Abstract
To elucidate the functions of rhabdovirus matrix (M) protein, we determined the localization of M in rabies virus (RV) and analyzed the properties of an M-deficient RV mutant. We provide evidence that M completely covers the ribonucleoprotein (RNP) coil and keeps it in a condensed form. As determined by cosedimentation experiments, not only the M-RNP complex but also M alone was found to interact specifically with the glycoprotein G. In contrast, an interaction of G with the nucleoprotein N or M-less RNP was not observed. In the absence of M, infectious particles were mainly cell associated and the yield of cell-free infectious virus was reduced by as much as 500,000-fold, demonstrating the crucial role of M in virus budding. Supernatants from cells infected with the M-deficient RV did not contain the typical bullet-shaped rhabdovirus particles but instead contained long, rod-shaped virions, demonstrating severe impairment of the virus formation process. Complementation with M protein expressed from plasmids rescued rhabdovirus formation. These results demonstrate the pivotal role of M protein in condensing and targeting the RNP to the plasma membrane as well as in incorporation of G protein into budding virions.
Matrix (M) proteins of negative-strand RNA viruses constitute major structural components of the virus and are believed to be required for virus assembly and budding. For rhabdoviruses, the first insight into the function of M protein was obtained by treating vesicular stomatitis virus (VSV) with nonionic detergent and analyzing intact VSV nucleocapsid-M complexes. Isolated subviral structures, which were named skeletons, resembled intact virions, despite their increased length and smaller diameter (3, 22). These results suggested that the viral assembly function is due to the ability of M to bind and probably condense the nucleocapsid (reviewed in reference 14). Most importantly, skeleton-like structures were also observed near the plasma membranes of VSV-infected cells, showing that these structures represent an intermediate between naked nucleocapsid and mature virions (24). VSV M has been also shown to bind to the plasma membrane even in the absence of other viral proteins, and such membrane-associated M was also able to bind to the nucleocapsid of VSV in vitro (4, 5). These properties of M protein make it likely that M functions in the viral assembly and budding process by acting as a bridge between the nucleocapsid and the plasma membrane.
In influenza virus, the matrix protein (M1) is proposed to surround the nucleocapsid (26), and it was suggested that M1 interacts with the membrane and perhaps also with the spike proteins. In contrast, VSV M protein was suggested to be inside the nucleocapsid coil and accessible to make contact with the lipid bilayer only at the extreme ends (1). This is in contrast to the more generally accepted view that M is responsible for the condensation of the nucleocapsid from outside and that it is localized between the nucleocapsid coil and the membrane (33). For both VSV and influenza virus, due to the lack of clear labeling with antibodies, no conclusive data on the localization of M have been obtained.
The genome organization and virion structure of rabies virus (RV), the prototype of the Lyssavirus genus within the family Rhabdoviridae, are highly similar to those of VSV. In both viruses, the nonsegmented negative-strand RNA together with the nucleoprotein (N), phosphoprotein (P), and polymerase (L) forms a helical ribonucleoprotein (RNP) complex. In the virion, the RNP is enwrapped by a lipid bilayer containing the single transmembrane spike glycoprotein (G) and, ostensibly, the matrix protein (M). Although the central role of M proteins of nonsegmented negative-strand RNA viruses in virus assembly and budding process appears to be obvious, direct experimental evidence for their functions has not been provided. This was mainly due to the lack of systems that would allow direct genetic manipulation of nonsegmented negative-strand RNA virus genomes. These technical difficulties have been overcome by the development of reverse genetics systems allowing recovery of viruses from cDNA, as first described for RV (31), which are now being employed for generation of nonsegmented negative-strand RNA virus mutants (reviewed in reference 9).
Recently, we have shown that spikeless rhabdovirus-shaped particles are released from cells infected with a G-deficient mutant, albeit at a 30-fold lower efficiency (21). This demonstrated that RNPs associated with M protein can be enwrapped with a membrane and bud at the cell surface. For VSV, it was shown that M is able to induce budding of vesicles in the absence of other VSV proteins (13, 15), indicating that M protein is able to pinch off membranes autonomously. Apart from M protein, rhabdovirus G proteins were also shown to contribute to the budding efficiency and even to be able to mobilize RNAs or RNPs (21, 25). Therefore, highly efficient budding of rhabdoviruses is achieved by a concerted action of both spikes and cores.
Another role of M protein in virion formation is its involvement in virion morphogenesis. It was shown that spherical particles were released from cells infected by a temperature-sensitive M mutant of VSV at the nonpermissive temperature (16). In addition, the involvement of M in determining the formation of spherical or filamentous particles was documented for influenza viruses (34), suggesting a pivotal role of M protein in virion morphogenesis.
Apart from their roles in virus assembly and budding, M proteins of rhabdoviruses, influenza viruses, and paramyxoviruses were reported to inhibit viral transcription (2, 6, 12, 18, 35, 37). Subgenomic mRNAs of these groups of viruses are transcribed from RNPs which also serve as templates for the replication of full-length RNAs. In an in vitro transcription assay with isolated RNPs and M protein of VSV, the transcription inhibition activity of the M protein was shown to be due to reassociation of M protein with RNP cores (2).
In this report, based on biochemical studies and reverse genetics approaches, we show the localization of M protein in mature RV and describe properties of an RV mutant lacking the entire M gene. The results demonstrate that M lies between the lipid bilayer and the RNP coil and that in the absence of M protein the virus assembly and budding process is severely impaired. Characterization by electron microscopy showed poor release of only rod-shaped particles or round vesicular structures but no bullet-shaped viruses. Subsequent expression of M protein from plasmid DNA in cells infected with the M-deficient mutant rescued high titers of infectivity and resulted in the release of typical bullet-shaped particles, demonstrating that RV M protein is responsible for the formation of virions with a bullet-like morphology. Moreover, we provide evidence for a direct interaction of G protein with M.
MATERIALS AND METHODS
Detergent treatment of virions and immunoelectron microscopy.
Approximately 106 and 107 BSR cells were infected with SAD L16 and SAD ΔM, respectively, at a multiplicity of infection (MOI) of 1. Approximately 5 × 108 SAD L16 virions were treated with either 0.05% Triton X-100 or 20 mM octyl-glucoside for 15 min at room temperature. Detergent-treated or untreated control samples were fixed with 0.5% glutaraldehyde for 30 min at 4°C and purified by centrifugation through 20% sucrose on a 60% sucrose cushion in a Beckman SW41 rotor at 27,000 rpm for 90 min. The interphase was then collected by side puncture. Carbon-coated nickel grids were deposited for 7 min on a drop of the virus suspension and washed four times with phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin. Samples were incubated for 45 min with anti-RV G monoclonal antibody (MAb) E543 (27), a monospecific anti-M polyclonal rabbit serum (S66), or a rabbit serum raised against purified RV RNP (S50), recognizing RV N and P proteins (21). Immunostaining was performed by incubating the samples with goat anti-mouse or goat anti-rabbit immunoglobulin G antibody (Biocell, Cardiff, United Kingdom) coupled to 10-nm gold particles. After being washed with PBS and distilled water, the samples were negatively stained with 1% uranyl acetate (unbuffered) and examined in a Zeiss 109 electron microscope. Micrographs were taken at an instrumental magnification of ×50,000 on Agfa Scientia 23D56 film.
