Summary
Background
Schistosomiasis is a disease that significantly impacts human health in the developing world. Effective diagnostics are urgently needed for improved control of this disease. CRISPR-based technology has rapidly accelerated the development of a revolutionary and powerful diagnostics platform, resulting in the advancement of a class of ultrasensitive, specific, cost-effective and portable diagnostics, typified by applications in COVID-19/cancer diagnosis.
Methods
We developed CRISPR-based diagnostic platform SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) for the detection of Schistosoma japonicum and S. mansoni by combining recombinase polymerase amplification (RPA) with CRISPR-Cas13a detection, measured via fluorescent or colorimetric readouts. We evaluated SHERLOCK assays by using 150 faecal/serum samples collected from Schistosoma-infected ARC Swiss mice (female), and 189 human faecal/serum samples obtained from a S. japonicum-endemic area in the Philippines and a S. mansoni-endemic area in Uganda.
Findings
The S. japonicum SHERLOCK assay achieved 93–100% concordance with gold-standard qPCR detection across all the samples. The S. mansoni SHERLOCK assay demonstrated higher sensitivity than qPCR and was able to detect infection in mouse serum as early as 3 weeks post-infection. In human samples, S. mansoni SHERLOCK had 100% sensitivity when compared to qPCR of faecal and serum samples.
Interpretation
These schistosomiasis diagnostic assays demonstrate the potential of SHERLOCK/CRISPR-based diagnostics to provide highly accurate and field-friendly point-of-care tests that could provide the next generation of diagnostic and surveillance tools for parasitic neglected tropical diseases.
Funding
Australian Infectious Diseases Research Centre seed grant (2022) and National Health and Medical Research Council (NHMRC) of Australia (APP1194462, APP2008433).
Keywords: Schistosomiasis, Schistosoma japonicum, Schistosoma mansoni, Diagnostics, CRISPR, Cas13a, SHERLOCK, Point-of-care
Research in context.
Evidence before this study
Parasitic helminths, including schistosomiasis, cause devastating diseases afflicting 1.5 billion people in the world and represent a significant public health and economic burden, especially in developing countries. Currently available diagnostic tools for helminth infections are neither sufficiently sensitive nor field-friendly for use in low-endemic or resource-poor settings where rebound infections can occur, leading to underestimation of true prevalence rates. Advanced tools are urgently needed for rapid mapping of helminthic diseases and monitoring control efforts as mass drug administration (MDA) programs are unsustainable long-term.
Since the award of the Nobel Prize for Chemistry (2020) for discovery of the CRISPR/Cas9 genetic scissors that revolutionized genome editing, interest in CRISPR technology has been accelerated in the scientific community and has led to the development of a powerful new generation CRISPR-based diagnostics platform. Utilization of the CRISPR-Cas12/13 system for nucleic acid detection has resulted in a powerful platform of ultra-sensitive, fast, specific, cost-effective and portable diagnostics without the need for specialized equipment or expertise, typified by applications in COVID-19 and cancer diagnosis. At present, the development of CRISPR-based platforms for parasite diagnosis has been limited to the detection of unicellular parasites (e.g., malaria, Cryptosporidium parvum and Enterocytozoon hepatopenaei) and the plant cyst nematode-Heterodera schachtii. This revolutionary technology has yet to be enlisted for detecting parasitic helminth infections by using field-derived samples.
Added value of this study
We have used the Schistosoma bloodfluke model to successfully establish diagnostic assays with the nucleic acid detection platform SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) by combining recombinase polymerase amplification (RPA) and CRISPR-Cas13a detection to diagnose schistosomiasis in humans and animals. We showed that these CRISPR-based assays are capable of ultra-sensitive detection and show great promise as a potential point-of-care diagnostic tool. They exhibit similar diagnostic sensitivity as qPCR-based assays, which are currently the most sensitive approach for the diagnosis of helminthic infections, but with significantly reduced requirements for trained personnel and expensive technical equipment. Our S. japonicum and S. mansoni SHERLOCK assays have the potential to fulfil key recommendations of the neglected tropical diseases (NTDs) 2021–2030 roadmap and the 2022 Guideline on the Control and Elimination of Human Schistosomiasis released recently by the World Health Organization.
Implications of all the available evidence
We have demonstrated that SHERLOCK is a sensitive, accurate, user-friendly nucleic acid detection platform that can be utilized for the detection of schistosomiasis. Although further optimisation is required before field-ready implementation, CRISPR-based nucleic acid detection shows great promise as the basis of a point-of-care (POC) diagnostic tool for clinical diagnosis and surveillance of schistosomiasis with potential extension to other helminthiases. Accurate and portable SHERLOCK POC diagnostics could play a crucial role in the prevention of severe NTDs by facilitating immediate and targeted treatment. It could also provide more accurate data on disease prevalence to policymakers and allow them to monitor transmission, identify new outbreaks of schistosomiasis, and sustain control efforts more effectively. Access to CRISPR-based POC diagnostics could be a game changer in the global fight against schistosomiasis and other helminthic diseases.
Introduction
Parasitic helminths are a global scourge but mainly impact low-income communities in developing tropical/subtropical countries. Asia and sub-Saharan Africa contain infection hotspots, with approximately 90% of the world's parasitic worm infestations occurring on these continents.1 Parasitic worms cause severe morbidity and mortality and result in chronic disease outcomes, with significant impacts on human health and economic development. Schistosomiasis is an acute and chronic parasitic disease caused by trematode blood flukes. Over 250 million people are infected and 800 million in 78 countries are at risk of infection.2 As the second most socioeconomically devastating parasitic disease after malaria,3 schistosomiasis is listed by the World Health Organisation (WHO) for control and elimination. One key roadblock to schistosomiasis elimination, identified in the new WHO neglected tropical diseases (NTDs) roadmap 2021–2030,4 is the lack of a diagnostic test that provides timely and accurate results while still being accessible, inexpensive, and able to be performed by local personnel with minimal training.5 As schistosomiasis often occurs in remote rural areas, a point-of-care (POC)-based diagnostic is particularly relevant.5 Deployment of a sensitive POC test will confer many benefits for control efforts by significantly reducing drug costs through targeted treatment of infected subjects rather than mass drug administration (MDA) with praziquantel, thereby reducing the risk of anthelminthic-drug resistance developing and increasing treatment compliance. Moreover, highly sensitive diagnostics will allow for better mapping of schistosome transmission dynamics and provide more accurate disease burden estimates. As elimination/control programs progress in schistosomiasis-endemic areas, ultra-sensitive and specific diagnostic procedures will be essential to accurately record the infection status of susceptible populations to prevent re-bound infections following the cessation of MDA programs.6
Currently, diagnosis of helminth infections (including schistosomiasis) in the field relies almost entirely on conventional diagnostics,7, 8, 9, 10 including stool-based Kato-Katz (KK) method, the miracidial hatching technique and antigen-based detection. These diagnostic tools lack sensitivity and/or demonstrate poor replicability with variability, and are often unreliable and labour-intensive, especially when applied in areas of low parasite prevalence/intensity.11,12 Currently, POC circulating cathodic antigen (POC-CCA) cassette is commercially available for detection of S. mansoni antigens, measured via lateral flow readout in 20 min by using urine samples, although sensitivity (70%) and specificity (57%) of the POC-CCA are low.13,14 Recently, diagnostics based on nucleic acid amplification (such as PCR-based assays and cell-free DNA detection), have achieved a much higher degree of sensitivity and specificity15; however the requirements for costly laboratory instrumentation, reagents, and trained personnel have significantly hampered the practicability of widespread use of these tests particularly in remote endemic areas in developing countries. Without the availability of a readily accessible, sensitive and affordable diagnostic test, there will be underestimations of active schistosomiasis cases, and errors in evaluation of transmission rates and in interpreting outcomes of MDA programs.12 A new generation of field-friendly schistosomiasis (and other helminth) diagnostics is urgently needed to improve control efforts and eventually eliminate the spread of parasitic worm infections globally.
Over the past 5 years, a new class of CRISPR-based diagnostics has emerged that has proven especially promising in virology (including COVID-19) and cancer diagnosis.16, 17, 18, 19 These systems utilise the programmable CRISPR-associated endonucleases Cas12 and Cas13, and have been shown to be ultra-sensitive, rapid, specific, cost-effective and portable, without the necessity for specialized equipment and expertise.18,20, 21, 22 The CRISPR-based diagnostic system typically involves amplification of target sequence using isothermal amplification methods, followed by CRISPR-Cas12/13-mediated detection that combines the binding specificity of programmable crRNAs with the collateral nonspecific enzymatic cleavage activities of Cas12 (targets dsDNA) and Cas13 (targets RNA) to cleave a reporter sensor23 that is measured via fluorescent or colorimetric readouts. Isothermal amplification methods include recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP), both of which can amplify target nucleic acids at a single temperature - usually 37 °C (RPA), or 65 °C (LAMP)- making the system especially suitable for field-based application. The Cas13a-based SHERLOCK platform has been shown to be able to clinically diagnose viral infections (Zika and dengue virus) at concentrations as low as 1 copy/μl,22 and with single-base mismatch specificity for detection of SARS-CoV-2 in less than 1 h.24 Recently, successful CRISPR-Cas12/13 mediated diagnostics have been developed only for the detection of unicellular protozoan parasites [including Plasmodium,25,26 Trypanosoma,27 Leishmania,28 Cryptosporidium parvum29,30 and Enterocytozoon hepatopenaei31] and the plant cyst nematode Heterodera schachtii.32 This revolutionary technology has the potential to fulfil the unmet and urgent needs for parasite detection recommended by WHO. Here, we describe the development of a new diagnostic tool for schistosomiasis using the RPA-based CRISPR-Cas13a nucleic acid detection platform (SHERLOCK) that enables ultrasensitive detection of two major schistosome species, Schistosoma japonicum and S. mansoni.
Methods
Ethics statement
All experiments undertaken in this study were approved by the Animal and Human Ethics Committee (ethics number P3706, P3705, P524, and P3700) of the QIMR Berghofer Medical Research Institute (QIMRB). The study was performed according to the guidelines of the National Health and Medical Research Council of Australia, which were published in the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 7th edition, 2004 (www.nhmrc.gov.au). All work related to live S. japonicum and S. mansoni life cycle stages was conducted in quarantine-approved facilities. The human research ethical approval for the study was obtained from the Institutional Review Board (IRB) of the Research Institute for Tropical Medicine (RITM), Manila, Philippines (IRB Number 2015–12) and Vector Control Division, Ministry of Health, Uganda (IRB number VCDREC160). Written consent was obtained from all participants (for children aged 15 years or under written consent was obtained from their legal guardians).
Mouse infection and faecal sample collection
S. japonicum: Two groups of six-week-old female ARC Swiss mice (8-week old, n = 10 mice/group, 5 mice/cage) were anesthetized and infected percutaneously with either a low dosage (30/mouse) and high dosage (70/mouse), respectively, of S. japonicum (Philippines strain) cercariae. Faeces were collected from individual mice weekly from week 1 to week 7 post-infection (p.i.) for DNA extraction. Egg counting of individual faecal samples was undertaken at 4–7 weeks p.i. as described.33 Mice were euthanized at 7 weeks p.i. and adult worms obtained by portal perfusion were counted. Uninfected mice were used as negative controls.
