Abstract
Background
Poor sleep is associated with many negative health outcomes, including multiple dimensions of psychopathology. In the past decade, sleep researchers have advocated for focusing on the concept of sleep health as a modifiable health behavior to mitigate or prevent these outcomes. Sleep health dimensions often include sleep efficiency, duration, satisfaction, regularity, timing, and daytime alertness. However, there is no consensus on how to best operationalize sleep health at the phenotypic and genetic levels. In some studies, specific sleep health domains were examined individually, while in others, sleep health domains were examined together (e.g., with an aggregate sleep health score).
Methods
Here, we compared alternative sleep health factor models using genomic structural equation modeling on summary statistics from previously published genome-wide association studies of self-reported and actigraphic sleep measures with effective sample sizes up to 452,633.
Results
Our best-fitting sleep health model had 6 correlated genetic factors pertaining to 6 sleep health domains: circadian preference, efficiency, alertness, duration, noninsomnia, and regularity. All sleep health factors were significantly correlated (|rgs| = 0.11–0.51), except for the circadian preference factor with duration and noninsomnia. Better sleep health was generally significantly associated with lower genetic liability for psychopathology (|rgs| = 0.05–0.48), yet the 6 sleep health factors showed divergent patterns of associations with different psychopathology factors, especially when controlling for covariance among the sleep health factors.
Conclusions
These results provide evidence for genetic separability of sleep health constructs and their differentiation with respect to associations with mental health.
Keywords: Chronotype, Factor, Genetics, Latent, Psychiatric, Structural equation model (SEM)
Poor sleep quantity and quality are problems associated with negative health outcomes, including psychopathology (1). More than a quarter of the adult population experiences sleep problems; thus, promoting sleep as a modifiable health behavior that can lead to positive outcomes is an important public health initiative (2). While research has traditionally focused on pathology and disordered sleep, sleep health is a construct that frames sleep measures in a positive light, identifying a typical range of healthy sleep as well as tangible targets for improving population health (3).
Sleep health includes 6 domains: timing, efficiency, alertness, duration, satisfaction, and regularity, although regularity is a more recent addition (3,4). Sleep efficiency reflects one aspect of sleep quality (e.g., percentage of time in bed asleep). Sleep duration is measured from the time of sleep onset to sleep offset. Sleep timing is typically assessed as bedtime or midpoint of sleep and is sometimes used as an estimate for circadian preference or an individual’s preference for being a morning versus evening person. An individual’s alertness reflects the level of sleepiness outside the sleep window. Sleep satisfaction is a measure of perception of sleep quality. Regularity indicates an individual’s consistency of sleep/wake times. Although all of these domains make up the sleep health concept, most studies referencing sleep health operationalize only one or a subset of these domains (3). However, it may be informative to use several measures of sleep health together because they can paint a fuller picture than one dimension on its own (5).
Two possible conceptualizations of sleep health exist. The first views sleep health as a unitary construct that captures variation across all sleep health domains (3) and suggests that the various sleep health domains would predict outcomes in similar direction and magnitude. This conceptualization is implicit when using a summary score or latent factor of measures across sleep domains to capture overall sleep health [e.g., Dalmases et al. (6)]. This view might predict that in a structural equation model (SEM), a single factor would explain the associations of various sleep health domains with health outcomes, i.e., the health outcomes would correlate with the sleep domains to the extent that the health outcomes are predicted by the common sleep health factor.
The second conceptualization views sleep health as a nonunitary construct, such that unique sleep health domains may relate differently to outcomes (7). If so, an aggregate sleep health measure may fail to capture crucial information. For example, Dalmases et al. (6) examined how 5 dimensions of sleep health related to self-reported poor health. In individual regressions, the sleep health dimensions showed variable odds ratios; sleep duration and timing did not significantly predict health, whereas alertness and satisfaction did. They then created an aggregate variable from those dimensions to predict a number of chronic diseases. Although this aggregate variable was significantly associated with a number of chronic diseases, the individual regressions suggest that not all components of the variable were contributing to that association. Thus, although an aggregate sleep health variable may be useful for characterizing how overall sleep health relates to outcomes, it may obscure differential associations of specific sleep health domains.
Sleep health dimensions are associated with psychiatric symptoms and disorders, but the nature of these relationships remains nebulous (8, 9, 10, 11, 12). One potential cause of poor sleep and psychopathology is shared genetic variants (pleiotropy). Genome-wide association studies (GWASs) demonstrate that sleep and psychiatric traits show genetic correlations (12, 13, 14). Genetic correlations based on GWAS data use millions of genetic variants to quantify whether genetic effects across genomes are similar across phenotypes. They are typically calculated using GWAS data from unrelated individuals to avoid conflating genetic similarity with environmental similarity. Furthermore, estimating genetic correlations based on GWAS summary statistics does not require the same individuals to be assessed on both traits (15). This advantage enables assessment of a much wider range of associations than is possible in a single study (16).
Recent genetic studies on sleep traits quantified the contributions of additive genetic effects from common variants on biological predispositions in sleep-psychopathology relationships. Using GWAS data, the morning chronotype was found to be negatively associated with schizophrenia and depression and positively associated with greater subjective well-being (13,17). Dashti et al. (18) found that sleep duration positively correlated with schizophrenia and bipolar disorder (BP) and that both short and long sleep positively genetically correlated with depressive symptoms. The first genetic study to look at multiple domains of objective sleep health with psychiatric disorders found that polygenic risk scores for depression significantly predicted measures of sleep quality, naps, and variability; polygenic risk scores for BP significantly predicted wake-up time and variability; and polygenic risk scores for schizophrenia significantly predicted wake-up time, sleep quality, naps, and variability (12).
The present study adds to this literature by incorporating a broader range of sleep health (objective and subjective) and psychopathology measures into genomic SEMs (19) to examine the genetic factor structure of sleep health and its relationship to psychopathology. It is important to account for objective and subjective traits because they often relate differentially to psychopathology at the phenotypic level (20). Furthermore, by analyzing sleep health as a whole, not just disordered sleep, and sleep health’s associations with psychopathology, we gain more knowledge about how sleep as a modifiable health behavior may be used to ameliorate the burden of psychiatric disorders associated with poor sleep health. Here, we tested multiple genetic structures using 12 sleep measures from published GWASs. Then, we analyzed the relationships of the sleep health latent genetic factors with psychopathology to better understand how sleep health is related to internalizing, externalizing, and 2 thought disorder psychopathology factors. We were motivated by 2 primary questions relevant to the conceptualizations of sleep health:
-
1.