Construction of a cDNA clone.
To delete the entire M protein-coding region from the RV genome, the following manipulations were performed with the full-length cDNA clone pSAD L16 (31), which possesses the authentic sequence of the RV strain SAD B19 (7). First, subclone pSKpm24, containing a 2.7-kb EcoRI-XhoI fragment of pSAD L16, representing SAD B19 nucleotides 1112 to 3823, was generated. By digestion of pSKpm24 with PflMI and XbaI, subsequent filling in by Klenow enzyme, and religation, a cDNA fragment comprising SAD B19 nucleotides 2435 to 3176 was removed. A BstBI fragment of the modified pSKpm24 (SAD B19 positions 1498 to 3739) was then isolated and used to replace the corresponding fragment of pSAD L16 to give rise to pSAD ΔM.
Recovery of M-deficient RV.
Transfection experiments were carried out as described previously (10). Approximately 106 BSR cells were infected with the recombinant vaccinia virus vTF7-3 (11) and then transfected with a plasmid mixture containing 5 μg of pT7T-N, 2.5 μg of pT7T-P, 2.5 μg of pT7T-L, 2 μg of pT7T-M, and 4 μg of pSAD ΔM by using the Stratagene mammalian transfection kit (CaPO4 protocol). Isolation of the transfectant virus and removal of vaccinia virus were carried out as described previously (31). For passage of SAD ΔM, which required complementation with M protein to be effective, cells were incubated with the clarified supernatant, infected with vTF7-3, and transfected with 2 μg of pT7T-M as described above. For detection of replicating RV, infected cells were fixed with 80% acetone and stained with a conjugate containing a mixture of MAbs directed to RV N protein (Centocor). Stocks of phenotypically complemented SAD ΔM were prepared after seven or eight serial passages. Infectious titers of RV and M-complemented SAD ΔM were determined by serial 10-fold dilution and staining of infected foci or syncytia with the N conjugate. For noncomplemented SAD ΔM, the undiluted supernatant was used for infection of cells, and fluorescent syncytia were counted at 24 h postinfection.
Analysis of RNA and RT-PCR.
Total RNA isolated from infected cells was electrophoresed on denaturing gels and analyzed by Northern hybridizations as described previously (8). RV N or M gene cDNA fragments were labeled with 32P by nick translation (Amersham nick translation kit). Reverse transcription-PCR (RT-PCR) was performed on 1 μg of total RNA from infected cells. Reverse transcription by avian myeloblastosis virus reverse transcriptase was primed by an RV P gene-specific oligonucleotide, NS1P (5′-GTCGAATCCGACAAGCTG-3′; SAD B19 nucleotides 2345 to 2362). DNA amplification was done with primer NS1P and a G gene-specific primer, G4M (5′-GGGTACAAACAGGACAGC-3′; SAD B19 nucleotides 3329 to 3346), or an M gene-specific primer, M4M (5′-TTGCAATCCGACGAACTC-3′). The PCR products resulting from 30 cycles (denaturation for 30 s at 94°C, annealing for 60 s at 45°C, and elongation for 120 s at 72°C) were analyzed on 1% agarose gels and used directly for sequencing.
Metabolic labeling and immunoprecipitation of proteins.
For surface immunoprecipitation, approximately 106 BSR cells were infected with recombinant RVs at an MOI of 1. After 16 h of infection, cells were labeled with 100 μCi of [35S]methionine (1,365 Ci/mmol; ICN) for 3 h. The labeled cells were then incubated with a MAb directed to RV G protein for 45 min at 4°C. Antigen extraction and immunoprecipitation were performed as described previously (21). Immunoprecipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and quantitated with a phosphorimager (Fuji BAS1500).
Sucrose gradient centrifugation and protein analysis.
For analysis of cell-free virions, approximately 106 and 107 cells were infected with SAD L16 and SAD ΔM, respectively, at an MOI of 1. For velocity centrifugation, supernatants were collected at 2 days postinfection and layered on 5 to 25% sucrose gradients prepared in TEN buffer (10 mM Tris [pH 7.4], 50 mM NaCl, 1 mM EDTA). The gradients were centrifuged at 31,500 rpm in an SW41 rotor for 30 min. To analyze the protein composition of cell-associated infectious particles, equal amounts of infected cells were washed five times with PBS and harvested by scraping the monolayer in TEN buffer, pH 7.4. Cells were disrupted by sonication for 10 s at 100 W with a Sonifier B12 (Branson Sonic Power Company), and the lysates were clarified by centrifugation at 5,000 × g for 5 min at 4°C. For isopycnic centrifugation, clarified cell lysates or supernatants containing released particles were layered over 10 to 70% continuous sucrose gradients and centrifuged in an SW41 rotor at 35,000 rpm for 18 h. Equal amounts of 12 fractions were collected, and an aliquot from each fraction was used to determine the refractive index and the titer of infectious particles by end point dilution. Virus proteins from gradient fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and incubated with a mixture of rabbit sera directed against RV proteins as described previously (21). Proteins were visualized after incubation with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Dianova) by using the ECL Western blot detection kit (Amersham) (1-min incubation) and exposure to X-ray films (Biomax MR; Kodak).
RESULTS
RV M is located beneath the lipid bilayer and surrounds the RNP.
