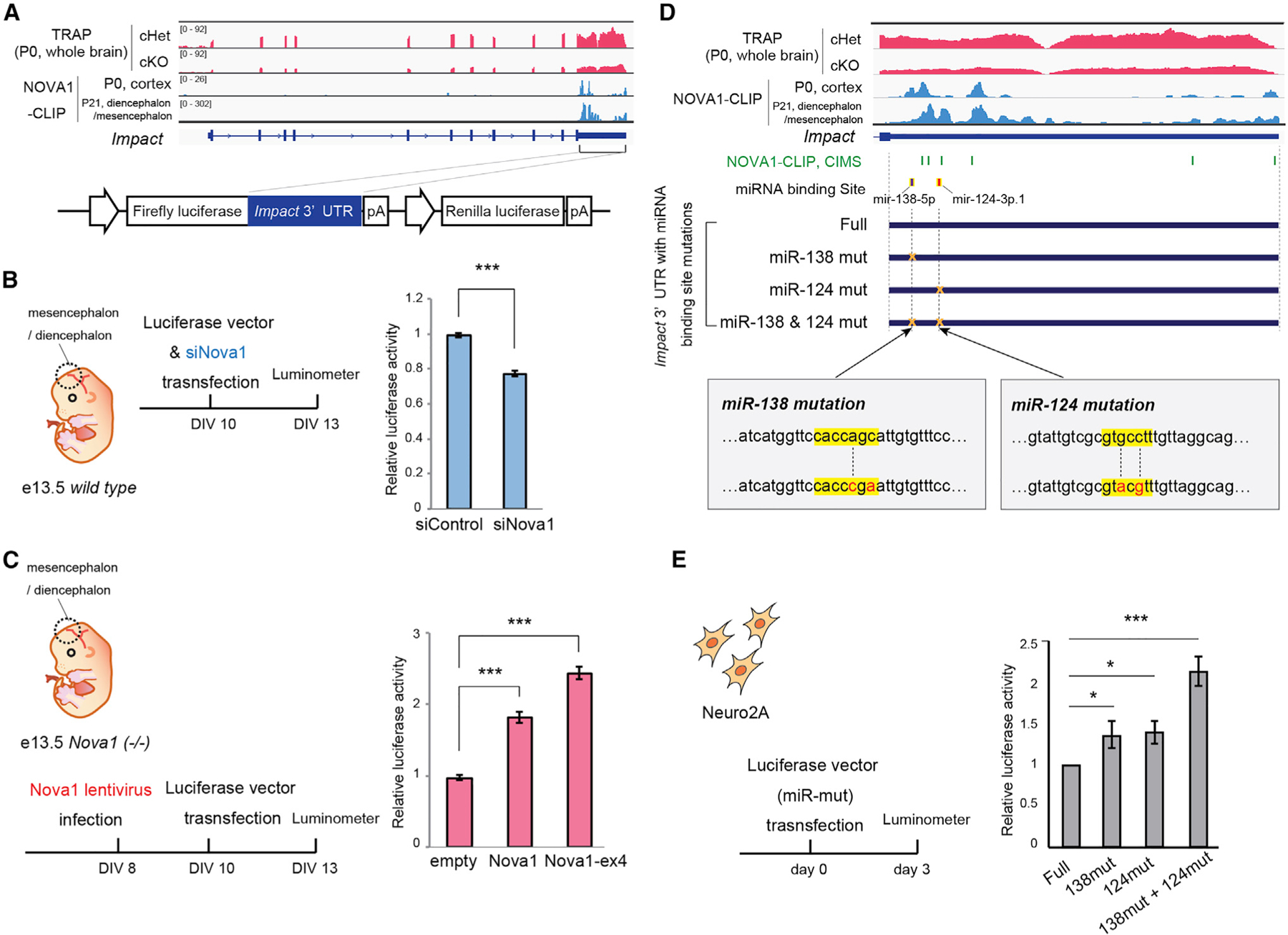
(A) IGV track of Impact locus/transcript. P0 TRAP data and NOVA1 CLIP data (with P0 cortex and P21 diencephalon/mesencephalon) are shown. The Impact 3′ UTR cloned into the luciferase vector (bottom) is indicated by the solid line.
(B) Luciferase assay examining the effect of Nova1 knockdown on expression of luciferase via the Impact-3′ UTR. A mixture of four siRNAs targeting Nova1 and Luciferase vector containing the Impact 3′ UTR was transfected into wild-type primary neurons from diencephalon/mesencephalon dissection. Luciferase activity was measured 3 days after transfection (mean ± SEM, representative data from more than three independent experiments, unpaired t test, ***p < 0.005).
(C) Luciferase assay examining the effect of Nova1 overexpression on mRNA expression via the Impact-3′UTR. A lentivirus expressing Nova1 (or the Nova1-ex4 isoform) was infected into primary neurons prepared from Nova1 KO mice, and then a luciferase vector containing the Impact-3′ UTR was transfected. After 3 days of transfection, Luciferase activity was measured (mean ± SEM, representative data from more than three independent experiments, unpaired t test, ***p < 0.005).
(D) IGV track of the Impact-3′ UTR. The positions of the miRNA binding sites (miR-138 and miR124) annotated in the TARGETSCAN and the constructs with mutations in the binding sites are shown below. The mutant sequences of each binding site are shown on the bottom; the seed sequence of miRNA is shown in yellow, and the mutated nucleotides are shown in red. Crosslinking-induced mutation sites (CIMSs) from NOVA1 CLIP analysis are shown in green.
(E) Luciferase assay using the Impact-3′ UTR with mutations in the miRNA binding sites (miR-mut). Each Luciferase construct was transfected into Neuro2a cells, and the luciferase activity was measured 3 days after transfection. Each value represents the ratio of firefly luciferase over Renilla luciferase activity. Values of the samples were normalized to that of the full-length Impact 3′ UTR-transfected sample (mean ± SEM, representative data from more than three independent experiments, unpaired t test, *p < 0.05, ***p < 0.005).
We confirmed that this same regulation could be reproduced in the Neuro-2a cell line (Figure S6A), suggesting a common regulatory mechanism within different neuronal cell types. Together, these results suggest that the presence of NOVA1, particularly the isoform missing the NOVA1 phosphorylation domain, regulates RNA expression through binding the Impact 3′ UTR, and this effect can be assessed in a synthetic reporter assay.
To further interrogate the regulatory elements within the Impact-3′ UTR, we divided the binding region of NOVA1 on the Impact 3′ UTR into three regions according to their NOVA1 CLIP binding intensity (peaks 1–3) and examined the effect of the loss of each region on expression of the luciferase reporter (Figure S6B). Comparison of each deletion construct revealed that the effect of NOVA1 to enhance luciferase expression was abolished in constructs lacking the most upstream NOVA1 binding region (peak 1) within the Impact-3′ UTR. This loss of regulation was observed to a lesser extent when peak 2 was deleted and abolished when peak 3 was deleted, highlighting the importance of NOVA1 binding, particularly to the peak 1 region. We further divided the region of peak 1 into two regions (p1a and p1b). Although deletion of the entire peak 1 region abolished NOVA1 regulation, deletion of p1a or p1b alone did not abolish this regulation (Figure S6C).