S. mansoni: Six-week-old female ARC Swiss mice (8-week old, n = 10 mice, 5 mice/cage) were infected percutaneously with 180 S. mansoni cercariae per mouse. Faeces were collected weekly from week 4 to week 7 p.i. and serum was collected at week 3, 5 and 7 p.i. from individual mice for DNA extraction. Mice were euthanized at week 7 p.i. and adult worms obtained by portal perfusion were counted. Uninfected mice were used as negative controls.
ARC Swiss mice were purchased from Australia Animal Resources Center (ARC). For all experiments, mice had free access to tap water and food (chow) and were kept in rooms with 20 °C and 50% humidity. All animals had standardised enrichment according to protocols of the animal facility.
Human sample (faeces and serum) collection
Human samples (n = 38 faeces and n = 37 sera) were collected from subjects residing in 18 barangays (villages) endemic for S. japonicum in Laoang and Palapag municipalities, Northern Samar, the Philippines, as previously described.34 Human samples (n = 57 faeces and n = 57 sera) were obtained from subjects in 6 villages endemic for S. mansoni in Mayuge and Kabale districts in Uganda (Kabale is a non-endemic area and the samples collected from Kabale were used as negative controls). Samples from different gender were selected randomly (self-reported by study participants). Briefly, serum samples were obtained by centrifuging blood samples in serum separation tubes at 1500 × g for 10 min after prior incubation at room temperature for 30 min. Age and gender information for each participant were recorded at the time of sampling. Individual stool (fixed in 80% ethanol) and serum samples, collected from each subject, were shipped with dry ice to QIMRB, Australia for DNA extraction and further analysis. Two faecal samples were collected from each individual and KK tests performed on 3 slides/faecal sample (total 6 slides/individual) with results presented as the average number of eggs per gram of faeces (EPG).35
DNA extraction from mouse and human samples
Genomic DNA was isolated from mouse faecal samples using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). Stool (200 mg/sample) was first washed with ddH2O, then 500 μl ROSE buffer was added and samples were manually homogenized using a toothpick as described.34 Homogenates were incubated at 95 °C for 10 min, centrifuged at 4000 × g for 5 min and 200 μl of the resulting supernatant was transferred into a new tube along with 25 μl of proteinase K. The remaining DNA isolation steps were carried out following the QIAamp protocol. DNA was isolated from mouse serum samples using the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek, Norcross, USA) using the whole blood and body fluids protocol. Genomic DNA was extracted from ethanol-fixed human faecal samples (200 mg/sample) using the Maxwell®16 Instrument (Promega, Wisconsin) and the Maxwell®16 LEV Plant DNA kit as previously described.34 For the human serum samples collected in the Philippines, DNA was extracted from 2 ml of each serum sample using the ChemagicTM360 instrument (PerkinElmer Inc., Massachusetts). For the human serum samples collected in Uganda, DNA was extracted from 250 μl serum using the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek).
RPA primer and crRNA design
RPA primers were designed to target recognised diagnostic genes for schistosomiasis. Two S. japonicum genes, the mitochondrial cox1 (cytochrome C oxidase subunit 1, Sjcox1, GenBank accession no: EU325891.1)36 and sap4 (coding saposin-like protein 4, Sjsap4, FN315320.1)37 were selected for detection of S. japonicum infection. Two S. mansoni genes including cox1 (Smcox1, AF216698.1)38 and the 121-bp tandem repeat sequence Sm1-7 (M61098)39 were specifically targeted for S. mansoni diagnosis. The NCBI Primer Basic Local Alignment Search Tool (BLAST) was used for RPA primer design according to the following criteria: 1) 30–35 nucleotides in length; and 2) amplicon length of ∼125 bp targeting a ∼28 nt crRNA binding site. A T7 promoter sequence (5′-GAAATTAATACGACTCACTATAGGG-3′) was incorporated into the 5′ end of the forward primer to enable in vitro transcription by T7 polymerase during SHERLOCK detection.19 Primers were synthesized by Integrated DNA Technologies (IDT, Singapore).
After RPA primer optimisation, crRNAs were designed to target Sjcox1, Sjsap4, Smcox1, and Sm1-7, synthesised by IDT, resuspended in nuclease-free water and stored at −20 °C. The full list of oligonucleotides (including the RPA primers, qPCR primers, crRNA) used in this study is provided in Supplementary Table S1.
Recombinase polymerase amplification (RPA)
RPA reactions were performed using the TwistAmp Basic (TwistDx, Maidenhead, UK) kit according to the manufacturer's instructions, with the following modifications. Magnesium acetate was added prior to sample input, and each 50 μl TwistAmp reaction was divided into 5 × 10 μl reactions, each containing 9 μl of the reconstituted RPA reaction and 1 μl DNA sample. Reactions were incubated at 37 °C in a thermal cycler for 1 h. After 7 min initial incubation, samples were removed, vortexed, centrifuged briefly, and then returned to the cycler for the remaining incubation time. In order to determine the best primer pairs for downstream Cas13 detection of S. japonicum, amplicons (produced using the RPA primers Sjcox1-F1R1, Sjcox1-F2R2, Sjsap4-F1R1 and Sjsap4-F2R2) were purified using the QIAquick PCR Purification Kit (Qiagen), which was only performed during RPA primer selection and then visualised on a 2% (w/v) agarose gel.
Detection using LwCas13a
Fluorescent detection assays were performed in triplicate in a 20 μl reaction volume containing 44 nM LwCas13a (GenScript, Piscataway, USA), 10 ng crRNA (IDT, Singapore), 125 nM fluorescent Poly-U reporter (IDT) (Supplementary Table S2), 1 mM rNTP mix (New England Biolabs; NEB, Ipswich, USA), 20 mM HEPES, 9 mM MgCl2, 0.6 μl Murine RNase inhibitor (ScienCell, Carlsbad, USA), 0.5 μl T7 RNA Polymerase (LGC Biosearch Technologies, Middlesex, UK) and 1 μl RPA product. Reactions were incubated at 37 °C for 1 h on a BioTek Synergy H4 multi-mode plate reader (Agilent, Santa Clara, USA) with kinetics measured every 5 min. For each sample, 2 biological replicates (separate RPA reactions) were tested. Each SHERLOCK run included a no-template ‘Blank’ (ddH2O replacing the RPA product), a no-template control (NTC; ddH2O as RPA input), a negative control (NC; serum or stool DNA extracted from uninfected mice or humans), and a positive control (PC; S. japonicum or S. mansoni DNA extracted from either eggs or adult worm pairs). Background-subtracted fluorescence was calculated by subtracting the absolute fluorescence values of no-template ‘Blank’ wells from sample well fluorescence values. To compare samples across runs, sample background-subtracted fluorescence values were normalised relative to positive control fluorescence values which were set to 40,000 a.u. Data are presented as the mean ± SE for 3 technical replicates. Some reactions were also visualised under UV light and the results were captured by smart phone camera.
Lateral flow assays were set up with the same reaction components as the fluorescent detection assays, substituting the fluorescent reporter with 250 nM FAM-Biotin Poly-U reporter (IDT) (Supplementary Table S2). After incubation at 37 °C for 1 h, 80 μl HybriDetect 1 assay buffer (Milenia Biotec, Gießen, Germany) was added to 20 μl of the Cas13a reaction and run on HybriDetect 1 lateral flow strips (Milenia Biotec, Gießen, Germany). The strips were incubated at room temperature for 2–3 min and the result was recorded by camera. Lateral flow strips were interpreted visually; a strong band at the test line indicated a positive result (see Supplementary Figure S1 for additional details on interpretation of lateral flow strips).
Analytical specificity
To evaluate the specificity of the SHERLOCK assays, Sjcox1 and Sm1-7 SHERLOCK assays were performed, respectively, using DNA (1 μl of 1 ng/μl) extracted from adult worms of a number of intestinal parasite species stored in our laboratory, including trematodes (Schistosoma haematobium, Fasciola gigantica), cestodes (Echinococcus granulosus, E. multiceps, Hymenolepis diminuta, Taenia multiceps, T. ovis, T. solium), and a nematode (Ascaris lumbricoides).
Real-time PCR
Real-time PCR (qPCR) was used as a gold standard to determine the sensitivity of the SHERLOCK system using identical samples as were tested using the SHERLOCK detection assays. Sequences of the qPCR primers used in this study are shown in Supplementary Table S1. Reaction mixtures (3 technical replicates) comprised: 10 μl QuantiNova SYBR Green PCR Master Mix (Qiagen), 200 nM each of the forward and reverse primers, 1–2 μl template DNA, and ddH2O to a final reaction volume of 20 μl. The following cycling conditions were used: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C (30 s), annealing at 59 °C (30 s), and extension at 72 °C (30 s).
The PCR was performed using the Mic qPCR cycler and software (Bio Molecular Systems, Upper Coomera, Australia). Detection of target sequence was indicated by cycle threshold (Ct) values as previously described.40,41 Each run included a NTC (ddH2O as input) and DNA extracted from either eggs or adult worm pairs of S. japonicum or S. mansoni as a positive control. A positive qPCR result was defined as successful amplification (positive Ct value) in >75% technical replicates. Data were analysed using a dynamic baseline correction with extensive exclusion parameters and manual exclusion of samples with an abnormal melt curve.
Statistical analysis
Statistical analyses were conducted using Prism GraphPad (Version 7, GraphPad Software, La Jolla, CA, USA). Each experiment was performed in duplicate and all data are presented as the mean ± SE of technical replicates. Comparisons between groups were performed to determine statistical significance using One-way ANOVA with Dunnett's correction for multiple comparisons. Sensitivity and specificity of the SHERLOCK assays were calculated from the 2 by 2 contingency tables comparing the SHERLOCK assay results versus qPCR (Sjcox1-F1R1 or Sm1-7-F1R2 primers) as the reference test. The confidence intervals were calculated using the Wilson Score method in JMP Pro (v17.1, SAS Institute, Cary, NC, USA). The receiver operating characteristic (ROC) curves were created in JMP Pro (v17.1, SAS Institute, Cary NC, USA) using simple logistic regression of the dichotomised qPCR result versus the quantitative SHERLOCK result. The thresholds were chosen to maximise the sum of sensitivity and specificity (Youden's Index). AUCs (area under the ROC curve), sensitivity and specificity are provided with 95% confidence intervals.
Role of funders
The funding sources were not involved in the study design, in the collection, analysis, and interpretation of data, in the writing of report or in the decision to submit this paper for publication.
