Abstract
Hepatocellular carcinoma (HCC) is a common malignant primary tumor that is usually diagnosed at an advanced stage; thus, there is an urgent need for efficient and sensitive novel diagnostic markers to determine the prognosis and halt disease progression in patients with HCC. Disulfidptosis is a recently discovered form of programmed cell death, essentially an abnormal accumulation of intracellular bisulfides. Therefore, our study aimed to investigate the role of disulfidptosis-related genes (DRGs) in the pathogenesis of HCC. Based on public databases, our work demonstrates the relationship between DRG and expression, immunity, mutation/drug sensitivity, and functional enrichment in HCC. We also revealed the significant heterogeneity of HCC in different DRGs sub-clusters and in differentially expressed genes (DEGs), respectively. Subsequently, the most relevant candidate gene, SLC7A11, was screened by machine learning to further validate the significance of SLC7A11 in the clinical features, prognosis, nomogram pattern, and immune infiltration of HCC. Our study, which elucidates the potential mechanisms of DRGs and HCC, reveals that SLC7A11 can serve as a novel prognostic biomarker and provides opportunities and challenges for individualized cancer immunotherapy strategies.
Keywords: Disulfidptosis, Hepatocellular carcinoma, Immune infiltration, Machine learning, SLC7A11
1. Introduction
Primary liver cancer is the fourth most common tumor worldwide. Hepatocellular carcinoma (HCC) is a common malignancy that occurs in the liver [1,2]. Since HCC accounts for over 90% of all liver cancer cases, chemotherapy and immunotherapy are currently the best treatment options for the condition. In addition, the use of natural compounds and/or nanotechnology can reduce systemic toxicity and side effects on patients, leading to better treatment outcomes [3]. Tislelizumab and sorafenib have been shown to be effective in improving the survival prognosis of previously treated patients with advanced HCC; however, it is not a complete cure [4,5]. Moreover, most patients are not eligible for radical treatment as they are usually diagnosed at an advanced stage.
Therefore, there is a need for efficient and sensitive novel diagnostic markers to determine the prognosis and halt disease progression, and an urgent need to investigate new treatments to improve the prognosis of HCC patients.
The current study proposes a new mode of cell death, disulfidptosis, which is a form of rapid death caused by disulfide stress due to excessive intracellular cystine accumulation, independent of the currently existing forms of programmed cell death such as apoptosis, ferroptosis, necroptosis, and cuproptosis [6]. Early studies found that under glucose-starvation conditions, NADPH was severely depleted in cells overexpressing SLC7A11, and disulfides such as cystine accumulated abnormally, causing disulfide stress and eventually leading to rapid cell death [7]. The formation of structural disulfide bonds is a catalytic process involving many proteins and small molecules. The mitochondria and endoplasmic reticulum of eukaryotic cells are capable of forming and transferring protein disulfide bonds [8,9]. Disulfide bond formation is found in cancer-related proteins, which means this metastable bond can be used as a target for new therapies [10]. Among them, disulfide isomerase, an endoplasmic reticulum protein, forms disulfide bonds in nascent proteins [11]. In addition, disulfide isomerases have been shown to be associated with tumorigenicity in a variety of tumors and are promising targets for cancer therapy [[12], [13], [14]]. Notably, protein disulfide isomerase family A member 3 is involved in immunogenic cell death, and it is associated with liver damage and hepatocellular carcinoma cell proliferation [15,16]. However, the role of disulfidptosis in the prognosis and immune infiltration of HCC is unclear and requires further study.
To explore possible pathogenic mechanisms, we explored comprehensive bioinformatics analyses of DRGs using the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. We searched for and explored the expression, immunity, mutations, and drug sensitivity of disulfidptosis-related genes (DRGs) in HCC. Sub-clusters of DRGs are constructed based on clinical features and gene expression, and relevant subgroups are explored for prognosis determination and differential gene and functional enrichment. Differential genes and DRGs were then extracted for crossover to find differentially expressed DRGs. In addition, machine learning algorithms and differential gene analyses were applied to find key DRGs. Finally, the strongest trait genes were identified in HCC and their association with prognosis determination and immune infiltration was further considered. This provides new insights into a better understanding of the molecular mechanisms of HCC's pathogenesis and its early clinical diagnosis.
2. Materials and methods
2.1. Identification of DRGs in HCC
We identified nine DRGs from previous studies [6]. The GeneMANIA Prediction Server is a biological network for gene biological relevance and function prediction [17]. We used the GeneMANIA website (http://www.genemania.org) to identify functionally similar genes, created 29 DRGs, and further evaluated the reliability of DRGs.
2.2. HCC datasets
We downloaded RNA sequencing data and related clinical information from TCGA (http://cancergenome.nih.gov) for 424 HCC tissues (374 tumoral and 50 normal tissues). In addition, the GSE25097 dataset was derived from GEO (https://www.ncbi.nlm.nih.gov/geo/.), which contains 268 HCC samples and 243 paired normal samples. Two datasets were used to study the DRG expression levels, sub-cluster type, differential gene analyses, machine learning, prognosis determination, immune infiltration, among others. Data were extracted in the TPM format, and further log2 (x+1) transformations were performed on each expression value. All data analyses were performed using R (version 4.2.1) and the associated bioinformatics analysis website.
2.3. Gene set and DEGs functional enrichment analysis
The KEGG enrichment analysis is a practical method of analyzing gene function and related genomic functional information. Gene Ontology (GO) is a widely used tool for functional gene annotation, specifically the Molecular Function (MF), Biological Pathway (BP), and Cellular Component (CC). For gene set functional enrichment, we used the KEGG API (https://www.kegg.jp/kegg/rest/keggapi.html) to obtain the latest KEGG Pathway gene annotations as the background, mapped 29 DRGs to the background set, and used the R package ClusterProfiler (version 3.14.3) [18] to perform the enrichment analysis to obtain the results of gene set enrichment. Similarly, we used the GO annotations of genes from the R package, org. Hs.eg.db (version 3.1.0), as a background, mapped the genes to the background set, and used the R package, clusterProfiler (version 3.14.3), to perform enrichment analyses to obtain DRG enrichment Results. P-values of <0.05 and FDR values of <0.1 were considered statistically significant. For DEG functional enrichment, we obtained enrichment results for differentially upregulated/downregulated genes of the KEGG pathway and enrichment results for differentially upregulated/downregulated genes of the GO term. P-values of <0.05 were considered statistically significant.
2.4. Gene Set Cancer Analysis
The Genomic Cancer Analysis (GSCA) is a platform for the expression, immunity, mutation, and drug sensitivity of pan-cancer at the gene set level [19]. In the GSCA, a convenient platform is provided to perform gene set genomic, including the expression, single nucleotide variation (SNV), copy number variation (CNV), methylation, and immune (24 immune cells) analysis. In addition, clinical information combined with small-molecule drugs can be mined for candidate biomarkers and valuable drugs to inform further clinical trials.