Is sleep health best represented by a single genetic factor or multiple distinct factors?
-
2.
At the genetic level, do sleep health domains differentially relate to psychopathology factors?
Methods and Materials
GWAS Summary Statistics
We obtained GWAS summary statistics from published GWASs. GWAS summary statistics are output files from GWAS analyses that typically contain genetic variant identifier (single nucleotide polymorphism), reference allele, effect size, standard error, and p values for the reference alleles. Here, summary statistics were limited to European ancestry because sample sizes for other populations were not large enough to be included. Table 1 provides detailed descriptions of the measures, which we briefly describe below.
Table 1.
Sample Sizes and Descriptions of Sleep and Psychopathology Phenotypes
| Trait | Sample/Effective N | Description/Ascertainment | Coding | Reference |
|---|---|---|---|---|
| Circadian Preference | Higher score = more morningness | |||
| Chronotype | UKB, 23andMe/449,734 | “Do you consider yourself to be: Definitely an evening person, More an evening than morning person, More a morning than an evening person, Definitely a morning person” | Ordinal scale: −2, −1, 1, 2 | Jones et al., 2019 (13) |
| Sleep midpointa | UKB/85,810 | Calculated for each sleep period as the midpoint between the start of the first detected sleep episode and the end of the last sleep episode | Clock times: continuous | Jones et al., 2019 (23) |
| Least active 5 hoursa | UKB/85,205 | Midpoint of the least active 5 hours of each day; the least active 5 hours was defined as the 5-hour period with the minimum average acceleration | Clock times: continuous | Jones et al., 2019 (23) |
| Most active 10 hoursa | UKB/85,670 | Midpoint of the most active 10 hours of each day; the most active 10 hours was defined as the 10-hour period with the maximum average acceleration | Clock times: continuous | Jones et al., 2019 (23) |
| Efficiency | Higher score = better sleep efficiency | |||
| Sleep efficiency | UKB/84,810 | Sleep duration divided by the time between the start and end of the first and last nocturnal inactivity period, respectively | Continuous | Jones et al., 2019 (23) |
| Number of sleep episodesa | UKB/84,810 | Periods of at least 5 min with no change larger than 5° associated with the z-axis of the activity-monitor | Continuous | Jones et al., 2019 (23) |
| Alertness | Higher score = more daytime alertness | |||
| Daytime sleepinessa | UKB/452,071 | “How likely are you to fall asleep when you don’t mean to?” | Never, sometimes, often, or all of the time | Wang et al., 2019 (21) |
| Diurnal inactivitya | UKB/84,757 | Total daily duration of estimated bouts of inactivity that fell outside of the sleep window; this measure captures very inactive states such as napping and wakeful rest but not inactivity such as sitting and reading or watching television, which are associated with a low but detectable level of movement. | Continuous | Jones et al., 2019 (23) |
| Nappinga | UKB/452,633 | “Do you nap during the day?” | Never/rarely, sometimes, usually | Dashti et al., 2021 (22) |
| Duration | Higher score = longer sleep duration | |||
| Self-reported sleep duration | UKB/446,118 | “On average how much do you sleep?” | Continuous | Dashti et al., 2019 (18) |
| Short sleep durationb | 346,794 | “On average how much do you sleep?” | Case (6 h or less), control | Dashti et al., 2019 (18) |
| Long sleep durationb | 135,283 | “On average how much do you sleep?” | Case (9 h or less), control | Dashti et al., 2019 (18) |
| Sleep duration actigraphyb | UKB/85,449 | Summed duration of all sleep episodes | Continuous | Jones et al., 2019 (23) |
| Noninsomnia | Higher score = no insomnia liability | |||
| Insomniaa | UKB/259,365 | “Do you have trouble falling asleep at night, or do you wake up in the middle of the night?” | Case (usually), control (never/rarely) | Lane et al., 2019 (14) |
| Regularity | Higher score = more sleep time regularity | |||
| Standard deviation sleep duration actigraphya | UKB/84,441 | Standard deviation of the summed duration of all actigraphy sleep episodes | Continuous | Jones et al., 2019 (23) |
| Internalizing | Higher score = internalizing psychopathology liability | |||
| Anxiety | UKB/259,365 | DSM-based anxiety disorders diagnoses | Case/control | Purves et al., 2019 (25) |
| MDD | PGC, UKB/424,616 | Self-report | Case/control | Howard et al., 2019 (26) |
| PTSD | Meta-analysis of 11 cohorts/30,273 | DSM-IV | Case/control | Nievergelt et al., 2019 (24) |
| Externalizing | Higher score = externalizing psychopathology liability | |||
| Problematic alcohol use | UKB/17,852 | AUDIT problematic use | Continuous | Sanchez-Roige et al., 2018 (34) |
| Cigarettes per day | GSCAN/337,334 | 1: 1–5, 2: 6–15, 3: 16–25, 4: 26–35, 5: 36+ cigarettes per day | Quasi-continuous | Liu et al., 2019 (33) |
| Cannabis use disorder | PGC, iPsych, deCode/384,032 | DSM-IV, DSM-III-R, ICD-10 | Case/control | Johnson et al., 2020 (28) |
| ADHD | PGC, iPsych/22,842 | ICD-10 | Case/control | Demontis et al., 2018 (27) |
| Psychosis Thought Disorders | Higher score = psychosis psychopathology liability | |||
| Schizophrenia | PGC/69,279 | DSM-IV, ICD-10, SCID | Case/control | Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014 (30) |
| Bipolar disorder | PGC/50,981 | DSM-IV, ICD-9, or ICD-10 | Case/control | Mullins et al., 2021 (29) |
| Compulsive Thought Disorders | Higher score = compulsive psychopathology liability | |||
| OCD | IOCDF-GC, OCGAS/3890 | DSM-IV | Case/control | International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS), 2018 (31) |
| Anorexia nervosa | PGC, ANGI/23,160 | DSM III-R, DSM-IV, ICD-8, ICD-9, ICD-10, and self-report | Case/control | Watson et al., 2019 (32) |
This table presents summary statistics from previously published genome-wide association studies. Sample size is the effective N, calculated as suggested by the Genomic SEM Wiki (Eff N = 4 × v × [1 − v] × [Ncases + Ncontrols]; v = sample proportion of cases) for case/control traits and full sample for continuous traits or meta-analyses.