Results
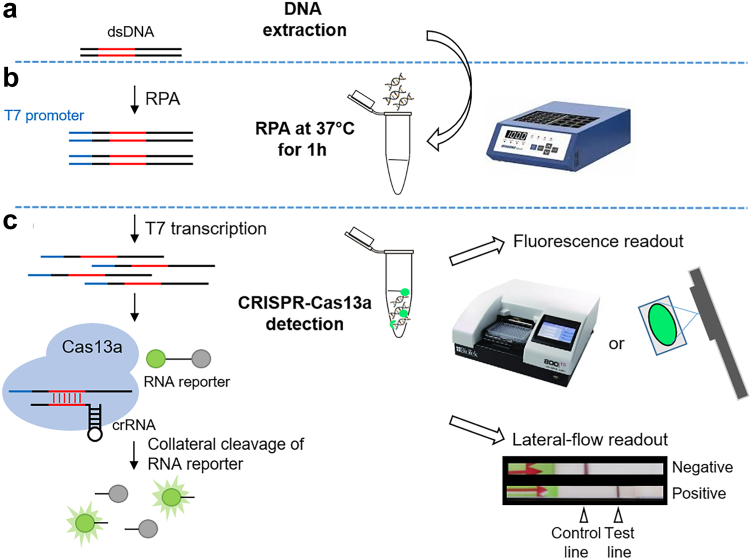
Here we describe the development of two SHERLOCK assays for the detection of S. japonicum and S. mansoni, evaluated using samples collected from experimentally infected mice and humans naturally infected with S. japonicum and S. mansoni, respectively. Our SHERLOCK assays combine RPA pre-amplification with CRISPR-Cas13a-mediated detection, visualised via a fluorescent or colorimetric readout using a reporter degradation strategy (Fig. 1). For fluorescent readout, this involves measuring an increase in fluorescence upon cleavage of a reporter when the gene target is present (Fig. 1c). For colorimetric lateral flow readout, the FITC/Biotin tagged reporter binds to the Streptavidin line (referred to as the control line), when intact. In the presence of target DNA, reporter cleavage occurs and cleaved reporter conjugates at the test line (Fig. 1c).
Fig. 1.
CRISPR-Cas13a SHERLOCK detection assay workflow. (a) DNA extraction from faecal or serum samples collected from human and animal; (b) Recombinase polymerase amplification (RPA) reaction performed at a single temperature (37 °C) for 1 h; (c) CRISPR-Cas13a detection via either fluorescence or lateral-flow readout. The S. japonicum/S. mansoni DNA target sequence is amplified by RPA. The RPA-amplified double-stranded DNA (dsDNA) is transcribed to single-stranded RNA (ssRNA) via T7 transcription. The Cas13a-crRNA complex is activated by crRNA specific binding to complementary ssRNA target sequences, triggering non-specific collateral cleavage of RNA reporters. Cleavage of quenched fluorophore-labelled ssRNA reporters is detectable by measuring fluorescence with a plate reader or qualitatively under a blue-light trans-illuminator (e.g., a handheld UV torch). Lateral flow-based detection can be read from strips with a coloured positive/negative band using a FAM/Biotin ssRNA reporter that conjugates to anti-FAM gold nanoparticles and accumulates at the control or test lines depending on whether the reporter is intact.
SHERLOCK assays for detection of S. japonicum infection
RPA primer selection and S. japonicum assay optimisation
Cox1 (cytochrome C oxidase subunit I) is a mitochondrial gene that has been broadly employed as a target for accurate PCR-based detection of S. japonicum, S. mansoni and other schistosome species.36 S. japonicum cox1 (Sjcox1) was selected as a target gene and incorporated into the design of a S. japonicum SHERLOCK detection assay. Two pairs of RPA primers were designed targeting Sjcox1, and the sensitivity of each primer pair was determined by RPA using DNA extracted from S. japonicum eggs as a template (20 pg/reaction). The RPA amplicons were visualised on a 2% (w/v) agarose gel (Supplementary Figure S2a) and a strong band at the expected size (∼125bp) was amplified by RPA using Sjcox1-F1R1 and Sjcox1-F2R2. It is noted that all RPA no-template controls (NTCs) appeared to show some smearing/banding in the agarose gels, though these were later shown to give negative results in the SHERLOCK Cas13a-detection assays. However, an extra smearing was observed in the NTC RPA product with the Sjcox1-F2R2 primer pair, possibly due to primer dimer (Supplementary Figure S2a). Thereby the primer pair Sjcox1-F1R1 was selected for downstream qPCR and SHERLOCK analysis.
Another target gene considered in developing the S. japonicum SHERLOCK detection assay was sap4 (S. japonicum saposin 4, Sjsap4). As reported, the Sjsap4 gene is highly expressed in schistosomula and adult worms of S. japonicum, but not in eggs, and saposin 4 has proven to be an excellent serological diagnostic marker for detecting early stages of S. japonicum infection.42,43 Two pairs of Sjsap4 RPA primers (Sjsap4-F1R1 and Sjsap4-F2R2) were designed and tested by RPA using DNA extracted from adult S. japonicum worms as a template (Supplementary Figure S2b). To determine the amplification efficiency of the Sjcox1 and Sjsap4 primer pairs in the detection of adult S. japonicum DNA, we undertook qPCR assays using the Sjcox1-F1R1, Sjsap4-F1R1 and Sjsap4-F2R2 qPCR primer pairs (Supplementary Figure 2c–f). The signal was detected in far fewer cycles (resulting in lower Ct values) from the reaction amplified by the Sjcox1-F1R1 primer pair, indicating a higher gene copy number of Sjcox1 compared to Sjsap4, and Sjcox1 represented a more sensitive target for DNA detection at early stages of S. japonicum infection. Therefore, the Sjcox1-F1R1 primer pair was employed in the subsequent development of the S. japonicum SHERLOCK system.
Sensitivity and specificity of the S. japonicum SHERLOCK system
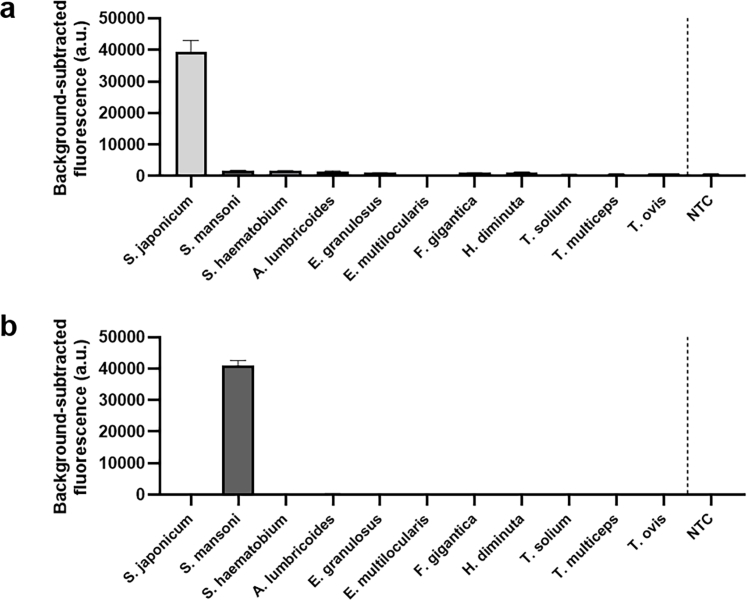
SHERLOCK and qPCR assays targeting Sjcox1 were performed using serially diluted S. japonicum egg DNA (20 pg/μl–0.02 pg/μl) as template (Fig. 2). The results obtained by qPCR (Fig. 2a), the SHERLOCK readout via lateral flow strip (Fig. 2b) and via BioTek fluorescence plate reader (Fig. 2c) all indicated an identical level of sensitivity. All three methods can detect S. japonicum egg DNA at a sensitivity of 0.2 pg/μl. It was noted that in the lateral flow readout, near complete reporter cleavage may occur in positive samples resulting in an extremely faint or absent ‘control’ band (as shown in the 20 pg/μl and 2 pg/μl DNA samples, Fig. 2b), an observation also found in other studies44 (see Supplementary Figure S1 for additional details on interpretation of lateral flow strips). Fluorescence-based SHERLOCK detection tests normally use a fluorescence plate reader (Fig. 2c) to read the fluorescence signal, providing real-time signal kinetics. We demonstrated that the fluorescence-based SHERLOCK assay readout could also be visualised clearly under UV light (Fig. 2d), a more field-friendly alternative. The specificity of the Sjcox1 SHERLOCK assay was determined using a panel of DNA (1 μl of 1 ng/μl) extracted from trematodes (S. japonicum, S. mansoni, S. haematobium and Fasciola gigantica), cestodes (Echinococcus granulosus, E. multiceps, Hymenolepis diminuta, Taenia multiceps, T. ovis, T. solium), and a nematode (Ascaris lumbricoides) (Supplementary Figure S3a). No cross-reactivity was observed among these different parasitic helminth species, including no cross reactivity with other Schistosoma species (S. mansoni and S. haematobium).
Fig. 2.
Diagnostic sensitivity of qPCR and SHERLOCK assays targeting Sjcox1 for the detection of S. japonicum, as determined using serially diluted S. japonicum egg DNA (20 pg/μl−0.02 pg/μl). (a) qPCR amplification plots; (b) Colorimetric SHERLOCK assay with lateral flow readout; Fluorescence-based SHERLOCK assay with signal detected using (c) the BioTek fluorescence plate reader (mean ± SEM of 3 technical replicates), and (d) UV light. H2O was used as a no-template control (NTC). All the tests were performed in biological duplicates.
S. japonicum SHERLOCK assay evaluation using mouse faecal samples
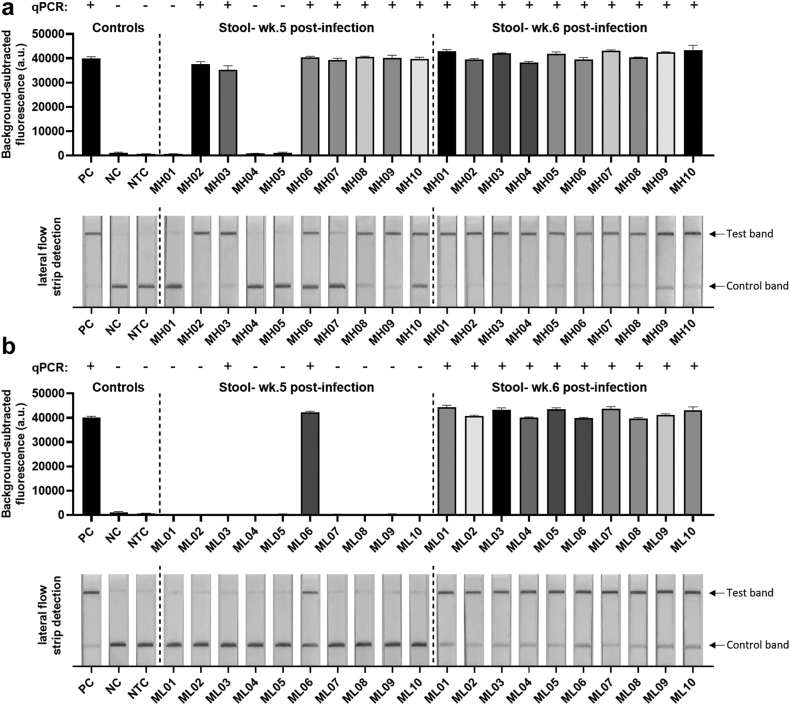
To evaluate the accuracy and efficacy of the SHERLOCK assay, we tested DNA isolated from faecal samples obtained at 1–7 weeks p.i. from individual Swiss mice infected with either a low (30/mouse) or high (70/mouse) exposure to S. japonicum (Philippines strain) cercariae. Faecal DNA samples were tested using qPCR (weeks 1–7 p.i.) and SHERLOCK detection assays (weeks 4–7 p.i.), both targeting the Sjcox1 mitochondrial gene. Adult worms, obtained from individual mice by portal vein perfusion at 7 weeks p.i., were counted and the faecal egg number of each animal was checked at 5–7 weeks p.i. (Supplementary Table S3). S. japonicum infection was not positively identified by qPCR or SHERLOCK at week 4 p.i., or by qPCR at weeks 1–3 p.i., in either the low-dose or high-dose group. However, in mice infected with higher cercarial exposure, 7/10 mice were diagnosed as S. japonicum-positive at week 5 p.i. both by qPCR and the fluorescence-based SHERLOCK assay (Fig. 3a). In mice that received the lower cercarial challenge exposure, infection was only identified at week 5 p.i. in 1/10 animals by SHERLOCK and 2/10 mice by qPCR (Fig. 3b). These outcomes are consistent with the fact that sexually mature female worms of S. japonicum commence egg-laying after pairing with mature males around week 4 p.i.,45 with oviposition occurring between 4 and 6 weeks p.i.,46 resulting in egg DNA being detectable around this time. In both the low-dose and high-dose groups, S. japonicum infection was positively diagnosed in all the mice at week 6 (10/10) and 7 (10/10) p.i., both by qPCR and the SHERLOCK assays (via either fluorescence-based or lateral-based detection). Overall, diagnosis with the Sjcox1 SHERLOCK assays matched well with the qPCR results with 100% agreement after 6 weeks p.i. using faecal DNA samples obtained from mice infected with 70 and 30 cercariae.