2.5. Sub-cluster analysis
The sub-cluster analysis can be up to six clusters using the ConsensusClusterPlus R package (v1.54 4.0) [20] and 80% of the total sample drawn 100 times, clusterAlg = “hc”, innerLinkage = “ward.D2.” The threshold for differential mRNA expression between the two clusters was set at “Adjusted p < 0.05 and |log2FC| > 2". Cluster heatmaps were performed using the R package pheatmap (v1.0.12). Gene expression heatmaps retained genes with an SD of >0.1.
2.6. Machine learning
The least absolute shrinkage and selection operator (LASSO) is a regression method used for regularization to improve prediction accuracy and models by selecting variables [21]. The Random Forest is a learning method that constructs many decision trees and outputs classes of individual trees with a high degree of accuracy, sensitivity, and specificity [22]. The log-rank test was used to determine differences in survival between the two groups via comparisons. The timeROC analysis was used to compare the predictive accuracy of DRGs and risk scores.
2.7. Statistical analysis
All statistical analyses were performed using the R package (version 4.2.1) and visualizations were plotted using the ggplot 2 package (version 3.3.6). Expression-related heatmaps for DRGs were analyzed and visualized using the igraph package (version 1.3.4) and the ggraph package (version 2.1.0). DEGs were calculated using the limma package (version 3.40.6). The LASSO regression and random forest analysis were performed using the R packages, “glmnet” [23] and “randomForest” [24]. The Kaplan–Meier survival analysis was carried out using the “survival R″ and “survminer R″ packages (version 3.3.1). The ROC analysis was performed using the qROC package (version 1.18.0). The “rms” package (version 6.4.0) was used to build and visualize the nomogram model. The Wilcoxon rank-sum test was used to determine differences between groups via comparisons. P-values of <0.05 were considered statistically significant (ns, p ≥ 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
3. Results
3.1. Preliminary exploration and definition of DRGs in HCC
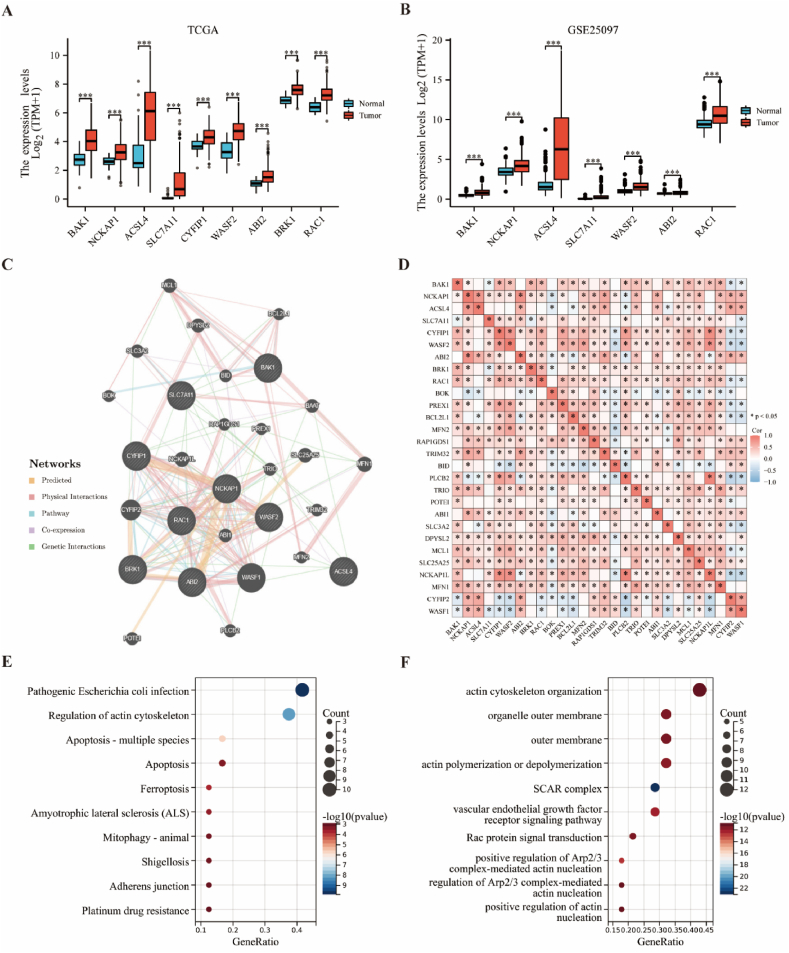
As previously mentioned, nine genes (BAK1, NCKAP1, ACSL4, SLC7A11, CYFIP1, WASF2, ABI2, BRK1, RAC1) were shown to be associated with disulfidptosis [6]. To confirm the expression of these genes in HCC, we downloaded expression data from the TCGA and GTEx databases of tumoral and normal tissues, which showed differences in the expression of nine genes, which were all upregulated and significant in tumor expression (Fig. 1A). Seven of these genes, including BAK1, NCKAP1, ACSL4, SLC7A11, WASF2, ABI2, and RAC1, also yielded consistent results in the GEO database (Fig. 1B). We used GeneMANIA to predict biologically similar genes in DRGs. We obtained 20 similar gene hub genes comprising WASF1, CYFIP2, MFN1, NCKAP1L, SLC25A25, MCL1, DPYSL2, BAAT, SLC3A2, ABI1, POTEI, TRIO, PLCB2, BID, TRIM32, RAP1GDS1, MFN2, BCL2L1, BOK and PREX1. The relationships between genes are based on five specific types, including Predicted, Physical Interactions, Pathway Co-expression, and Genetic Interactions (Fig. 1C). The heat map of the correlation analysis showed that the expression of most DRGs was positively correlated (Fig. 1D). The enrichment analysis of DRGs in the KEGG dataset identified some cellular processes and human diseases such as pathogenic Escherichia coli infection, actin cytoskeleton regulation, apoptosis-multiple species, ferroptosis, amyotrophic lateral sclerosis (ALS), shigellosis, and so on (Fig. 1E). Further enrichment analyses of these genes on the GO dataset indicated that certain related actin nucleation items, such as the actin cytoskeleton organization, organelle outer membrane, outer membrane, vascular endothelial growth factor receptor signaling pathway, positive regulation of Arp2/3 complex-mediated actin nucleation, actin polymerization or depolymerization, regulation of Arp2/3 complex-mediated actin nucleation, positive regulation of actin nucleation, and so on (Fig. 1F). The above preliminary analysis showed that the correlation between DRG and HCC progression has some reliability.
Fig. 1.
Reasonable definition of DRGs in HCC (A).