ANGI, Anorexia Nervosa Genetics Initiative; AUDIT, Alcohol Use Disorders Identification Test; GSCAN, GWAS & Sequencing Consortium of Alcohol and Nicotine use; GWAS, genome-wide association study; IOCDF-GC, International OCD Foundation Genetics Collaborative; MDD, major depressive disorder; OCD, obsessive-compulsive disorder; OCGAS, OCD Collaborative Genetics Association Study; PGC, Psychiatric Genomics Consortium; PTSD, posttraumatic stress disorder; SCID, Structured Clinical Interview for DSM; UKB, UK Biobank.
Summary statistics were reverse coded by multiplying the z-statistic by −1 before being used in analyses.
Summary statistics were not used in the final sleep health model. See Figure S3 for full correlation matrix with all sleep health and psychopathology traits.
Sleep Phenotypes
Subjective (self-reported) sleep phenotypes were insomnia (14), chronotype (13), sleep duration (18), daytime sleepiness (21), and napping (22). Objective sleep phenotypes were collected via actigraphic data (AX3; Axivity, worn for 7 days) and consisted of sleep midpoint, most active 10 hours of the day, least active 5 hours of the day, sleep efficiency, sleep episodes, diurnal inactivity, and the standard deviation of actigraphy sleep duration (23). We reverse coded the summary statistics of episodes, daytime sleepiness, diurnal inactivity, napping, insomnia, midpoint, most active 10 hours of the day, least active 5 hours of the day, and standard deviation of sleep duration so that the factors would all indicate better health. See the Supplement for a discussion of the measures that were not included in the models, particularly short and long sleep variables derived from the self-reported sleep duration measure as well as actigraphy duration.
Psychopathology Phenotypes
Case-control psychopathology phenotypes were posttraumatic stress disorder (24), anxiety (25), major depressive disorder (26), attention-deficit/hyperactivity disorder (27), cannabis use disorder (28), BP (29), schizophrenia (30), obsessive-compulsive disorder (31), and anorexia nervosa (32). Quasi-continuous psychopathology phenotypes were cigarettes per day (33) and problematic alcohol use (34).
Statistical Analyses
Genomic SEM (19) is a flexible R package that enables SEM on genetic covariances derived from GWAS summary statistics. It is an extension of linkage disequilibrium score regression (35), which calculates bivariate genetic covariances by using a weighted regression of the product of GWAS summary statistics on linkage disequilibrium scores (a measure of how much genetic variation is tagged by the candidate single nucleotide polymorphism).
We munged summary statistics with the linkage disequilibrium score regression munge sumstats function in Python (version 3.7.3) and then used the Genomic SEM ldsc() function in R (version 4.1.2) to create 2 matrices: an S matrix that contained the genetic variances and covariances of the traits, and a V sampling matrix that contained the squared standard errors of the estimates from S on the diagonal and the covariances between the heritability and covariances on the off diagonal. The V matrix takes into account sample overlap between traits by assessing dependencies between estimation errors (19).
Genomic SEM uses lavaan syntax to specify models of interest and estimates models using the diagonally weighted least squares estimation with the S and V matrices. Model fit was evaluated using the χ2 test, confirmatory fit index (CFI), and standard root mean residual (SRMR). In Genomic SEM, the χ2 is often uninterpretable because it is a product of the sample size, which can be quite large in these analyses. Thus, we used CFI > 0.95 and SRMR < 0.08 as the criteria for good fit (36) and CFI > 0.90 as a criterion for acceptable fit. We calculated χ2 difference tests (Δχ2) to compare nested models.
Sleep Health Models
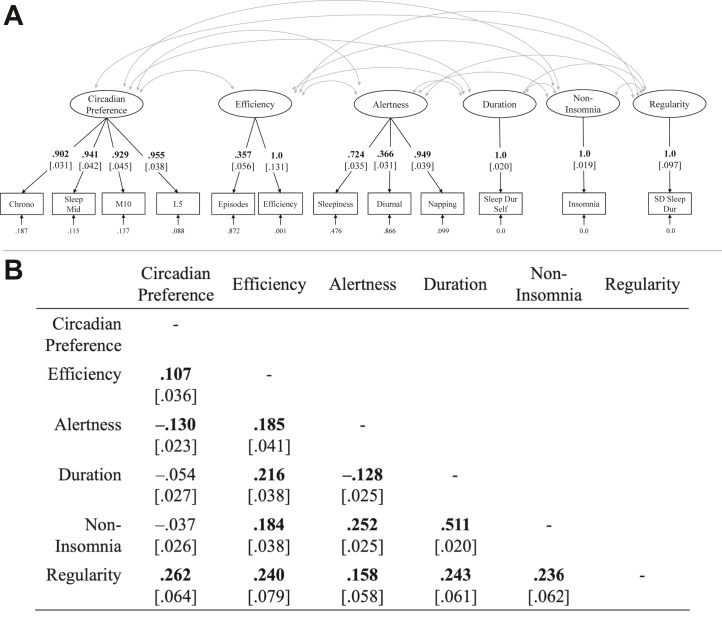
We fitted 3 sleep health models based on prior literature (Figure 1 and Figure S1) (3,4,6,7).
-
1.
Model 1: If the specific facets of sleep health are genetically unique, the data should be well represented by a correlated factors model. We fitted a latent model with 6 factors to indicate these unique facets. Circadian preference (indicated by chronotype, sleep midpoint, least active 5 hours of the day, and most active 10 hours of the day), efficiency (indicated by episodes and efficiency), alertness (indicated by napping, diurnal inactivity, and daytime sleepiness), and duration (indicated by self-reported sleep duration) were based on the original sleep health framework proposed by Buysse (3). The fifth factor, regularity, indicated by the standard deviation of actigraphic sleep duration, is a more recent addition to sleep health (37). Finally, we included a sixth factor, noninsomnia, indicated by insomnia because it is highly comorbid with psychopathology. We reverse-coded sleep midpoint, least active 5 hours, most active 10 hours, episodes, daytime sleepiness, napping, diurnal inactivity, standard deviation sleep duration, and insomnia so that the factors would reflect better health. Because noninsomnia, duration, and regularity were single-indicator factors, we constrained their loadings to be 1 and residual variances to be 0 to identify the factors. See the Supplement for more details on how these factors were chosen.