Fig. 3.
Fluorescence-based (upper panel) and lateral flow-based (bottom panel) SHERLOCK detection of S. japonicum (targeting the Sjcox1 gene) in mouse faecal samples. Faecal DNA (50 ng/reaction) of individual mice (n = 10) infected with (a) 70 S. japonicum cercariae and (b) 30 S. japonicum cercariae (Philippines strain) per mouse, was used for SHERLOCK and qPCR assays. Stools were collected at week 5 (wk.5) and week 6 (wk.6) post-infection. S. japonicum DNA (2 pg/μl) extracted from eggs (Philippines strain) was used as a positive control (PC), uninfected mouse stool as a negative control (NC) and H2O as a no-template control (NTC). qPCR test result of each sample was shown on the top panel (+, infected; -, uninfected). Background-subtracted fluorescence values are given as the mean ± SEM of 3 technical replicates. All the tests were performed in biological duplicates.
S. japonicum SHERLOCK assay validation using human faecal and serum samples
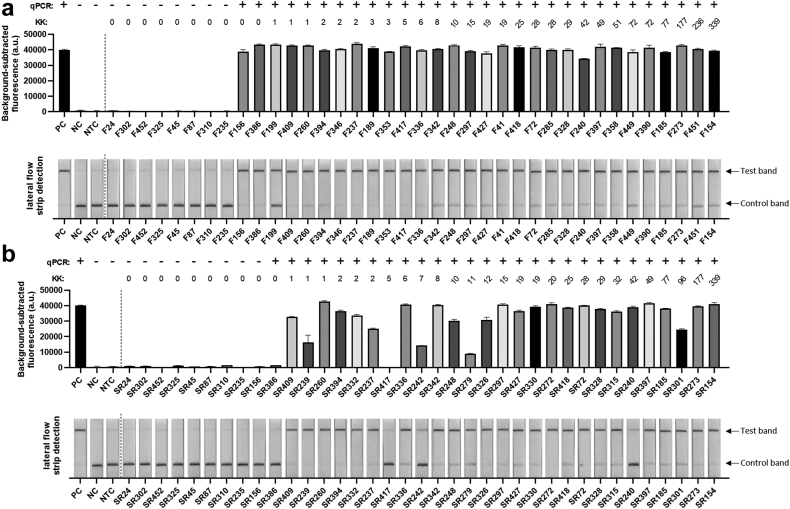
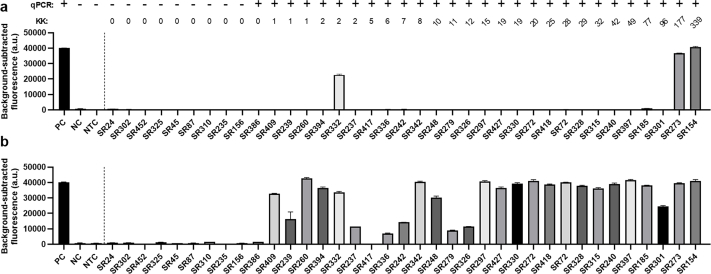
To further validate the clinical diagnostic capabilities of the SHERLOCK assay, samples (serum and stool) collected from human subjects living in an S. japonicum endemic region in Northern Samar, the Philippines, were tested by qPCR (using Sjcox1-F1R1 primers) and the SHERLOCK assay (using Sjcox1-F1R1 primers/crRNA). A detailed summary of results is presented in Supplementary Table S4. We found that the Sjcox1 SHERLOCK assay successfully detected S. japonicum in the faecal DNA of 100% (30/30) of qPCR-positive samples and 0% (0/8) of qPCR-negative samples, using both fluorescence and lateral flow readout methods (Fig. 4a). Both SHERLOCK detection methods showed 100% sensitivity [95% confidence interval (CI) (88.6%, 100%)] and 100% specificity [95% CI (67.6%, 100%)] relative to qPCR in testing clinical faecal samples. Indeed, two samples that were identified as negative for S. japonicum infection by the KK procedure (i.e. 0 EPG) tested positive in both the qPCR and SHERLOCK assays, emphasising the high sensitivity of the qPCR and SHERLOCK assays when testing stool samples.
Fig. 4.
Fluorescence-based (upper panel) and lateral flow-based (bottom panel) SHERLOCK detection of S. japonicum (targeting the Sjcox1 gene) in human (a) faecal (50 ng DNA/reaction) samples (n = 38) and (b) serum (7.5 ng DNA/reaction) samples (n = 37) collected from 18 barangays endemic forS. japonicuminfection in Laoang and Palapag municipalities, Northern Samar, the Philippines.S. japonicum DNA (2 pg/μl) extracted from eggs (Philippines strain) was used as a positive control (PC), human stool/serum from a non-endemic area as a negative control (NC) and H2O as a no-template control (NTC). Background-subtracted fluorescence values are given as the mean ± SEM of 3 technical replicates. All the tests were performed in biological duplicates. qPCR test result of each sample was shown on the top panel (+, infected; -, uninfected). Faecal egg counts obtained using the stool-based Kato-Katz (KK) procedure are presented as eggs per gram of faeces (EPG).
We also assessed the capacity of SHERLOCK to detect S. japonicum DNA in human serum using a panel of qPCR-positive (n = 28) and qPCR-negative (n = 9) serum samples (using Sjcox1-F1R1 primers with crRNA) (Fig. 4b). Using fluorescence-based detection, the Sjcox1 SHERLOCK assay successfully detected S. japonicum in the DNA of 26/28 qPCR-positive serum samples and 0/9 qPCR-negative serum samples, showing 93% sensitivity [95% CI (77.4%, 98.0%)] and 100% specificity [95% CI (70.1%, 100%)]. A lower fluorescence value (<50% of the positive control) was detected in some serum samples, which indicates the RPA pre-amplification did not reach the saturation point, likely due to very low target copy numbers present in the samples. Nevertheless, samples with low mean fluorescence were still considered ‘positive’ as they provided a fluorescence readout that was significantly higher than the negative and no-template controls (∗∗∗∗, P < 0.0001, one way ANOVA).
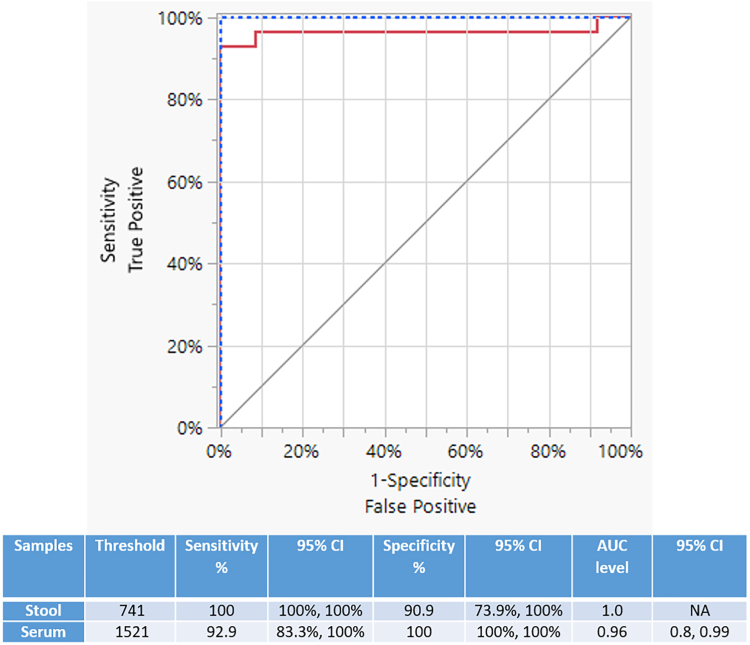
The ROC analysis (Fig. 5) revealed that the fluorescence-based Sjcox1 SHERLOCK assay in detecting S. japonicum, had an AUC level of 1.00 [95% CI (NA)] for stool samples (threshold = 741) and an AUC level of 0.96 [95 CI (0.80, 0.99)] for serum samples (threshold = 1521). The ROC AUC values (0.96 and 1.00) indicate perfect and nearly perfect separation for stool and serum samples, respectively.
Fig. 5.
The receiver operating characteristic (ROC) curve analysis of fluorescence-based Sjcox1 SHERLOCK in detecting S. japonicum. The ROC was generated to assess the capability of the fluorescence-based Sjcox1 SHERLOCK assay in discriminating the negative controls (n = 8–9) and qPCR (+) subjects (n = 28–30) of S. japonicum infections. The red line represents the serum samples and the blue-dotted line indicates the stool samples. Thresholds and AUC levels of clinical samples were determined using the ROC curve analysis and represented in the Table.
Lateral flow-based SHERLOCK presented 86% sensitivity [95% CI (68.5%, 94.3%)] and 100% specificity [95% CI (70.1%, 100%)] in testing the same clinical serum samples. It is notable that 2 of 26 serum samples that tested positive by the fluorescence-based SHERLOCK assay presented a faint test line in the lateral-based detection assay (Fig. 4b, SR242 and SR240). This could be considered as an ambiguous or open to interpretation result, and was also recorded with one mouse faecal sample (MH07; at 5 weeks p.i., Fig. 3a). Lateral flow-based detection had reduced sensitivity (86%) compared with fluorescence-based SHERLOCK detection (93%), the latter of which can provide a clearer and more easily interpretable result.
Although Sjcox1 was initially selected over Sjsap4 as a sensitive S. japonicum SHERLOCK assay gene target, we did perform an alternative fluorescence-based SHERLOCK assay, incorporating Sjsap4-F2R2 RPA primers/crRNA, to test the same panel of human serum from the Philippines (Supplementary Figure S4). However, the Sjsap4 SHERLOCK only identified 3 of the 28 qPCR-positive serum samples as positives, demonstrating its low sensitivity compared with Sjcox1 SHERLOCK. In light of a previous study of ours that found sap4 is not transcribed in S. japonicum eggs,43 we did not test the Sjsap4 SHERLOCK assay using stool samples. Clearly, the SHERLOCK assay targeting Sjcox1-F1R1 provided a considerably higher level of diagnostic sensitivity and accuracy than the Sjsap4-F2R2 primer pair achieved.