The NOVA1-binding peak 1 region contains two highly conserved neuronally expressed microRNA (miRNA) target sites (miR-138 and miR-124), leading us to test whether NOVA1 might compete with these miRNAs in the Impact 3′ UTR. Indeed, cross-link-induced mutagenesis site (CIMS) analysis54 on NOVA1 CLIP data in the diencephalon/mesencephalon of P21 mice revealed three CIMSs in peak 1 surrounding the miRNA binding sites (Figure 6D). This strongly suggested that the effect of NOVA1 was mediated through competition against miRNA binding. To experimentally validate this model, we inserted mutations into these miRNA seed-binding sites and analyzed their effects on regulation (Figure 6D). The results showed that mutations in miR-138 and miR-124 target sites resulted in increased expression of upstream transcripts in the absence of NOVA1 (Figure 6E). This effect was more pronounced when mutations were introduced in both sites. Moreover, mutation of the two miRNA binding sites eliminated the effect of ectopic NOVA1 on enhancing the luciferase activity (Figure S6D). These results suggest that NOVA1 regulates the expression of Impact through the 3′ UTR by competing with miRNAs such as miR-138 and miR-124.
DISCUSSION
We describe a human with NOVA1 haploinsufficiency and a series of neuro-behavioral deficits consistent with the proposed role for NOVA1 in inhibitory motor and cognitive functions seen in paraneoplastic opsoclonus-myoclonus syndrome in which NOVA1 is targeted. In the present work, we examined the function of NOVA1 in adult inhibitory neurons in mice. We found that Nova1 in Gad2+ neurons controls behaviors (i.e., motor abnormalities, working memory, anxiety) consistent with the phenotype of human NOVA1 haploinsufficiency. We also discovered a role of NOVA1 relating to hypothalamic functions, including thermogenesis, growth, and feeding behavior, and the possibility that NOVA1 may play a similar role in regulating human hypothalamic functions. Molecularly, we identified a widespread action of NOVA1 to suppress expression of the components of the translational machinery. This led us to identify IMPACT, a translational regulatory protein, as a direct target of NOVA1. We uncovered the mechanism by which NOVA1 regulates Impact expression: through antagonism of 3′ UTR miRNA binding and subsequent RNA stabilization. This work provides a clear instance of close integration of RNA regulation, from nuclear splicing factors to translational control in neurons.
NOVA1-IMPACT pathway
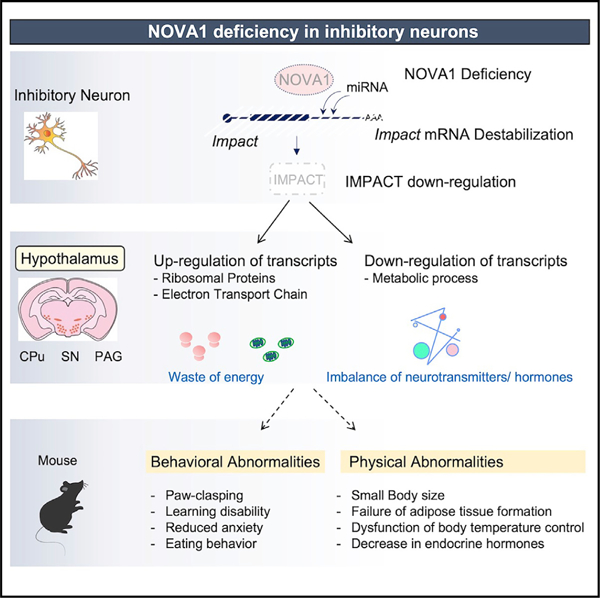
Based on our findings, we propose the following model for some of NOVA1’s normal physiologic actions in the brain (Figure 7). In the steady state, NOVA1 binds to the 3′ UTR of Impact, antagonizing miRNA-induced transcript destabilization and supporting IMPACT stability and protein expression. NOVA1-IMPACT-expressing neurons are distributed throughout the hypothalamus. Within these neurons, IMPACT suppresses expression of ribosomal proteins and the electron transport system, leading to reduced wasteful energy consumption in the neuron. The NOVA1-IMPACT pathway also has a role in supporting intracellular metabolic pathways such as cholesterol and lipids.
Figure 7. NOVA1-IMPACT pathway in mice.
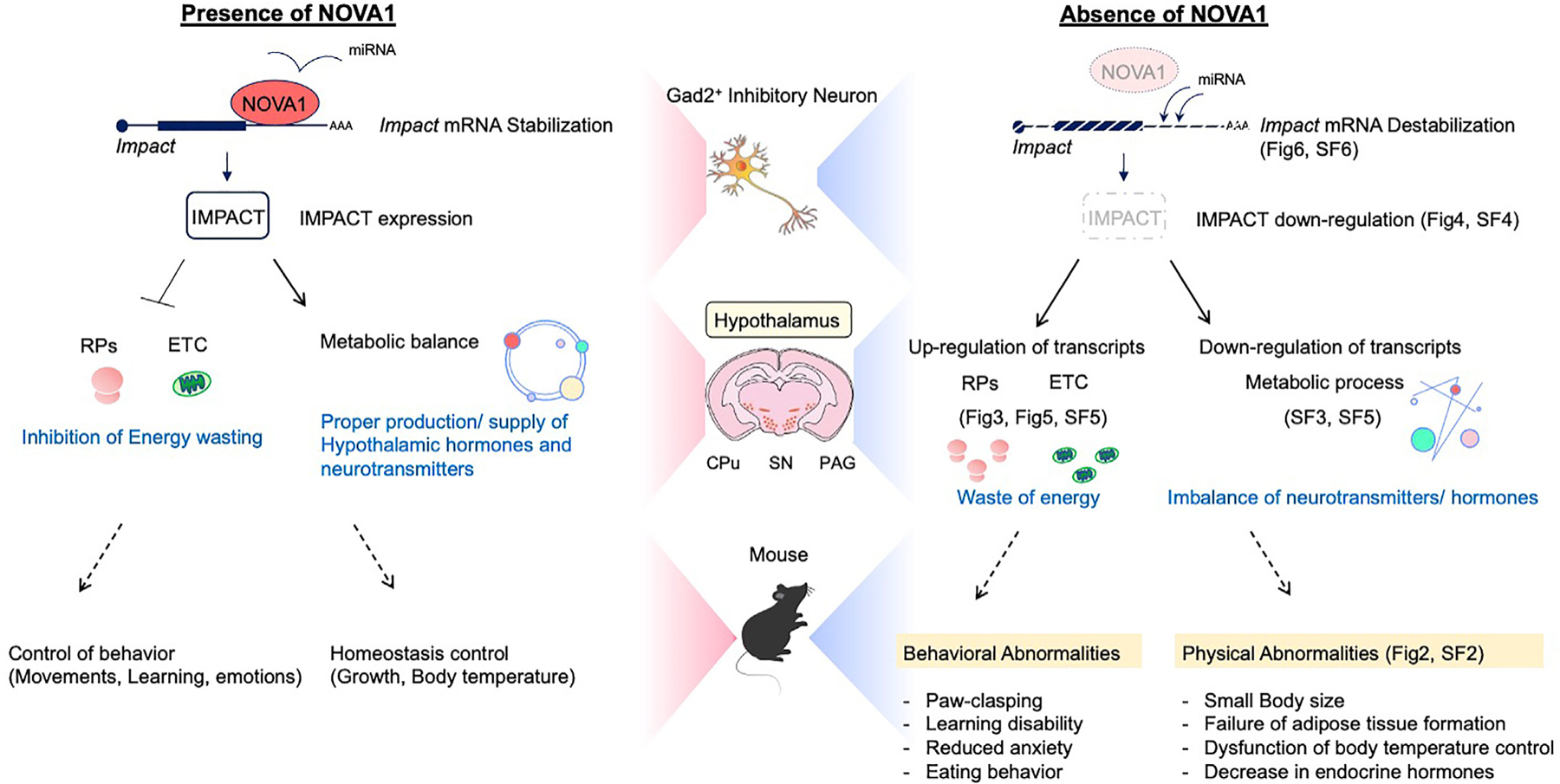
Proposed models of NOVA1 function in Gad2+ inhibitory neurons in this study (see Discussion). RP, ribosomal protein; ETC, electron transport chain. Relevant figures or supplemental figures (SFs) are indicated.