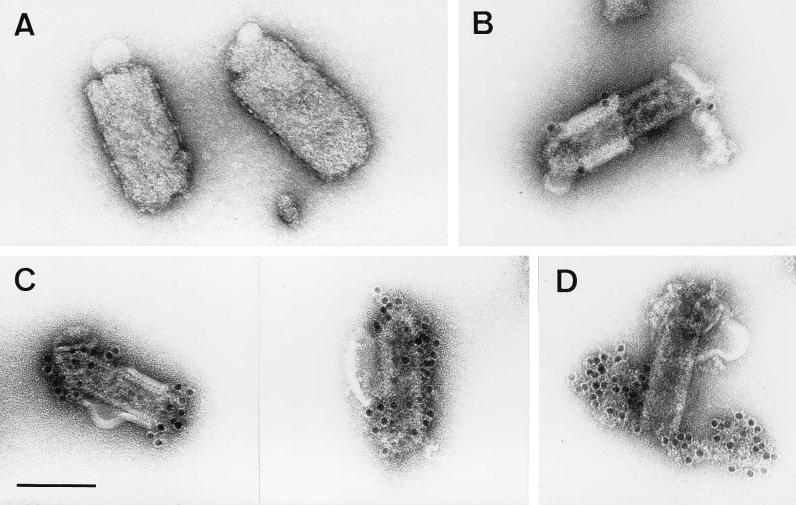
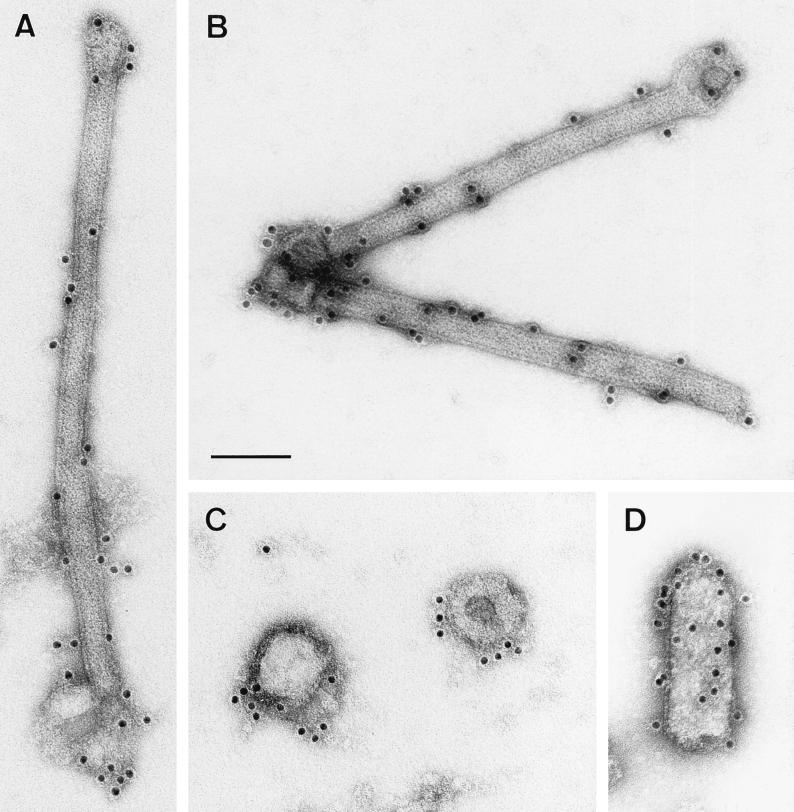
The virtual dogma that matrix proteins underlie the viral lipid bilayer was recently challenged by results obtained in electron microscopic studies of VSV, the prototype rhabdovirus. It was suggested that VSV M is inside the RNP coil and that only the part of M that protrudes from the RNP ends is able to contact the lipid bilayer (1). To determine the localization of M in RV virions, supernatants from infected-cell cultures were harvested 24 h after infection, treated with detergent, and fixed with glutaraldehyde. Virions were then purified over sucrose gradients. Electron microscopic examination of untreated control samples which were incubated with a monospecific anti-M polyclonal rabbit serum did not show labeling on the outer surface, confirming that M protein does not protrude through the virus membrane (Fig. 1A). Treatment of virions with 0.05% Triton X-100 removed the lipid bilayer together with the surface transmembrane glycoprotein and resulted in subviral structures with the same lengths as intact RV but with smaller diameters (50 to 60 nm) than intact RV (90 to 100 nm) (Fig. 1). Similar structures of VSV that was treated with octyl-glucoside have been named skeletons (3), and we also use this term for the subviral RV structures obtained after treatment with Triton X-100. The shapes of the structures observed ranged from intact skeletons with residual lipid bilayers (Fig. 1B) to partially disassembled ones in which RNP extended out of the skeleton (Fig. 1D). This heterogeneity helped us to determine the locations of virus proteins in the different structures. By using an anti-G MAb, no gold labeling of skeletons was detected, except for areas where the lipid bilayer was not completely removed (Fig. 1B). To analyze the localization of M protein in the subviral structure, the treated particles were labeled with the monospecific anti-M serum and examined by electron microscopy. Intensive labeling of intact skeletons was observed in areas completely devoid of membranes, demonstrating that RV M lies beneath the lipid bilayer and covers the nucleocapsid (Fig. 1C). In partially disassembled skeletons, M labeling was not detected on the uncoiled RNP, except at the unravelling ends of skeletons (not shown). Incubation of such partially disassembled skeletons with a rabbit serum raised against purified RNP resulted in intense labeling of the unwound nucleocapsid, but no gold labeling was observed on the surface of the intact skeleton (Fig. 1D). Obviously, RV M protein covers the entire surface of condensed RNPs. Even upon removal of the membrane, M protein is associated with the nucleocapsid and maintains it in a compact form. Upon partial removal of M, the RNP coil is released at one or both ends of the skeleton (Fig. 1D). Intensive treatment with the detergent resulted in a complete disassembly of the skeleton structure (not shown), demonstrating that M protein imposes a definite shape by maintaining the condensation of the RNP coil.
FIG. 1.
RV M protein is positioned between the lipid bilayer and the RNP coil. Virions treated with Triton X-100 (B to D) and untreated control samples (A) were purified as described in Materials and Methods and analyzed by electron microscopy. The absence of anti-M labeling on the outer membranes of untreated virions (90 to 100 nm in diameter) (A) and an intense anti-M labeling on the surfaces of the RNP coil or skeletons (50 to 60 nm in diameter) of detergent-treated virions (C) demonstrate that M surrounds the RNP coil. Only areas where the lipid bilayer was not completely removed (60 to 70 nm in diameter) are weakly labeled with anti-G MAb (B). In partially disassembled skeletons (D), only the uncoiled nucleocapsid, and not the surface of the skeleton, is labeled with anti-RNP serum. Bar, 100 nm.
Generation of an M-deficient RV mutant.
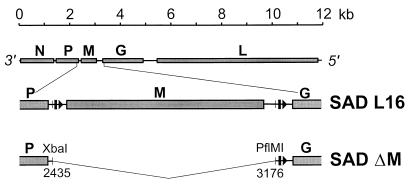
Analysis of genetically engineered viruses that are completely devoid of a particular gene and the ability to trans complement the essential functions have proved useful in investigating the roles of viral proteins. In order to address the functions of RV M protein in virion formation, we constructed a cDNA in which the entire RV M gene sequence (SAD B19 positions 2435 to 3176) was deleted (Fig. 2). To rescue the modified cDNA into virus, we used a previously described system (31) in which T7 RNA polymerase transcripts corresponding to RV antigenome RNA are assembled into transcriptionally active RNPs in cells that express RV N, P, and L proteins. Not unexpectedly, it was not possible to recover infectious M-deficient virions by using the standard protocol, indicating severe impairment of virus formation in the absence of M protein. We therefore complemented the defects during recovery experiments by expression of M protein. In addition to the plasmids encoding RV N, P, and L proteins and the antigenome of the M-deficient mutant (SAD ΔM), cell cultures were transfected with pT7T-M, encoding the SAD B19 M protein (10). After incubation for 2 days, the presence of virus was detected by direct immunofluorescence with an anti-N conjugate. Stocks of cell-free phenotypically complemented SAD ΔM were prepared after seven or eight passages in cells transiently expressing RV M protein from transfected plasmids. The recombinant vaccinia virus vTF7-3 expressing T7 RNA polymerase was then removed from the resulting supernatant by filtration.