SHERLOCK assays for detection of S. mansoni infection
RPA primer selection for S. mansoni assays
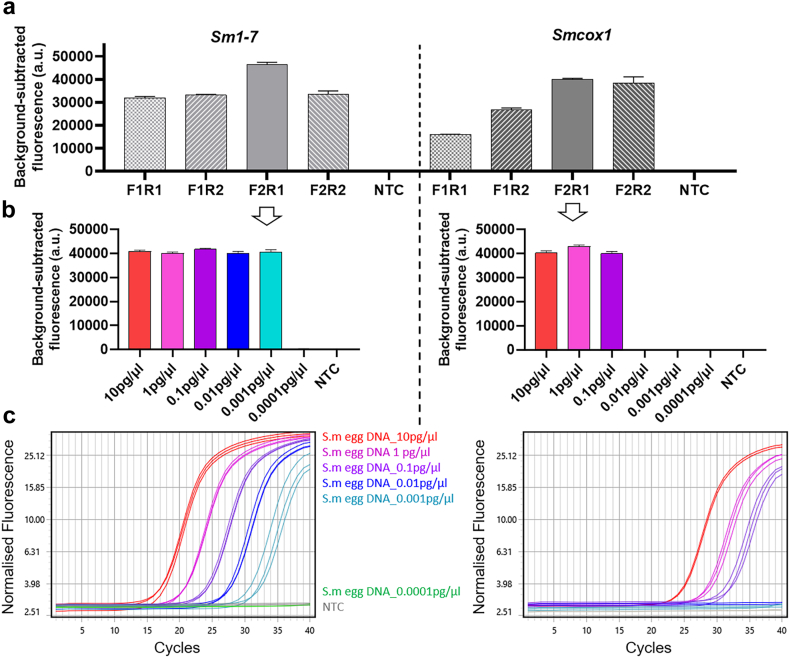
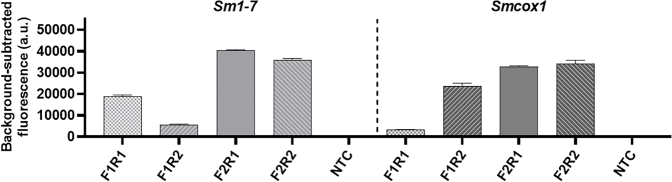
We then developed SHERLOCK assays for the detection of S. mansoni by targeting two S. mansoni specific genes; cox1- Smcox1,38 and Sm1-7, a high-copy tandem repeat sequence commonly used for nucleic acid-based detection assays for S. mansoni.39 For each gene target, RPA primers (two forward and two reverse) were designed to amplify a region surrounding a crRNA target site. RPA forward and reverse primer combinations were tested by fluorescence-based SHERLOCK detection assays using S. mansoni egg DNA as a template (0.1 ng/reaction) (Fig. 6a). The highest fluorescent signal was observed in the Smcox1 SHERLOCK assay using Smcox1-F2R1 and in the Sm1-7 SHERLOCK assay using Sm1-7-F2R1. Similar results were also obtained by the assays when using adult worm S. mansoni DNA (1 ng/reaction) as a template (Supplementary Figure S5). Therefore, these two primer pairs (Smcox1-F2R1 and Sm1-7-F2R1) were used in all subsequent S. mansoni assays. No fluorescence signal was detected in the NTC reactions (ddH2O as RPA template).
Fig. 6.
Optimisation of RPA primers and assessment of diagnostic sensitivity of qPCR and SHERLOCK assays for detection of S. mansoni DNA targeting Sm1-7 (left panel) and Smcox1 (right panel). (a) Optimisation of RPA primers for detection of S. mansoni DNA targets using fluorescence-based SHERLOCK detection with the BioTek fluorescence plate reader (mean ± SEM of 3 replicates). To amplify gene Sm1-7, RPA primer pairs Sm1-7-F1R1, -F1R2, F2R1 –F2R2 (right panel) were paired with Sm1-7 crRNA1. For gene Smcox1, the primer pairs Smcox1-F1R1, -F1R2, F2R1 –F2R2 (left panel) were paired with Smcox1 crRNA1. DNA (1 μl of 0.1 ng/μl) extracted from S. mansoni eggs was used as the positive control template, H2O was used as a no-template control (NTC); Diagnostic sensitivity of SHERLOCK and qPCR assays as determined using serially diluted S. mansoni egg DNA samples (10 pg/μl −0.0001 pg/μl); (b) Fluorescence-based SHERLOCK assay with signal detected using the BioTek fluorescence plate reader (mean ± SEM of 3 technical replicates); (c) qPCR amplification plots. All the tests were performed in biological duplicates.
Sensitivity and specificity of the S. mansoni SHERLOCK system
The sensitivity of the SHERLOCK assays targeting Sm1-7 and Smcox1 was then assessed using 10-fold serial dilutions of S. mansoni egg DNA (10 pg/μl - 0.0001 pg/μl) as a template. We found the Smcox1-F2R1 SHERLOCK assay was able to detect S. mansoni egg DNA at a sensitivity of 0.1 pg/μl (Fig. 6b), which was similar to the level of DNA detectable by the SHERLOCK assay targeting Sjcox1 (0.2 pg/μl of S. japonicum egg DNA). The Sm1-7-F2R1 SHERLOCK assay was capable of detecting S. mansoni egg DNA at a sensitivity of 0.001 pg/μl (Fig. 6b), a 100-fold increase in sensitivity compared to the Smcox1 SHERLOCK assay. In addition, qPCR assays that targeted a similar gene region were developed for Smcox1 and Sm1-7, respectively, and presented the same level of sensitivity as their corresponding SHERLOCK assays (Fig. 6c). The specificity of the Sm1-7 SHERLOCK assay was also determined using a panel of DNA (1 μl of 1 ng/μl) isolated from 10 different parasitic helminth species (Supplementary Figure S3b). No cross-reactivity was observed for other helminth parasites, including two Schistosoma species (S. japonicum and S. haematobium).
S. mansoni SHERLOCK assay evaluation using mouse faecal and serum samples
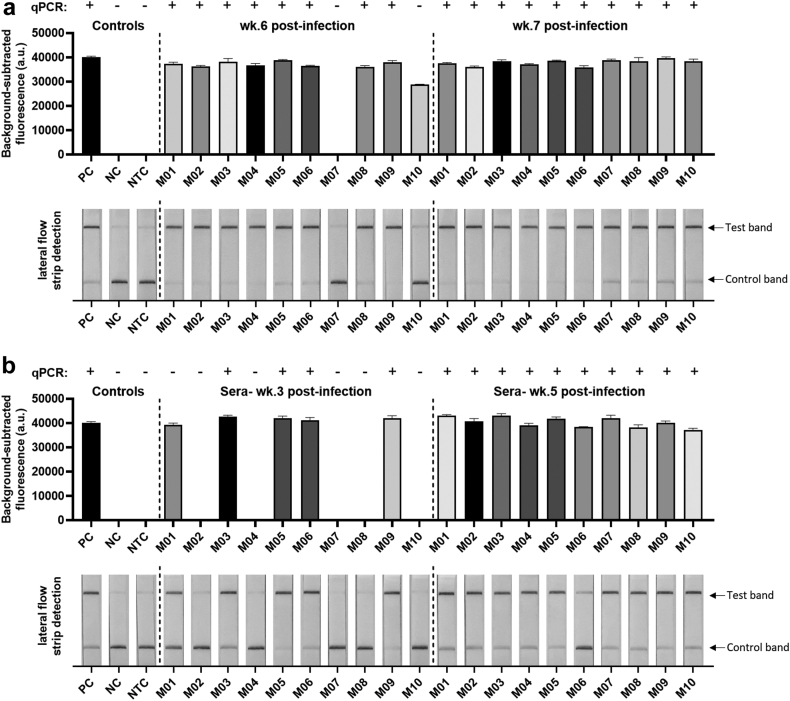
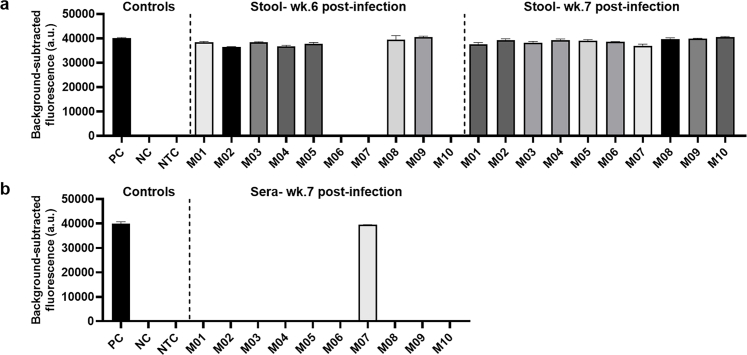
The S. mansoni SHERLOCK assays were evaluated with serum and faecal samples obtained from S. mansoni-infected Swiss mice. This cercarial dosage produced a similar infection intensity (measured by the adult worm count) to that observed in mice infected with a high exposure of S. japonicum (Philippines strain) (70 cercariae/mouse). Serum was collected from mice at weeks 3, 5 and 7 p.i. and stool obtained at 4–7 weeks p.i. (see Supplementary Table S5 for detailed results summary). At 4–5 weeks p.i., S. mansoni infection was not able to be positively identified in mouse faecal DNA by either Smcox1 or Sm1-7 SHERLOCK assays. The earliest infection phase that could be detected in the stool DNA of S. mansoni infected mice was week 6 p.i., where 9/10 mice tested positive using the fluorescence-based Sm1-7 SHERLOCK assay (Fig. 7a), and 8/10 mice tested positive by Sm1-7 qPCR detection. The fluorescence-based Smcox1 SHERLOCK assay detected infection in only 7/10 mice at week 6 p.i. (Supplementary Figure S6a), exhibiting a lower sensitivity than that of the Sm1-7 SHERLOCK assay. As observed in mice infected with S. japonicum infection, detection of faecal DNA corresponds with egg-laying, which occurs one week later in S. mansoni than in S. japonicum.47 At week 7 p.i., both S. mansoni SHERLOCK assays and qPCR detected infection in 10/10 mice.
Fig. 7.
Fluorescence-based (upper panel) and lateral flow-based (bottom panel) SHERLOCK detection of S. mansoni DNA (targeting the Sm1-7 gene) in samples collected from individual mice (n = 10) infected with 180 S. mansoni cercariae. (a) Stool (50 ng/reaction) collected at week 6 (wk.6) and week 7 (wk.7) post-infection (p.i.) and (b) serum (1.5 ng/reaction) collected at week 3 (wk.3) and week 5 (wk.5) p.i. S. mansoni DNA (1 ng/μl) extracted from adult worms was used as a positive control (PC), uninfected mouse stool/serum as a negative control (NC) and H2O as a no-template control (NTC). qPCR test result of each sample was shown on the top panel (+, infected; -, uninfected). Background-subtracted fluorescence values are given as the mean ± SEM of 3 technical replicates. All the tests were performed in biological duplicates.