Disruption of the NOVA1-IMPACT pathway (Figure 7) causes intracellular energy wasting and abnormal metabolic circuitry, resulting in an imbalance in the production and supply of neurotransmitters and hormones. NOVA1 cKO mice show physical and behavioral phenotypes in which the hypothalamus plays a central role, such as growth failure, thermoregulatory dysfunction, and abnormal feeding behavior.
Phenotypes of cKO mice
While conventional Nova1-null mice are neonatal lethal, 75% of the cKO mice survive to adulthood (25% die for unknown reasons before and after weaning). This suggests that the lethality observed in conventional Nova1-null mice involves NOVA1 action in other cell types (i.e., excitatory neurons that do not express Gad2). In contrast, the behavioral and physical abnormalities in cKO mice persisted throughout life, suggesting ongoing importance of NOVA1 in Gad2 neurons in the adult. Importantly, some of these phenotypes resembled the symptoms of patients with NOVA1 haploinsufficiency, underscoring the utility of linking studies of cKO mice to human neurology. The cKO mice showed severe growth retardation despite an increase in daily food intake, consistent with elevated levels of peptide neurotransmitters such as Npy and Agrp in the hypothalamus, which are known to promote food intake. In the hypothalamus of cKO mice, increased expression of somatostatin (Sst) was also detected. Somatostatin is a hormone that regulates the endocrine system and suppresses the secretion of many secondary hormones, such as growth hormone and gonadotropin.47,48 In fact, expression of these hormones was greatly reduced in cKO mice. We suspect that such a hormonal imbalance in cKO mice may be responsible for the seemingly conflicting phenotypes of the mice.
Translation and working memory
Numerous reports have linked translational dysregulation to human neurological disorders such as Alzheimer’s disease and autism, underscoring its clinical importance.55,56 In learning experiments using the Y maze, cKO mice showed reduced working memory compared with control mice. In recent years, other groups have reported that enhanced translation in inhibitory neurons expressing Gad2 or Sst leads to enhanced long-term memory in mice.57,58 One possible explanation for this discrepancy is that the region analyzed in detail in this study was the diencephalon/mesencephalon, where NOVA1 expression is particularly high, as our immunostaining results demonstrate. NOVA1 expression is also found in scattered neurons and regions in the cerebral cortex, including the retrosplenial region and hippocampus (Figure 1D), important limbic regions for learning and emotional control. Thus, neurons (non-Gad2-Cre expressing but otherwise undefined) may contribute to the Nova-dependent learning phenotype. Given that the expression of IMPACT, a direct NOVA1 target identified in this study, is particularly enriched in the hypothalamus, it is also possible that other key NOVA1 targets are present in a region-specific (cortex, limbic, or hippocampus) manner.
IMPACT, a direct target of NOVA1
IMPACT, a direct NOVA1 target mRNA identified in this study, has been reported to be a repressor of the eIF2α kinase GCN2.51 eIF2α phosphorylation represses general translation and upregulates some specific gene expression in cells. Our result suggests the hypothesis that NOVA1 may regulate translational control by eIF2α phosphorylation of GCN2 through upregulation of IMPACT expression. However, a series of ribosomal proteins whose expression was upregulated by loss of Nova1 was repressed by induction of Impact (Figures 5C–5E). This result is seemingly contradictory to the observation that IMPACT is a repressor of GCN2 (which would suggest that elevated IMPACT would be permissive for general translational activation). Puromycin incorporation into newly translated proteins was increased in Nova1-null cells (Figure 3D). Moreover, when we analyzed the relevance of this IMPACT-GCN2 pathway by crossing cKO mice with Gcn2 KO mice, no apparent improvement of phenotypes such as growth retardation or paw clasping was observed. This is consistent with the description of Impact-KO mice, which also showed that their lean phenotype was not rescued by crossing with Gcn2 KO mice.53 There are several reports that suggest the existence of GCN2-independent pathways downstream of IMPACT, particularly in neurons.59,60 IMPACT has an intramolecular RWD domain (domain in RING finger and WD repeat containing proteins and DEXDc-like helicases subfamily related to the UBCc domain) and an “ancient” domain.61 The RWD domain has been reported to be important for inhibition of GCN2 activity but has also been suggested to be important for the interaction with actin and the cell cycle regulator CDC28, suggesting that IMPACT may have GCN2-independent functions. Moreover, the function of the IMPACT “ancient” domain is unknown. These reports, together with our data, suggest that there are likely other pathways downstream of IMPACT that do not involve GCN2 (and these may have opposite effects on translational regulation).
Association between NOVA1 and miRNAs
In this study, we suggest competition between NOVA1 and miR-NAs as a mechanism by which Impact expression is regulated. Interestingly, specific deletion of Dicer in proopiomelanocortin (POMC) neurons, a population of cells involved in regulation of feeding in the hypothalamus, results in the opposite phenotype of Nova1-cKO mice, such as decreased expression of several ribosomal proteins as well as increased body weight.62,63 This may indicate that competition between NOVA1 and miRNAs is part of an important molecular mechanism in homeostatic networks, including control of feeding behavior and regulation of energy metabolism in the body. Identification of upstream factors that control the expression and function of NOVA1 will be important for future studies. It has been reported that NOVA1 directly interacts with Argonaute (Ago), a major component of the miRNA-induced silencing complex (miRISC), to regulate miRNA function.64 However, it is not clear whether the regulation of Impact expression by NOVA1 is mediated by Ago. Interestingly, this study also reported that knockdown of Nova1 in primary neuronal cultures attenuates the repressive effect of mir138 on mRNA.64 This conflicts with the finding that KO of Nova1 in our diencephalon/mesencephalon primary neurons downregulates expression of a reporter linked to the Impact 3′ UTR. One possible explanation for this is that we studied the diencephalon/mesencephalon, where NOVA1 expression is high, versus studies in a culture system from the cerebral cortex. Factors that are differentially expressed in these cells may affect NOVA1 function on its targets. Moreover, our results may be specific to Impact regulation, which is particularly enriched in the hypothalamus.