FIG. 2.
Organization of the M-deficient RV genome. The entire RV genome with its five open reading frames (top), a detailed M cistron region (middle), and the region encompassing a deletion (bottom) are shown. Arrowheads and bars represent transcriptional start and stop signals, respectively. In SAD ΔM, the entire M cistron (SAD L16 nucleotides 2435 to 3176) was removed by a deletion starting within the nontranslated region of the P gene and ending close to the transcriptional stop signal of the M gene.
To first confirm the absence of the M gene in SAD ΔM, total RNA was isolated from cells infected with phenotypically complemented SAD ΔM virions, and RT-PCR was performed. By using the P gene-specific primer NS1P and the G gene-specific primer G4M, DNA fragments of ∼990 and ∼250 bp were obtained from the genomes of SAD L16 and SAD ΔM, respectively (not shown). The size difference of ∼740 bp corresponded to the deletion introduced into the cDNA. In addition, by using primer NS1P and an M gene-specific primer, M4M, a DNA fragment of ∼600 bp was obtained from the genome of SAD L16, but no product from the SAD ΔM genome was detected. The identity of the recombinant SAD ΔM virus was further verified by sequencing the PCR products and by Northern blotting of total RNA from cells infected with SAD ΔM or SAD L16. With a probe spanning the entire coding region of the RV M gene, no hybridization of SAD ΔM RNAs was observed in Northern blots, once again confirming the absence of the M gene (not shown). As determined with a phosphorimager, approximately twofold more N mRNA was reproducibly present in SAD ΔM-infected cells than in SAD L16-infected cells, which might in part reflect alleviation of the transcription-inhibitory activity of rhabdovirus M protein (2, 6, 12).
SAD ΔM induces increased cell-cell fusion.
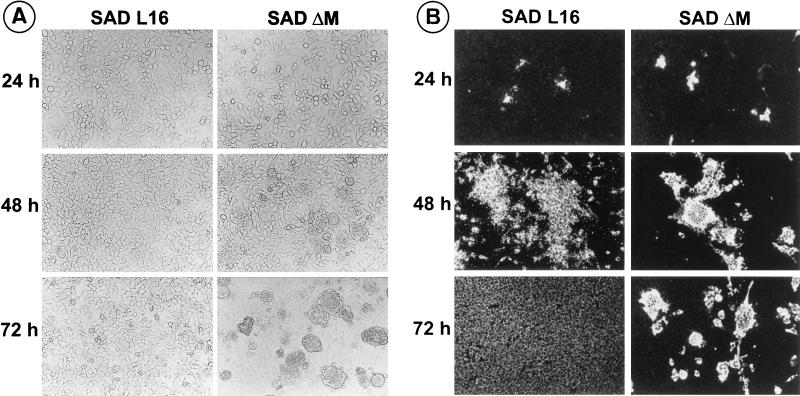
To investigate the growth characteristics of SAD ΔM in the absence of RV M protein, cells were infected with phenotypically complemented SAD ΔM virions at an MOI of 1 and examined by immunofluorescence at various times after infection. Interestingly, cells infected with SAD ΔM showed a marked cell-cell fusion activity already after 24 h postinfection. In contrast, cells infected in parallel with SAD L16 appeared unchanged even until 72 h of infection. To analyze this unexpected effect of SAD ΔM, infection was done at a lower MOI (0.005), and both live and fixed cells were examined for morphological changes and viral protein expression. As shown in Fig. 3A, increased cell-cell fusion was observed in SAD ΔM-infected cells at 48 h postinfection. Fused cells started to detach from the monolayer and float in the supernatant at 72 h postinfection. In contrast, cells infected with the wild-type virus did not show much alteration even at 72 h of infection.
FIG. 3.
SAD ΔM induces increased cell-cell fusion and cell death. (A) Following infection of cultures with an MOI of 0.005, live cells were examined microscopically for morphological changes at the indicated times. Cell fusion is detected in SAD ΔM-infected cells after 48 h, and large syncytia are present at 72 h postinfection. (B) In cells infected in parallel, the spread of infection was monitored at the indicated times by direct immunofluorescence with a conjugate directed against RV N protein. Secondary infection of cells, as indicated by the appearance of fluorescing granules, is virtually absent in SAD ΔM. After 48 h, SAD ΔM N expression is found in large syncytia.
To compare the spreads of SAD ΔM and SAD L16, infected-cell cultures were fixed with acetone and examined by direct immunofluorescence with an anti-N conjugate (Fig. 3B). In cultures incubated with SAD L16, primary infection of single cells spread to neighboring cells within 24 h, as indicated by the appearance of small fluorescent granules. Large foci of approximately 50 to 100 fluorescing cells were observed at 48 h postinfection, and the entire cell monolayer was infected at 72 h. In contrast, the typical small fluorescent granules of early RV infection were entirely absent at 24 h postinfection in cultures incubated with SAD ΔM. At 48 h after infection, cells expressing RV N protein were found to fuse with neighboring cells and to form large multinucleated syncytia, which is not a characteristic property of a standard RV infection. At 72 h, these syncytia start to break into small pieces and either remain on the monolayer or float in the supernatant (Fig. 3).
G protein accumulates to high levels on the surfaces of cells infected with SAD ΔM virus.
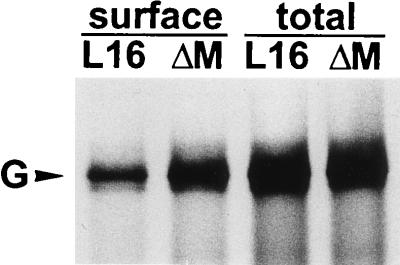
Previous results obtained with a G-deficient RV mutant showed that infection of cells with supernatant virus and cell-to-cell spread of RV absolutely depend upon the presence of the G protein (21). Since the rate of intracellular transport and accumulation of the G protein at the cell surface may affect the growth characteristics of RV, cells infected with SAD ΔM and SAD L16 were analyzed for the amount of G protein expressed at the cell surface. After 16 h of infection, cells were labeled with 100 μCi of [35S]methionine for 3 h, incubated with an anti-G MAb, and analyzed by surface immunoprecipitation (Fig. 4). As quantitated with a phosphorimager, the total amount of G protein in SAD ΔM-infected cells was only 3% more than that in SAD L16-infected cells. However, the amount of G protein present on the surfaces of infected cells was 2.6-fold more for SAD ΔM-infected cells. The cell surface G was 13 and 33% of the total G content for SAD L16 and SAD ΔM, respectively. Since G is the only RV protein that has a fusion activity, the enhanced cell surface accumulation of G protein may be responsible for the increased cell-cell fusion in SAD ΔM-infected cells.
FIG. 4.
Analysis of cell surface expression of G proteins. Approximately 106 BSR cells were infected at an MOI of 1 and after 16 h were labeled with 100 μCi of [35S]methionine for 3 h. Anti-G MAb was incubated with live cells to bind G proteins expressed at the cell surface or with cell lysates containing total G protein. Phoshorimager quantitation revealed a 2.6-fold-higher level of G protein on the surfaces of cells infected by SAD ΔM.
