Summary
Finding cancer-driver genes has been a central theme of cancer research. We took a different perspective; instead of considering normal cells, we focused on cancerous cells and genes that maintained abnormal cell growth, which we named cancer-keeper genes (CKGs). Intervening CKGs may rectify aberrant cell growth, making them potential cancer therapeutic targets. We introduced control-hub genes and developed an efficient algorithm by extending network controllability theory. Control hub are essential for maintaining cancerous states and thus can be taken as CKGs. We applied our CKG-based approach to bladder cancer (BLCA). All genes on the cell-cycle and p53 pathways in BLCA were identified as CKGs, showing their importance in cancer. We discovered that sensitive CKGs — genes easily altered by structural perturbation — were particularly suitable therapeutic targets. Experiments on cell lines and a mouse model confirmed that six sensitive CKGs effectively suppressed cancer cell growth, demonstrating the immense therapeutic potential of CKGs.
Subject areas: Therapeutics, Drugs, Cancer
Graphical abstract

Highlights
-
•
Focus on genes that maintain aberrant cell growth, named cancer-keeper genes (CKGs)
-
•
Develop a novel approach for finding CKGs
-
•
Apply the new method to bladder cancer and validate CKGs in cancer cells and mice
Therapeutics; Drugs; Cancer
Introduction
A primary objective of cancer research is to identify cancer-driver genes (CDGs) that may trigger tumorigenesis or promote aberrant cell growth.1,2 CDGs can help elucidate cancer etiology3 and be used as diagnostic biomarkers and therapeutic targets.4 Most existing methods for CDG discovery look for genes with substantial mutations that can separate cancer subjects and normal controls.5,6,7 Despite many efforts,1,2,8 CDGs that can be discovered seem saturated with increasing sample sizes.9
Deviated from the prevalent mutation-based methods for CDG finding is a recent approach based on network structural controllability.10,11 Structural controllability specifies how a networked system can be driven from any state to the desired state in finite time by exerting stimuli on the driver nodes defined by a control scheme (i.e., a minimal set of non-overlapping paths covering all nodes) for the network.10 Network controllability has been applied to various biological networks,12,13,14 including protein-protein interaction networks,15 gene regulatory networks,16,17 and metabolic networks.18,19 Instead of analyzing individual genes in isolation, this approach organizes all genes of interest in a network and identifies the network’s driver nodes (i.e., genes). When applied to cancer regulatory networks, it identifies driver nodes regarded as CDGs and therapeutic targets for cancer treatment.20
However, two critical issues hinder network controllability from becoming a practical method for finding CDGs. First, given a network, the control scheme is often not unique, but numerous control schemes and different sets of driver nodes exist.21,22 It is difficult, if not infeasible, to determine the most effective control scheme for the network. All control schemes may be compared to select the best, e.g., one with the shortest control trajectory to the desired state. However, finding all control schemes, a #P-hard problem,23 is computationally prohibitive. In addition, one control scheme may contain a substantial number of driver genes,21 which may need to be stimulated together to control the cell. It is challenging to manipulate many genes at once for disease treatment. Second, a fundamental but mostly neglected assumption underlying structural controllability methods is that the biological network is a model describing both cancerous and normal cell states so that mutations in some genes can drive the cell from a normal state to a cancerous state, resulting in cancer. However, little is known about which states are normal and which other states are cancerous, so a driver node in the network may not necessarily be a CDG.
We adopt a different perspective on cancer and cancer treatment. Instead of considering mutated genes that may transform normal cells into abnormal ones, we are interested in genes that maintain cancerous cell states, which we call cancer-keeper genes (CKGs). We postulate that intervention to such genes may terminate or prevent aberrant cell differentiation and proliferation and hypothesize that CKGs are ideal candidate biomarkers for diagnosis and therapeutic targets for therapy. To this end, we focused on homogeneous networks that model cancer cells and utilized and extended the network structural controllability theory. We went beyond one control scheme and considered the overall structural controllability governed by all network control schemes. We introduced control hubs in the middle of a control path of every control scheme. We developed a polynomial-time algorithm for finding all control hubs without computing all control schemes. We were particularly interested in those control hubs that were sensitive to changes to network structures, so they were better suited as therapeutic targets. In a case study, we applied our approach to bladder cancer (BLCA) and identified 35 sensitive CKGs (sCKGs) for the disease. One important finding was that the genes on the cell cycle and p53 pathways were sCKGs. This result showed that these genes were critical for maintaining BLCA and demonstrated the power of our new method. Using small-interferencing RNA (siRNA) knockdown, we experimentally validated six sCKGs in vitro by examining their effects on the proliferation of two BLCA cell lines. We also validated RPS6KA3, the only sensitive CKG on the p53 pathway, in vivo in a mouse model of BLCA by showing its prohibitive function on tumor progression. Additional results on cervical and head-and-neck cancer cells demonstrated the rigor of our approach to other types of cancer.
Results
The new CKG-based approach consists of four steps: (1) Construct a biological network describing gene-gene interactions/associations for diseases of interest; (2) Identify control hubs in the network and from which, identify CKGs; (3) Analyze the functions of selected CKGs; and (4) Experimentally characterize the cancer suppression functions of sCKGs in cell lines and animal models. In the sequel, we present these major components along with the rationale and key ideas of the new approach. We focus on BLCA as an application and show that the new method is general and applicable to cancers like cervical cancer and head-and-neck carcinoma.
Construction of cancer gene regulatory network
We developed a novel method for constructing gene regulatory networks for cancer study (Figure 1A). Such a network consists of cancer-related genes and interactions of the ten most important and common types of interactions and signaling pathways (Table S1) from five well-curated, disease- and cancer-related pathway databases, including the NCI Pathway Interaction Database,24 PhosphoSite Kinase-substrate information,25 HumanCyc,26 Reactome,27 and PANTHER Pathways28 (see STAR Methods). These databases were created from disease studies containing information on mutated genes and disease- and cancer-related pathways. For example, Reactome comprises the KEGG databases with disease and drug information. PhosphoSite contains data on missense mutations from UniProtKB, TCGA, and many other sources, collected from over 2,000 diseases and syndromes, and has data on polymorphisms associated with hundreds of cancers.
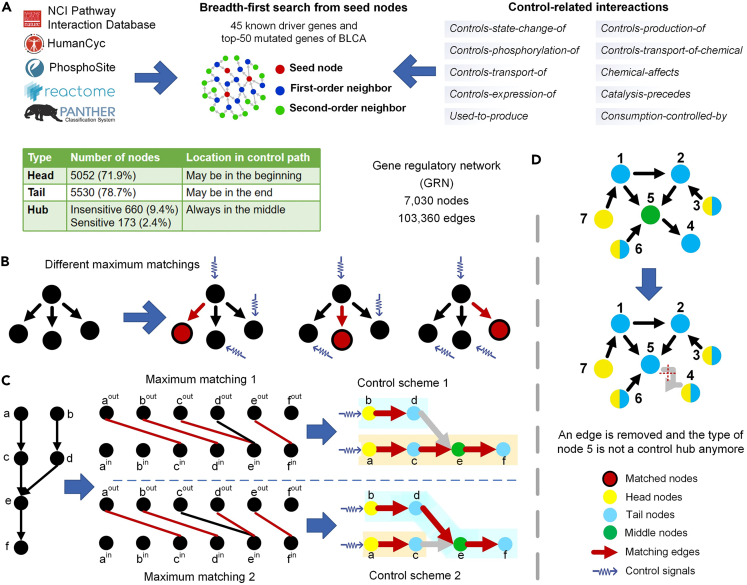
Figure 1.