3.2. Comprehensive analysis of the expression, immunity, mutation, and drug sensitivity of twenty-nine DRGs in HCC
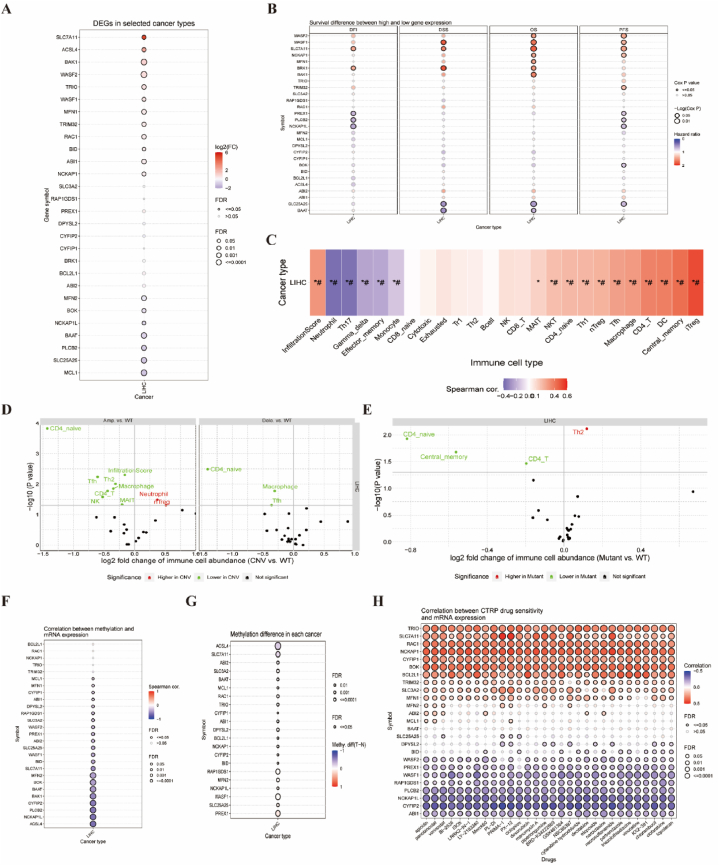
To gain a comprehensive understanding of the role and relevance of DRGs in cancer diagnosis, we further correlated the analysis of four modules: expression, immunity, mutation, and drug sensitivity using Gene Set Cancer Analysis (GSCA). In the expression module, we summarize the DEGs between tumoral and normal samples in HCC (Fig. 2A). Among them, upregulated genes include SLC7A11, ACSL4, BAK1, WASF2, and so on, compared with downregulated genes, including MCL1, SLC25A25, PLCB2, BAAT, and so on. After that, we analyzed the survival difference between the high and low DRG-expressing groups. Specifically, SLC7A11 is the only gene that has significant associations with the DFI, DSS, OS, and PFS (Fig. 2B). In the immune cell type, gene sets were most significantly and positively correlated with iTreg (p < 0.05, #FDR <0.05) and most negatively correlated with neutrophils (p < 0.05, #FDR <0.05; Fig. 2C). Interestingly, Fig. 2D summarizes the difference in immune infiltration between gene set CNV groups. iTreg and neutrophils are significantly highly expressed between amplification and wild-type in HCC. Fig. 2E summarizes the difference in immune infiltration between gene set SNV groups. Th2 is significantly highly expressed between mutant and wild-type in HCC. However, methylation affected the extensive downregulation of their mRNA expression in DRGs (Fig. 2F). Among them, ACSL4 and SLC7A11 have a significant negative correlation with methylation (Fig. 2G). In addition, there was a strong correlation between DRGs and CTRP drug sensitivity (top 30) in pan-cancer (Fig. 2H). In summary, the analysis of the multiple modules described above leads to a strong correlation between the expression, immunity, mutation, and drug sensitivity of DRGs in HCC.
Fig. 2.
Comprehensive analysis of 29 DRGs in HCC.
3.3. Analysis of two disulfidptosis sub-cluster and their sub-cluster differential genes
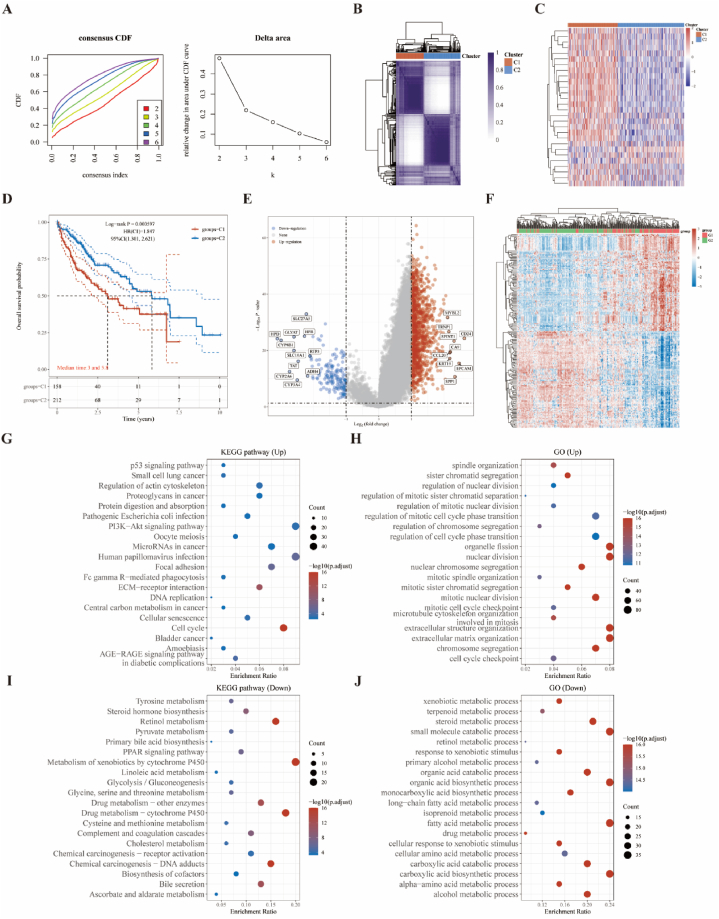
We performed consistent unsupervised clustering on a sample of 370 HCC patients in the TCGA database, in which the relative stability under two clusters (cluster 1 [n = 158], cluster 2 [n = 212]) was selected for the distribution (Fig. 3A and B). The high expression of DRGs in the C1 sub-cluster compared to the C2 sub-cluster (Fig. 3C) was associated with a poorer prognosis on the survival curve (Fig. 3D). To explore this further, we used the Limma analysis of the volcano map (Fig. 3E) and heat map (Fig. 3F) to show differential genes between the two subgroups. A total of 1083 upregulated genes such as MYBL2, TRNP1, SPINT1, CD24, and 158 downregulated genes were identified in DRG-high group, such as SLC27A5, HPD, GLYAT, and HPR, were identified in the DRG-low group (C1) compared to DRG-high group (C2) (Fig. 3E). Enrichment analyses of the upregulated genes in the KEGG dataset are shown in the cell cycle pathway (Fig. 3G), compared to the downregulated genes in the metabolism of xenobiotics by cytochrome P450, drug metabolism–cytochrome P450, and retinol metabolism pathways (Fig. 3I). Further enrichment analyses of the upregulated genes in the GO dataset are shown in organelle fission, nuclear division, extracellular structure organization, and extracellular matrix organization (Fig. 3H), compared to the downregulated genes in small-molecule catabolic process, organic acid biosynthetic process, fatty acid metabolic process, and carboxylic acid biosynthetic process (Fig. 3J).
Fig. 3.
Two disulfidptosis sub-clusters are shown.