M is the driving force in virus assembly and budding.
The characteristic pattern of spread observed for SAD ΔM in cell culture and the high-level accumulation of G protein on the surfaces of SAD ΔM-infected cells suggested a defect in the virus assembly and budding process. To determine the efficiency of virus formation and release, noncomplementing cells were infected at an MOI of 1 with phenotypically complemented SAD ΔM virions. The supernatants as well as cell extracts were titrated at 24, 48, and 72 h after infection. Compared to that of SAD L16, the total amount of infectious SAD ΔM, including both cell-associated and cell-free virus, was reduced by factors of approximately 600 at 24 h and 10,000 at 48 h postinfection (Table 1). When only the titers of cell-free infectious virus were compared, the yield was reduced by as much as 5 × 105-fold. In agreement with this, 98% of SAD ΔM infectious particles were found to be cell associated, compared to fewer than 10% for the wild-type virus, at 48 h postinfection. This indicated severe defects in the virus assembly and budding mechanism in the absence of M protein (Table 1).
TABLE 1.
Titers of released and cell-associated RV
| Virus | h post-infection | Focus-forming units/mla in:
|
% Cell-associated virusb | |
|---|---|---|---|---|
| Supernatant | Cell lysate | |||
| SAD L16 | 24 | 3.5 × 106 | 4.5 × 106 | 56 |
| SAD ΔM | 24 | 6 × 101 | 1.2 × 104 | 99.5 |
| SAD L16 | 48 | 1.6 × 108 | 1.5 × 107 | 8.6 |
| SAD ΔM | 48 | 3 × 102 | 1.5 × 104 | 98 |
| SAD L16 | 72 | 8 × 107 | 1 × 106 | 1.3 |
| SAD ΔM | 72 | 1.8 × 102 | 7.5 × 103 | 97.7 |
Titer of infectious particles as determined by end point dilution in cell culture.
Percentage of total infectious virions in the cell-associated form.
Physical characteristics of infectious SAD ΔM particles.
To analyze the sedimentation rate of the infectious particles, supernatants from cells infected with SAD ΔM were layered on a 5 to 25% continuous sucrose gradient and centrifuged at 31,500 rpm in an SW41 rotor for 30 min. Twelve equal fractions were collected from the gradient, and the infectivities of individual fractions were determined by end point dilution. For wild-type RV, the peak of infectivity was in fractions 9 and 10 (numbering is from the top to the bottom of the gradient), whereas for SAD ΔM, the majority of infectious particles were found in two distinct peaks, encompassing fractions 7 and 8 and fraction 11. This result indicated that the size of SAD ΔM particles differs considerably from that of wild-type RV. To determine the density of the infectious SAD ΔM particles, samples were sedimented to equilibrium on 10 to 70% continuous sucrose gradients, and the infectivities of 12 equal fractions were determined as described above. The densities of the fractions containing the majority of infectious SAD ΔM and SAD L16 were found to be 1.17 and 1.16 g/cm3, respectively.
RV M interacts with G.
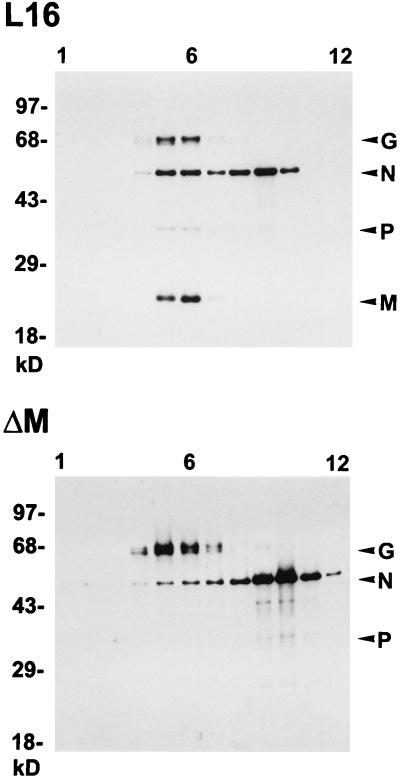
An interaction between G and M and/or N has been postulated to be required for efficient budding of rhabdoviruses (17, 21). However, there was little direct evidence in support of this central dogma. The analysis of the SAD ΔM mutant was therefore expected to give clues as to whether G interacts with M, N, or the RNP. Since most SAD ΔM infectious particles are cell associated, we analyzed the protein composition and infectivity of cell-bound particles after equilibrium centrifugation on sucrose gradients. Total cell lysates were prepared as described in Materials and Methods, layered on a 10 to 70% sucrose gradient, and fractionated as described above. The locations of virus proteins were determined by Western blotting, and the infectivities and refractive indices of individual fractions were determined. The majority of cell-associated infectious SAD L16 virions were localized in fraction 6 (2 × 106/ml), which also contained the largest amount of virus proteins (Fig. 5). The infectivity of cell-bound SAD ΔM also peaked in fraction 6 (1.5 × 103/ml), which has a density of 1.14 g/cm3. Another protein peak in both gradients in which mainly N was detected was localized in fraction 9 for SAD L16 and in fractions 9 and 10 for SAD ΔM. The density of 1.21 g/cm3 for fraction 9 in both gradients is identical to the density of RV RNPs obtained after detergent treatment of purified virions (21).
FIG. 5.
Protein composition of cell-associated SAD L16 and SAD ΔM virions. Cell extracts from approximately 106 infected cells were prepared as described in Materials and Methods and purified by 10 to 70% sucrose gradient centrifugation. Twelve equal gradient fractions (numbered from top to bottom) were analyzed by Western blotting for the presence of viral proteins. The sucrose density was determined from the refractive index of each fraction. For both preparations, the peak of infectivity was present in fraction 6, which has a density of 1.14 g/cm3.
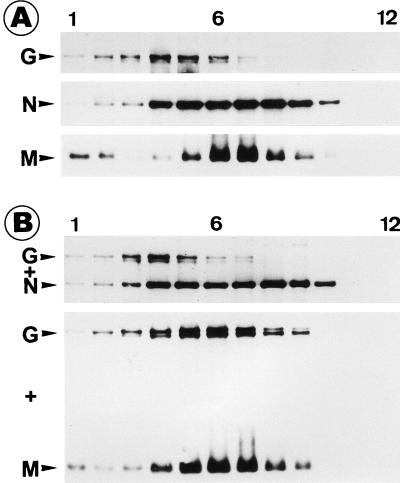
A striking difference between the two gradients was in the proportions of N protein localized in the two peaks (Fig. 5). While the N protein in fractions 5 and 6, where G is also colocalized, in the SAD L16 gradient constitutes 36% of the total N protein, only 6% of the SAD ΔM N protein is localized in the corresponding fractions. The lack of colocalization of the SAD ΔM RNP with G protein indicated that only those RNPs enwrapped by M protein are able to specifically interact with the spike protein. To investigate whether M alone can interact with G or whether formation of an M-RNP complex is necessary, we expressed RV M, N, and G proteins either alone or in combinations and analyzed cell lysates by sedimentation on sucrose gradients. When M protein was expressed in the absence of other viral proteins, about 10% of M remained within the top three fractions of the gradient, whereas more than 90% moved towards the bottom of the gradient and peaked in fractions 6 and 7 (Fig. 6). When G was expressed alone, the majority of G was localized in fractions 3 to 5. Most interestingly, when M and G were coexpressed, the localization of M remained unaltered, but G followed M and colocalized with it throughout the gradient, except at the top. In contrast, the distribution pattern of G remained unchanged when it was coexpressed with N protein, suggesting the absence of a direct G-N interaction. These results indicated that M is able to interact with G even in the absence of other viral components and also in the form of an M-RNP complex (Fig. 5). Obviously, the interaction between G and the RNP-M complex is crucial for efficient budding of infectious progeny virions.