-
2.
Model 2: If all sleep health domains are highly genetically correlated, they might reflect a single factor. Thus, we fitted a single-factor model with loadings for all 12 sleep traits (Figure S1).
-
3.
Model 3: Even if a single-factor model does not fit well, there is potential for a hierarchical model to fit the data if the sleep health factors share genetic variance. Thus, we fitted a model in which the 6 sleep factors estimated in model 1 loaded on a single higher-order factor (Figure S1).
Figure 1.
Genomic structural equation model sleep health model. (A) Genomic structural equation model of 6 correlated sleep health factors. (B) Factor correlations. Boldface font for factor loadings indicates p < .05 (standard errors in brackets). Boldface font for factor correlations indicates significance after false discovery rate correction. Dur, duration; L5, least active 5 hours of the day; M10, most active 10 hours of the day; Mid, midpoint; SD, standard deviation of sleep duration; Self, self-report.
After running models and assessing fit statistics and residual correlations between indicators, we made slight model modifications and ran 2 model comparisons: 6-factor sleep health compared with single-factor sleep health and 6-factor sleep health compared with hierarchical sleep health (Table 2). The residual variance of the efficiency indicator was negative, leading to nonconvergence; therefore, we constrained the residual variance to be positive (>0.001), allowing the model to converge. See the Supplement for details on model modifications.
Table 2.
Fit of Genomic Structural Equation Models of Sleep Health
| Model | χ2 (df) | CFI | SRMR | Compared With | Δχ2 (df) |
|---|---|---|---|---|---|
| Sleep Models | |||||
| Six-factor sleep healtha | 977.931 (41)b | 0.914 | 0.065 | – | – |
| Single factor | 7838.253 (54)b | 0.284 | 0.163 | Sleep model 1 | 6860.321 (13)b |
| Hierarchical factor | 1514.193 (50)b | 0.865 | 0.114 | Sleep model 1 | 536.262 (9)b |
| Psychopathology Models | |||||
| Four correlated factors | 169.389 (38)b | 0.964 | 0.083 | – | – |
| Combined Models | |||||
| Sleep model 1 and Psych model 1a | 4864.961 (188)b | 0.921 | 0.067 | Combined model 3 | 5.789 (1)c |
| Sleep model 3 and Psych model 1 | 12,187.550 (217)b | 0.798 | 0.091 | – | – |
This table presents models of sleep health and psychopathology fit.
CFI, confirmatory factor index; SRMR, standard root mean residual.
Models for which main analyses were performed.
Indicates χ2 values and χ2 difference test p values < .001.
Indicates χ2 difference test p values < .05.
Psychopathology Model
We specified a psychopathology model based on prior work (38, 39, 40). The model was largely based on the work by Caspi et al. (38), but considerations from more recent models were also factored in. We focused on the correlated factors model proposed by Caspi et al., which contained 3 factors: internalizing psychopathology, externalizing psychopathology, and thought disorders, rather than their p-factor model, given the lack of utility for a p-factor at the genetic level shown by Grotzinger et al. (41). The correlated factors model by Caspi et al. is easily interpretable, and the broad categorization into internalizing, externalizing, and thought disorders is well accepted in the literature and consistent with most phenotypic and genetic correlational patterns (42). However, we separated the thought disorders factor into 2 factors, compulsive thought disorders and psychosis thought disorders, based on recent literature (40). Although recent genetic work has also included Tourette syndrome and/or autism spectrum disorder (39, 40, 41), with the latter as part of a neurodevelopmental factor, those models vary in terms of other disorders that are clustered in the factors that explain these disorders and cross-paths that are needed in the model to accommodate them; autism spectrum disorder also seems to show divergent patterns of genetic association with external correlates compared with other psychiatric disorders or even other neurodevelopmental disorders (41). Because testing alternative psychopathology factor structures was not the emphasis of this investigation, we focused on the more typically examined psychiatric disorders that serve as indicators for the main factors examined by Caspi et al. (38). Our model contained 4 correlated factors: internalizing psychopathology, indicated by anxiety, major depressive disorder, and posttraumatic stress disorder; externalizing psychopathology, indicated by cigarettes per day, problematic alcohol use, cannabis use disorder, and attention-deficit/hyperactivity disorder; psychosis thought disorders, indicated by schizophrenia and BP; and compulsive thought disorders, indicated by anorexia nervosa and OCD.
Combined Model
Finally, we fitted a model with both sleep health and psychopathology structures and allowed all latent factors to correlate. Then, to determine whether the sleep health factors showed distinct patterns of associations with psychopathology, controlling for each other, we let all sleep health factors associate with all psychopathology factors in a multiple regression framework. We used the p.adjust function in R to false discovery rate (FDR)–corrected p values for all correlations and regression coefficients. p.adjust (method=‘FDR’) uses the Benjamini-Hochberg method to control for the expected number of false discoveries, and all p values per model were corrected at once. The genetic correlations and betas we present as significant in the results are significant after FDR correction.