Targets other than Impact
While this study focused on Impact as a target of NOVA1 in inhibitory neurons, we also found that Ahi1 and Dzip1 are repressed by loss of Nova1. In particular, Ahi1 has been reported as a promising candidate gene for neuropsychiatric disorders such as Joubet syndrome, schizophrenia, and autism. A detailed examination of the molecular mechanisms of Ahi1 expression control by Nova1 should be beneficial from the viewpoint of therapeutic targets.
Overall, we uncovered the biological importance of Nova1 function in adult mice and identified direct target transcripts with regulatory mechanisms beyond splicing control. The function and downstream pathway of NOVA1 is a clinically significant research topic in view of its association with human psychiatric disorders and various cancers. The biological relevance of the newly identified targets as well as the cellular responses by Nova1, such as enhanced translation-related factors, downregulation of cholesterol pathways, or, more generally, a physiologic role in hypothalamic function, in NOVA-related disorders will be an important topic to be explored.
Limitations of the study
One of the striking findings in the work is the discovery that aspects of the NOVA1 haploinsufficient human phenotype, most clearly those relating to failure of motor inhibition and cognitive challenges, are phenocopied in a mouse in which Nova1 is specifically lost in inhibitory neurons. This correlation is tempered by confounding factors, including sample size (one human clinical case versus statistically significant findings across multiple mice), age (the human was medically followed for a decade, while the mouse was examined at 6 weeks), and variables in loss-of-function parameters (Nova1 cKO mice have homozygous loss but only in when and where Gad-2 is expressed). We recognize that such discrepancies may account for differences in phenotypes seen in the two instances. In addition, while we demonstrate a role for NOVA1 in the hypothalamus and metabolism, it will require future work to demonstrate a direct role for NOVA1 in these aspects of biology; for example, by genetic rescue of phenotype in Nova1 Gad2-Cre-null mice with cell-specific rescue of NOVA1 in the hypothalamus (and ultimately in a relevant subset of hypothalamic neurons). Finally, one specific predicted phenotype, rescue of growth retardation in Gcn2 KO mice by Nova1 KO, yielded a negative result (see above “IMPACT, a direct target of NOVA1”), which was consistent with similar findings observed after crossing Impact KO mice with Gcn2 mice,53 suggesting yet-to-be discovered, GCN2-independent actions of NOVA1 and IMPACT on translational control pathways.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Robert B. Darnell (darnelr@rockefeller.edu).
Materials availability
Nova1-flox mouse line generated in this study have not been deposited. The line is available from the lead contact with a completed Material Transfer Agreement.
Data and code availability
RNA-seq data (Gene expression omnibus (GEO): GSE212473) and NOVA1-CLIP data (GEO: GSE212547) reported in this paper have been deposited at GEO and are publicly available as of the data of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Subject
The patient was a 9-year-old boy. The patient’s information can be found in the first section of the Result. The patient was seen on a protocol approved by the Rockefeller University IRB (RDA-747) after written consent was obtained from the parents and assent was obtained from the patient.
Animal experiments
All procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) at Rockefeller University. C57BL/6J (stock no. 000664), ACTB-FLPe (stock no. 005703), tdTomato reporter (stock no. 007914), GAD2-Cre (stock no. 019022) mice were obtained from the Jackson Lab. Nova1-KO mice were generated in Robert Darnell lab32 and back-crossed to C57BL/6J strain at least 10 times. Mice were housed in a 12-h light/dark cycle, up to 5 mice per cage. Male or female mice aged 8–12 weeks were used for animal experiments unless otherwise stated. Littermates of the same-sex were randomly assigned to experimental groups.
Generation of Nova1-cKOGad2 mice line
Nova1-cKO targeting vectors were generated by standard restriction cloning; Nova1-cKO targeting vector contains FRT-NEO-FRT cassette and Nova1 exons 1 and 2. Constructs were electroporated into Bruce 4 ES cells. Correctly targeted ES cells were injected into C57BL/6J blastocysts and screened for chimeras. Chimera males were crossed with C57BL/6J females to generate heterozygous Nova1-cKO. The FRT-NEO-FRT cassette was removed by crossing heterozygous Nova1-cKO to ACTB-FLPe mice. Nova1-cKO mice were crossed with GAD2-IRES-Cre knock-in mice (The Jackson Laboratory #019022) to generate GAD2-positive cell lineage-specific Nova1 knockout mice (Nova1-cKOGad2). Within the viable homozygous cKO mice, 5–10% animals showed malocclusion phenotype. Although these mice with malocclusion grew similar to other cKO mice, we excluded them from behavior experiments.
METHOD DETAILS
Behavior test
Male mice aged 8–12 weeks were used for the experiments and were moved to the behavior room 1 h prior to the start of the experiment to acclimate to the environment. After each trial, all apparatus were cleaned with super hypochlorous water to prevent a bias based on olfactory cues.
Y-maze test
The Y-maze tests were conducted according to the described procedure.38 The test is performed in a Y-maze with three arms of equal length at 120° angles to each other. The mouse is placed in the center of the maze and has free access to all three arms. If the animal chooses an arm different from the arm it arrived in, this choice is called an alteration. This is considered a correct response; conversely, returning to the previous arm is considered an error. The number of times and the order in which the animals entered the arms are recorded and used to calculate the alternation rate. The behavior of the mice was recorded for 8 min.
Open field test
Open field tests were conducted according to the procedure described.39 The test apparatus consisted of an enclosed plastic square box (50 × 50 × 40 cm). 10 cm × 10 cm grid lines were drawn on the floor of the box, with the squares on the wall side as the outer zone and the inner squares as the inner zone. Mice were placed in the central part of the box at the start of the experiment and their behavior was recorded for 5 min. The number of times the mice moved across the grid lines and the number of times they entered and the duration of stay in the inner zone were measured.
Light/dark transition test
Light/dark transition tests were conducted according to the procedure described.40 The apparatus consisted of a cage (21 × 42 × 25 cm) divided into two chambers of equal size by a partition with door. One chamber is an enclosed ‘dark’ half and the other an open ‘light’ half. Mice are initially placed in a “dark” room and are free to move between the two rooms throughout the experiment. Time spent in either half is measured over a 5-min period.
Food intake measurement
Mice were fed pre-weighed solid feed and water under normal rearing conditions; feed was collected after 24 h and the amount of reduction was calculated. The amount of feed reduction was measured for each mouse for three consecutive days, and the average value was defined as the amount of feed consumed per day.
Thermogenesis assay in newborn mice
For thermogenesis assays of Nova1-cKOGad2 mice, body temperatures of newborn pups (P5) were measured with a thermography camera (FLIR system). An average of three thermal images per litter were taken at 1min and 10min after pup isolation from the mother. Images were taken by placing the newborn pups into six-well plates at the time of recording.
Primary neuron culture
For the primary neuron culture of diencephalon/mesencephalon, we followed the method of Gaven et al. in 2014.66 Briefly, ventral diencephalon/mesencephalon were isolated from e13.5 embryos and plated on laminin-coated plates in culture medium (1xDMEM; 5 mM HEPES; 2 mM L-Glutamine; 0.6% glucose) with each hormone (apo-transferrin, insulin, putrescine dihydrochloride, sodium selenite, progesterone) for 15 days. At each culture day, cells were collected and used for Western blotting, immunostaining, or luciferase assay.