Encouragingly, we found the Sm1-7 SHERLOCK assay was able to detect S. mansoni infection as early as week 3 p.i. in serum DNA, with infection positively identified in 5/10 mice using Sm1-7 SHERLOCK (both fluorescence and lateral flow readout methods) and in 4/10 mice using Sm1-7 qPCR (Fig. 7b). SHERLOCK and qPCR assays targeting Sm1-7 also detected S. mansoni DNA in all mice at week 5 (10/10) and week 7 (10/10) post-infection. However, the Smcox1 SHERLOCK assay was poor at detecting S. mansoni infection in infected mouse serum, with only 1/10 mice tested S. mansoni-positive at week 7 p.i. (Supplementary Figure S6b). Compared to Smcox1, Sm1-7 proved to be a more sensitive target for the detection of S. mansoni DNA, particularly when testing infected mouse serum, therefore we decided to focus on Sm1-7 for further validation in human samples.
S. mansoni SHERLOCK assay validation using human faecal and serum samples
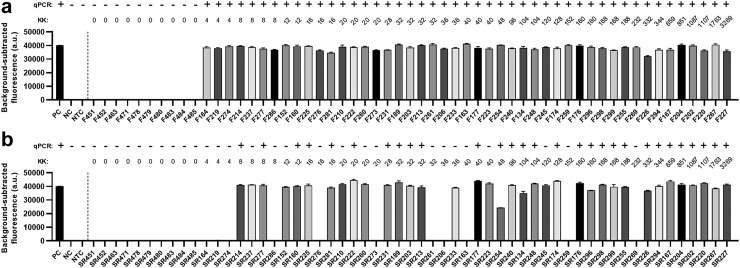
The diagnostic capabilities of the S. mansoni SHERLOCK assay targeting Sm1-7 were further validated using human samples (faeces and blood) collected from subjects residing in 6 villages in the Mayuge and Kabale districts in Uganda. The panel tested comprised of 47 kK-positive individuals recruited from a S. mansoni endemic region (EPG >1), along with 10 kK-negative individuals from a non-endemic region. Individual stool and serum DNA samples were tested by qPCR (using Sm1-7-F1R2 primers) and the fluorescence-based SHERLOCK assay (using Sm1-7-F2R1 primers/crRNA). A detailed summary of results is shown in Supplementary Table S6. The Sm1-7 SHERLOCK assay successfully detected S. mansoni infection in the stool DNA of 100% (47/47) of KK/qPCR-positive samples and 0% (0/10) of KK/qPCR-negative samples (Fig. 8a), showing 100% sensitivity [95% CI (92.4%, 100%)] and 100% specificity [95% CI (72.2%, 100%)] relative to qPCR. However, in the 47 kK-positive human serum samples, 36 tested positive using the Sm1-7 SHERLOCK assay and only 29 were identified as positive by the Sm1-7 qPCR assay (Fig. 8b). Compared to qPCR, Sm1-7 SHERLOCK presented 100% sensitivity [95% CI (88.3%, 100%)] and 75% specificity [95% CI (56.6%, 87.3%)]. It is worth noting that although specificity of SHERLOCK detection in serum samples is 75% when qPCR is used as a reference test, the same individuals tested positive for S. mansoni in stool samples using both KK and Sm1-7 SHERLOCK/qPCR assays. It was observed that SHERLOCK and qPCR assays detected positives in the serum of only 18/24 (75%) and 12/24 (50%) individuals, respectively, that had a KK count of 8–48 EPG, and no individuals (0/3) with EPG <8. In individuals with higher faecal egg counts (EPG >96), the detection rate in serum was increased to 18/20 (90%) and 17/20 (85%) detectable by SHERLOCK and qPCR assays, respectively, indicating the detection rate of SHERLOCK/qPCR is improved with the increasing of faecal egg burden These results further implied that the Sm1-7 SHERLOCK assay is actually more sensitive than the Sm1-7 qPCR assay in testing serum samples, which was also observed in mouse samples at early stages of infection. We found a greater number of positives were detected in faecal DNA than in serum DNA, a similar outcome as observed in the Sjcox1 SHERLOCK assay.
Fig. 8.
Fluorescence-based (upper panel) and lateral flow-based (bottom panel) SHERLOCK detection of S. mansoni DNA (targeting the Sm1-7 gene) in human (a) faecal (50 ng/reaction) samples (n = 57) and (b) serum (1.5 ng/reaction) samples (n = 57) collected in 6 villages endemic for S. mansoni infection in Mayuge and Kabale districts in Uganda.S. mansoni DNA (1 ng/μl) extracted from adult worms was used as a positive control (PC), human stool/serum from a non-endemic country as a negative control (NC) and H2O as a no-template control (NTC). Background-subtracted fluorescence values are given as the mean ± SEM of 3 technical replicates. All the tests were performed in biological duplicates. qPCR test result of each sample was shown on the top panel (+, infected; -, uninfected). Faecal egg counts obtained using the stool-based Kato-Katz (KK) procedure are presented as eggs per gram of faeces (EPG).
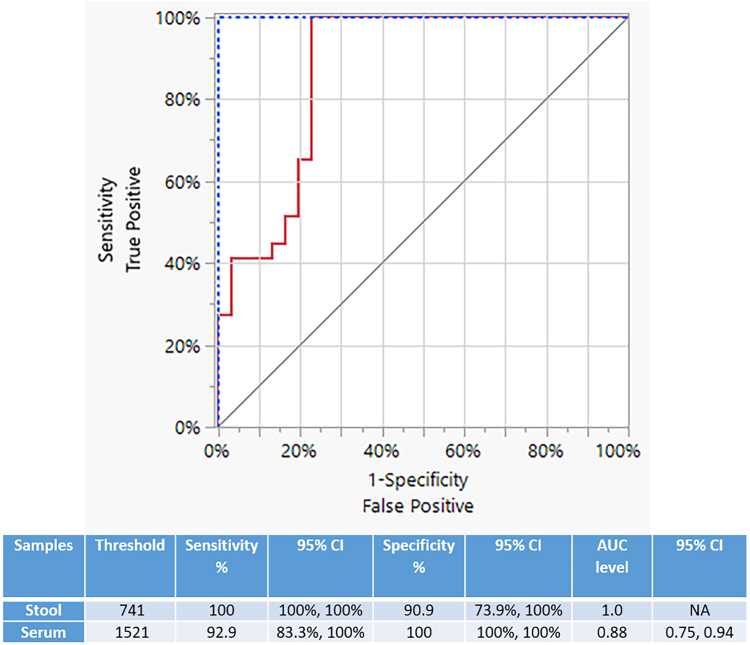
Our ROC analysis also showed that the fluorescence-based Sm1-7 SHERLOCK assay in detecting S. mansoni (Fig. 9) had an AUC level of 1.00 [95% CI (NA)] for stool samples (threshold = 741) and an AUC level of 0.88 [95% CI (0.75, 0.94)] for serum samples (threshold = 1521), indicating perfect and near perfect separation in stool and serum samples, respectively.
Fig. 9.
The ROC curve analysis of fluorescence-based Sm1-7 SHERLOCK in detecting S. mansoni. The ROC analysis was performed to determine the capability of the Sm1-7 SHERLOCK assay in discriminating the negative controls (n = 10) and qPCR (+) subjects (n = 47) of S. mansoni infections. The red line represents the serum samples and the blue-dotted line indicates the stool samples. Thresholds of clinical samples and AUC level were determined using the ROC curve analysis and represented in the Table.
Discussion
CRISPR-based technology represents a powerful new generation diagnostics platform that has not been previously applied for the detection of any helminth infections, Schistosoma spp. or any other helminth diseases of humans or animals by using samples collected from infected animals or clinical samples obtained from endemic regions. We demonstrate here the capacity of the CRISPR-Cas13a SHERLOCK platform to detect schistosome infections, achieving a level of sensitivity similar to qPCR-based assays (Figs. 2 and 6), as has also been clearly evidenced in previous CRISPR-based diagnostics developed for viral diseases.18,48 Our SHERLOCK diagnostic assays achieved high sensitivity when applied to samples obtained from mice experimentally-infected with S. japonicum/S. mansoni, and human samples collected from S. japonicum-endemic areas in the Philippines and S. mansoni-endemic regions in Uganda. The results we present here reinforce the potential of SHERLOCK and the CRISPR diagnostic platform to fulfill the urgent requirements for new diagnostic tools to better assess and assist in the control and elimination of schistosomiasis and other parasitic NTDs globally.
The most commonly utilised readout methods of SHERLOCK are fluorescence-based detection via a plate reader and colorimetric detection via a lateral flow strip. Lateral flow-based detection methods have the benefit of being portable and easy to use. For the most part, the interpretation of results is straight-forward, but we found that some of the analysed samples occasionally produced a very faint test line when we analysed samples with very low infection intensities. Such anomalies may cause difficulties in interpreting these as definitively positive, an observation that has also been noted in other CRISPR-based and POC-CCA diagnostic studies.44,49 Meanwhile, fluorescence-based detection using a plate reader is more suitable for mass-screening as the interpretation of positive and negative results can be clearly defined; however the requirement of a fluorescence plate reader means this method is not convenient to use in the field, especially in low resource, rural environments. To make the fluorescence-based SHERLOCK readout more user-friendly, we showed that the fluorescent signal could also be visualised in reaction tube, under blue-light trans-illuminator or UV light, by the naked eye (Fig. 2d). This detection method provided the same level of sensitivity as the fluorescence plate reader (Fig. 2c). To reduce user bias when interpreting results, a potential solution could be to incorporate a handhold UV torch or an UV reader device/app that can be added to a standard smartphone24,50 to aid in result analysis.
In developing the SHERLOCK assay for the detection of S. japonicum, we considered two key target genes, cox1 and sap4. Cox1 is a haploid-inherited mitochondrial gene that is repetitively represented in schistosomes,36 and has been employed to identify and differentiate infections among S. mansoni, S. haematobium, S. japonicum, S. mekongi and S. bovis.38,51 We showed here that Sjcox1 was also a robust diagnostic target for S. japonicum identification using SHERLOCK, resulting in 100% and 93% consensus with qPCR, respectively, targeting human stool and serum samples. The sap4 gene encodes the saposin-like protein SAP4 which is an excellent target for serological diagnosis of S. japonicum infection,42,43 being consistently released in worm vomitus into host blood,52 thereby inducing a strong host immune response resulting in high levels of anti-SAP4 antibody. However, SHERLOCK targeting Sjsap4 showed poor sensitivity when applied to human serum samples (Supplementary Figure S4), which may reflect a low transcript level of Sjsap4 present in patient serum, given that gene abundance levels are frequently not reflected in protein abundance levels.53
In optimising the SHERLOCK assay for the detection of S. mansoni, we compared two key target genes, Smcox1 and Sm1-7 54. The latter was found to be a better target, as the Sm1-7 SHERLOCK assay showed a 100-fold increase in sensitivity in the detection of S. mansoni egg DNA compared with the Smcox1 SHERLOCK assay (Fig. 6b). Encouragingly, the fluorescence-based Sm1-7 SHERLOCK assay was able to detect early S. mansoni infection in mice at week 3 p.i. using serum DNA (Fig. 7b). When we tested human serum samples collected from S. mansoni-endemic villages in Uganda, 36/47 and 29/47 of the KK-positive serum samples tested positive by fluorescence-based Sm1-7 SHERLOCK and Sm1-7 qPCR, respectively (Fig. 8b). This shows that Sm1-7 SHERLOCK assays may be more sensitive than Sm1-7 qPCR; this result was supported by a recent study showing that a subspecies-specific SHERLOCK tool for diagnosis of trypanosomiasis had 10 to 100-fold more sensitivity than that of qPCR.27 However, this also indicates that for some individuals that tested positive for S. mansoni infection by SHERLOCK/qPCR/KK testing in stool samples, infection was not detectable in serum DNA. Fluorescence-based SHERLOCK assays for the detection of faecal egg DNA always showed 100% consensus with qPCR/KK results (Figs. 4a and 8a) for both S. japonicum and S. mansoni infections, suggesting that SHERLOCK detection using faecal samples can provide more reliable and consistent results than when using serum sample. Indeed, most studies using PCR for schistosomiasis detection have found a higher number of positives are detected using faecal DNA than in serum DNA.34
Several factors could be responsible for this outcome.