FIG. 6.
RV M interacts with G. Approximately 106 BSR cells were first infected with vTF7-3 and then transfected with 5 μg of plasmid DNA. After 16 h of infection, cell lysates were prepared, centrifuged to equilibrium, and analyzed as described for Fig. 5. (A) Density gradients from cells expressing G, N, or M protein individually; (B) cells coexpressing G and N proteins or G and M proteins. When G and M proteins were coexpressed, the G protein cosedimented with M protein (numbering is from the top to the bottom of the gradient).
M renders RV bullet shaped.
As shown above, the infectious particles produced in SAD ΔM-infected cells varied in many respects from wild-type RV. To analyze the morphology of the infectious particles released in the absence of M protein, supernatant samples were fixed with 0.5% glutaraldehyde and purified over a sucrose gradient. Samples were incubated with a MAb directed against RV G protein, stained with a secondary antibody coupled to colloidal gold, and examined by immunoelectron microscopy. Although control samples prepared in parallel from SAD L16-infected cells contained almost exclusively typical bullet-shaped particles, we repeatedly failed to detect even a single bullet-shape particle in samples prepared from SAD ΔM-infected cells. Instead, the latter samples contained either unique long, rod-shaped particles with smaller diameters or rounded, vesicular structures (Fig. 7A to C). The long, rod-shaped particles that were found exclusively in the supernatants of cells infected by SAD ΔM varied greatly in size, ranging from 500 to 1,000 nm by 50 nm, compared to 200 to 300 nm by 95 nm for wild-type RV. Both types of particles were labeled with anti-G MAb, showing that they contain spike G protein on their surfaces. Due to the high accumulation of G protein on the surfaces of SAD ΔM infected cells, the particles may have acquired G protein nonspecifically. Attempts were also made to visualize SAD ΔM virions budding from the surfaces of virus-infected cells by examining thin sections in the electron microscope. However, we failed to detect any virus-like particle budding at the plasma membrane, further confirming that in the absence of M, RNPs alone are not competent for budding, even at G-containing membranes.
FIG. 7.
Morphology of particles released from SAD ΔM-infected cells. SAD ΔM was grown in the absence of RV M protein (A to C) or in cells that express RV M protein from transfected plasmids (D). Purified supernatants from approximately 107 SAD ΔM-infected BSR cells were immunostained with a MAb specific for the RV G protein and analyzed by electron microscopy. In the absence of RV M protein, either long rod-shaped particles (A and B) or round vesicular structures (C), but no bullet-shaped particles, were detected. After expression of RV M protein, the majority of particles had the typical bullet-shaped rhabdovirus morphology (D). Bar, 100 nm.
To confirm the possibility of correcting the observed defects in the assembly and budding process of the SAD ΔM RV mutant, we complemented the M deficiency by expressing RV M protein in infected cells. The resulting supernatant was analyzed for infectivity by end point dilution and for particle morphology by electron microscopy as described above. After complementation of SAD ΔM, the infectious particle yield at 24 h posttransfection was increased approximately 10,000 fold, from 102/ml (SAD ΔM) to 106/ml (SAD ΔM plus M), thus reaching 1% of the maximal SAD L16 titers. This indicated alleviation of the major defects by M protein provided in trans. Moreover, the increase in titer in the supernatant was accompanied by the formation of typical bullet-shaped particles (Fig. 7D). Although round vesicular structures were still present in smaller number, the long rod-shaped particles were virtually absent in the supernatants after complementation with M protein from transfected plasmids. Taken together, these results show the absolute requirement of rhabdovirus M protein for efficient virus assembly and budding and for the bullet-like morphology.
DISCUSSION
In the assembly pathway of negative-strand RNA viruses, the steps following encapsidation of the RNA genome by nucleoprotein and finally resulting in the budding of mature viral particles are believed to involve the matrix (M) protein. However, knowledge about the biological functions of M protein is restricted mainly to membrane- or nucleocapsid-binding properties (reviewed in reference 14), and more direct evidence that M protein does execute virus assembly and budding was not available. Recently, the M protein of VSV, the prototype rhabdovirus, was reported to be localized within the RNP coil (1), challenging the general view of how M proteins contribute to virus assembly. In this work, we show the localization of M protein in RV virions and describe a recombinant mutant RV that lacks the entire M gene. Characterization of this helper virus-free, M-deficient mutant provided greater insight into the role of M protein in the different steps of virus formation, including virus protein assembly, morphogenesis, and release of mature virions.
To determine the localization of M protein in mature RV in order to understand how it exerts its action in the various steps of virion formation, the membranes of mature virions were peeled off by using a nonionic detergent. After immunogold labeling of the resulting subviral structures, an intense anti-M gold labeling was seen on the surface of the intact coiled nucleocapsid, the so-called skeleton. In contrast, anti-RNP label was bound only to uncoiled RNP extending from skeletons and not to the surfaces of intact skeletons. Both the intense M labeling of intact skeletons and their inaccessibility to N and P antibodies indicate that the RNP coil is completely covered with M protein. Removal of M protein from skeletons apparently results in uncoiling of RNP, which then can be labeled by anti-RNP antibodies. Thus, the M protein layer surrounding skeletons is responsible for keeping the RNP in the condensed form. The integrity and shape of the RV skeleton are therefore defined by the condensing effect of the M protein.
The finding that RV M lies beneath the lipid bilayer and surrounds the RNP contradicts the results published for VSV (1). M protein of VSV was suggested to be only inside the nucleocapsid coil and to be able to interact with the membrane only at the extreme ends. The absence of anti-M labeling on the surfaces of VSV skeletons may be due to conformational differences between VSV M inside the RNP coils and VSV M that might surround the RNP, so that epitopes of the latter perhaps were not accessible to the antibodies used. Alternatively, M protein outside the RNP coil may be rapidly removed under the conditions used, while that inside may be more resistant and sufficient to keep the skeletons condensed. However, a significant difference in the structural organizations of the two rhabdoviruses cannot be excluded (see below). In fact, in contrast to the case for RV, we could also observe central structures resembling the described cigar-shaped core (1) in partially uncoiled VSV RNPs (not shown).