Gene regulatory network construction and control hubs of complex networks
(A) Construction of a gene regulatory network, GRN. Ten control-related gene/protein interaction types were extracted from five high-quality pathway databases. A breadth-first search was then performed, starting from 45 known BLCA driver genes and the 50 most highly mutated genes of BLCA. The traversed nodes and edges constitute GRN.
(B) An example of a simple network with three different maximum matchings. The unmatched nodes are driver nodes, and the red edges are matched edges.
(C) Two different control schemes of a simple network. A node may lie at the head, tail, or middle of a control path. Node e resides in the middle of some control paths of the two control schemes, becoming a control hub of the network.
(D) Sensitive control hubs of a simple network. After removing edge e(5,4), node 5 changes from a control hub to a tail node, making it a sensitive control hub of the network.
To construct a regulatory network relevant to the types of cancer of interest (e.g., BLCA), we used some cancer driver genes as seeds to traverse the ten disease- and cancer-related interaction databases. From the data of TCGA Research Network (https://www.cancer.gov/tcga), we chose 45 known BLCA CDGs29 and 50 most mutated genes in BLCA as seed nodes (Table S2). A breadth-first search from the 95 seed genes was used to traverse the databases’ ten major types of control-related interactions. The traversed genes and the interactions formed the gene regulatory network (GRN) with 7,030 nodes (genes) and 103,360 directed edges (Figure 1A; see Data S1). Although we initially utilized BLCA-related seed nodes to obtain GRN, we found that the interactions in the databases consist of a vast connected component (including 99.2% nodes and 99.9% edges) and 14 small connected components. Consequently, regardless of which cancer we choose the seed nodes from, the resulting GRN will remain almost unchanged, implying that the GRN is a pan-cancer network with high generalizability. The network was built upon protein interaction data and signaling pathway information describing regulatory relationships among genes (i.e., proteins), aligning well with the notion of network controllability.
Control hub nodes of a network
Our new method hinges upon the concept of control hubs of a network. They are the most vulnerable spots for structural controllability, so intervention on any of them may render the network uncontrollable by external stimuli. When applied to network models of cancer cells, interventions on control hubs may terminate aberrant cell differentiation and proliferation so that they may be taken as therapeutic targets.
A key observation supporting structural controllability is that a node in a directed network can control one of its outgoing neighbors,30 the controlled neighbor can control a neighbor of its own and so forth. These nodes collectively form a control path with a head node at the beginning, also called control node12 or driver node,11 a tail node at the end of the path, and middle nodes, if any (Figures 1B and 1C, STAR Methods). The set of the smallest number of control paths and their driver nodes constitute a control scheme of the network (Figure 1C). Notably, the control scheme is not unique to a complex network.21,22 A node may take different positions and thus play distinct roles in various control schemes (e.g., nodes c and d in Figure 1C). It is difficult to determine the best control scheme. We may compute all control schemes to select the best. However, it is computationally prohibitive to derive all control schemes because the problem is #P-hard,23 meaning no polynomial algorithm is known for the problem. Furthermore, given all control schemes, it is nontrivial to determine the best because little is known about the network dynamics, and different optimality criteria (e.g., the fewest nodes to be controlled versus the fewest steps to reach the desirable state) will lead to distinct control schemes. Moreover, many nodes may serve as driver nodes in different control schemes,21 making it costly to control the network. All these unfavorable factors cast doubts on the utility of structural controllability for finding cancer driver genes.
We go beyond one control scheme and consider the overall structural controllability of a network by considering all control schemes. To ameliorate the computational burden, we avoid computing all control schemes and focus on all control hubs that are middle nodes of some control paths of every control scheme (e.g., node e in Figure 1C). A prominent feature of the control hub is that it is essential for maintaining the overall structural controllability of the network. A perturbation to or blockade of any control hub may void all control schemes and consequently take apart the overall controllability of the network. Therefore, it is critical to protect all control hubs in order to maintain structural controllability and the validity of the network control model, meaning that control hubs are the most vulnerable spots for structural controllability. Here, we explicitly explore and exploit this property of control hubs.
However, identifying all control hubs of a large network is technically nontrivial. We developed a novel polynomial-time algorithm for finding all control hubs without computing all control schemes.31 The algorithm is based on previous work enumerating all driver nodes.21,32 It first identifies the head and tail nodes of the control paths of all control schemes and subsequently identifies control hubs. It has a complexity of O(n×m) on a network with n nodes and m edges.
Sensitive control hub nodes as cancer-keeper genes
We consider a model representing cancerous cells, focusing on the overall network controllability using control hubs. When a change to the network structure turns a control hub into a non-control hub, the network is no longer controllable by any control scheme. As a result, the cell may transition from the current cancerous state to the normal state. Therefore, we name control hub genes in a cancer regulatory network CKGs because they maintain the network controllability of the model.
We expect that some control hubs are more sensitive and vulnerable to external perturbations than others and may be better therapeutic targets. We are particularly interested in those control hubs that can be turned into non-control hubs when a single edge is removed from the network as a perturbation (Figure 1D), which we call sensitive control hubs or sensitive CKGs (sCKGs). All sCKGs can be identified by removing every edge of the network one at a time (see STAR Methods).
Cancer-keeper genes are essential in the cancer gene regulatory network
We applied our novel CKG approach to the cancer gene regulatory network GRN containing 7,030 genes and 103,360 interactions (Figures 1A and 2A, Data S1). One control scheme has 3,115 driver genes (44.3% of all 7,030 genes), and all control schemes contain 5,052 driver genes (71.9%, Figure 2B). So many control schemes and driver genes made choosing a suitable control scheme to adopt structural controllability infeasible.
Figure 2.
Control hubs or cancer-keeper genes (CKGs) in the gene regulatory network (GRN)
(A) Topological structure of GRN. Node size is proportional to node degree. The network contains 7,030 genes and 103,360 directed interactions.
(B) Proportions of the driver nodes, control hubs, and sensitive control hubs in GRN.
(C) Node degrees of different types of genes in GRN. Control hubs have a significantly higher average degree than the other genes.
(D) Control hubs of GRN are enriched in the context of essentiality, evolutionary conservation, cell signaling, protein abundance, posttranslational modifications (PTMs), regulators of cell proliferation, diseases, virus targets, drug targets, and immune regulation.
(E) Proportions of sensitive CKGs and CKGs in ten key cancer signaling pathways with significant genetic variations. Most of the genes in these pathways are CKGs.
(F) Two TCGA-analyzed signaling pathways of BLCA. FGFR3 and RPS6KA3, two control hubs, reside upstream of the most mutated genes in the pathways. Removing the edge from FGFR3 to RPS6KA3 will change their node types in the control scheme and make them sensitive CKGs.