Lentivirus production
6 μg amounts of lentiviral plasmids (pUltra-eGFP, pUltra-eGFP-P2A-Cre, pUltra-eGFP-P2A-Cre-T2A-Impact) were transfected together with 3 μg of GAG-Pol and VSVG-Rev plasmids into 293T cells in 10 cm plates using polyethyleneimine (MW25000, Polyscience Inc. #23966). 10 μM forskolin (SIGMA #F3917) was added to the culture 16 h after transfection, and supernatant was collected at 48 h after transfection. Pooled viral supernatant was centrifuged and concentrated.
SUnSet
Immunohistochemical SUnSET were conducted according to the procedure described.49 Briefly, primary neurons of Nova1cKOf/f mouse diencephalon/mesencephalon were infected with control (empty) or Cre-expressing lentivirus on day 7 of culture. On day 14 of culture, samples were incubated for 30 min in culture medium containing 1 μg/mL puromycin, fixed in 2% PFA for 10 min, then washed in 0.1M glycine. Samples were incubated with anti-mouse IgG Fab for 1 h to remove background signal. Then reacted with anti-puromycin (Clone 12D10) for 2 h and incubated with Alexa 647-labeled anti-mouse IgG2a for 1 h. After washing, samples were mounted and observed under a Keyence microscope.
Luciferase assay
For the luciferase assay, the Dual-Luciferase Reporter Assay system (E1910, Promega) was used and analyzed according to the manufacturer’s instructions. The vector used was pmirGLO Dual-Luciferase (E1330, Promega), with each 3′UTR region cloned downstream of luc2. Vectors were transfected into cells using Lipofectamine 3000 Transfection Reagent (L3000001, Thermo Fisher Scientific), and cells were collected after 3 days and reporter activity was measured by luminometer.
Antibodies
Primary antibodies used for immunohistochemistry and western blotting were as follows; rabbit anti-NOVA1 [EPR13847] (ab183024, abcam), human anti-pan NOVA (anti-Nova paraneoplastic human serum), mouse anti-Impact (ab72444, abcam), rabbit Anti-Impact (NBP1–42678, Novus Biologicals), rabbit anti-Impact (PA5–59805, Thermo Fisher Scientific), rabbit anti-beta Actin (ab8227, abcam), mouse anti-Puromycin (MABE343, Millipore Sigma).
Immunohistochemistry
Twelve-week-old mice were perfused with PBS and 4% paraformaldehyde and further fixed overnight by 4% PFA at 4°C. They were sequentially replaced with 15% sucrose/PBS and 30% sucrose/PBS, embedded with OCT compound, and stored at −80°C until use. Frozen brains were sliced into 30 μm thick sections in a cryostat (CM3050S, LEICA). Slices were washed three times with PBS at room temperature (RT), incubated in 0.2% Triton X-100/PBS for 15 min at RT, blocked in 1.5% normal donkey serum (NDS)/PBS for 1 h at RT, incubated overnight at 4°C with primary antibody in 1.5% NDS/PBS, then incubated in Alexa Incubated with 488, 555 or 647 conjugated donkey secondary antibody (1:1000). Images of specimens were collected with a BZ-X700 (KEYENCE) microscope.
Protein extraction and immunoblotting
As samples, we used each dissected region of P21 mouse brain, the excised pituitary gland, and primary neurons from each day of culture (one well of a 12-well plate). Each sample was lysed in RIPA buffer (50mM Tris-HCl; 150mM NaCl; 0.1% SDS; 0.5% sodium deoxycholate; 1% NP-40), and the lysates were separated in NuPAGE™ 4–12% Bis-Tris Protein Gel (Invitrogen; NP0323BOX) and blotted onto a nitrocellulose blotting membrane (GE Healthcare Life science; 10600002). Antibodies used were as described above. Western blots were quantified by normalizing each lane with an Actb signal to control for differences in loading.
RNA-seq and TRAP-seq library preparation and analysis
For the TRAP method, we followed the procedure reported in.67 For the mice used in the analysis, Nova1-cKOf/f mice were crossed with RPL22-HA knock-in and GAD2-IRES-Cre mice to create RiboTag (Rpl22-HA) GAD2-Cre Nova1cKO mice. P0 RiboTag whole brain from Nova1-cKOGad2 (cKO) and control (cHet) littermates were rapidly dissected and treated with ice-cold polysome buffer (20 mM HEPES pH7.4, 150 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 0.1 mg/mL cycloheximide). After removal of insoluble material by centrifugation at 20,000 xg for 10 min at 4°C, the cells were precleared by incubation with Protein A Dynabeads (Invitrogen) for 45 min at 4°C, and then incubated with anti-HA (ab9110, abcam) for 2 h at 4°C with rotation. The antibody-polysome complexes were immunoprecipitated by adding Protein A Dynabeads and rotating at 4°C for 1 h. Protein A Dynabeads were washed with polysome buffer containing 1% NP-40 and high salt buffer containing 50 mM Tris pH 7.5, 500 mM KCl, 12 mM MgCl2, 1% NP-40, 1 mM DTT and 0.1 mg/mL cycloheximide. Polysomes were eluted from the beads by incubating with Trizol (Invitrogen) for 5 min at room temperature with occasional vortexing. RNA was extracted from the Trizol reagent according to the manufacturer’s instructions.
For RNA-seq, samples included dissected hypothalamus, substantia nigra, periaqueductal gray, caudate putamen, and pituitary gland from 3-week-old Nova1cKOGad2 (cKO) and control (cHet) mice, or primary neurons from e13.5 diencephalon/mesencephalon on day 14 in culture. The mRNA-seq library was prepared from RNA extracted with Trizol following the Illumina TruSeq protocol of polyA selection, fragmentation, and adapter ligation. Multiplex libraries were sequenced as 125 nt paired-end runs on the HiSeq-2500 platform at Rockefeller University Genomic Core. These raw datasets and processed data files have been deposited with Gene Expression Omnibus. Reads were aligned to mm10 builds of the mouse genome using OLego,68 and AS and gene expression were analyzed using the Quantas pipeline.69 mRNA abundance changes were assessed by differential analysis of raw sequence counts in edgeR65 using the TMM technique.