3.4. Machine learning to identify DRGs in the TCGA dataset
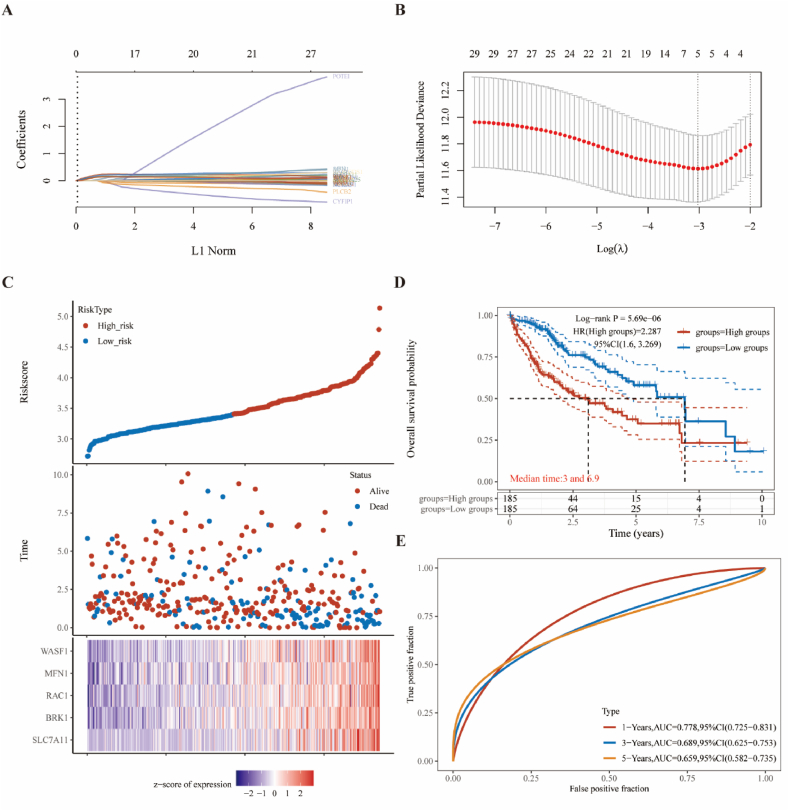
To identify new prognostic markers for HCC, we performed a LASSO regression analysis based on 29 DRGs for patients with HCC in the TCGA database. The LASSO regression algorithm used 10-fold cross-validation for feature selection, and all genes, except POTEI, showed consistency (Fig. 4A). Finally, five genes were identified as prognostic markers, including WASF1, MFN1, RAC1, BRK1, and SLC7A11 (Fig. 4B and C). We also confirmed that OS was significantly shorter in the high-risk group than in the low-risk group (HR = 2.287, 95%CI = 1.6–3.269, P < 0.001), comparing median durations of 3 years–6.9 years, respectively (Fig. 4D). Meanwhile, ROC time-dependent curves showed that the accuracy of the 14 genetic traits was close to or greater than 0.70 for 1-, 3- and 5-year survival (AUC >0.7 indicates higher accuracy; Fig. 4E).
Fig. 4.
Machine learning to identify disulfidptosis-signatures in the TCGA dataset.
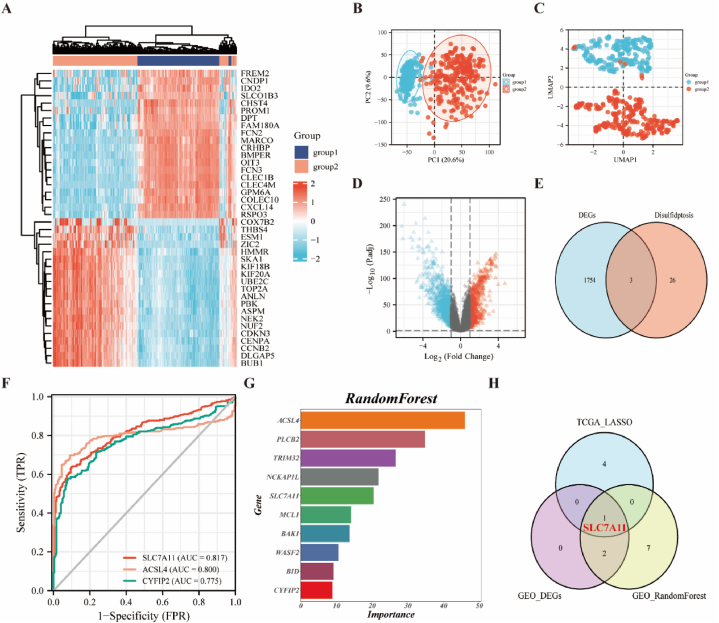
3.5. Differential gene analysis and machine learning to identify trait genes in the GEO dataset
To validate the reliability of the above analysis based on TCGA data, further differential gene analyses and machine learning were performed on HCC patients based on the GEO database. First, we used Limma analysis heat maps (showing top 20 upregulated/downregulated genes, respectively; Fig. 5A) and volcano maps (Fig. 5D) to demonstrate differential genes in HCC patients in GSE25097. For the PCA plot (Fig. 5B) and UMAP plot (Fig. 5C), there were significant differences between groups. A total of 592 upregulated genes and 1165 downregulated genes were identified, three of which were associated with DRGs (Fig. 5E). Second, we analyzed the ROC curves for the three trait genes described above. Notably, the AUC values for SLC7A11, ACSL4, and CYFIP2 were 0.817, 0.800, and 0.775, respectively (Fig. 5F). Next, we used the RandomForest algorithm to screen DRGs and construct potential genes based on the GSE25097 dataset. We show the top 10 genes in order, including ACSL4, PLCB2, TRIM32, NCKAP1L, SLC7A11, MCL1, BAK1, WASF2, BID, and CYFIP2 (Fig. 5G). Ultimately, SLC7A11 was selected as the only candidate gene (Fig. 5H).
Fig. 5.
Differential gene analyses and machine learning to identify trait genes in the GEO dataset.
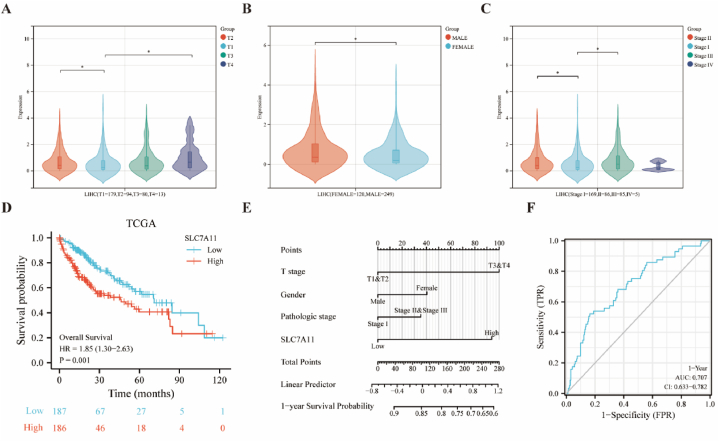
3.6. Clinical diagnosis and prognostic value analysis of SLC7A11
In the current study, we assessed SLC7A11 expression according to the T stage, sex, and pathologic stage in HCC and found that high SLC7A11 expression was significantly associated with T4, the female sex, and stage III, compared to T2, the male sex, and stage II, respectively (Fig. 6A–C). Survival analyses of HCC patients suggest the reliability of SLC7A11 as a poor prognostic factor (HR = 1.85, P = 0.001; Fig. 6D). SLC7A11 was combined with three other markers to construct a new nomogram to predict the probability of survival one year after the clinical diagnosis of HCC according to the patient's T stage, sex, and histological type (Fig. 6E). The one-year ROC time curve indicates AUC >0.7 in HCC (Fig. 6F).