Our CKG approach identified 660 CKGs in GRN, 9.4% of all 7,030 genes and 13.1% of all 5,052 driver genes in the network (Figure 2B). These CKGs have several characteristics. First, they have higher connectivity, with a degree of 96.4 on average, than the other nodes, which have an average degree of 29.4 (Figure 2C), reflecting the greater controlling power of the CKGs. Next, we characterized the CKGs, head and tail genes by essentiality, evolutionary conservation, and regulation at the levels of translational and posttranslational modifications (PTMs) as done previously.33 The CKGs were significantly enriched in most of the above datasets (Figures 2D and S2–S4). They were enriched considerably with pathogenic mutants and targets of viruses, drugs, and immunotherapies (Figures 2D, S5, and S6). Altogether, these results revealed that CKGs play crucial regulatory roles.
We further analyzed the CKGs against the potential targets of tumor immunotherapy, including core cancer-intrinsic CTL-evasion genes (coreCTL),34 CAR therapy targets,35 and immune checkpoints.36,37 Twenty-seven CKGs were known targets of tumor immunotherapy, of which 20 CKGs were coreCTL genes, and 7 were CAR genes or resided at immune checkpoints. Surprisingly, although 437 CKGs were not directly immune-related, their direct neighbors in the network were immune regulatory genes (Data S2). These results suggested that the 660 CKGs were involved in multiple regulatory processes of tumor immunity, reflecting their functional importance.
To assess the potential oncogenic functions of the 660 CKGs, we examined their involvement in ten key cancer-related signaling pathways with genes with extensive genetic variations in 33 types of cancer analyzed by TCGA.38 Interestingly, 70.4% of the genes in four cancer signaling pathways (i.e., the cell-cycle, p53, TGFβ, and RTK-RAS pathways) were CKGs (Figures 2E and 2F, Table S4). Remarkably, all the genes in the cell-cycle and p53 pathways were CKGs (Figure 2E), which were key signaling pathways for cell differentiation and proliferation. Many CKGs (e.g., ACVR1B in the TGFβ pathway and FGFR3, KIT, NTRK2, and BRAF in the RTK-RAS pathway) were located upstream of the cell-cycle and p53 pathways. Compared to nine popular node/gene identification methods (Figure S7), such as that based on node connectivity and centrality, more CKGs are in the cell-cycle, p53, TGF β, and Myc pathways, which are all known to regulate cell differentiation and proliferation.38 These results indicated that CKGs were tightly regulated and critical in cancer.
We compared CKGs with housekeeping genes (HKGs), known for their stable expression and function in all cells of an organism at all times.39 Although 112 of the 660 CKGs were HKGs, a random sampling analysis (see STAR Methods) revealed that HKGs were significantly under-represented in CKGs but greatly enriched in the head node genes (Figure S8A). Notably, the CKGs had significantly lower expression levels than the HKGs (Figures S8B and S8C). These results implied a fundamental difference in the expression patterns and potential functions between CKGs and HKGs.
Sensitive cancer-keeper genes have oncogenic functions and clinical importance
To select a small subset of the 660 CKGs as potentially druggable targets,40 we looked for sCKGs in GRN. The rationale for edge removal as network perturbation is that many cancer drugs are kinase receptor inhibitors, thus blocking protein-protein interactions and acting as edge removal. One example is an irreversible fibroblast growth factor inhibitor futibatinib41 for BLCA. We name an edge a sensitive edge if removing it makes a CKG become an sCKG. An sCKG has at least one sensitive edge associated; the more sensitive edges that an sCKG is associated with, the more sensitive and druggable it is. Of the 660 CKGs, 173 (26.2%) were sCKGs, among which more than 35.8% were associated with more than one sensitive edge (Figure 3A). All sensitive edges had high confidence scores42 (Table S5). The 173 sCKGs had several characteristics. They predominantly had moderate or low connectivity; their average connectivity was less than half of the other CKGs (Figure 2C). Notably, the direct neighboring genes of sCKGs in the GRN exhibited significantly higher genetic alterations (including mutations, copy number variations, and homozygous deletions) and mRNA expression (Figure 3B, Data 2), so sCKGs might not be detected by conventional frequency-based43,44 and network-based methods.43,45 These findings suggested that these sCKGs could regulate genes with high genetic alteration and mRNA expression.
Figure 3.
Characterizing sensitive CKGs (sCKGs) in GRN
(A) The distribution of the number of edges that could change an sCKG. Most sCKGs have less than three edges that may change their node types, indicating they are robust to random structural perturbations.
(B) Differential analysis of the genetic alteration frequencies and mRNA expression between the sCKGs and their target neighbors in GRN. SCKGs had lower rates of genetic alteration and mRNA expression level, but there were significantly highly altered and mRNA-expressed genes around them. All the samples are from the cBioPortal database.
(C) The subnetwork of sCKGs and their surrounding cancer driver genes.
(D) Characteristics of 35 sensitive CKGs. Twenty-eight sCKGs are not cancer-driver genes, and the rest 7 are. Most CKGs are directly connected to drug targets, cancer-driver genes, and/or immune genes in GRN. The CKGs with names in red are experimentally studied (Figure 4).
(E) Survival analysis of the 35 sensitive CKGs in BLCA. Simultaneous alterations to more than one CKG will significantly increase the survival rate of patients from 59.31 months (one sCKG altered) to 104.65 months (three or more sCKGs changed).
We mapped the 173 sCKGs to the indispensable genes of the human PPI network,33 resulting in 35 sCKGs that might be better therapeutic targets (Figure 3D). Most of these sCKGs were directly connected to the known cancer driver genes,29 cancer therapeutic targets46 or immune genes35,36,37 in GRN (Figures 3C, 3D, and S9). They were enriched with regulatory genes (Figure S10). For instance, 26 of the 35 sCKG had cancer driver genes as direct neighbors (Figure 3D, Table S6), 30 (85.7%) were directly connected to cancer therapeutic targets (Figures 3D and S11A, and Table S7), and 27 (77.1%) were directly connected to immune genes (Figures 3D and S11B, and Table S8).
Many of the 35 sCKGs had beneficial epistatic relations. Consider BLCA as an example. Increased mutations to more than one of these genes significantly increased the overall survival rates of many BLCA subjects based on the TCGA PanCancer clinical data (see STAR Methods). The median survival time of BLCA patients would increase from 28.41 months (for the unaltered group) to 59.31 months (for the one sCKG altered group), 66.41 months (for the two sCKGs altered group), and 104.65 months (for the three and more sCKGs altered group) under the stringent criterion of the Log -rank Test p value <0.015 (Figure 3E). This result strongly suggested that the sCKGs were excellent diagnostic and prognostic biomarkers and therapeutic targets for BLCA.
Two sCKGs (FGFR3 and RPS6KA3) deserved further scrutiny in BLCA. FGFR3 is a well-characterized CDG for BLCA,47 and FGFR3 and RPS6KA3 are involved in two critical pathways of BLCA, i.e., RTK-RAS-PI(3)K and p5348 (Figure 2F). FGFR3 is upstream of PIK3CA in the RTK-RAS-PI(3)K pathway, and RPS6KA3 is upstream of TP53 in the p53 pathway, suggesting that FGFR3 and RPS6KA3 are upstream drivers of two key CDGs. Furthermore, RPS6KA3 is a substrate of FGFR3, so they are related and share many functions. FGFR3 can phosphorylate Y529 and Y707 of RPS6KA3 to assist ERK1/2 in connecting to RPS6KA3 and keep RPS6KA3 active.49 The interaction between FGFR3 and RPS6KA3 was critical; removing the link between the two in GRN would change them from control hubs to head nodes and subsequently invalidate all control schemes for GRN, suggesting that the two genes must play critical roles in BLCA.