HITS-CLIP
NOVA1 HITS-CLIP was performed in the wild-type diencephalon/mesencephalon of P21 using three biological replicates. Tissues were dissected in PBS, triturated using 20G needle and crosslinked three times on ice for 400 mJ/cm2 using Stratalinker. Crosslinked material was collected by centrifugation, resuspended in wash buffer (1X PBS, 0.5% NP-40, 0.5% deoxycholate and 0.1% SDS with protease inhibitor), and subjected to DNase (RQ1 DNase: Promega) and RNase (RNase A: Affymetrix) treatment at a final dilution of 1:20,000 for 5 min. The lysate was clarified by centrifugation at 20,000 x g for 20 min. The supernatant was used for immunoprecipitation with 200 μL of Protein A Dynabeads (Invitrogen) loaded with 18 μg anti-Nova1 antibody (abcam) for 2 h at 4°C. The samples were washed as follows: twice with wash buffer, twice with Nelson stringent wash buffer (15mM Tris pH 7.4, 5mM EDTA, 2.5 mM EGTA, 1% Triton X-100, 1% Sodium deoxycholate, 0.1% SDS, 120 mM NaCl, 25mM KCl), twice with Nelson high salt buffer (15mM Tris pH 7.4, 5mM EDTA, 2.5 mM EGTA, 1% Triton X-100, 1% Sodium deoxycholate, 0.1% SDS, 1M NaCl), twice with Nelson low salt buffer (15mM Tris pH 7.4, 5mM EDTA), and twice with PNK wash buffer (50mM Tris pH 7.4, 10 mM MgCl2, 0.5% NP-40). RNA fragments were dephosphorylated using FastAP Alkaline phosphatase (Thermo Fisher Scientific) and subjected to 3′ ligation overnight at 16°C with a pre-adenylated linker (preA-L32) using truncated KQ T4 RNA Ligase2 (NEB). The RNA-protein complexes were labeled with 32P-γ-ATP using T4 PNK (NEB), and subjected to SDS-PAGE and transfer to nitrocellulose membrane. Appropriate regions of the membrane were cut out and RNA was extracted according to the following conditions: 100mM Tris PH7.5, 50mM NaCl, 10mM EDTA, 7M Urea with proteinase K. RNA was purified by phenol-chloroform extraction method. Cloning was performed using the BrdU-CLIP protocol. Briefly, the reverse transcription reaction was performed using Superscript III (Thermo Fisher Scientific), and the cDNA was BrdU-labeled by including BrdU in the reaction solution. Immunoprecipitation was performed with 5 μg anti-BrdU antibody (abcam) and 25 μg protein G Dynabeads per reaction (45 min at room temperature), followed by washing with the following solutions (including Denhardt’s solution): once with IP buffer (0.3x SSPE, 1mM EDTA, 0.05% Tween 20), twice with Nelson low salt buffer, twice with Nelson stringent wash buffer, twice with IP buffer. After eluting the cDNA, BrdU-immunoprecipitation was performed again under the same conditions. cDNA was circularized on beads using CircLigase II (Epicentre) and PCR was performed using Accuprime Pfx supermix (Thermo Fisher Scientific) and Syber Green until RFU 250–500. PCR products were purified using Agencourt AMPure XP (Beckman Coulter) and concentrations were measured by TapeStation. High-throughput sequencing was performed at the Rockefeller University Genome Resource Center. Sequence tags were aligned to the mouse genome (mm10) by novoalign. Unique tags were collected by eliminating PCR duplicates.
Analysis of CLIP data
Analysis of CLIP data was performed as in previous reports.70 To reduce misalignment due to sequencing errors, reads were first filtered based on quality score (≥20 in the degenerate linker region; average of ≥20 in the remaining read). Exact sequences were collapsed to remove PCR duplicates and demultiplexed. The degenerate barcode was removed and the 3′ linker was trimmed. CLIP reads were mapped by novoalign (www.novocraft.com) on the mouse genome mm10 build. Only unique tags were used for subsequent analysis. All scripts and detailed information used in the analysis, including peak-finding algorithms, are available at http://zhanglab.c2b2.columbia.edu/index.php/Resources.
QUANTIFICATION AND STATISTICAL ANALYSIS
Information of statistical details and methods are indicated in the figure legends. Data are represented as mean ± standard error of the mean (s.e.m.) and analyzed using two-tailed unpaired Student’s t-test or Mann-Whitney test. p values are shown in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Rabbit anti-Nova1 | Abcam | Cat# ab183024; RRID:AB_2632587 |
| Mouse anti-Impact | Abcam | Cat# ab72444; RRID:AB_1269162 |
| Rabbit anti-Impact | Thermo Fisher Scientific | Cat# PA5–59805; RRID:AB_2642711 |
| Mouse anti-hCG alpha | Nnovus biochlogicals | Cat# NBP2–52442 |
| Goat anti-Doublecortin | Santa cruz | Cat# sc-271390; RRID:AB_10610966 |
| Chicken anti-Map2 | Novus biological | Cat# NB300–213; RRID:AB_2138178 |
| Human anti-pan NOVA | Yang et al.4 | N/A |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| pUltra | Addgene | Plasmid #24129 |
| pUltra-Cre | This study | N/A |
| pUltra-mNova1 | This study | N/A |
| pUltra-mNova1-ex4 | This study | N/A |
| pUltra-Cre-Impact | This study | N/A |
|
| ||
| Critical commercial assays | ||
|
| ||
| Dual-Luciferase Reporter Assay System | Promega | E1910 |
| Dynabeads mRNA Purification Kit | Ambion | Cat# 61006 |
| Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) | Illumina | Cat# MRZH116 |
| TruSeq RNA Sample Preparation Kit v2 | Illumina | Cat# RSS-122–2001 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-seq | This paper | GEO: GSE212473 |
| NOVA1-CLIP | This paper | GEO: GSE212547 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Neuro-2A | ATCC | CCL-131 |
|
| ||
| Experimental models: Organisms/Strains | ||
|
| ||
| Mouse: cKO: GAD2cre-Nova1f/f | This paper | N/A |
| Mouse: cHet: GAD2cre-Nova1f/w | This paper | N/A |
| Mouse: conditional Nova1 KO: Nova1f/f | This paper | N/A |
| Mouse: GAD2-IRES-Cre: B6N.Cg-Gad2tm2(cre)Zjh/J | The Jackson Labpratory | Strain# 019022 |
| Mouse: Nova1 KO: Nova1 knock out | Robert B. Darnell Lab. | Neuron. 2000 Feb; 25(2):359–71. https://doi.org/10.1016/s0896-6273(0080900-9)). |
|
| ||
| Oligonucleotides | ||
|
| ||
| siGENOME Mouse Nova1 siRNA SMARTpool | horizon | M-161826-00–0005 |
| siGENOME Non-Targeting siRNA Pool #1 | horizon | D-001206-13-05 |
| qPCR, mNova1, Fwd: 5′-catctgaccccatgaccac-3′ | This paper | N/A |
| qPCR, mNova1, Rvs: 5′-tgctgtgctgttgggaacta-3′ | This paper | N/A |
| qPCR, mImpact, Fwd: 5′-ccttcttgcaggactgtgaa-3′ | This paper | N/A |
| qPCR, mImpact, Rvs: 5′-caccaccaccatgacattct-3′ | This paper | N/A |
| qPCR, mGapdh, Fwd: 5′-aagagggatgctgcccttac-3′ | This paper | N/A |
| qPCR, mGapdh, Rvs: 5′-ccattttgtctacgggacga-3′ | This paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pcDNA3.1(+) | Thermo Fisher | Cat# V79020 |
| pcDNA3.1(+)-mNova1 | This paper | N/A |
| pcDNA3.1(+)-mNova1-ex4 | This study | N/A |
| pmirGLO Dual-Luciferase miRNA Target Expression Vector | Promega | Cat# E1330 |
| pmirGLO-Impact 3′UTR | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak1 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak2 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak3 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak1&2 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak1&3 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak2&3 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆peak1&2&3 | This study | N/A |
| pmirGLO-Impact 3′UTR_∆p1a | This study | N/A |
| pmirGLO-Impact 3′UTR_miRNAmut | This study | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| CLIP Tool Kit (CTK) | Zhang lab | https://zhanglab.c2b2.columbia.edu/index.php/CTK_Documentation |
| OLego | Zhang lab | https://zhanglab.c2b2.columbia.edu/index.php/OLego_Documentation |
| Quantas | Zhang lab | https://zhanglab.c2b2.columbia.edu/index.php/Quantas_Documentation |
| edgeR | Robinson et al.65 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| IGV (2.13.0) | Thorvaldsdottir et al.50 | https://software.broadinstitute.org/software/igv/2.13.x |
Highlights.