Results
Sleep Health Latent Genetic Structure
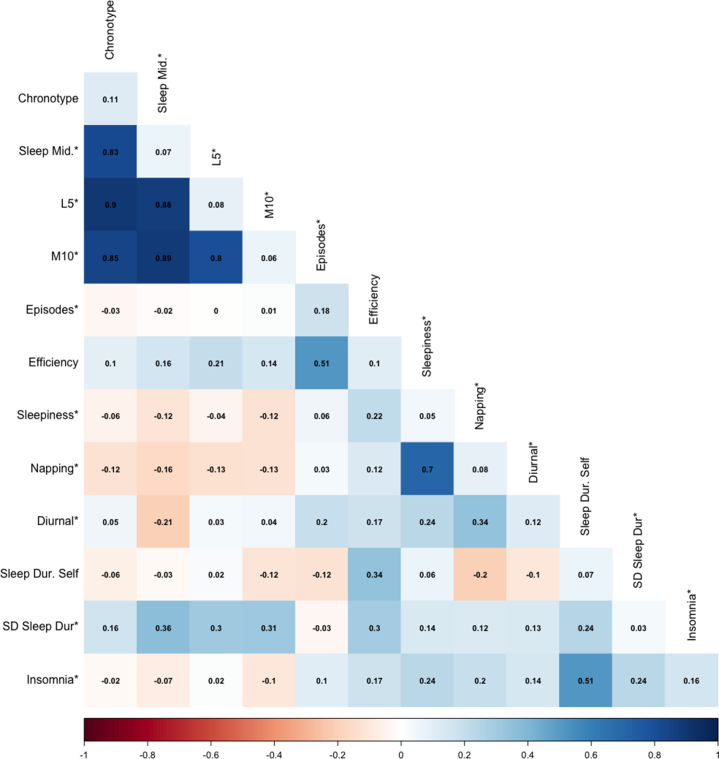
Genetic correlations between all sleep health indicators are presented in Figure 2, and model fit statistics are shown in Table 1. A sleep health model with 6 factors (model 1) (Figure 1A) fitted acceptably, with χ242 = 977.93, CFI = 0.914, and SRMR = 0.065. All factor correlations were significant except those between circadian preference and duration and circadian preference and noninsomnia (Figure 1B). Otherwise, factor correlations ranged from |rg| = 0.11–0.51. In contrast, a single-factor sleep health model (model 2) did not fit well, with χ254 = 7838.253, CFI = 0.284, and SRMR = 0.163. A model with a higher-order sleep health factor (model 3) also did not fit well, with χ251 = 1628.48, CFI = 0.855, and SRMR = 0.117 and provided a significantly poorer fit to the data than the 6-factor sleep health model (model 1) (Table 2). All loadings on the higher-order sleep health factor were significant (λs = 0.27–0.78, ps < .001) except for circadian preference.
Figure 2.
Genetic correlations between sleep traits with heritability on the diagonal. Trait heritability and bivariate genetic correlations calculated from linkage disequilibrium score regression. ∗indicates that summary statistics were reverse coded. Acti., actigraphy; Dur, duration; L5, least active 5 hours of the day; M10, most active 10 hours of the day; Mid., midpoint; SD, standard deviation of sleep duration.
Sleep Health and Psychopathology Latent Genetic Correlations
The psychopathology model fitted well, with χ238 = 169.39, CFI = 0.964, and SRMR = 0.083, and latent correlations were mostly positive and significant. Externalizing psychopathology correlated positively with the psychosis thought disorder factor but negatively with the compulsive thought disorder factor, showing some genetic divergence in the relationships of the 2 thought factors (Figure S2).
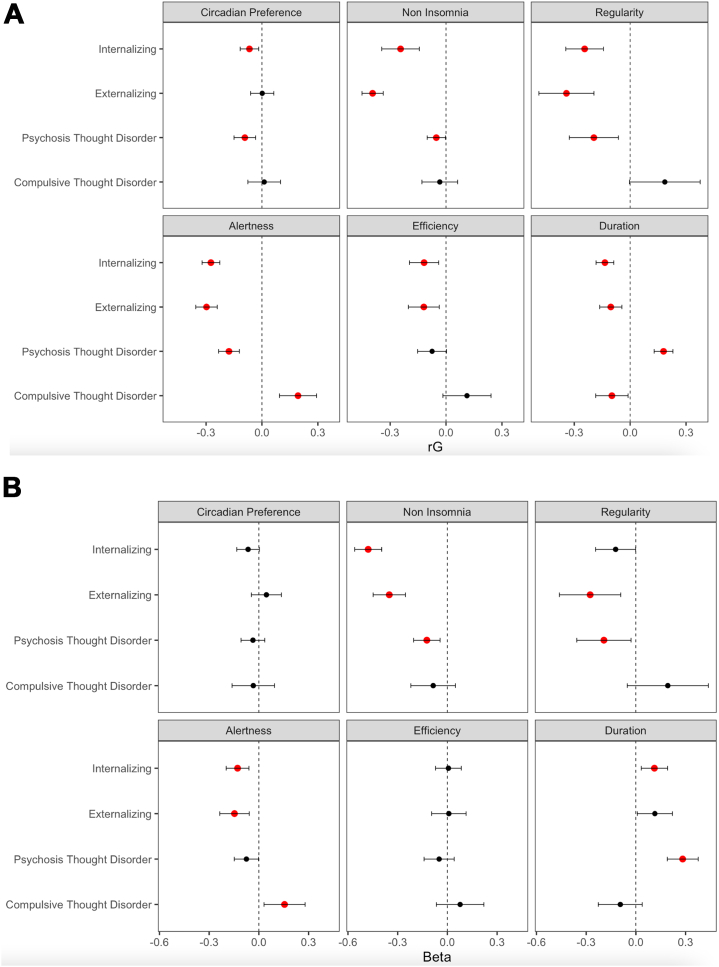
The combined model with the 6-factor sleep health structure and psychopathology structure fitted acceptably, with χ2188 = 4864.96, CFI = 0.921, and SRMR = 0.067. See Figure S3 for all sleep and psychopathology indicator genetic correlations. The latent variable correlations for the full model are shown in Figure 3A. Their directions were as expected in most cases; better sleep health (alertness, noninsomnia, longer duration, higher efficiency, morning circadian preference, and greater regularity) was associated with lower genetic risk for psychopathology, but we did find one significant exception to this trend. The compulsive thought disorder factor positively correlated with alertness (rg = −0.18, p < .001), indicating that being more alert during the day is associated with a genetic liability for compulsive thought disorders.
Figure 3.
Genetic correlations and partial regression coefficients of sleep health factors and psychopathology factors. (A) Latent correlations between sleep health and psychopathology factors. (B) Regression coefficients of sleep health factors predicting psychopathology factors (i.e., statistically controlling for covariances among sleep health factors). Error bars are 95% confidence intervals, and red coloring indicates statistical significance after false discovery rate correction in both panels.