-
1.
Parasite DNA is present in host blood/serum in the form of cell-free DNA (cfDNA), short nucleic acid fragments that can be challenging to extract due to their low concentration in blood and short half-life.54 Many factors can affect cfDNA yield, including sample volume, storage conditions and extraction method.55 For human samples collected in Uganda, due to the smaller volume of blood obtained from each individual we used 250 μl serum for DNA extraction. This was considerably lower than the 2 ml serum used for DNA extraction from subjects in the Philippines, and the majority of studies looking at cfDNA detection in humans used a minimum serum volume of 1 ml, though higher volumes are often recommended.55,56
-
2.
DNA extracted from Uganda serum samples had reduced S. mansoni detection rates relative to stool samples particularly in individuals with a lower faecal egg burden detected by KK method, which is not sufficiently sensitive in samples with low egg density,57 while frequently leading to overestimation of egg counts.49,58 In individuals with higher faecal egg counts, the detection rate in serum was improved from 75% (with 8–48 EPG) to 90% (with EPG>96) tested by SHERLOCK and from 50% (with 8–48 EPG) to 85% (with EPG>96) detected by qPCR assay.
-
3.
Biological/environmental factors in endemic communities could complicate the detection of schistosomiasis in human serum DNA, such as presence/absence of drug-treatment, reinfection rates, differences in infection stage, and co-infection with other parasites and diseases.59 Other potential confounding factors could cause a false negative result, such as DNA degradation in serum samples during transport from Uganda/Philippines to our laboratory in Australia.
In addition, we acknowledge the limitations of our CRISPR-Cas13a based assay and the improvements required before its application in resource-limited settings as a clinical POC test. Based on the CRISPR-based diagnostics literature and our study results, we have identified several strategies to develop our assay towards the goal of its field-deployment and testing its diagnostic effectiveness in the future.
-
1.
Further improvements to the sensitivity of the SHERLOCK assay, particularly during the early stages of an infection, could be made by testing additional gene targets and applying a wider panel of crRNA/RPA primers. Potential target genes include the mitochondrial NADH dehydrogenase 1 (nad1) and the SjR2 retrotransposon for S. japonicum,60 and the mitochondrial minisatellite DNA region (SmMIT) for S. mansoni.61
-
2.
The SHERLOCK assay procedure could be streamlined by combining the RPA reaction with Cas13a detection (Cas13a protein, crRNA and reporter) in one single reaction (one-pot detection). This would simplify the protocol, improving ease-of-use and reducing processing time, as well as reducing the risk of introducing contaminants. In our two-step protocol we used an extended incubation time for the RPA and CRISPR-Cas13a detection, but these could be reduced to as little as 5 min for each step as described.19 One-pot CRISPR-Cas13a detection has been successfully implemented for the detection of the Zika, Dengue viruses, and malaria.20,25 One-pot CRISPR-Cas detection can also be lyophilized, which increases thermostability and reagent shelf life as well as enabling convenient field deployment.63 We are currently working on developing one-pot versions of our S. japonicum and S. mansoni SHERLOCK assays in our laboratory and preliminary results are promising.
-
3.
Protocol changes, such as using LAMP-based CRISPR-Cas12 mediated detection, could be considered. Both RPA and LAMP are commonly used in CRISPR-Cas detection protocols as pre-amplification of the target gene significantly enhances sensitivity.63 Although LAMP primer design is more complex, it exhibits greater specificity,63 and there is some evidence that LAMP may have greater sensitivity compared to RPA and a greater tolerance of inhibitory substances,64 which may facilitate rapid DNA extraction methods. LAMP is more commonly coupled with CRISPR-Cas12 mediated detection and although it requires a higher amplification temperature, one-pot detection coupling LAMP and Cas12 b is possible.65
The SHERLOCK assays developed in this study are capable of detecting S. japonicum and S. mansoni infections at a similar level of sensitivity as gold-standard PCR-based testing, and this technology should be readily adaptable to detect other schistosome and helminth species. When considering its ease of access, our SHERLOCK assay has many benefits over qPCR. SHERLOCK can be performed at a constant temperature of 37 °C, which is easily achievable using a simple, portable heat block and bypasses the requirement of a thermal cycler. Indeed, no specialized equipment is required and, with multiple field-friendly readout methods available including lateral-flow or in-tube fluorescence detection by a handheld UV torch, the interpretation of results is simple and straight-forward. Our current cost of reagents used for SHERLOCK was estimated to $4.66–5.02 AUD per test (Supplementary Table S7). Since the major cost of the test comes from using commercially available products (including reporter, protein and inhibitor), bulk enzyme purchase or in-house production (i.e. of Cas13a protein and crRNAs) could significantly lower the price. Further optimization of the SHERLOCK detection process could also reduce cost and enhance field applicability as development of a thermostable, lyophilized SHERLOCK reaction would allow for cold-chain independence and ease of operation without specialised training, while also reducing preparation time and user error.66 The field of CRISPR-Cas based diagnostics is growing rapidly and collective research efforts have already led to many improvements in assay design including one-pot detection, DNA extraction from crude samples, and increased accuracy or affordability at the POC and in the field.62 The technology is readily adaptable, and capable of incorporating multiplex assays that can distinguish between individual species, specific to 1–2 single nucleotide polymorphisms (SNPs).24 As development continues, CRISPR based diagnostics show great promise and move closer to POC application in resource-limited settings. It will provide the ammunition to further extend this sensitive, field-friendly diagnostics approach for future wide-scale application to other parasitic NTDs.
With its versatility, sensitivity, and its potential to be an inexpensive, portable POC tool, SHERLOCK can become a major and dominant player as a next-generation diagnostic platform that can be optimised for the detection of a range of NTDs. The CRISPR-Cas based diagnostics platform could be the game-changer needed to address the unmet and urgent requirements outlined by the WHO5 for rapid mapping of helminthic diseases, monitoring helminth control programs and assessing elimination targets. Future investigations are needed to optimise performance of SHERLOCK in field settings but accelerated research will open new opportunities for CRISPR diagnostics in clinical, surveillance and research applications. The S. japonicum and S. mansoni SHERLOCK assays presented in this study highlight the potential of CRISPR diagnostics and demonstrate the vital role they may play in the global control efforts to eliminate schistosomiasis and other NTDs.
Contributors
HY, HS, JDF, and DPM contributed to study design; SRM, NC and HY performed all experiments; HY, SRM, HS, JDF contributed to development of methodology; SRM, MGD, TGE, MA, CAG, PC, RMO, GH and MKJ provided reagents, contributed to sample collection and data analysis; SRM, HY, DPM, JDF, CGT and MKJ, drafted and revised the manuscript. HY, DPM, and PC are responsible for funding acquisition and project administration. All authors read and approved the final version of the manuscript. SRM and HY have accessed and verified the data, and HY was responsible for the decision to submit the manuscript.
Data sharing statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
We thank the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) providing Oncomelania hupensis quadrasi snails (NIH-NIAID Contract HHSN272201700014I) for distribution through BEI Resources. We thank all research participants and the local Philippines field and clinical staff for their kind assistance allowing collection of the clinical samples used in this study. This work received support from an Australian Infectious Disease Research Centre Seed Grant and Grants (APP1194462, APP2008433) from the National Health and Medical Research Council of Australia.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104730.
Appendix A. Supplementary data
Supplementary Figure S1.
Supplementary Figure S2.
Supplementary Figure S3.
Supplementary Figure S4.
Supplementary Figure S5.
Supplementary Figure S6.