Immunotherapy has been adopted in treating BLCA, and several drugs (e.g., atezolizumab and avelumab) have been developed to target immune checkpoint inhibitors (e.g., PD-1 and PD-L1).50 Among the six well-known genes targeted by BLCA drugs, five (CD274/PD-L1, CTLA4, IL12B, PTGS2, and TFDP1) were head or tail nodes except FKBP1A (Data S2). Nevertheless, all six genes were well connected with CKGs (i.e., control hubs) in GRN. For example, CTLA4 has seven neighbors, six of which were CKGs. Such a close connection with CKGs suggested these genes as potential drug targets for BLCA treatment.
Sensitive CKGs as potential therapeutic targets for bladder, cervical and head-and-neck cancers
We experimentally analyzed the potential therapeutic values of some CKGs in three types of cancer, BLCA, cervical cancer (CC) and head-and-neck cancer (HNC). We used cancer cell lines in our experiments, i.e., T24 and UMUC3 for BLCA, SiHa for CC, and FaDu for HNC. We also experimented in vivo with a mouse model of BLCA. When choosing CKGs for experimental validation and analysis, we focused on genes or proteins that are less characterized or not CDGs for these cancers. We included a few known CDGs as positive controls for comparison.
We adopted small-interferencing RNAs (siRNAs) to knock down these six sCKGs in two BLCA cell lines (T24 and UMUC3) and one sCKG (RPS6KA3) in a mouse model of BLCA. The siRNA knockdown experiments were repeated three times for every gene, and every cell line, and the experiments were also repeated on ten mice. For each experiment, the expression of the sCKG was measured by quantitative PCR. The results of the repeated experiments were consistent (Figure 4A). We subjected six sCKGs to in vitro and one sCKG to in vivo analyses to assess their impact on BLCA cell proliferation and migration, thus evaluating and confirming their function in maintaining cancerous cell states and their potential therapeutic values. We chose four sCKGs (RPS6KA3, N-cadherin (CDH2), caspase-1, and FN1) that are not CDGs for BLCA or other malignancies to assess their potential function in BLCA. We also selected two sCKGs (FGFR3 and EP300), known CDGs for BLCA,47 for comparison and validation (Figure 3D, gene names marked in red). The absence of RPS6KA3, FGFR3, and CDH2 significantly decreased the proliferation of the two BLCA cells (Figures 4A and 4B). In contrast, the knockdown of EP300 and FN1 significantly promoted the proliferation of UMUC3 cells (Figure 4B) but had little effect on the proliferation of T24 cells (Figure S12). Interestingly, the loss of caspase-1, which was thought to play an important role in inflammation and tumor microenvironment, promoted the proliferation and survival of BLCA cells (Figures 4A and 4B). As expected, the deletion of FGFR3, RPS6KA3, EP300, FN1, and CDH2 significantly reduced the migration ability of bladder tumor cells (Figures 4C and 4D). In contrast, the absence of caspase-1 promoted the migration of BLCA cells (Figures 4C and 4D).
Figure 4.
Experimental validation of representative sensitive CKGs (sCKGs) in bladder cancer
(A and B) CCK-8 assay and corresponding quantitative detection of the effect of siRNA-mediated knockdown sCKGs on the proliferation of T24 (A) and UMUC3 (B) BLCA cells.
(C and D) Transwell migration assay (upper) and corresponding quantitative detection (down) of the effect of siRNA-mediated knockdown sCKGs on the migration ability of T24 (C) and UMUC3 (D) cells. Scale bar represents 100 μm.
(E) RPS6KA3 promoted tumor proliferation in vivo. BALB/c nude mice were injected with UMUC3 cells stably transfected with RPS6KA3 knockdown plasmids or control vector in the xenotransplantation model. After 4 weeks, the tumor size (left) and weight (right) in the RPS6KA3 knockdown group were significantly lower than in the vector group.
Furthermore, we used a nude mouse model with transplanted BLCA tumors to examine the function of a typical sCKG, RPS6KA3, which has Serine/Threonine kinase activities and is vital in the proliferation of some solid tumors.49,51 Consistent with the in vitro results, the knockdown of RPS6KA3 in the mouse model significantly inhibited the growth of BLCA xenografts in the mice (Figure 4E). All in vitro and in vivo experimental results confirmed the functional roles of some of the sCKGs for maintaining the validity of cancer cells. They validated the feasibility of our novel network control hub-based method.
The six sCKGs evaluated in T24 and UMUC3 BLCA cells were examined in SiHa CC and FaDu HNC cells. Except for FGFR3, siRNA-mediated knockdown assay did not adequately detect the expression of the other five sCKGs in SiHa cells (Figure S13A), which might be plausibly not expressed in SiHa in vitro. Nevertheless, experiments on FGFR3 and two sCKGs (AP2M1 and BCAR1) added to the investigation showed that the down-regulation of these sCKGs had prohibitive effects on SiHa cell proliferation (Figure S13A). Two of the six sCKGs (RPS6KA3 and CDH2) were detected in FaDu cells. The knockdown of RPS6KA3 and CDH2 led to reduced migration of FaDa cells (Figure S13B).
Discussion
One of our primary contributions was a novel perspective on viewing and treating cancer. It fundamentally deviates from the prevalent concept of CDGs that are defined by high mutation rates and may transform normal cells into abnormal ones with uncontrolled cell growth. In contrast, we introduced a new notion of CKGs defined on cancerous cells, which may terminate or prevent abnormal cell differentiation and proliferation when intervened. To utilize the new concept of CKGs, we extended the well-developed theory of network structural controllability,11 which has been adopted to identify CDGs and cancer therapeutic targets.33,52 Going beyond one control scheme in structural controllability, we introduced control hubs over all control schemes. We adopted control hubs as candidates of CKGs defined for abnormal cells.
There are typically fewer control hubs (i.e., CKGs) than driver nodes (i.e., driver genes) in a gene regulatory network. For example, GRN has 5,052 driver genes out of 7,030 genes but, in contrast, only 660 CKGs which can be further reduced to 173 sCKGs. Most CKGs are involved in critical tumor signaling pathways. Remarkably, the genes in the cell-cycle and p53 signaling pathways, reported by the TCGA project,38 are all CKGs (Figure 2F). Dysregulation of cell-cycle control is a hallmark of cancer.53 Many CKGs in the cell-cycle pathway, including CDKN1A,54 CDK2,55 and E2F1,56 have been considered potential targets of anticancer drugs. The p53 pathway is tightly regulated in BLCA,57 and the genetic variations of the genes in the pathway have been an attractive topic for BLCA treatment.58,59 CDKN1A in the cell-cycle pathway and p53 in the TP53 pathways have been taken as therapeutic targets for BLCA.60 The enrichment of the CKGs in TGFβ and receptor-tyrosine kinase (RTK)/RAS/MAP-Kinase signaling pathways is also thought-provoking. These two pathways are known to be essential for BLCA. TGFβ can act as a critical tumor suppressor,61 and the dysregulation of the TGFβ pathway may increase the risk of BLCA. Sixteen CKGs also involve other cancer-related signaling pathways, including PI3K, Myc, Wnt, Notch, Hippo, and Nrf2 signaling pathways (Table S4).