Sleep Health and Psychopathology Latent Genetic Multiple Regressions
To test whether the relationships of the sleep health domains with psychopathology were independent of each other, we ran a multiple regression model in Genomic SEM. In this model, all psychopathology factors were regressed on all the sleep health latent factors. Because the models were statistically equivalent, the model fit was identical to that of the correlational model. As shown in Figure 3B, several associations became nonsignificant when controlled for correlations among sleep health factors: circadian preference with internalizing and psychosis thought disorders, regularity with internalizing, alertness with psychosis thought disorders, efficiency with internalizing and externalizing disorders, and duration with externalizing. Finally, the association of duration with internalizing reversed direction when controlled for other sleep health factors, but the effect size was small. Together, sleep health factors explained 28% of the internalizing psychopathology factor variance, 26% of the externalizing psychopathology factor variance, and 11% and 9%, respectively, of the psychosis and compulsive thought disorder factor variances.
Discussion
Although recent sleep GWASs (13,14,18,21) provide insight into the molecular underpinnings of specific sleep measures, genetic analyses of the overall sleep health construct are lacking. We addressed this gap by modeling the genetic correlations of multiple sleep phenotypes to create a multidimensional sleep health structure. Our results support a genetic sleep health model that parsed 12 sleep traits into 6 correlated factors. Further demonstrating divergence, the sleep health factors did not relate uniformly to psychopathology factors and were associated with independent variance in some of those psychopathology factors. These results suggest that sleep health may be best conceptualized as a family of genetically correlated but separable domains, which is important for contextualizing sleep health as a public health initiative that may ease the burden of comorbid psychiatric disorders.
As expected, our sleep health factors were generally negatively associated with psychopathology at the genetic level. Sleep health factors collectively explained the most genetic variance in the internalizing and externalizing factors, consistent with prior observations that have linked insomnia to anxiety, depression, and substance use and chronotype to depression (11, 12, 13, 14,43,44). Noninsomnia, daytime alertness, sleep duration, sleep regularity, and sleep efficiency all significantly correlated with internalizing and externalizing psychopathology such that poorer sleep health was genetically associated with higher psychopathology risk. Morning circadian preference was associated with lower internalizing and psychosis thought disorder liability, also consistent with prior literature (13).
However, we also found that some sleep health factors related differently and unexpectedly to psychopathology latent factors. Alertness showed significant and distinct patterns of associations with the psychosis and compulsive thought disorder factors. Alertness negatively correlated with psychosis thought disorders, such that lower daytime alertness was genetically associated with risk for schizophrenia or BP as expected, but positively correlated with the compulsive thought disorder factor. Post hoc model tests reported in the Supplement suggest that this relationship was driven by OCD because higher alertness was genetically associated with risk for OCD but not anorexia nervosa.
To our knowledge, an association between better sleep health and higher liability for compulsive disorders has not been observed before. In fact, prior research indicates that OCD is associated with eveningness and sleep disturbances (45, 46, 47). Our result implies the existence of common genetic factors that influence higher daytime alertness and OCD. It is possible that something specific to OCD is mediating the relationship with daytime alertness, such as a heightened cognitive awareness to focus on compulsions regardless of sleep quality. Irrespective of the explanation, these findings suggest poorer sleep health does not uniformly relate negatively to outcomes. It is also important to keep in mind that this finding reflects shared genetic risk for OCD and higher daytime alertness, not necessarily that individuals diagnosed with OCD show higher daytime alertness (i.e., a phenotypic correlation). A genetic correlation in the context of a null or opposing phenotypic correlation can be observed, such as autism and intelligence showing a small positive genetic correlation, although phenotypically, autism is associated with lower intelligence (48). Thus, genetic risk factors can diverge from environmental or phenotypic associations.
Another instance of sleep health relating differentially to psychopathology was that sleep duration positively correlated with the psychosis thought disorder factor, such that longer duration related to higher genetic liability for psychosis disorders, but negatively correlated with the compulsive thought disorder factor (and the other psychopathology factors), such that longer duration was associated with lower genetic liability for compulsive disorders (and internalizing and externalizing disorders). These findings are difficult to parse because we used a linear sleep duration conceptualization. Many psychopathologies are associated with both short and long sleep, and here, our results indicated at the genetic level that liability for psychosis disorders is associated with longer duration, while liability for compulsive disorders is associated with shorter duration.
If sleep health factors independently associate with psychopathology, it would further support the conclusion that sleep health consists of distinct domains. Thus, we ran a genomic SEM multiple regression to assess whether the sleep health factors remained associated with psychopathology factors, controlling for their intercorrelations. We found that several sleep health factors were independently associated with psychopathology. This pattern is inconsistent with a general unitary concept of sleep health. In particular, regularity was more strongly related to externalizing than internalizing, and noninsomnia, duration, and alertness divergently related to psychosis and compulsive thought disorders. These findings are notable given the genetic overlap among the 2 thought disorder factors (rg = 0.43) and among internalizing and externalizing psychopathology (rg = 0.67).
Sleep disturbance has been proposed as a transdiagnostic risk factor for psychopathology (12,49). Our results partially support this proposal but suggest that the genomic relationships between sleep health domains and psychopathology are more complex than would be predicted by a simple model in which better cumulative sleep health relates to lower risk of psychopathology. While most sleep health domains were significantly associated with more than one domain of psychopathology, the patterns differed across sleep health domains and psychopathology factors, leading to a poor fit for the model and associating a higher-order sleep health factor with psychopathology factors. However, poorer sleep health is still related to many negative outcomes and targeting improvements in sleep health will likely benefit other areas of health. The finding that sleep health domains show different patterns of association with different aspects of psychopathology suggests that sleep interventions tailored to specific types of psychopathology merit investigation.
Limitations and Future Directions
Our results support the conclusion that sleep health and psychopathology share genetic variance, but not necessarily that improving sleep health will reduce psychopathology symptoms (or vice versa). Quasi-experimental designs such as co-twin control and Mendelian randomization (15), as well as the gold standard randomized controlled clinical trials could be used to examine directionality and causality.
Modeling genetic correlations is informative but is limited in that it does not explore genetic pathways involved in these traits. We plan to leverage this work by performing GWASs on our sleep health factors to investigate genetic variants common to the factor that may not have reached significance in GWASs for each sleep measure individually.