Fig. 6.
Construction of a nomogram for OS prediction based on TCGA.
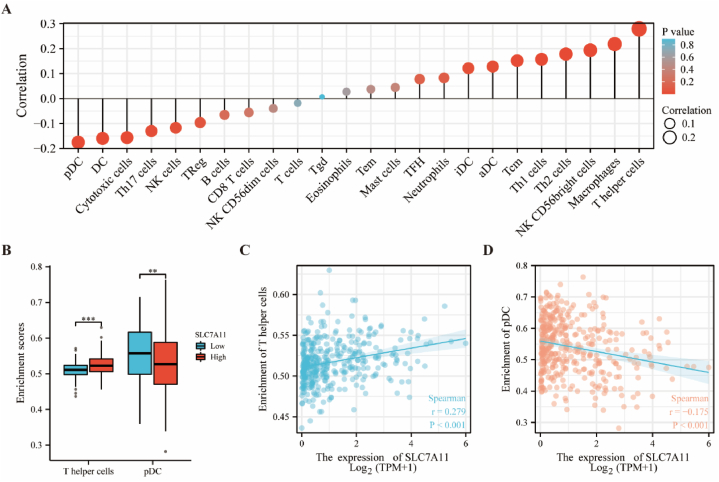
3.7. Immune infiltration analysis of SLC7A11 in HCC
By performing the ssGSEA algorithm on 24 immune cells, we analyzed the results of the correlation between SLC7A11 and immune infiltration and presented them as a lollipop plot (Fig. 7A). Specifically, SLC7A11 was positively correlated with most immune cells such as T helper cells, macrophages, and NK CD56 bright cells, and negatively correlated with pDC, DC, and cytotoxic T cells. In the analysis of high and low SLC7A11 expression in HCC, we showed a statistically significant correlation between T helper cells and NK CD56 bright cells (Fig. 7B). SLC7A11 expression levels were significantly positively and negatively correlated with the enrichment scores of T helper cells (Fig. 7C) and pDC (Fig. 7D), respectively.
Fig. 7.
SLC7A11 expression is associated with immune infiltration in HCC based on TCGA.
4. Discussion
The study of cell death mechanisms has not only contributed to a fundamental understanding of internal cellular stability and homeostasis but has also provided important new ideas for cancer treatment [25]. Recent studies have identified a novel form of disulfide-induced cell death in human cells, a phenomenon known as disulfidptosis. This study suggests that disulfidptosis induced by GLUT inhibitors may be an effective tumor treatment strategy [6].
People have long been looking for better ways of treating HCC, especially since early-stage HCC is relatively less malignant and can be cured by the aggressive treatment of some who suffer from the disease and are more hopeful [26,27]. It is difficult to avoid tumor resistance and progression through traditional surgical resection combined with chemotherapy and stereotactic body radiation therapy [28,29]. In addition, first-line targeted drugs such as tislelizumab and sorafenib do not cure advanced HCC [4,5]. Therefore, it has become crucial to evaluate HCC for early diagnosis and to investigate new drugs that target specific functional pathways. Our study links disulfidptosis to the pathogenicity of HCC, identifies possible key genes through bioinformatic analyses, and further analyzes the prognosis and immune infiltration.
In the present study, the expression of nine DRGs in HCC tumors and normal tissues showed a significant consistent upregulation of tumor expression based on the TCGA database. Seven of these genes, including BAK1, NCKAP1, ACSL4, SLC7A11, WASF2, ABI2, and RAC1, also yielded consistent results in the GEO database. We obtained 29 functionally positively correlated DRGs using GeneMANIA. The defined gene set is scientifically valid and credible. The gene set KEGG enrichment analysis revealed apoptosis and disease-related pathways, confirming the intrinsic pathway correlation between disulfidptosis and various forms of cell death (such as ferroptosis and apoptosis). In GO terms studies, we found that DRGs are involved in actin cytoskeleton organization. Disruptions in the organization and dynamics of the actin cytoskeleton lead to age-related symptoms and diseases ranging from cancer to neurodegeneration [30]. This explains, at least to some extent, the oncogenic effect of disulfidptosis in HCC cells. At an aggregate level, we have conducted a preliminary exploration of DRGs that are closely associated with HCC expression, immune infiltration, mutation, and drug sensitivity. As an example, solute carrier family 7 member 11 (SLC7A11), which ranks first in the list of differential genes for positive correlation, is a significant prognostic risk factor (including DFI, DSS, OS, and PFS) for HCC. SLC7A11 (also commonly referred to as xCT), which functions to import cysteine for glutathione biosynthesis and antioxidant defense, is overexpressed in a variety of human cancers [31]. SLC7A11 has been documented to be associated with efferocytosis, ferroptosis, and cancer [32,33]. Notably, SLC7A11 was positively correlated with 30 CTRP drug sensitivities, which contributed to further clinical drug trials. In addition, other genes such as TRIO, RAC1, NCKAP1, CYFIP1, BOK, and BCL2L1 also showed consistent results. Interestingly, the expression of SLC7A11 was significantly negatively correlated with methylation. RNA methylation and its associated downstream signaling pathways are involved in several biological processes, including cell differentiation, sex determination, and stress responses. There is growing evidence that this process is closely linked to cancer cell proliferation, cellular stress, metastasis, and immune responses [34].
Based on consensus clustering, we identified two HCC sub-clusters (C1 and C2) by the expression of DRGs and found that the C2 sub-cluster was associated with a poor prognosis. Sub-clusters of differential genes were then further explored and, finally, an enrichment analysis of upregulated genes in the KEGG pathway was found to observe the cell cycle and the GO term is mainly enriched in DNA-damaging processes such as organelle fission, nuclear division, extracellular structure organization, and extracellular matrix organization. Cell division is tightly regulated by a variety of evolutionarily conserved cell cycle control mechanisms to ensure the production of two genetically identical cells. Cancer, a group of diseases in which cells continue to divide excessively, is associated with a greater propensity to accumulate DNA damage. Cancer-associated mutations that disrupt cell cycle control allow continuous cell division primarily by disrupting the ability of cells to exit the cell cycle [35]. Besides, abnormal DNA replication may lead to genetic mutations which, in turn, may lead to the growth of cancer cells [36]. This provides further evidence for a potential molecular mechanism for the triggering of disulfidptosis in the pathogenesis of HCC.