We would like to highlight that CKGs are network-structure-based, fundamentally different from mutation-based CDGs. Most known CDGs have high mutation rates or greater connectivity in gene regulatory networks. In contrast, CKGs, particularly sCKGs, have average mutation rates and thus are unlikely to be detected by the existing mutation-based methods. On the other hand, most CKGs are directly connected to known CDGs and drug targets in GRN, forming gene pairs essential for the network’s function. Many CKGs are also located upstream in tumor-related signaling pathways and thus can control CDGs. Therefore, CKGs provide alternatives to CDGs, and many CKGs can be viewed or adopted as latent master regulators of CDGs. Indeed, manipulating CKGs can affect cancer cell proliferation and migration and suppress tumor growth, as we showed in our experiments using four cancer cell lines and a mouse model of BLCA (Figures 4 and S13). The six sCKGs (RPS6KA3, FGFR3, N-cadherin (CDH2), EP300, caspase-1, and FN1) that we experimentally analyzed are neither known CDGs of BLCA nor drug targets for BLCA and have not been well studied for BLCA. Combined, our analytic and experimental results strongly suggest CKGs be a class of novel regulatory elements that, when perturbed, can affect the states of the underlying cells. In particular, the six experimentally analyzed sCKGs are novel putative drug targets for BLCA treatment.
sCKGs have different characteristics from the conventional network hub nodes defined by various structure-based methods, such as the nine popular methods we analyzed (Figure S7). sCKGs are not typical network hubs and have lower centrality. Therefore, the existing hub-based methods would not be able to detect them. The sCKGs are essential to network control, and any variation to them may change overall network states. Meanwhile, the lower centrality of sCKGs makes variations to them less impact the network (or cell) stability than the other methods. Therefore, they are excellent candidates for therapeutic targets.
CKGs are also fundamentally different from housekeeping genes; the former typically have a reduced expression (Figures S8B and S8C) and are located in the middle of the control paths of a regulatory network. In contrast, housekeeping genes tend to posit at the heads of the control paths (Figure S8A). While these two types of genes are non-exclusive, most housekeeping genes are not CKGs (Figure S8A, Data S1).
Our approach can be improved by incorporating gene expression in network construction. The regulatory network we used in the study was built from 10 types of important interactions and signaling pathways, so it was a pan-cancer gene regulatory network suitable for applying network controllability. In addition to using some seed genes specific to one type of cancer, the network can be revised to a one-cancer-specific network by removing nodes (i.e., genes or proteins) and links (i.e., interactions) unsupported by gene expression data for that specific cancer. In other words, we may use gene expressions and their correlations as evidence to retrofit the network to be specific to a particular cancer by removing incompatible interactions detected in different conditions.
In summary, the extension to network structural controllability is thought-provoking and is expected to inspire future work. It can be extended in multiple dimensions to meet the urgent demands for innovative approaches for analyzing large quantities of biological data. Our novel control hub–based approach can be directly applied to various types of cancer and be readily extended to other complex diseases, such as neurodegenerative disorders and metabolic diseases.
Limitations of the study
Our gene regulatory network was built upon cancer-related pathways and protein interaction data, which may not capture all the potential protein interactions. Additionally, the network was designed for pan-cancer studies, so it might retain protein interactions occurring in different conditions that might obscure specific regulatory interactions in particular cancer. This limitation suggested using cancer-specific gene regulatory networks. Our approach did not fully consider biological factors that could influence CKGs, such as mRNA expression, posttranslational and epigenetic modifications, non-coding RNA regulation, and the influence of tumor microenvironments, which may play pivotal roles in cancer biology. Lastly, further experimental validations are needed to elucidate molecular mechanisms of CKGs in cancer progression. Despite these limitations, the new control hub-based concept of CKGs and the results presented in this paper provided a starting point in understanding CKGs and their potential as therapeutic targets and, more importantly, inspiring further exploration beyond the prevalent concept of CDGs.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Xizhe Zhang (zhangxizhe@njmu.edu.cn).
Materials availability
The study did not generate new unique reagents.
Experimental model and study participant details
Xenograft mouse model
UMUC3 cells expressing control shRNA or PRS6KA3 shRNA (2×106) were subcutaneously injected into the dorsal flank of 4-week-old male athymic nude mice (n=5 mice per group, Shanghai SLAC Laboratory AnimalCo. Ltd.). Mice were sacrificed after 3 weeks, and tumors were excised and weighed. Mice were used in the experiment at random. During testing the tumors' weight, the researchers were blinded to the information and shape of tumor tissue masses. Animal studies were conducted with approval from the Animal Research Ethics Committee of China Medical University.
Cell culture
Human bladder cancer-derived UMUC3 and T24 cells were purchased from the Chinese Academy of Sciences (Shanghai, China). UMUC3 and T24 cells were cultured in DMEM (11995-065, Gibco, USA) and DMEM-F12 (10565-018, Gibco), respectively. These mediums were supplemented with 10% heat-inactivated fetal bovine serum (FBS; 1750114, Gibco). Cells were maintained at 37°C in an incubator containing 5% CO2.
Method details
Gene regulatory network construction and data used
Five well-curated disease- and cancer-related pathway databases were used to construct a gene regulatory network (GRN). In particular, the ten most important and common types of interactions (Table S1) were used to link genes. The five databases included the National Cancer Institute (NCI) Nature Pathway Interaction Database,24 PhosphoSite Kinase-substrate information,25 HumanCyc26 (https://humancyc.org), the Reactome27 (https://www.reactome.org) and PANTHER Pathways28 (http://www.pantherdb.org). Disease studies partly motivated these database projects, so they contained information on mutated genes and cancer-related pathways. For example, Reactome consists of the KEGG databases, including disease and drug databases. Phosphosite contains data on missense mutations from UniProtKB, TCGA, and many other sources, collected from over 2,000 diseases and syndromes, and has data on polymorphisms associated with hundreds of cancers.
Structural controllability of cancer gene regulatory network
According to control theory, a dynamic networked system can be driven from any initial state to any final state in a finite time if influenced by external control signals.11 Structural controllability was introduced by Lin10 and further developed by Liu et al.11 It maps the problem of finding the minimum inputs or driver nodes into identifying a maximum matching of the network. Although most biological networks are inherently nonlinear, the experimentally obtained network, however, these networks can be assumed to capture linear effects around homeostasis. Therefore, we used the concept of local structural controllability11 and the analysis tools based on linear controllability to investigate the controllability of the cancer gene regulatory network.
Consider a directed network . The network can be modeled as a linear time-invariant system whose states are determined by the following Equation 1:
| (Equation 1) |
where the state denotes the state value of all nodes at time ; is the transpose of the adjacency matrix of ; is the input signal; is the input matrix that defines how control signals are inputted into the network. The nodes receiving external signals are called driver nodes. The minimum set of driver nodes to fully control the network is called the Minimum Driver node Set (MDS). Based on the minimum input theorem present by Liu et al.,11 the MDS can be obtained by computing any maximum matching of an equivalent undirected bipartite graph .62 The set of unmatched nodes of are the MDS. While solving the maximum matching process as mentioned above, the network is transformed into a bipartite graph by splitting the node set V into two node sets and and the maximum matching is obtained by finding the augmented path.