Although the recent increase in large-scale GWASs and public summary data allowed us to investigate numerous sleep and psychopathology traits, the number of independent GWASs was also a limiting factor. Because many of the sleep phenotypes were derived from the same measurements (efficiency and actigraphy sleep duration, self-reported sleep duration, and long and short sleep duration), their inclusion in the same SEM model would lead to problems. The limited number of phenotypes also required us to include multiple single-indicator factors in our data (noninsomnia, duration, and regularity). While these single-indicator variables do not add much beyond the original GWASs, considering them in conjunction with the other sleep health domains and psychopathology is nevertheless informative about the genetic structure of sleep health.
Future studies could also implement more robust psychopathology factors once more data are available. Similarly, because some of the clinical phenotypes are case/control GWASs, we could not assess the relationship between severity of symptoms and sleep health.
Finally, owing to the complex structure of genetic data, we were able to relate findings only to those of European ancestry. Future GWASs that include more diverse samples are needed to ensure that these results are representative and benefit all individuals (50).
Conclusions
Sleep is a crucial and modifiable health behavior related to a host of negative outcomes. Thus, it is important to understand the etiology and nature of these relationships. Our results show that sleep health is best represented by multiple distinct genetic factors and that these sleep health factors differentially relate to psychopathology factors. Incorporating the many unique aspects of sleep health may aid in disentangling the relationship sleep has with mental health and other health outcomes.
Acknowledgments and Disclosures
This research was supported by grants from the National Institutes of Health: CLM is supported by Grant No. MH016880; EAW is supported by Grant Nos. MH015442 and DA017637; NPF is supported by Grant Nos. DA046064, DA046413, DA051018, DA042742, MH117131, HD078532, and AG046938.
KPW reports research support/donated materials from DuPont Nutrition & Biosciences, Grain Processing Corporation, and FrieslandCampina Innovation Centre. KPW reports consulting with or without receiving fees and/or serving on the advisory boards for Circadian Therapeutics, Circadian Biotherapies, Inc., and the United States Army Medical Research and Development Command–Walter Reed Army Institute of Research. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2022.07.002.
Supplementary Material
References
- 1.Grandner M.A. In: Sleep and Health. Grandner M.A., editor. Academic Press; Cambridge: 2019. Social-ecological model of sleep health; pp. 45–53. [Google Scholar]
- 2.Murawski B., Wade L., Plotnikoff R.C., Lubans D.R., Duncan M.J. A systematic review and meta-analysis of cognitive and behavioral interventions to improve sleep health in adults without sleep disorders. Sleep Med Rev. 2018;40:160–169. doi: 10.1016/j.smrv.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Buysse D.J. Sleep health: Can we define it? Does it matter? Sleep. 2014;37:9–17. doi: 10.5665/sleep.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ravyts S.G., Dzierzewski J.M., Perez E., Donovan E.K., Dautovich N.D. Sleep health as measured by RU SATED: A psychometric evaluation. Behav Sleep Med. 2021;19:48–56. doi: 10.1080/15402002.2019.1701474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koinis-Mitchell D., Kopel S.J., Boergers J., McQuaid E.L., Esteban C.A., Seifer R., et al. Good sleep health in urban children with asthma: A risk and resilience approach. J Pediatr Psychol. 2015;40:888–903. doi: 10.1093/jpepsy/jsv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalmases M., Benítez I.D., Mas A., Garcia-Codina O., Medina-Bustos A., Escarrabill J., et al. Assessing sleep health in a European population: Results of the Catalan Health Survey 2015. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace M.L., Yu L., Buysse D.J., Stone K.L., Redline S., Smagula S.F., et al. Multidimensional sleep health domains in older men and women: An actigraphy factor analysis. Sleep. 2021;44:zsaa181. doi: 10.1093/sleep/zsaa181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory A.M., Sadeh A. Sleep, emotional and behavioral difficulties in children and adolescents. Sleep Med Rev. 2012;16:129–136. doi: 10.1016/j.smrv.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Benham G. Sleep: An important factor in stress-health models. Stress Health. 2010;26:204–214. [Google Scholar]
- 10.Coulombe J.A., Reid G.J., Boyle M.H., Racine Y. Concurrent associations among sleep problems, indicators of inadequate sleep, psychopathology, and shared risk factors in a population-based sample of healthy ontario children. J Pediatr Psychol. 2010;35:790–799. doi: 10.1093/jpepsy/jsp097. [DOI] [PubMed] [Google Scholar]
- 11.Tkachenko O., Olson E.A., Weber M., Preer L.A., Gogel H., Killgore W.D.S. Sleep difficulties are associated with increased symptoms of psychopathology. Exp Brain Res. 2014;232:1567–1574. doi: 10.1007/s00221-014-3827-y. [DOI] [PubMed] [Google Scholar]
- 12.Wainberg M., Jones S.E., Beaupre L.M., Hill S.L., Felsky D., Rivas M.A., et al. Association of accelerometer-derived sleep measures with lifetime psychiatric diagnoses: A cross-sectional study of 89,205 participants from the UK Biobank. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S.E., Lane J.M., Wood A.R., van Hees V.T., Tyrrell J., Beaumont R.N., et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10:343. doi: 10.1038/s41467-018-08259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane J.M., Jones S.E., Dashti H.S., Wood A.R., Aragam K.G., van Hees V.T., et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51:387–393. doi: 10.1038/s41588-019-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman N.P., Banich M.T., Keller M.C. Twin studies to GWAS: There and back again. Trends Cogn Sci. 2021;25:855–869. doi: 10.1016/j.tics.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Loughlin J., Casanova F., Jones S.E., Hagenaars S.P., Beaumont R.N., Freathy R.M., et al. Using Mendelian Randomisation methods to understand whether diurnal preference is causally related to mental health. Mol Psychiatry. 2021;26:6305–6316. doi: 10.1038/s41380-021-01157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dashti H.S., Jones S.E., Wood A.R., Lane J.M., van Hees V.T., Wang H., et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10:1100. doi: 10.1038/s41467-019-08917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grotzinger A.D., Rhemtulla M., de Vlaming R., Ritchie S.J., Mallard T.T., Hill W.D., et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav. 2019;3:513–525. doi: 10.1038/s41562-019-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armitage R., Trivedi M., Hoffmann R., Rush A.J. Relationship between objective and subjective sleep measures in depressed patients and healthy controls. Depress Anxiety. 