To identify new prognostic markers for HCC, we performed a LASSO regression analysis based on 29 DRGs from the TCGA database. We screened for five genes with a disulfidptosis signature, including WASF1, MFN1, RAC1, BRK1, and SLC7A11. We also confirmed that survival durations were significantly shorter in the high-risk group than in the low-risk group. Also, ROC time-dependent curves showed an accuracy of greater than or close to 0.70 for 1-, 3- and 5-year survival rates. To verify the reliability of the above data analysis, further differential gene analyses and machine learning were performed on HCC patients based on the GEO database. First, we identified a total of 592 upregulated and 1165 downregulated genes by the Limma analysis, three of which were associated with DRGs, including SLC7A11, ACSL4, and CYFIP2. Meanwhile, we evaluated the DRGs using the RandomForest algorithm, which showed the top 10 genes.
Ultimately, SLC7A11 was selected as the only candidate gene, a result that provides further evidence of its oncogenic role in HCC. In recent studies, SLC7A11 has been shown to be highly expressed in certain types of cancer, such as lung cancer [33], breast cancer [37], and bladder cancer [38], and to promote cell progression and invasion. SLC7A11 can be used as a therapeutic target. In this paper, a survival analysis of HCC patients demonstrates the reliability of SLC7A11 as a poor prognostic factor. SLC7A11 ultimately establishes the role of the nomogram as an aid in the early clinical diagnosis of HCC. At the same time, we are concerned that ACSL4 (Acyl-CoA Synthetase Long-Chain Family Member 4) is an enzyme that catalyzes the activation of long-chain fatty acids by attaching them to coenzyme A (CoA), a process known as fatty acid activation or esterification [39]. In cancer, ACSL4 has been shown to promote tumor growth and metastasis by modulating lipid metabolism, promoting epithelial-mesenchymal transition, and enhancing cellular resistance to ferroptosis and oxidative stress [40]. ACSL4 has also been identified as a potential therapeutic target for cancer treatment, as the inhibition of ACSL4's activity or expression can sensitize cancer cells to chemotherapy and radiation therapy [41,42].
Accelerated tumor progression is not only associated with intracellular dysregulation but is also influenced by the tumor microenvironment [43]. As their understanding of the tumor microenvironment increases, scholars are exploring relevant immune cells and their roles in the tumor microenvironment to guide immunotherapy [44,45]. Among immune cell types, DRGs were most significantly positively correlated with Inducible regulatory T (iTreg) cells and most significantly negatively correlated with neutrophils. These iTreg cells, also known as peripherally induced Treg (pTreg) cells, occur in secondary lymphoid tissues and play an important role in immune suppression [46]. Neutrophils are no longer seen as innate immune cells with a single function; thus, they are less likely to be bystanders in the pathogenesis of cancer. Neutrophils have an important role and mechanism in the development, progression, metastasis, and recurrence of cancer [47]. We also analyzed the results of the correlation between SLC7A11 and immune infiltration. SLC7A11 was positively correlated with T helper cells (CD4+ T cells) and negatively correlated with plasmacytoid pre-dendritic cells (pDC). T helper cells (CD4+ T cells) are essential for host defense but are also important drivers of immune-mediated disease [48]. There is literature highlighting the significance of T cells in the tumor microenvironment and identifying unique candidate prognostic genetic markers for CD4+ T cells in patients with colorectal cancer [49]. pDCs are involved in the initiation of antiviral immune responses through interactions with innate and adaptive immune cell populations [50]. pDC levels correlate with immune cell infiltration and patient survival in triple-negative breast cancer [51]. Hence, studies focusing on one or more immune cells may help to identify potential mechanisms of disulfidptosis and SLC7A11 and demonstrate that SLC7A11 is a promising diagnostic biomarker for HCC that is involved in immune regulation. Ultimately, SLC7A11-related studies and new targeted immunotherapies may help improve poor patient prognosis and provide the latest insights for physicians to diagnose and intervene early in the treatment of HCC.
This study has some limitations. Firstly, we explored the genes associated with disulfidptosis, using public databases such as GEO and TCGA but did not have our own clinical data. Secondly, human tissue validation was not performed in this study, and further precise validation needs to be performed through experiments.
Our study reveals the relationship between DRGs and the expression, immunity, mutation, and drug sensitivity of HCC, as well as significant heterogeneity among different sub-clusters of DRGs and differential gene analyses. In conclusion, we found that in HCC, SLC7A11 may play a key role in predicting poor outcomes and correlates with the level of immune infiltration. Thus, SLC7A11 could serve as a new prognostic biomarker, providing directions and identifying challenges for the development of new immunotherapies. This is a new brave and scientific attempt.
Expression distributions of nine DRGs between cancer and normal tissues in the TCGA dataset. (B) Expression distributions of seven DRGs between cancer and normal tissues in the GEO dataset. (C) Identify genes with similar functions and establish 29 DRGs by GeneMANIA website. The five line colors represent the types of gene interactions. (D) Expression correlations of DRGs. (E) KEGG and (F) GO concentrated air bubble diagram. *p < 0.05, ***p < 0.001.
(A) The DEGs between tumoral and normal samples in HCC. (B) The survival difference between high and low DRG expression groups. (C) The association between the GSVA score and the activity of cancer-related immune cell types in HCC. (D) The difference in immune infiltration between gene set CNV groups. (E) The difference in immune infiltration between gene set SNV groups. (F) The profile of correlations between methylation and the mRNA expression of DRGs in HCC. (G) The methylation difference between tumoral and normal samples of DRGs in HCC. (H) CTRP drug sensitivity (top 30) in pan-cancer. DFI: disease-free interval; DSS: Disease Specific Survival; OS: Overall Survival; PFI: Progression-free survival; GSVA score: gene set expression score; CNV: Copy Number Variation; SNV: Single Nucleotide Variation; Amp: amplification; Dele: deletion; WT: wild-type; CTRP: The Cancer Therapeutics Response Portal. *p < 0.05, #FDR <0.05.
(A) The cumulative distribution function (CDF) curve of K [[2], [3], [4], [5], [6]]. The relative change in area under the CDF curve of K [[2], [3], [4], [5], [6]]. (B) Appropriate unsupervised clustering analysis (k = 2). (C) Heat map showing the relationship between DRG expressions in subgroups. (D) Survival curve analysis revealing differences in OS between the two subgroups. Two groups were tested using the log-rank test, with the 95% CI representing the HR confidence interval; The median time represents the time in years corresponding to survival in the different groups at 50%. (E) Differential analyses of subgroups. (F) Heatmap showing differential gene expressions. (G-J) The KEGG pathway and GO air bubble diagram. In the enrichment result, P-values of <0.05 are considered statistically significant (enrichment score with −log 10 (P) of more than 1.3). OS: overall survival; HR: hazard ratio.
(A) Construction of disulfidptosis-signatures using the LASSO regression. (B) Determining the appropriate number of genes by confidence intervals of lambda. (C) The risk score, survival time, and five-gene signature expression in HCC. (D) The OS (per the Kaplan–Meier survival analysis) was compared between low-risk-score and high-risk-score groups in HCC. (E) ROC curves over time at 1, 3, and 5 years, respectively.