References
- 1.Hotez P.J., Fenwick A., Savioli L., Molyneux D.H. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373:1570–1575. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 2.Toor J., Alsallaq R., Truscott J.E., et al. Are we on our way to achieving the 2020 goals for schistosomiasis morbidity control using current world health organization guidelines? Clin Infect Dis. 2018;66:S245–S252. doi: 10.1093/cid/ciy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundamaraju R. Novel antipathy for schistosomiasis-the most lethal ailment of the tropical region. Asian Pac J Trop Biomed. 2014;4:S43–S45. doi: 10.12980/APJTB.4.2014C831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO . World Health Organization; 2021. Ending the neglect to attain the sustainable development goals: a road map for neglected tropical diseases 2021–2030. 28 January 2021. [Google Scholar]
- 5.WHO . World Health Orgnization; 2021. Public consultation: target product profiles for diagnostic tests to meet Schistosomiasis and Soil-transmitted helminth programme needs. 9 February 2021. [Google Scholar]
- 6.Ogongo P., Nyakundi R.K., Chege G.K., Ochola L. The road to elimination: current state of Schistosomiasis research and progress towards the end game. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amoah A.S., Hoekstra P.T., Casacuberta-Partal M., et al. Sensitive diagnostic tools and targeted drug administration strategies are needed to eliminate schistosomiasis. Lancet Infect Dis. 2020;20:e165–e172. doi: 10.1016/S1473-3099(20)30254-1. [DOI] [PubMed] [Google Scholar]
- 8.Gordon C.A., Kurscheid J., Williams G.M., et al. Asian schistosomiasis: current status and prospects for control leading to elimination. Trop Med Infect Dis. 2019;4:40. doi: 10.3390/tropicalmed4010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He P., Gordon C.A., Williams G.M., et al. Real-time PCR diagnosis of Schistosoma japonicum in low transmission areas of China. Infect Dis Poverty. 2018;7:8. doi: 10.1186/s40249-018-0390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CF V.A.N.D., Gordon C.A., Li Y., et al. Rodents, goats and dogs - their potential roles in the transmission of schistosomiasis in China. Parasitology. 2017;144:1633–1642. doi: 10.1017/S0031182017000907. [DOI] [PubMed] [Google Scholar]
- 11.Graeff-Teixeira C., Favero V., Pascoal V.F., et al. Low specificity of point-of-care circulating cathodic antigen (POCCCA) diagnostic test in a non-endemic area for schistosomiasis mansoni in Brazil. Acta Trop. 2021;217 doi: 10.1016/j.actatropica.2021.105863. [DOI] [PubMed] [Google Scholar]
- 12.Song J., Liu C., Bais S., Mauk M.G., Bau H.H., Greenberg R.M. Molecular detection of schistosome infections with a disposable microfluidic cassette. PLoS Negl Trop Dis. 2015;9 doi: 10.1371/journal.pntd.0004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin X., N’Goran E.K., Ouattara M., et al. Comparison of POC-CCA with kato-katz in diagnosing Schistosoma mansoni infection in a pediatric L-praziquantel clinical trial. Front Trop Dis. 2021;2 [Google Scholar]
- 14.Peralta J.M., Cavalcanti M.G. Is POC-CCA a truly reliable test for schistosomiasis diagnosis in low endemic areas? The trace results controversy. PLoS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weerakoon K.G., McManus D.P. Cell-free DNA as a diagnostic tool for human parasitic infections. Trends Parasitol. 2016;32:378–391. doi: 10.1016/j.pt.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Palaz F., Kalkan A.K., Tozluyurt A., Ozsoz M. CRISPR-based tools: alternative methods for the diagnosis of COVID-19. Clin Biochem. 2021;89:1–13. doi: 10.1016/j.clinbiochem.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palaz F., Kalkan A.K., Can O., et al. CRISPR-Cas13 system as a promising and versatile tool for cancer diagnosis, therapy, and research. ACS Synth Biol. 2021;10:1245–1267. doi: 10.1021/acssynbio.1c00107. [DOI] [PubMed] [Google Scholar]
- 18.Gootenberg J.S., Abudayyeh O.O., Lee J.W., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J.S., Ma E., Harrington L.B., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myhrvold C., Freije C.A., Gootenberg J.S., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Arizti-Sanz J., Freije C.A., Stanton A.C., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee R.A., Puig H., Nguyen P.Q., et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci U S A. 2020;117:25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham C.H., Hennelly C.M., Lin J.T., et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. eBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sima N., Dujeancourt-Henry A., Perlaza B.L., Ungeheuer M.N., Rotureau B., Glover L. SHERLOCK4HAT: a CRISPR-based tool kit for diagnosis of Human African Trypanosomiasis. eBioMedicine. 2022;85 doi: 10.1016/j.ebiom.2022.104308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duenas E., Nakamoto J.A., Cabrera-Sosa L., et al. Novel CRISPR-based detection of Leishmania species. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.958693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Deng F., Hall T., Vesey G., Goldys E.M. CRISPR/Cas12a-powered immunosensor suitable for ultra-sensitive whole Cryptosporidium oocyst detection from water samples using a plate reader. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117553. [DOI] [PubMed] [Google Scholar]
- 30.Yu F., Zhang K., Wang Y., et al. CRISPR/Cas12a-based on-site diagnostics of Cryptosporidium parvum IId-subtype-family from human and cattle fecal samples. Parasit Vectors. 2021;14:208. doi: 10.1186/s13071-021-04709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanitchinda S., Srisala J., Suebsing R., Prachumwat A., Chaijarasphong T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol Rep (Amst) 2020;27 doi: 10.1016/j.btre.2020.e00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao K., Peng D., Jiang C., et al. Rapid and visual detection of Heterodera schachtii using recombinase polymerase amplification combined with Cas12a-mediated technology. Int J Mol Sci. 2021;22:12577. doi: 10.3390/ijms222212577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You H., Gobert G.N., Duke M.G., et al. The insulin receptor is a transmission blocking veterinary vaccine target for zoonotic Schistosoma japonicum. Int J Parasitol. 2012;42:801–807. doi: 10.1016/j.ijpara.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Weerakoon K.G., Gordon C.A., Williams G.M., et al. Droplet digital PCR diagnosis of human schistosomiasis: parasite cell-free DNA detection in diverse clinical samples. J Infect Dis. 2017;216:1611–1622. doi: 10.1093/infdis/jix521. [DOI] [PubMed] [Google Scholar]
- 35.Keller L., Patel C., Welsche S., Schindler T., Hurlimann E., Keiser J. Performance of the Kato-Katz method and real time polymerase chain reaction for the diagnosis of soil-transmitted helminthiasis in the framework of a randomised controlled trial: treatment efficacy and day-to-day variation. Parasit Vectors. 2020;13:517. doi: 10.1186/s13071-020-04401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He P., Song L.G., Xie H., et al. Nucleic acid detection in the diagnosis and prevention of schistosomiasis. Infect Dis Poverty. 2016;5:25. doi: 10.1186/s40249-016-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mu Y., Gordon C.A., Olveda R.M., et al. Identification of a linear B-cell epitope on the Schistosoma japonicum saposin protein, SjSAP4: potential as a component of a multi-epitope diagnostic assay. PLoS Negl Trop Dis. 2022;16 doi: 10.1371/journal.pntd.0010619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato-Hayashi N., Kirinoki M., Iwamura Y., et al. Identification and differentiation of human schistosomes by polymerase chain reaction. Exp Parasitol. 2010;124:325–329. doi: 10.1016/j.exppara.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Hamburger J., Turetski T., Kapeller I., Deresiewicz R. Highly repeated short DNA sequences in the genome of Schistosoma mansoni recognized by a species-specific probe. Mol Biochem Parasitol. 1991;44:73–80. doi: 10.1016/0166-6851(91)90222-r. [DOI] [PubMed] [Google Scholar]
- 40.Cnops L., Tannich E., Polman K., Clerinx J., Van Esbroeck M. Schistosoma real-time PCR as diagnostic tool for international travellers and migrants. Trop Med Int Health. 2012;17:1208–1216. doi: 10.1111/j.1365-3156.2012.03060.x. [DOI] [PubMed] [Google Scholar]
- 41.Cnops L., Soentjens P., Clerinx J., Van Esbroeck M. A Schistosoma haematobium-specific real-time PCR for diagnosis of urogenital schistosomiasis in serum samples of international travelers and migrants. PLoS Negl Trop Dis. 2013;7:e2413. doi: 10.1371/journal.pntd.0002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S., Zhou X., Piao X., et al. Saposin-like proteins, a multigene family of schistosoma species, are biomarkers for the immunodiagnosis of schistosomiasis japonica. J Infect Dis. 2016;214:1225–1234. doi: 10.1093/infdis/jiw188. [DOI] [PubMed] [Google Scholar]
- 43.Cai P., Weerakoon K.G., Mu Y., et al. A parallel comparison of antigen candidates for development of an optimized serological diagnosis of schistosomiasis japonica in the Philippines. eBioMedicine. 2017;24:237–246. doi: 10.1016/j.ebiom.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broughton J.P., Deng X., Yu G., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashton P.D., Harrop R., Shah B., Wilson R.A. The schistosome egg: development and secretions. Parasitology. 2001;122:329–338. doi: 10.1017/s0031182001007351. [DOI] [PubMed] [Google Scholar]
- 46.Weerakoon K.G.A.D., Gobert G.N., Cai P., McManus D.P. Advances in the diagnosis of human schistosomiasis. Clin Microbiol Rev. 2015;28:939–967. doi: 10.1128/CMR.00137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheever A.W., Eltoum I.A., Andrade Z.A., Cox T.M. Biology and pathology of Schistosoma mansoni and Schistosoma japonicum infections in several strains of nude mice. Am J Trop Med Hyg. 1993;48:496–503. doi: 10.4269/ajtmh.1993.48.496. [DOI] [PubMed] [Google Scholar]
- 48.Patchsung M., Jantarug K., Pattama A., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 49.Lindholz C.G., Favero V., Verissimo C.M., et al. Study of diagnostic accuracy of Helmintex, Kato-Katz, and POC-CCA methods for diagnosing intestinal schistosomiasis in Candeal, a low intensity transmission area in northeastern Brazil. PLoS Negl Trop Dis. 2018;12 doi: 10.1371/journal.pntd.0006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fozouni P., Son S., Diaz de Leon Derby M., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333.e9. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webster B.L., Rollinson D., Stothard J.R., Huyse T. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate Schistosoma haematobium and S. bovis. J Helminthol. 2010;84:107–114. doi: 10.1017/S0022149X09990447. [DOI] [PubMed] [Google Scholar]
- 52.Hall S.L., Braschi S., Truscott M., Mathieson W., Cesari I.M., Wilson R.A. Insights into blood feeding by schistosomes from a proteomic analysis of worm vomitus. Mol Biochem Parasitol. 2011;179:18–29. doi: 10.1016/j.molbiopara.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H., Park C., Na W., Park K.H., Shin S. Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics. NPJ Precis Oncol. 2020;4:3. doi: 10.1038/s41698-019-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murugesan K., Hogan C.A., Palmer Z., et al. Investigation of preanalytical variables impacting pathogen cell-free DNA in blood and urine. J Clin Microbiol. 2019;57:e00782. doi: 10.1128/JCM.00782-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wichmann D., Panning M., Quack T., et al. Diagnosing schistosomiasis by detection of cell-free parasite DNA in human plasma. PLoS Negl Trop Dis. 2009;3:e422. doi: 10.1371/journal.pntd.0000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berhe N., Medhin G., Erko B., et al. Variations in helminth faecal egg counts in Kato-Katz thick smears and their implications in assessing infection status with Schistosoma mansoni. Acta Trop. 2004;92:205–212. doi: 10.1016/j.actatropica.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 58.Menezes D.L., Santos C.T.J., Oliveira Y., et al. Accuracy study of kato-katz and helmintex methods for diagnosis of schistosomiasis mansoni in a moderate endemicity area in sergipe, northeastern Brazil. Diagnostics (Basel) 2023;13:527. doi: 10.3390/diagnostics13030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Assefa A., Erko B., Gundersen S.G., Medhin G., Berhe N. Co-Infections and comorbidities of multiple parasites and hepatitis B virus infections in the lowland area of western Ethiopia: implications for integrated approaches. J Multidiscip Healthc. 2021;14:3369–3383. doi: 10.2147/JMDH.S341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weerakoon K.G., Gordon C.A., Cai P., et al. A novel duplex ddPCR assay for the diagnosis of schistosomiasis japonica: proof of concept in an experimental mouse model. Parasitology. 2017;144:1005–1015. doi: 10.1017/S003118201700021X. [DOI] [PubMed] [Google Scholar]
- 61.Mesquita S.G., Lugli E.B., Matera G., Fonseca C.T., Caldeira R.L., Webster B. Development of real-time and lateral flow recombinase polymerase amplification assays for rapid detection of Schistosoma mansoni. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaminski M.M., Abudayyeh O.O., Gootenberg J.S., Zhang F., Collins J.J. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5:643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- 63.Oliveira B.B., Veigas B., Baptista P.V. Isothermal amplification of nucleic acids: the race for the next “gold standard”. Front Sensor. 2021;2 [Google Scholar]
- 64.Howson E.L.A., Kurosaki Y., Yasuda J., et al. Defining the relative performance of isothermal assays that can be used for rapid and sensitive detection of foot-and-mouth disease virus. J Virol Methods. 2017;249:102–110. doi: 10.1016/j.jviromet.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li L., Li S., Wu N., et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 66.Heithoff D.M., Barnes L., Mahan S.P., et al. Assessment of a smartphone-based loop-mediated isothermal amplification assay for detection of SARS-CoV-2 and influenza viruses. JAMA Netw Open. 2022;5 doi: 10.1001/jamanetworkopen.2021.45669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.