Next, we will introduce some concepts in graph theory. A set of edges in is called a matching M if no two edges in M have a node in common.63 A node is said to be matched by M if an edge of M is linked to ; otherwise, is unmatched.62 The maximum matching is a matching with the maximum number of edges of the graph. A path P is said to be M-alternating if the edges of P are alternately in and not in M. An M-alternating path P that begins and ends at the unmatched nodes is called an M-augmenting path. The maximum matching is a matching with the maximum number of edges of the graph.
Control schemes and control hubs of cancer gene regulatory network
Based on structural controllability theory, for a directed network , the matching edges of a maximum matching form the cactus structures in the network, which are the basic control structure of the network. Therefore, the matched edges form a set of edge-independence paths in the directed network . We call these paths control paths.30 The control paths start with driver nodes and end with tail nodes. The driver nodes (unmatched nodes) and the corresponding control path in the network are called a control scheme.31
Considering a node in the network, based on its position on the control path, we can classify it into three types. A node is called a head node if it is at the beginning of a control path of at least one control scheme. Similarly, a node is a tail node if it is at the end of a control path of at least one control scheme. A node is a middle node if it is in the middle of a control path of at least one control scheme.31
Since maximum matching is not unique,21,22 a network often has multiple control schemes. A node may change its type in different control schemes, i.e., a head node in one control scheme may become a middle node in another control scheme. Therefore, we consider a special type of node, which always lies in the middle of a control path in all control schemes. We call these nodes control hubs.31
We were interested in identifying control hubs, which are middle nodes in all control schemes. If a node is not a head node or tail node in all control schemes, it must be in the middle of a control path in all control schemes, and it must be a control hub. Therefore, we need first to find all the head and tail nodes in all control schemes, and then the rest nodes are the control hub nodes. To find head or tail nodes, we have the following claims:
Property 1. For a given network and all its control schemes, the head nodes are the union of all driver nodes of all control schemes.
Proof: Based on the definition of the driver nodes, the external control signals are inputted into the network from the driver nodes. Therefore, the driver nodes lie at the beginning of the control paths of a control scheme. Thus, the head nodes are the union of all the driver nodes of all control schemes of a network.
Consider a network . We say a network is the transpose network of if it has the same nodes set, but the directions of all edges are reversed compared to the corresponding edges in . To identify tail nodes, we have the following property:
Property 2. For a given network and its transpose network , the head nodes of are the tail nodes of .
Proof: For the directed network , let be the corresponding undirected bipartite graph. Consider the transpose network ; the only difference between and is that all edges' direction is reversed (see Figure S1 for an example). Therefore, the undirected bipartite graph of must be . Because and have the same edges set, a maximum matching of must be a maximum matching of . Therefore, the control paths of all control schemes of are the same as that of except they have reversed directions, making the head nodes of to be the tail nodes of , which completes the proof.
Based on the above properties, we can obtain all control hubs by computing all driver nodes of network G and its transpose network G'. Our previous work31,32 has proved that all driver nodes of a network can be found in polynomial time. Therefore, we can identify all control hubs in polynomial time without computing all control schemes. More details about all driver nodes and control hubs can be found in our previous work.21,31,32
Identification of sensitive control hub nodes or sensitive cancer-keeper genes
While control hubs are vulnerable spots for the structural controllability of a network, they may have different resilience to external stimuli. A control hub is considered sensitive if it is no longer a control hub after removing any edge from the network. All sensitive control hubs of a network can be identified by removing one edge at a time and examining whether the set of control hubs of every control scheme remains intact. The complexity of this algorithm is O(n0.5m2) on a network with n nodes and m links in the worst.
Functional enrichment analysis of cancer-keeper genes
A function enrichment analysis was adopted to assess the biological functions of CKGs using the information in 33 gene function datasets (Table S3). These 33 gene function datasets include essential and conserved gene datasets, protein datasets that regulate translation and posttranslational modifications (PTMs), and datasets of pathogenic mutations and targets of viruses, drugs, and immunotherapies. All datasets used for this study’s enrichment analysis are listed in Data S1. The enrichment for a particular function was reflected by those CKGs that were annotated with that function. A sample of N genes was randomly chosen from the regulatory network GRN with N CKGs. The functional enrichment of this random sample was the overlapping of the random set with the given functional dataset. Random samplings were chosen 1,000 times, and their enrichments constituted an empirical enrichment distribution of CKGs.
Map sensitive CKGs with cancer driver genes and therapeutic targets
The CKGs were further analyzed using several datasets, including datasets of CDGs, therapeutic targets, and immune genes (Table S3). The CDG dataset29 includes 739 genes from 9,423 tumor exome sequencing data from TCGA. Cancer therapeutic target dataset46 consists of 628 genes obtained from genome-scale CRISPR-Cas9 screening of 324 human cancer cell lines from 30 cancer types. Immune-related genes include 358 genes obtained by coreCTL immune-related gene dataset,30 CAR immune-related gene datasets,31 and immune checkpoints.36,37 We analyzed the overlapping genes among the three datasets with the CKGs.
Survival analysis
Survival analysis was performed on 35 sCKGs using the clinical data of 411 samples of Bladder Urothelial Carcinoma (TCGA PanCancer analysis) from the cbioportal website (http://www.cbioportal.org). We counted the mutated genes in 35 sCKG across all 411 samples and selected 4 subsets based on the number of simultaneously mutated genes. Whether a gene was mutated depends on the common alterations, including somatic mutation, gene fusion, copy number amplification, or homozygous deletion. These subsets had no mutated genes, at least one, two, or three mutated genes in the 35 sCKGs. The survival analysis was performed using the Kaplan-Meier estimator64 from the cbioportal website (http://www.cbioportal.org).
Transient siRNA-mediated gene knockdown
The negative control (siRNA-NC) and siRNAs targeting the CKGs to be tested were purchased from Shanghai Genechem Co., Ltd. (China). For transient transfection, a designated siRNA was introduced into cells in the presence of Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, US) according to the manufacturer’s instructions. The siRNA was diluted to the final concentration of 20 μM following the manufacturer’s instructions. Cancer cells were transfected using Lipofectamine TM RNAiMAX (Invitrogen, Carlsbad, CA, US) and opti-MEM (Gibco). Twenty-four hours after transfection, cells were collected and subjected to subsequent experiments. The siRNA sequences tested are listed in Tables S9 and S10. Three siRNAs were designed to target different regions of each target gene tested, and the knockdown efficiency was tested. The siRNA with a significant knockdown effect was selected for subsequent functional verification, and the rest siRNAs were discarded.
Cell counting Kit 8 (CCK-8) assay
The effect of knocking down candidate CKGs on cell proliferation was examined by CCK-8 assays. Twenty-four hours after transfection, cells were collected and maintained in 96-well plates (1,500 cells/well). Four hours after incubation, 10 μl of CCK-8 reagent (Vazyme, Nanjing, China) was added to each well, and the reaction mixtures were incubated for 2 h at 37°C under a 5% CO2 atmosphere. CCK-8 assay was performed at the same time every day for 4 consecutive days, and a total of 96h of cell activity was recorded. The absorbance (OD) value at 450 nm wavelength was then measured using a Microplate reader (Bio-Rad, CA).