NOVA1 haploinsufficiency impairs motor inhibition behavior and learning in humans
Nova1 deletion in Gad2+ neurons causes abnormalities referable to the hypothalamus
NOVA1 regulates translation- and electron transport chain-related gene expression
NOVA1 stabilizes Impact mRNA via 3′ UTR binding and miRNA competition
ACKNOWLEDGMENTS
The authors wish to thank members of the Darnell lab for thoughtful discussions and review of the manuscript. This project was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Rockefeller University, grant UL1 TR001866. This project was also supported by a Japan Society for the Promotion of Science postdoctoral fellowship for research abroad (JSPS) (to Y.T.), Simons Foundation research award DFARI 240432, and NIH awards NS081706 and R35NS097404 (to R.B.D.). R.B.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112050.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Darnell RB, and Posner JB (2003). Paraneoplastic syndromes involving the nervous system. N. Engl. J. Med 349, 1543–1554. [DOI] [PubMed] [Google Scholar]
- 2.Luque FA, Furneaux HM, Ferziger R, Rosenblum MK, Wray SH, Schold SC Jr., Glantz MJ, Jaeckle KA, Biran H, and Lesser M (1991). Anti-Ri: an antibody associated with paraneoplastic opsoclonus and breast cancer. Ann. Neurol 29, 241–251. [DOI] [PubMed] [Google Scholar]
- 3.Buckanovich RJ, Posner JB, and Darnell RB (1993). Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron 11, 657–672. [DOI] [PubMed] [Google Scholar]
- 4.Yang YY, Yin GL, and Darnell RB (1998). The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc. Natl. Acad. Sci. USA 95, 13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckanovich RJ, Yang YY, and Darnell RB (1996). The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J. Neurosci 16, 1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckanovich RJ, and Darnell RB (1997). The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol. Cell Biol 17, 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, and Burley SK (2000). Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell 100, 323–332. [DOI] [PubMed] [Google Scholar]
- 8.Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, and Patel DJ (2011). Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1. Structure 19, 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito Y, Yuan Y, Zucker-Scharff I, Fak JJ, Jereb S, Tajima Y, Licatalosi DD, and Darnell RB (2019). Differential NOVA2-mediated splicing in excitatory and inhibitory neurons regulates cortical development and cerebellar function. Neuron 101, 707–720.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dredge BK, and Darnell RB (2003). Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol. Cell Biol 23, 4687–4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggiu M, Herbst R, Kim N, Jevsek M, Fak JJ, Mann MA, Fischbach G, Burden SJ, and Darnell RB (2009). Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc. Natl. Acad. Sci. USA 106, 3513–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito Y, Miranda-Rottmann S, Ruggiu M, Park CY, Fak JJ, Zhong R, Duncan JS, Fabella BA, Junge HJ, Chen Z, et al. (2016). NOVA2-mediated RNA regulation is required for axonal pathfinding during development. Elife 5, e14371. 10.7554/eLife.14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, and Darnell RB (2003). CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215. [DOI] [PubMed] [Google Scholar]
- 14.Ule J, Jensen K, Mele A, and Darnell RB (2005). CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386. [DOI] [PubMed] [Google Scholar]
- 15.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dredge BK, Stefani G, Engelhard CC, and Darnell RB (2005). Nova autoregulation reveals dual functions in neuronal splicing. EMBO J 24, 1608–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eom T, Zhang C, Wang H, Lay K, Fak J, Noebels JL, and Darnell RB (2013). NOVA-dependent regulation of cryptic NMD exons controls synaptic protein levels after seizure. Elife 2, e00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Racca C, Gardiol A, Eom T, Ule J, Triller A, and Darnell RB (2010). The neuronal splicing factor Nova Co-localizes with target RNAs in the dendrite. Front. Neural Circuits 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, and Darnell RB (2010). Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, and Darnell RB (2006). An RNA map predicting Nova-dependent splicing regulation. Nature 444, 580–586. [DOI] [PubMed] [Google Scholar]
- 21.Darnell RB (2013). RNA protein interaction in neurons. Annu. Rev. Neurosci 36, 243–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fryssira H, Tsoutsou E, Psoni S, Amenta S, Liehr T, Anastasakis E, Skentou C, Ntouflia A, Papoulidis I, Manolakos E, and Chaliasos N (2016). Partial monosomy14q involving FOXG1 and NOVA1 in an infant with microcephaly, seizures and severe developmental delay. Mol. Cytogenet 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi M, Zhao Y, Wang Y, and Li T (2013). A new small supernumerary marker chromosome involving 14pter / q12 in a child with severe neuro-developmental retardation: case report and literature review. Gene 531, 457–461. [DOI] [PubMed] [Google Scholar]
- 24.Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta L.d.l.T., Huang J, et al. (2016). Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gimenez M, Marie SKN, Oba-Shinjo S, Uno M, Izumi C, Oliveira JB, and Rosa JC (2015). Quantitative proteomic analysis shows differentially expressed HSPB1 in glioblastoma as a discriminating short from long survival factor and NOVA1 as a differentiation factor between low-grade astrocytoma and oligodendroglioma. BMC Cancer 15, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhi F, Wang Q, Deng D, Shao N, Wang R, Xue L, Wang S, Xia X, and Yang Y (2014). MiR-181b-5p downregulates NOVA1 to suppress proliferation, migration and invasion and promote apoptosis in astrocytoma. PLoS One 9, e109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon SO, Kim EK, Lee M, Jung WY, Lee H, Kang Y, Jang Y-J, Hong SW, Choi SH, and Yang WI (2016). NOVA1 inhibition by miR-146b-5p in the remnant tissue microenvironment defines occult residual disease after gastric cancer removal. Oncotarget 7, 2475–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EK, Yoon SO, Jung WY, Lee H, Kang Y, Jang Y-J, Hong SW, Choi SH, and Yang WI (2017). Implications of NOVA1 suppression within the microenvironment of gastric cancer: association with immune cell dysregulation. Gastric Cancer 20, 438–447. [DOI] [PubMed] [Google Scholar]
- 29.Shen B, Zhang Y, Yu S, Yuan Y, Zhong Y, Lu J, and Feng J (2015). MicroRNA-339, an epigenetic modulating target is involved in human gastric carcinogenesis through targeting NOVA1. FEBS Lett 589, 3205–3211. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y-A, Zhu J-M, Yin J, Tang W-Q, Guo Y-M, Shen X-Z, and Liu T-T (2014). High expression of neuro-oncological ventral antigen 1 correlates with poor prognosis in hepatocellular carcinoma. PLoS One 9, e90955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim EK, Yoon SO, Kim SH, Yang WI, Cho YA, and Kim SJ (2016). Upregulated neuro-oncological ventral antigen 1 (NOVA1) expression is specific to mature and immature T- and NK-cell lymphomas. J. Pathol. Transl. Med 50, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, and Darnell RB (2000). Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25, 359–371. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quina LA, Walker A, Morton G, Han V, and Turner EE (2020). GAD2 expression defines a class of excitatory lateral habenula neurons in mice that project to the raphe and pontine tegmentum. eNeuro 7, ENEURO. 0527, 19.2020. 10.1523/ENEURO.0527-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. (2011). Brown adipose tissue activity controls triglyceride clearance. Nat. Med 17, 200–205. [DOI] [PubMed] [Google Scholar]