To elucidate the importance of RV M protein in virion formation, we generated the RV mutant SAD ΔM, which lacks the entire M gene. The mode of spread of SAD ΔM in cell culture was significantly different from that of wild-type RV. SAD ΔM-infected cells showed an increased cell-cell fusion and enhanced cell death, in contrast to the noncytopathic type of growth of wild-type RV in cell culture. When infection was done at a low MOI (0.005), the formation of giant multinucleated cells was more evident. Such syncytia reach the maximum size at 48 h postinfection and then start to disintegrate. Although RV is a rather noncytopathogenic virus, higher accumulation of RV G protein in infected cells was reported to be associated with increased cytopathic effects in fibroblasts and RV-induced apoptosis in lymphocytes (23, 36). Thus, the 2.6-fold higher level of G protein expression on the surfaces of cells infected by SAD ΔM compared to SAD L16 may explain the increased cell-cell fusion and cytopathic effects observed in SAD ΔM-infected cultures. In addition, the high-level accumulation of G on the surfaces of SAD ΔM-infected cells strongly suggests low-level depletion of surface G by budding virions.
The findings that the total amount of infectious SAD ΔM particles was decreased by up to 10,000-fold and that of cell-free particles was decreased by as much as 5 × 105-fold revealed severe defects in the virus formation process if M protein is not present. As shown by the density gradient centrifugation of cell-associated infectious particles, the middle peak in the SAD L16 gradient accounted for 36% of the total RNP, compared to only 6% of the corresponding SAD ΔM RNP (Fig. 5). Fraction 6 in both gradients contained the majority of infectious particles and presumably the majority of condensed RNP-M cores in the SAD L16 gradient. Such condensed RV RNP-M cores were demonstrated to be able to bud at the cell surface and mature into spikeless particles (21). Thus, the considerable drop in SAD ΔM infectious particle formation is mainly due to the absence of RNP-M cores, which may represent the minimum budding-competent subviral structure. Presumably, condensed RNPs, i.e., RNPs surrounded by M protein, already assume the skeleton-like structure intracellularly and not much change in shape is required during envelopment and budding. Thus, the way that RNPs of negative-strand RNA viruses are condensed by their M proteins may be the basis for morphological variation and efficiency of virion release.
Interestingly, more than 98% of SAD ΔM infectious particles were cell associated, compared to fewer than 10% for SAD L16. This suggests that the assembly intermediates found in fraction 6 of SAD ΔM-infected cells were not competent for envelopment with the plasma membrane. The released SAD ΔM infectious particles, although heterogenous in size, have a density that is similar to that of wild-type RV, indicating that they are also composed of enveloped RNPs but in an uncondensed form. Such uncondensed RNPs might be enwrapped in G-containing membranes and released into the supernatant. Since rhabdovirus G proteins were shown to autonomously mobilize even unrelated RNAs or RNPs (21, 25), this process of SAD ΔM particle release does not represent the normal virus budding mechanism. We assume that the long rod-shaped particles contain uncondensed RNPs and constitute all free infectious SAD ΔM particles. However, we cannot rule out the possibility that some of the G protein-containing round vesicles may also contain RNP-like structures, since particles both with and without electron-dense content were observed (Fig. 7C). The complete absence of bullet-shaped particles in supernatants of SAD ΔM-infected cells, however, shows the absolute requirement of M protein for the characteristic rhabdovirus morphology. Furthermore, after transient expression of RV M protein in cells that were infected by SAD ΔM, the majority of released particles had the typical bullet-shape morphology. The titer in the supernatant was also raised 10,000-fold, clearly showing the central role of rhabdovirus M proteins in both virus morphogenesis and the budding process.
For efficient budding of RV, a concerted action of both the core (i.e., RNPs surrounded by M protein) and the spike protein was demonstrated to be essential (21). In addition, the RV G cytoplasmic tail was shown to contain a signal to direct efficient incorporation of not only G but also chimeric proteins with foreign transmembrane domains and ectodomains into the envelopes of budding virions (19–21). These data suggested that the RV G cytoplasmic tail specifically interacts with one or more internal virus proteins to facilitate both sorting of surface proteins into virus envelopes and efficient budding. The present results indicate that M is the only internal virus protein with which the G cytoplasmic tail may interact. The RV nucleocapsid skeleton is completely covered with M, rendering it inaccessible to antibodies and probably also to the G cytoplasmic tail. The lack of colocalization of M-less RNPs and G protein in SAD ΔM gradients further supports the idea that the G cytoplasmic tails interact solely with RV M localized between the membrane and the nucleocapsid. This is further supported by the finding of colocalization between M-RNP and G in the wild-type RV (Fig. 5) as well as of cosedimentation of M and G in the absence of any other viral protein (Fig. 6). Obviously, the interaction between G and the RNP complex is mediated by the M layer, allowing efficient release of infectious progeny virions.
In striking contrast to RV, the closely related VSV appears to utilize a nonspecific way of incorporating VSV G protein (28) or heterologous surface proteins (29, 32) into the envelope. For instance, the human transmembrane protein CD4 was found to be incorporated into the virus envelope with the same efficiency as a chimeric CD4 containing the transmembrane and cytoplasmic domains of VSV G (29, 32). In contrast to the case for VSV, CD4 and the human chemokine receptor CXCR4 proteins were efficiently incorporated into an RV-derived envelope only when the autologous G tail was present (20, 30). We have shown here that the M protein of RV interacts with G and is thus responsible for recruiting G protein into the virus. We are now identifying the residues of the cytoplasmic tail responsible for this interaction (unpublished data). A direct interaction of VSV M and VSV G has so far not been shown unequivocally (4, 17). If the localization of VSV M is considered to be RV-like, the compelling differences in spike protein incorporation by VSV and RV might be explained by a less stringent and weaker interaction between M and G. If VSV M is considered to be only inside the RNP, as suggested recently (1), a basic difference in the assembly mechanisms of RV and VSV has to be postulated. In such a situation, M-G interactions would perhaps not be relevant, and this might be reflected by the observed nonselective incorporation of VSV surface proteins. Only some M protein protruding from the ends of RNPs would remain able to contact G-containing membranes. Moreover, a localization of M protein in VSV inside the RNP coil would require a substantially modified model of how efficient budding is achieved.
ACKNOWLEDGMENTS
We thank K. Kegreiss, V. Schlatt, and K. Mildner for their perfect technical assistance and W. Kramer for the photographic work.
This work was supported by grant BEO/0311171 from the Bundesministerium für Forschung und Technologie.