Transwell assay
Transwell assays were employed to determine the effect of candidate cancer-keeper genes on cancer cells' migration and invasion abilities. Twenty-four hours after transfection, cells were collected and subjected to transwell assay. In brief, the upper compartment of the filters (Corning, NY, USA) was coated with (invasion) or without (migration) 55 μl of the basal membrane matrix (1:7 dilution, Corning). Cells were diluted with serum-free medium to 1× 105 cells per ml. Cell suspension (200 μl) was added to the upper compartment, and 600 μl of RPMI-1640 containing 10% FBS was added to the lower compartment. Twenty-four hours after incubation at 37°C under 5% CO2, filters were fixed with 4% formaldehyde for 30 min, then stained with 0.1% crystal violet at room temperature for 30 min. Filters were then rinsed 3 times with PBS, and the unmigrated cells were removed with cotton swabs. Finally, cells were photographed and counted in a 5-independent microscopic field.
Quantification and statistical analysis
One-way ANOVA was adopted to measure the degree difference between control hubs and other genes, followed by multiple comparison tests with a 0.05 significance level. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. A one-sided t-test was adopted to measure the statistical significance of sCKGs in bladder cancer. Data represent mean±s.d. from three replicate cultures. A z-test, using Z = , was adopted to measure the difference between the function enrichment of CKGs () and the empirical distribution (), along with the statistical significance of the two-tailed p-value.
Acknowledgments
This work was partly supported by the National Natural Science Foundation of China (grant numbers 62176129 and 81672523), the Scientific Research Foundation of Liaoning Province (LJKZ1134), the Hong Kong Global STEM Professorship Scheme, the Hong Kong Jockey Club Charities Trust, and a grant from the Hong Kong Health and Medical Fund (grant number 10211696).
Author contributions
X.Z., W.Z., and Y.Z. conceived and designed the project. X.Z. supervised the work, designed the algorithm, analyzed its correctness and complexity, performed data analysis, and drafted the manuscript. W.Z. conceived the main conceptual ideas and wrote the manuscript. Y.Z. and Y.Y. designed and supervised the biological experiments. Y.Z. analyzed the results and helped with writing the manuscript. C.P. and X.W. implemented the algorithm and collected and analyzed the data. Y.M. was involved in the design of in vivo experiments and related data analysis. S.L., J.A., and J.Y. conducted the in vitro and in vivo experiments and collected data. B.W. and W.H. analyzed the data and results from the in vitro and in vivo experiments.
Declaration of interests
No potential conflicts of interest were disclosed.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107296.
Contributor Information
Xizhe Zhang, Email: zhangxizhe@njmu.edu.cn.
Yang Yao, Email: yaoyang1972@gmail.com.
Yuyan Zhu, Email: yyzhu@cmu.edu.cn.
Weixiong Zhang, Email: weixiong.zhang@polyu.edu.hk.
Supplemental information
Data and code availability
-
•
DATA: The data for establishing gene regulatory networks in this study have been deposited at Github: https://github.com/network-control-lab/control-hubs and are publicly available as of the date of publication. Additionally, in vivo experimental data from our mouse model, as reported in this study, are available upon request from the lead contact.
-
•
CODE: All original code for finding control hubs and cancer-keeper genes are freely available at GitHub: https://github.com/network-control-lab/control-hubs, and it is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sayılgan J.F., Haliloğlu T., Gönen M. Protein dynamics analysis identifies candidate cancer driver genes and mutations in TCGA data. Proteins. 2021;89:721–730. doi: 10.1002/prot.26054. [DOI] [PubMed] [Google Scholar]
- 3.Lee G.Y., Haverty P.M., Li L., Kljavin N.M., Bourgon R., Lee J., Stern H., Modrusan Z., Seshagiri S., Zhang Z., et al. Comparative Oncogenomics Identifies PSMB4 and SHMT2 as Potential Cancer Driver Genes. Cancer Res. 2014;74:3114–3126. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z., Zhou J., Jiang J., Yuan J., Zhang Y., Wei X., Loo N., Wang Y., Pan Y., Zhang T., et al. Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment. Nat. Commun. 2019;10:733. doi: 10.1038/s41467-019-08554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.-P., Nickerson E., Auclair D., Li L., Place C., et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dees N.D., Zhang Q., Kandoth C., Wendl M.C., Schierding W., Koboldt D.C., Mooney T.B., Callaway M.B., Dooling D., Mardis E.R., et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamborero D., Gonzalez-Perez A., Perez-Llamas C., Deu-Pons J., Kandoth C., Reimand J., Lawrence M.S., Getz G., Bader G.D., Ding L., Lopez-Bigas N. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsiehchen D., Hsieh A. Nearing saturation of cancer driver gene discovery. J. Hum. Genet. 2018;63:941–943. doi: 10.1038/s10038-018-0481-4. [DOI] [PubMed] [Google Scholar]
- 10.Ching-Tai, Lin Structural controllability. IEEE Trans. Automat. Contr. 1974;19:201–208. doi: 10.1109/TAC.1974.1100557. [DOI] [Google Scholar]
- 11.Liu Y.-Y., Slotine J.-J., Barabási A.L. Controllability of complex networks. Nature. 2011;473:167–173. doi: 10.1038/nature10011. [DOI] [PubMed] [Google Scholar]
- 12.Gu S., Pasqualetti F., Cieslak M., Telesford Q.K., Yu A.B., Kahn A.E., Medaglia J.D., Vettel J.M., Miller M.B., Grafton S.T., Bassett D.S. Controllability of structural brain networks. Nat. Commun. 2015;6:8414. doi: 10.1038/ncomms9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barabási A.L., Gulbahce N., Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., Li M., Wang J.-X., Wu F.-X. Controllability and Its Applications to Biological Networks. J. Comput. Sci. Technol. 2019;34:16–34. doi: 10.1007/s11390-019-1896-x. [DOI] [Google Scholar]
- 15.Kanhaiya K., Czeizler E., Gratie C., Petre I. Controlling Directed Protein Interaction Networks in Cancer. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-10491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolouri H. World Scientific Publishing Company; 2008. Computational Modeling of Gene Regulatory Networks-A Primer. [Google Scholar]
- 17.Qian X., Ivanov I., Ghaffari N., Dougherty E.R. Intervention in gene regulatory networks via greedy control policies based on long-run behavior. BMC Syst. Biol. 2009;3:61. doi: 10.1186/1752-0509-3-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D.-H., Motter A.E. Slave nodes and the controllability of metabolic networks. New J. Phys. 2009;11 doi: 10.1088/1367-2630/11/11/113047. [DOI] [Google Scholar]
- 19.Asgari Y., Salehzadeh-Yazdi A., Schreiber F., Masoudi-Nejad A. Controllability in cancer metabolic networks according to drug targets as driver nodes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W.-F., Zhang S.-W., Zeng T., Akutsu T., Chen L. Network control principles for identifying personalized driver genes in cancer. Brief. Bioinform. 2020;21:1641–1662. doi: 10.1093/bib/bbz089. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Lv T., Pu Y. Input graph: the hidden geometry in controlling complex networks. Sci. Rep. 2016;6 doi: 10.1038/srep38209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia T., Liu Y.Y., Csóka E., Pósfai M., Slotine J.J., Barabási A.L. Emergence of bimodality in controlling complex networks. Nat. Commun. 2013;4:2002. doi: 10.1038/ncomms3002. [DOI] [PubMed] [Google Scholar]