1997;5:97–102. doi: 10.1002/(sici)1520-6394(1997)5:2<97::aid-da6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang H., Lane J.M., Jones S.E., Dashti H.S., Ollila H.M., Wood A.R., et al. Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat Commun. 2019;10:3503. doi: 10.1038/s41467-019-11456-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashti H.S., Daghlas I., Lane J.M., Huang Y., Udler M.S., Wang H., et al. Genetic determinants of daytime napping and effects on cardiometabolic health. Nat Commun. 2021;12:900. doi: 10.1038/s41467-020-20585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones S.E., van Hees V.T., Mazzotti D.R., Marques-Vidal P., Sabia S., van der Spek A., et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun. 2019;10:1585. doi: 10.1038/s41467-019-09576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nievergelt C.M., Maihofer A.X., Klengel T., Atkinson E.G., Chen C.Y., Choi K.W., et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558. doi: 10.1038/s41467-019-12576-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purves K.L., Coleman J.R.I., Meier S.M., Rayner C., Davis K.A.S., Cheesman R., et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2020;25:3292–3303. doi: 10.1038/s41380-019-0559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard D.M., Adams M.J., Clarke T.K., Hafferty J.D., Gibson J., Shirali M., et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson E.C., Demontis D., Thorgeirsson T.E., Walters R.K., Polimanti R., Hatoum A.S., et al. A large-scale genome-wide association study meta-analysis of cannabis use disorder [published correction appears in Lancet Psychiatry 2022; 9:e12] Lancet Psychiatry. 2020;7:1032–1045. doi: 10.1016/S2215-0366(20)30339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullins N., Forstner A.J., O’Connell K.S., Coombes B., Coleman J.R.I., Qiao Z., et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–829. doi: 10.1038/s41588-021-00857-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Obsessive Compulsive Disorder Foundation Genetics Collaborative (IOCDF-GC) and OCD Collaborative Genetics Association Studies (OCGAS) Revealing the complex genetic architecture of obsessive–compulsive disorder using meta-analysis. Mol Psychiatry. 2018;23:1181–1188. doi: 10.1038/mp.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson H.J., Yilmaz Z., Thornton L.M., Hübel C., Coleman J.R.I., Gaspar H.A., et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51:1207–1214. doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu M., Jiang Y., Wedow R., Li Y., Brazel D.M., Chen F., et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Roige S., Palmer A.A., Fontanillas P., Elson S.L., 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, Adams MJ, et al. Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176:107–118. doi: 10.1176/appi.ajp.2018.18040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu L., Bentler P.M. Cutoff criteria for fit indexes in covariance structure analysis: Conventional criteria versus new alternatives. Struct Equ Model. 1999;6:1–55. [Google Scholar]
- 37.Fritz J., Phillips A.J.K., Hunt L.C., Imam A., Reid K.J., Perreira K.M., et al. Cross-sectional and prospective associations between sleep regularity and metabolic health in the Hispanic Community Health Study/Study of Latinos. Sleep. 2021;44:zsaa218. doi: 10.1093/sleep/zsaa218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caspi A., Houts R.M., Belsky D.W., Goldman-Mellor S.J., Harrington H., Israel S., et al. The p factor: One general psychopathology factor in the structure of psychiatric disorders? Clin Psychol Sci. 2014;2:119–137. doi: 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldman I.D., Poore H.E., Luningham J.M., Yang J. Testing structural models of psychopathology at the genomic level. World Psychiatry. 2020;19:350–359. doi: 10.1002/wps.20772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross-Disorder Group of the Psychiatric Genomics Consortium Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179:1469–1482.e11. doi: 10.1016/j.cell.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grotzinger A.D., Mallard T.T., Akingbuwa W.A., Ip H.F., Adams M.J., Lewis C.M., et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat Genet. 2022;54:548–559. doi: 10.1038/s41588-022-01057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smoller J.W., Andreassen O.A., Edenberg H.J., Faraone S.V., Glatt S.J., Kendler K.S. Psychiatric genetics and the structure of psychopathology [published correction appears in Mol Psychiatry 2019; 24:471. Mol Psychiatry. 2019;24:409–420. doi: 10.1038/s41380-017-0010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson M., Munafò M.R., Taylor A.E., Treur J.L. Evidence for genetic correlations and bidirectional, causal effects between smoking and sleep behaviors. Nicotine Tob Res. 2019;21:731–738. doi: 10.1093/ntr/nty230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong M.M., Brower K.J., Zucker R.A. Childhood sleep problems, early onset of substance use and behavioral problems in adolescence [published correction appears in Sleep Med 2010; 11:110–111] Sleep Med. 2009;10:787–796. doi: 10.1016/j.sleep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox R.C., Olatunji B.O. Circadian rhythms in obsessive–compulsive disorder: Recent findings and recommendations for future research. Curr Psychiatry Rep. 2019;21:54. doi: 10.1007/s11920-019-1033-0. [DOI] [PubMed] [Google Scholar]
- 46.Cox R.C., Olatunji B.O. Sleep disturbance and obsessive–compulsive symptoms: Results from the national comorbidity survey replication. J Psychiatr Res. 2016;75:41–45. doi: 10.1016/j.jpsychires.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Cox R.C., Tuck B., Olatunji B.O. The role of eveningness in obsessive–compulsive symptoms: Cross-sectional and prospective approaches. J Affect Disord. 2018;235:448–455. doi: 10.1016/j.jad.2018.04.060. [DOI] [PubMed] [Google Scholar]
- 48.Crespi B.J. Autism as a disorder of high intelligence. Front Neurosci. 2016;10:300. doi: 10.3389/fnins.2016.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harvey A.G., Murray G., Chandler R.A., Soehner A. Sleep disturbance as transdiagnostic: Consideration of neurobiological mechanisms. Clin Psychol Rev. 2011;31:225–235. doi: 10.1016/j.cpr.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harden K.P., Koellinger P.D. Using genetics for social science. Nat Hum Behav. 2020;4:567–576. doi: 10.1038/s41562-020-0862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.