REFERENCES
- 1.Barge A, Gaudin Y, Coulon P, Ruigrok R W. Vesicular stomatitis virus M protein may be inside the ribonucleocapsid coil. J Virol. 1993;67:7246–7253. doi: 10.1128/jvi.67.12.7246-7253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll A R, Wagner R R. Role of the membrane (M) protein in endogenous inhibition of in vitro transcription by vesicular stomatitis virus. J Virol. 1979;29:134–142. doi: 10.1128/jvi.29.1.134-142.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartwright B, Smale C J, Brown F. Dissection of vesicular stomatitis virus into the infective ribonucleoprotein and immunizing components. J Gen Virol. 1970;7:19–32. doi: 10.1099/0022-1317-7-1-19. [DOI] [PubMed] [Google Scholar]
- 4.Chong L D, Rose J K. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–414. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong L D, Rose J K. Interactions of normal and mutant vesicular stomatitis virus matrix proteins with the plasma membrane and nucleocapsids. J Virol. 1994;68:441–447. doi: 10.1128/jvi.68.1.441-447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinton G M, Little S P, Hagen F S, Huang A S. The matrix (M) protein of vesicular stomatitis virus regulates transcription. Cell. 1978;15:1455–1462. doi: 10.1016/0092-8674(78)90069-7. [DOI] [PubMed] [Google Scholar]
- 7.Conzelmann K K, Cox J H, Schneider L G, Thiel H J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 8.Conzelmann K K, Cox J H, Thiel H J. An L (polymerase)-deficient rabies virus defective interfering particle RNA is replicated and transcribed by heterologous helper virus L proteins. Virology. 1991;184:655–663. doi: 10.1016/0042-6822(91)90435-e. [DOI] [PubMed] [Google Scholar]
- 9.Conzelmann K K, Meyers G. Genetic engineering of animal RNA viruses. Trends Microbiol. 1996;4:386–393. doi: 10.1016/0966-842X(96)10062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conzelmann K K, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito Y, Nishizono A, Mannen K, Hiramatsu K, Mifune K. Rabies virus M protein expressed in Escherichia coli and its regulatory role in virion-associated transcriptase activity. Arch Virol. 1996;141:671–683. doi: 10.1007/BF01718325. [DOI] [PubMed] [Google Scholar]
- 13.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Luo L, Schubert M, Wagner R R, Kang C Y. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J Virol. 1993;67:4415–4420. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyles D S, McKenzie M, Kaptur P E, Grant K W, Jerome W G. Complementation of M gene mutants of vesicular stomatitis virus by plasmid-derived M protein converts spherical extracellular particles into native bullet shapes. Virology. 1996;217:76–87. doi: 10.1006/viro.1996.0095. [DOI] [PubMed] [Google Scholar]
- 17.Lyles D S, McKenzie M, Parce J W. Subunit interactions of vesicular stomatitis virus envelope glycoprotein stabilized by binding to viral matrix protein. J Virol. 1992;66:349–358. doi: 10.1128/jvi.66.1.349-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marx P A, Portner A, Kingsbury D W. Sendai virion transcriptase complex: polypeptide composition and inhibition by virion envelope proteins. J Virol. 1974;13:107–112. doi: 10.1128/jvi.13.1.107-112.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mebatsion T, Conzelmann K K. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc Natl Acad Sci USA. 1996;93:11366–11370. doi: 10.1073/pnas.93.21.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mebatsion T, Finke S, Weiland F, Conzelmann K K. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell. 1997;90:841–847. doi: 10.1016/s0092-8674(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 21.Mebatsion T, König M, Conzelmann K. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell. 1996;84:941–951. doi: 10.1016/s0092-8674(00)81072-7. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb W W, Brown J C. Role of the vesicular stomatitis virus matrix protein in maintaining the viral nucleocapsid in the condensed form found in native virions. J Virol. 1981;39:295–299. doi: 10.1128/jvi.39.1.295-299.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni Y, Iwatani Y, Morimoto K, Kawai A. Studies on unusual cytoplasmic structures which contain rabies virus envelope proteins. J Gen Virol. 1996;77:2137–2147. doi: 10.1099/0022-1317-77-9-2137. [DOI] [PubMed] [Google Scholar]
- 24.Odenwald W F, Arnheiter H, Dubois-Dalcq M, Lazzarini R A. Stereo images of vesicular stomatitis virus assembly. J Virol. 1986;57:922–932. doi: 10.1128/jvi.57.3.922-932.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolls M M, Webster P, Balba N H, Rose J K. Novel infectious particles generated by expression of the vesicular stomatitis virus glycoprotein from a self-replicating RNA. Cell. 1994;79:497–506. doi: 10.1016/0092-8674(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 26.Ruigrok R W, Calder L J, Wharton S A. Electron microscopy of the influenza virus submembranal structure. Virology. 1989;173:311–316. doi: 10.1016/0042-6822(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 27.Schneider L, Meyers S. Antigenic determinants of rabies virus as demonstrated by monoclonal antibody. In: Bishop D H L, Compans R W, editors. The replication of negative strand RNA viruses. Amsterdam, The Netherlands: Elsevier; 1981. pp. 947–953. [Google Scholar]
- 28.Schnell M J, Buonocore L, Botitz E, Ghosh H P, Chernish R, Rose J K. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 1998;17:1289–1296. doi: 10.1093/emboj/17.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnell M J, Buonocore L, Kretzschmar E, Johnson E, Rose J K. Foreign glycoproteins expressed from recombinant vesicular stomatitis viruses are incorporated efficiently into virus particles. Proc Natl Acad Sci USA. 1996;93:11359–11365. doi: 10.1073/pnas.93.21.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnell M J, Johnson J E, Buonocore L, Rose J K. Construction of a novel virus that targets HIV-1-infected cells and controls HIV-1 infection. Cell. 1997;90:849–857. doi: 10.1016/s0092-8674(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 31.Schnell M J, Mebatsion T, Conzelmann K K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert M, Joshi B, Blondel D, Harmison G G. Insertion of the human immunodeficiency virus CD4 receptor into the envelope of vesicular stomatitis virus particles. J Virol. 1992;66:1579–1589. doi: 10.1128/jvi.66.3.1579-1589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simons K, Garoff H. The budding mechanisms of enveloped animal viruses. J Gen Virol. 1980;50:1–21. doi: 10.1099/0022-1317-50-1-1. [DOI] [PubMed] [Google Scholar]
- 34.Smirnov Y A, Kuznetsova M A, Kaverin N V. The genetic aspects of influenza virus filamentous particle formation. Arch Virol. 1991;118:279–284. doi: 10.1007/BF01314038. [DOI] [PubMed] [Google Scholar]
- 35.Suryanarayana K, Baczko K, ter Meulen V, Wagner R R. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J Virol. 1994;68:1532–1543. doi: 10.1128/jvi.68.3.1532-1543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoulouze M I, Lafage M, Montano-Hirose J A, Lafon M. Rabies virus infects mouse and human lymphocytes and induces apoptosis. J Virol. 1997;71:7372–7380. doi: 10.1128/jvi.71.10.7372-7380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zvonarjev A Y, Ghendon Y Z. Influence of membrane (M) protein on influenza A virus virion transcriptase activity in vitro and its susceptibility to rimantadine. J Virol. 1980;33:583–586. doi: 10.1128/jvi.33.2.583-586.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]