- 23.Valiant L.G. The complexity of computing the permanent. Theor. Comput. Sci. 1979;8:189–201. doi: 10.1016/0304-3975(79)90044-6. [DOI] [Google Scholar]
- 24.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H. PID: the Pathway Interaction Database. Nucleic Acids Res. 2008;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornbeck P.V., Chabra I., Kornhauser J.M., Skrzypek E., Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 26.Romero P., Wagg J., Green M.L., Kaiser D., Krummenacker M., Karp P.D. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2005;6:R2–R17. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H., Thomas P. Springer; 2009. PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B., et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruths J., Ruths D. Control profiles of complex networks. Science. 2014;343:1373–1376. doi: 10.1126/science.1242063. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X., Pan C., Zhang W. 2022. Control Hubs of Complex Networks and a Polynomial-Time Identification Algorithm. arXiv:2206.01188. [DOI] [Google Scholar]
- 32.Zhang X., Han J., Zhang W. An efficient algorithm for finding all possible input nodes for controlling complex networks. Sci. Rep. 2017;7:10677–10678. doi: 10.1038/s41598-017-10744-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinayagam A., Gibson T.E., Lee H.-J., Yilmazel B., Roesel C., Hu Y., Kwon Y., Sharma A., Liu Y.-Y., Perrimon N., Barabási A.L. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc. Natl. Acad. Sci. USA. 2016;113:4976–4981. doi: 10.1073/pnas.1603992113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawson K.A., Sousa C.M., Zhang X., Kim E., Akthar R., Caumanns J.J., Yao Y., Mikolajewicz N., Ross C., Brown K.R., et al. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature. 2020;586:120–126. doi: 10.1038/s41586-020-2746-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKay M., Afshinnekoo E., Rub J., Hassan C., Khunte M., Baskaran N., Owens B., Liu L., Roboz G.J., Guzman M.L., et al. The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat. Biotechnol. 2020;38:233–244. doi: 10.1038/s41587-019-0329-2. [DOI] [PubMed] [Google Scholar]
- 36.Wei S.C., Duffy C.R., Allison J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.Cd-18-0367. [DOI] [PubMed] [Google Scholar]
- 37.He X., Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660–669. doi: 10.1038/s41422-020-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 41.Sootome H., Fujita H., Ito K., Ochiiwa H., Fujioka Y., Ito K., Miura A., Sagara T., Ito S., Ohsawa H., et al. Futibatinib Is a Novel Irreversible FGFR 1–4 Inhibitor That Shows Selective Antitumor Activity against FGFR-Deregulated Tumors. Cancer Res. 2020;80:4986–4997. doi: 10.1158/0008-5472.CAN-19-2568. [DOI] [PubMed] [Google Scholar]
- 42.Damian S., Gable A.L., Nastou K.C., David L., Rebecca K., Sampo P., Doncheva N.T., Marc L., Fang T., Peer B. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2020;D1 doi: 10.1093/nar/gkaa1074. [DOI] [Google Scholar]
- 43.Dimitrakopoulos C.M., Beerenwinkel N. Wiley Interdisciplinary Reviews: Systems Biology; 2017. Computational Approaches for the Identification of Cancer Genes and Pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajendran B.K., Deng C.X. Characterization of potential driver mutations involved in human breast cancer by computational approaches. Oncotarget. 2017;8:50252–50272. doi: 10.18632/oncotarget.17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan A., Huang H., Zhang P., Li S. Network-based cancer precision medicine: A new emerging paradigm. Cancer Lett. 2019;458:39–45. doi: 10.1016/j.canlet.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 46.Behan F.M., Iorio F., Picco G., Gonçalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M., et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 47.Sanli O., Dobruch J., Knowles M.A., Burger M., Alemozaffar M., Nielsen M.E., Lotan Y. Bladder cancer. Nat. Rev. Dis. Primers. 2017;3 doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. Akbani R., Broom B.M., Wang W., Verhaak R., Al E. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang S., Dong S., Gu T.L., Guo A., Cohen M.S., Lonial S., Khoury H.J., Fabbro D., Gilliland D.G., Bergsagel P.L., et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasekar M., Degraff D., Joshi M. Immunotherapy in Bladder Cancer. Curr. Mol. Pharmacol. 2016;9:242–251. doi: 10.2174/1874467208666150716120945. [DOI] [PubMed] [Google Scholar]
- 51.Zucman-Rossi J., Villanueva A., Nault J.C., Llovet J.M. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology. 2015;149:1226–1239.e4. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 52.Guo W.F., Zhang S.W., Feng Y.H., Liang J., Zeng T., Chen L. Network controllability-based algorithm to target personalized driver genes for discovering combinatorial drugs of individual patients. Nucleic Acids Res. 2021;49:e37. doi: 10.1093/nar/gkaa1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vermeulen K., Van Bockstaele D.R., Berneman Z.N. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. doi: 10.1046/j.1365-2184.2003.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stivala L.A., Cazzalini O., Prosperi E. The cyclin-dependent kinase inhibitor p21CDKN1A as a target of anti-cancer drugs. Curr. Cancer Drug Targets. 2012;12:85–96. doi: 10.2174/156800912799095126. [DOI] [PubMed] [Google Scholar]
- 55.Chohan T.A., Qian H., Pan Y., Chen J.-Z. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr. Med. Chem. 2015;22:237–263. doi: 10.2174/0929867321666141106113633. [DOI] [PubMed] [Google Scholar]
- 56.Bertino J.R., Banerjee D. E2F-1 as an anticancer drug target. Oncol. Rev. 2009;3:207–214. doi: 10.1007/s12156-009-0028-1. [DOI] [Google Scholar]
- 57.Mitra A.P., Birkhahn M., Cote R.J. p53 and retinoblastoma pathways in bladder cancer. World J. Urol. 2007;25:563–571. doi: 10.1007/s00345-007-0197-0. [DOI] [PubMed] [Google Scholar]
- 58.Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 59.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y., Kwiatkowski D.J. Combined CDKN1A/TP53 mutation in bladder cancer is a therapeutic target. Mol. Cancer Ther. 2015;14:174–182. doi: 10.1158/1535-7163.MCT-14-0622-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L. TGFß, a potent regulator of tumor microenvironment and host immune response, implication for therapy. Curr. Mol. Med. 2010;10:374–380. doi: 10.2174/156652410791317039. [DOI] [PubMed] [Google Scholar]
- 62.Hopcroft J.E., Karp R.M. An nˆ5/2 algorithm for maximum matchings in bipartite graphs. SIAM J. Comput. 1973;2:225–231. doi: 10.1137/0202019. [DOI] [Google Scholar]
- 63.Lovász L., Plummer M. American Mathematical Soc); 2009. Matching Theory. [Google Scholar]
- 64.Peterson A.V., Jr. Expressing the Kaplan-Meier estimator as a function of empirical subsurvival functions. J. Am. Stat. Assoc. 1977;72:854–858. doi: 10.1080/01621459.1977.10479970. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
DATA: The data for establishing gene regulatory networks in this study have been deposited at Github: https://github.com/network-control-lab/control-hubs and are publicly available as of the date of publication. Additionally, in vivo experimental data from our mouse model, as reported in this study, are available upon request from the lead contact.
-
•
CODE: All original code for finding control hubs and cancer-keeper genes are freely available at GitHub: https://github.com/network-control-lab/control-hubs, and it is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.