Differential genes in GSE25097 showing (A) a heat map and (D) a volcano map (|log2(FC)| > 1, p. adj <0.05). (B) Principal component analysis (PCA) in GSE25097. (C) Uniform Manifold Approximation and Projection (UMAP) analysis in GSE25097. (E) Trait genes shared by DRGs and differential genes of GSE25097. (F) ROC curves of three trait genes. (G) Selection of the most relevant DRGs based on GSE25097 using RandomForest (top 10). (H) Venn diagram showing the overlap of candidate genes between the two databases.
SLC7A11 expression among different groups of HCC based on the (A) T stage, (B) sex, and (C) pathologic stage. (D) Kaplan–Meier survival analysis of HCC in the high-risk and low-risk groups. (E) Nomogram combining SLC7A11 and clinicopathological features of HCC. (F) ROC curves over time at one year in HCC.
(A) Correlation between SLC7A11 and multiple immune cells. (B) SLC7A11 was associated with T helper cells and pDC. Correlation between enrichment scores and (C) SLC7A11 in T helper cells and (D) pDC. **p < 0.01, ***p < 0.001.
Author contributions
ZX and XL conceived and designed the experiments; SL and YL performed the experiments; XM and SZ analyzed and interpreted the data; ZX and SZ contributed reagents, materials, analysis tools or data; SL and XL wrote the paper.
Funding
This work was supported by the National Natural Science Foundation of China, China (No.81001340), the Medical Scientific Research Foundation of Guangdong Province, China (A2021474), the Medical and Health Science and Technology Project of Shantou, Guangdong, China (210526156491330), the Medical and Health Science and Technology Project of Shantou, Guangdong, China (221117126497682), the 2021 Guangdong Clinical Teaching Base Teaching Reform Research Project (2021JD065), and the 2022 Teaching Reform and Research Project of Shantou Medical College (2022–19).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Shao-bo Zhang, Email: 1652192302@qq.com.
Ze-feng Xie, Email: lsp13119@163.com.
References
- 1.Villanueva A. Hepatocellular carcinoma. N. Engl. J. Med. 2019;380(15):1450–1462. doi: 10.1056/NEJMra1713263. https://10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 2.Gravitz L. Liver cancer. Nature. 2014;516(7529):S1. doi: 10.1038/516S1a. https://10.1038/516S1a [DOI] [PubMed] [Google Scholar]
- 3.Anwanwan D., Singh S.K., Singh S., Saikam V., Singh R. Challenges in liver cancer and possible treatment approaches. Biochem. Biophys. Acta. Rev. Cancer. 2020;1873(1) doi: 10.1016/j.bbcan.2019.188314. https://10.1016/j.bbcan.2019.188314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren Z., Ducreux M., Abou-Alfa G.K., Merle P., Fang W., Edeline J., et al. Tislelizumab in patients with previously treated advanced hepatocellular carcinoma (RATIONALE-208): a multicenter, non-randomized, open-label, phase 2 trial. Liver Cancer. 2023;12(1):72–84. doi: 10.1159/000527175. https://10.1159/000527175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu L., Liu L., Yang S., Ren J., Lai P.B.S., Chen G.G. New insights into sorafenib resistance in hepatocellular carcinoma: responsible mechanisms and promising strategies. Biochem. Biophys. Acta. Rev. Cancer. 2017;1868(2):564–570. doi: 10.1016/j.bbcan.2017.10.002. https://10.1016/j.bbcan.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Nie L., Zhang Y., Yan Y., Wang C., Colic M., et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat. Cell Biol. 2023;25(3):404–414. doi: 10.1038/s41556-023-01091-2. https://10.1038/s41556-023-01091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X., Olszewski K., Zhang Y., Lim E.W., Shi J., Zhang X., et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020;22(4):476–486. doi: 10.1038/s41556-020-0496-x. https://10.1038/s41556-020-0496-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bien M., Longen S., Wagener N., Chwalla I., Herrmann J.M., Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell. 2010;37(4):516–528. doi: 10.1016/j.molcel.2010.01.017. https://10.1016/j.molcel.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 9.Sevier C.S., Kaiser C.A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3(11):836–847. doi: 10.1038/nrm954. https://10.1038/nrm954 [DOI] [PubMed] [Google Scholar]
- 10.Hogg P.J. Targeting allosteric disulphide bonds in cancer. Nat. Rev. Cancer. 2013;13(6):425–431. doi: 10.1038/nrc3519. https://10.1038/nrc3519 [DOI] [PubMed] [Google Scholar]
- 11.Depuydt M., Messens J., Collet J.F. How proteins form disulfide bonds. Antioxidants Redox Signal. 2011;15(1):49–66. doi: 10.1089/ars.2010.3575. https://10.1089/ars.2010.3575 [DOI] [PubMed] [Google Scholar]
- 12.Chen T.Y., Yang C.Y., Yang M.T., Kuo T.F., Chang C.L., Chen C.L., et al. Protein disulfide isomerase a4 promotes lung cancer development via the Stat 3 pathway in stromal cells. Clin. Transl. Med. 2022;12(2) doi: 10.1002/ctm2.606. https://10.1002/ctm2.606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S., Jackson C., Karapetyan E., Dutta P., Kermah D., Wu Y., et al. Roles of protein disulfide isomerase in breast cancer. Cancers. 2022;14(3) doi: 10.3390/cancers14030745. https://10.3390/cancers14030745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S., Sankar S., Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov. Today. 2014;19(3):222–240. doi: 10.1016/j.drudis.2013.10.017. https://10.1016/j.drudis.2013.10.017 [DOI] [PubMed] [Google Scholar]
- 15.Clement C.C., Osan J., Buque A., Nanaware P.P., Chang Y.C., Perino G., et al. PDIA3 epitope-driven immune autoreactivity contributes to hepatic damage in type 2 diabetes. Sci. Immunol. 2022;7(74) doi: 10.1126/sciimmunol.abl3795. https://10.1126/sciimmunol.abl3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo R., Ishino K., Wada R., Takata H., Peng W.X., Kudo M., et al. Downregulation of protein disulfide-isomerase A3 expression inhibits cell proliferation and induces apoptosis through STAT3 signaling in hepatocellular carcinoma. Int. J. Oncol. 2019;54(4):1409–1421. doi: 10.3892/ijo.2019.4710. https://10.3892/ijo.2019.4710 [DOI] [PubMed] [Google Scholar]
- 17.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. https://10.1093/nar/gkq537 Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. https://10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C.J., Hu F.F., Xie G.Y., Miao Y.R., Li X.W., Zeng Y., et al. GSCA: an integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Briefings Bioinf. 2023;24(1) doi: 10.1093/bib/bbac558. https://10.1093/bib/bbac558 [DOI] [PubMed] [Google Scholar]
- 20.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. https://10.1093/bioinformatics/btq170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibshirani R., Tibshirani R. 1996. Regression Shrinkage via the Lasso. [Google Scholar]
- 22.Breiman L.J., Cm Jo. Random forests. Mach. Learn. 2001;45(2):199–228. [Google Scholar]
- 23.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 24.Liaw A., Wiener M.J.R.N. Classification and Regression by randomForest. 2002;23(23) [Google Scholar]