- 36.Townsend KL, and Tseng Y-H (2014). Brown fat fuel utilization and thermogenesis. Trends Endocrinol. Metab 25, 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalonde R, and Strazielle C (2011). Brain regions and genes affecting limb-clasping responses. Brain Res. Rev 67, 252–259. [DOI] [PubMed] [Google Scholar]
- 38.Kraeuter A-K, Guest PC, and Sarnyai Z (2019). The Y-maze for assessment of spatial working and reference memory in mice. Methods Mol. Biol 1916, 105–111. [DOI] [PubMed] [Google Scholar]
- 39.Seibenhener ML, and Wooten MC (2015). Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. J. Vis. Exp., e52434. [DOI] [PMC free article] [PubMed]
- 40.Takao K, and Miyakawa T (2006). Light/dark transition test for mice. J. Vis. Exp 104, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams G, Bing C, Cai XJ, Harrold JA, King PJ, and Liu XH (2001). The hypothalamus and the control of energy homeostasis: different circuits, different purposes. Physiol. Behav 74, 683–701. [DOI] [PubMed] [Google Scholar]
- 42.Clarke IJ (2015). Hypothalamus as an endocrine organ. Compr. Physiol 5, 217–253. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Z-D, Yang WZ, Gao C, Fu X, Zhang W, Zhou Q, Chen W, Ni X, Lin J-K, Yang J, et al. (2017). A hypothalamic circuit that controls body temperature. Proc. Natl. Acad. Sci. USA 114, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison SF (2016). Central control of body temperature. F1000Res 5. 10.12688/f1000research.7958.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beck B (2006). Neuropeptide Y in normal eating and in genetic and dietary-induced obesity. Philos. Trans. R. Soc. Lond. B Biol. Sci 361, 1159–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ilnytska O, and Argyropoulos G (2008). The role of the Agouti-Related Protein in energy balance regulation. Cell. Mol. Life Sci 65, 2721–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koerker DJ, Ruch W, Chideckel E, Palmer J, Goodner CJ, Ensinck J, and Gale CC (1974). Somatostatin: hypothalamic inhibitor of the endocrine pancreas. Science 184, 482–484. [DOI] [PubMed] [Google Scholar]
- 48.Eigler T, and Ben-Shlomo A (2014). Somatostatin system: molecular mechanisms regulating anterior pituitary hormones. J. Mol. Endocrinol 53, R1–R19. [DOI] [PubMed] [Google Scholar]
- 49.Goodman CA, and Hornberger TA (2013). Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc. Sport Sci. Rev 41, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira CM, Sattlegger E, Jiang H-Y, Longo BM, Jaqueta CB, Hinnebusch AG, Wek RC, Mello LEAM, and Castilho BA (2005). IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J. Biol. Chem 280, 28316–28323. [DOI] [PubMed] [Google Scholar]
- 52.Cambiaghi TD, Pereira CM, Shanmugam R, Bolech M, Wek RC, Sattlegger E, and Castilho BA (2014). Evolutionarily conserved IMPACT impairs various stress responses that require GCN1 for activating the eIF2 kinase GCN2. Biochem. Biophys. Res. Commun 443, 592–597. [DOI] [PubMed] [Google Scholar]
- 53.Pereira CM, Filev R, Dubiela FP, Brandão BB, Queiroz CM, Ludwig RG, Hipolide D, Longo BM, Mello LE, Mori MA, and Castilho BA (2019). The GCN2 inhibitor IMPACT contributes to diet-induced obesity and body temperature control. PLoS One 14, e0217287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C, and Darnell RB (2011). Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol 29, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sossin WS, and Costa-Mattioli M (2019). Translational control in the brain in Health and disease. Cold Spring Harb. Perspect. Biol 11, a032912. 10.1101/cshperspect.a032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skariah G, and Todd PK (2021). Translational control in aging and neurodegeneration. Wiley Interdiscip. Rev. RNA 12, e1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma V, Sood R, Khlaifia A, Eslamizade MJ, Hung T-Y, Lou D, Asgarihafshejani A, Lalzar M, Kiniry SJ, Stokes MP, et al. (2020). eIF2α controls memory consolidation via excitatory and somatostatin neurons. Nature 586, 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Q, Bestard-Lorigados I, and Song W (2021). Cell-type-specific memory consolidation driven by translational control. Signal Transduct. Target. Ther 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roffé M, Hajj GNM, Azevedo HF, Alves VS, and Castilho BA (2013). IMPACT is a developmentally regulated protein in neurons that opposes the eukaryotic initiation factor 2a kinase GCN2 in the modulation of neurite outgrowth. J. Biol. Chem 288, 10860–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silva RC, Dautel M, Di Genova BM, Amberg DC, Castilho BA, and Sattlegger E (2015). The Gcn2 regulator Yih1 interacts with the cyclin dependent kinase Cdc28 and promotes cell cycle progression through G2/M in budding yeast. PLoS One 10, e0131070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sattlegger E, Barbosa JARG, Moraes MCS, Martins RM, Hinnebusch AG, and Castilho BA (2011). Gcn1 and actin binding to Yih1: implications for activation of the eIF2 kinase GCN2. J. Biol. Chem 286, 10341–10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneeberger M, Altirriba J, García A, Esteban Y, Castaño C, García-Lavandeira M, Alvarez CV, Gomis R, and Claret M (2012). Deletion of miRNA processing enzyme Dicer in POMC-expressing cells leads to pituitary dysfunction, neurodegeneration and development of obesity. Mol. Metab 2, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greenman Y, Kuperman Y, Drori Y, Asa SL, Navon I, Forkosh O, Gil S, Stern N, and Chen A (2013). Postnatal ablation of POMC neurons induces an obese phenotype characterized by decreased food intake and enhanced anxiety-like behavior. Mol. Endocrinol 27, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Störchel PH, Thümmler J, Siegel G, Aksoy-Aksel A, Zampa F, Sumer S, and Schratt G (2015). A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. EMBO J 34, 2237–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaven F, Marin P, and Claeysen S (2014). Primary culture of mouse dopaminergic neurons. J. Vis. Exp., e51751. [DOI] [PMC free article] [PubMed]
- 67.Heiman M, Kulicke R, Fenster RJ, Greengard P, and Heintz N (2014). Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc 9, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J, Anczuków O, Krainer AR, Zhang MQ, and Zhang C (2013). OLego: fast and sensitive mapping of spliced mRNA-Seq reads using small seeds. Nucleic Acids Res 41, 5149–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charizanis K, Lee K-Y, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, et al. (2012). Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 75, 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, and Darnell RB (2014). Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc 9, 263–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data (Gene expression omnibus (GEO): GSE212473) and NOVA1-CLIP data (GEO: GSE212547) reported in this paper have been deposited at GEO and are publicly available as of the data of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