- 25.Bertheloot D., Latz E., Franklin B.S. Necroptosis, pyroptosis and apoptosis: an intricate game of cell death. Cell. Mol. Immunol. 2021;18(5):1106–1121. doi: 10.1038/s41423-020-00630-3. https://10.1038/s41423-020-00630-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7(1):6. doi: 10.1038/s41572-020-00240-3. https://10.1038/s41572-020-00240-3 [DOI] [PubMed] [Google Scholar]
- 27.Li X., Ramadori P., Pfister D., Seehawer M., Zender L., Heikenwalder M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat. Rev. Cancer. 2021;21(9):541–557. doi: 10.1038/s41568-021-00383-9. https://10.1038/s41568-021-00383-9 [DOI] [PubMed] [Google Scholar]
- 28.Llovet J.M., De Baere T., Kulik L., Haber P.K., Greten T.F., Meyer T., et al. Locoregional therapies in the era of molecular and immune treatments for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021;18(5):293–313. doi: 10.1038/s41575-020-00395-0. https://10.1038/s41575-020-00395-0 [DOI] [PubMed] [Google Scholar]
- 29.Couri T., Pillai A. Goals and targets for personalized therapy for HCC. Hepatol. Int. 2019;13(2):125–137. doi: 10.1007/s12072-018-9919-1. https://10.1007/s12072-018-9919-1 [DOI] [PubMed] [Google Scholar]
- 30.Lai W.F., Wong W.T. Roles of the actin cytoskeleton in aging and age-associated diseases. Ageing Res. Rev. 2020;58 doi: 10.1016/j.arr.2020.101021. https://10.1016/j.arr.2020.101021 [DOI] [PubMed] [Google Scholar]
- 31.Koppula P., Zhuang L., Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi: 10.1007/s13238-020-00789-5. https://10.1007/s13238-020-00789-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maschalidi S., Mehrotra P., Keçeli B.N., De Cleene H.K.L., Lecomte K., Van der Cruyssen R., et al. Targeting SLC7A11 improves efferocytosis by dendritic cells and wound healing in diabetes. Nature. 2022;606(7915):776–784. doi: 10.1038/s41586-022-04754-6. https://10.1038/s41586-022-04754-6 [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Sun Y., Bai L., Zhi L., Yang Y., Zhao Q., et al. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11. J. Clin. Invest. 2021;131(22) doi: 10.1172/JCI152067. https://10.1172/jci152067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B., Wang J.Q., Tan Y., Yuan R., Chen Z.S., Zou C. RNA methylation and cancer treatment. Pharmacol. Res. 2021;174 doi: 10.1016/j.phrs.2021.105937. https://10.1016/j.phrs.2021.105937 [DOI] [PubMed] [Google Scholar]
- 35.Matthews H.K., Bertoli C., de Bruin R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022;23(1):74–88. doi: 10.1038/s41580-021-00404-3. https://10.1038/s41580-021-00404-3 [DOI] [PubMed] [Google Scholar]
- 36.Basu A.K. DNA damage, mutagenesis and cancer. Int. J. Mol. Sci. 2018;19(4) doi: 10.3390/ijms19040970. https://10.3390/ijms19040970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav P., Sharma P., Sundaram S., Venkatraman G., Bera A.K., Karunagaran D. SLC7A11/xCT is a target of miR-5096 and its restoration partially rescues miR-5096-mediated ferroptosis and anti-tumor effects in human breast cancer cells. Cancer Lett. 2021;522:211–224. doi: 10.1016/j.canlet.2021.09.033. https://10.1016/j.canlet.2021.09.033 [DOI] [PubMed] [Google Scholar]
- 38.Shen L., Zhang J., Zheng Z., Yang F., Liu S., Wu Y., et al. PHGDH inhibits ferroptosis and promotes malignant progression by upregulating SLC7A11 in bladder cancer. Int. J. Biol. Sci. 2022;18(14):5459–5474. doi: 10.7150/ijbs.74546. https://10.7150/ijbs.74546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soupene E., Fyrst H., Kuypers F.A. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Nat. Acad. Sci. U.S.A. 2008;105(1):88–93. doi: 10.1073/pnas.0709737104. https://10.1073/pnas.0709737104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Li J., Kang R., Klionsky D.J., Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17(9):2054–2081. doi: 10.1080/15548627.2020.1810918. https://10.1080/15548627.2020.1810918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai G., Wang D., Ma S., Hong S., Ding K., Tan X., et al. ACSL4 promotes colorectal cancer and is a potential therapeutic target of emodin. Phytomedicine : Int. J. Phytother. Phytopharmacol. 2022;102 doi: 10.1016/j.phymed.2022.154149. https://10.1016/j.phymed.2022.154149 [DOI] [PubMed] [Google Scholar]
- 42.Hou J., Jiang C., Wen X., Li C., Xiong S., Yue T., et al. ACSL4 as a potential target and biomarker for anticancer: from molecular mechanisms to clinical therapeutics. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.949863. https://10.3389/fphar.2022.949863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson N.M., Simon M.C. The tumor microenvironment. Curr. Biol. : CB (Curr. Biol.) 2020;30(16):R921. doi: 10.1016/j.cub.2020.06.081. https://10.1016/j.cub.2020.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng S., Xiao F., Chen M., Gao H. Tumor-microenvironment-responsive nanomedicine for enhanced cancer immunotherapy. Adv. Sci. 2022;9(1) doi: 10.1002/advs.202103836. https://10.1002/advs.202103836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Y., Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Therapeut. 2021;221 doi: 10.1016/j.pharmthera.2020.107753. https://10.1016/j.pharmthera.2020.107753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao F., Tian M., Zhang X., Jin X., Jiang Y., Sun X., et al. Butyrate enhances CPT1A activity to promote fatty acid oxidation and iTreg differentiation. Proc. Nat. Acad. Sci. U.S.A. 2021;118(22) doi: 10.1073/pnas.2014681118. https://10.1073/pnas.2014681118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong S., Dong L., Cheng L. Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 2021;14(1):173. doi: 10.1186/s13045-021-01187-y. https://10.1186/s13045-021-01187-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Shea J.J., Paul W.E. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science (New York, N.Y.) 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. https://10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toor S.M., Sasidharan Nair V., Saleh R., Taha R.Z., Murshed K., Al-Dhaheri M., et al. Transcriptome of tumor-infiltrating T cells in colorectal cancer patients uncovered a unique gene signature in CD4(+) T cells associated with poor disease-specific survival. Vaccines. 2021;9(4) doi: 10.3390/vaccines9040334. https://10.3390/vaccines9040334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alculumbre S., Raieli S., Hoffmann C., Chelbi R., Danlos F.X., Soumelis V. Plasmacytoid pre-dendritic cells (pDC): from molecular pathways to function and disease association. Semin. Cell Dev. Biol. 2019;86:24–35. doi: 10.1016/j.semcdb.2018.02.014. https://10.1016/j.semcdb.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 51.Oshi M., Newman S., Tokumaru Y., Yan L., Matsuyama R., Kalinski P., et al. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC) Cancers. 2020;12(11) doi: 10.3390/cancers12113342. https://10.3390/cancers12113342 [DOI] [PMC free article] [PubMed] [Google Scholar]