Graphical abstracts
Keywords: DC-SIGN, Oligomannoside ligand, Candida albicans mannan, Glycoarray, NMR binding and conformational studies
Abstract
The dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) is an innate immune C-type lectin receptor that recognizes carbohydrate-based pathogen associated with molecular patterns of various bacteria, fungi, viruses and protozoa. Although a range of highly mannosylated glycoproteins have been shown to induce signaling via DC-SIGN, precise structure of the recognized oligosaccharide epitope is still unclear. Using the array of oligosaccharides related to selected fragments of main fungal antigenic polysaccharides we revealed a highly specific pentamannoside ligand of DC-SIGN, consisting of α-(1 → 2)-linked mannose chains with one inner α-(1 → 3)-linked unit. This structural motif is present in Candida albicans cell wall mannan and corresponds to its antigenic factors 4 and 13b. This epitope is not ubiquitous in other yeast species and may account for the species-specific nature of fungal recognition via DC-SIGN. The discovered highly specific oligosaccharide ligands of DC-SIGN are tractable tools for interdisciplinary investigations of mechanisms of fungal innate immunity and anti-Candida defense. Ligand- and receptor-based NMR data demonstrated the pentasaccharide-to-DC-SIGN interaction in solution and enabled the deciphering of the interaction topology.
Introduction
The dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) is a receptor of the immune system, which plays an important role in host immune recognition of many pathogens. DC-SIGN has a C-type lectin domain which recognizes carbohydrate epitopes on the surface of various bacteria, fungi, viruses and protozoa (Erwig and Gow, 2016, Garcia-Vallejo and van Kooyk, 2013, Geijtenbeek et al., 2009, Švajger et al., 2010, Viljoen et al., 2023). Since DC-SIGN is a key pathogen recognition receptor of innate immunity, understanding of its interaction with carbohydrate ligands is of great importance in immunobiology.
It is known that DC-SIGN binds to L-fucose (Fuc) and D-mannose (Man) containing oligosaccharide chains in a calcium-dependent manner (Lee et al., 2011, van Liempt et al., 2006), including fungal mannans (Cambi et al., 2008). It has been documented that their recognition is dependent on the origin and structure of such mannan components. For example, the mannan from C. albicans is well recognized by DC-SIGN while the mannan from Saccharomyces cerevisiae (Cambi et al., 2008) is not. Candida is a major fungal pathogen of humans that, each year, causes over 400,000 life-threatening infections with high levels of mortality (46–75%) (Brown et al., 2012). Furthermore, 70–75% of women suffer at least one episode of vulvovaginal candidiasis during their lifetime (Bongomin et al., 2017). DC-SIGN plays an important role in anti-Candida defenses by promoting binding and phagocytosis of the fungus by human dendritic cells (Cambi et al., 2008).
Several studies of DC-SIGN carbohydrate specificity have been performed to date that have been focused on the investigation of non-mannan oligosaccharide ligands (Geissner et al., 2019, Valverde et al., 2019a) or heterosaccharides with oligomannose fragments which relate mainly to mammalian glycoprotein oligomannoside chains (Guo et al., 2004). These analyses did not explore fungi–specific mannan structures to investigate the nature of the DC-SIGN binding ligand. Fungal mannan and glycoprotein oligomannose fragments include a number of unique carbohydrate signatures. Therefore, it was of principle interest to identify oligosaccharide ligands of DC-SIGN that are components of the complex of N-linked and O-linked fungal mannans (Fig. 1). Present work is dedicated to addressing this gap and describes our results on the investigation of direct interaction of several forms of DC-SIGN and distinct oligosaccharides which are structurally related to determinant fragments of Candida α- and β-mannan chains as well as to other types of polysaccharides of fungal cell wall.
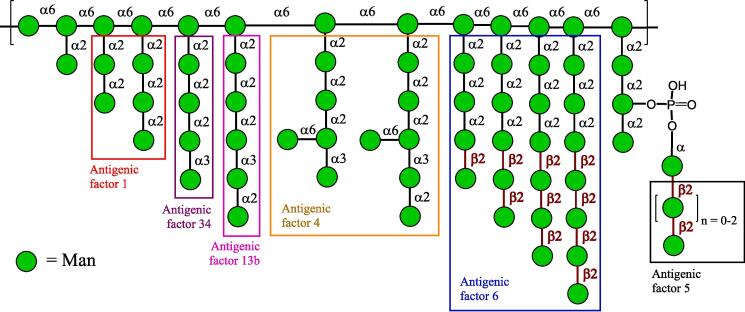
Fig. 1.
Structure of the cell wall mannan of C. albicans with marked structures of main antigenic factors. The carbohydrate sequences are represented according to symbol carbohydrate nomenclature. The carbohydrate sequences are represented according to symbol carbohydrate nomenclature (Neelamegham et al. 2019).
Materials and methods
Glycoarray
200 pmol/mL solutions of biotinylated oligosaccharides (obtained via synthetic routes described earlier (Argunov et al., 2019, Argunov et al., 2015, Karelin et al., 2010, Karelin et al., 2007, Kazakova et al., 2020, Komarova et al., 2015, Komarova et al., 2018, Krylov et al., 2018, Yashunsky et al., 2016a, Yashunsky et al., 2016b) in PBS-BSA-Tween buffer (phosphate buffered saline, Sigma Aldrich, supplemented with 0.1% bovine serum albumin, Sigma-Aldrich, and 0.05% Tween 20) were added to Thermo Scientific Pierce streptavidin-coated plates (100 μL per well). The plates were incubated for 2 h at 37 °C and after that the wells were washed three times with PBS-BSA-Tween buffer. For the rest of the assay Tris-Ca2+ buffer was used for dilutions and washing (150 mM NaCl; 10 mM Tris-HCl pH = 7.4; 10 mM CaCl2; 0.4% BSA; 0.05% Tween 20). The wells were washed one more time with Tris-Ca2+ buffer and after that human DC-SIGN with hFc tag (Sino Biological) was added (2.0 or 0.4 μg/mL, 100 μL per well) and incubated for 1 h at 37 °C. Then the wells were washed three times with Tris-Ca2+ buffer and 0.13 μg/mL of anti-human IgG rabbit IgG-horseradish peroxidase conjugate solution was added (100 μL per well) and incubated for 30 min at 37 °C. After washing three times, the color was developed using TMB mono-component substrate (100 μL) for 10 min, and the reaction was stopped with 50 μL of 1 M sulfuric acid. The absorbance was measured at 450 nm using a MultiSkan GO plate reader (Thermo Fisher Scientific, USA). All measurements were independently repeated twice in triplicate. Results are presented as the means ± s.d.
DC-SIGN lectin expression and purification
The extracellular domain of DC-SIGN (ECD DC-SIGN, residues 70–404) was obtained as a tetramer following well established methodologies (Daniel Martínez et al., 2019, Srivastava et al., 2021). The carbohydrate recognition domain of DC-SIGN (CRD DC-SIGN) in its 15N labelled form was obtained as previously described (Valverde et al., 2019a).
NMR experiments
The total volume for the NMR samples was 350 μL, using a precision NMR Shigemi tube with 5 mm outer diameter (SHIGEMI Co., LTD, Nakano Sanno, Tokyo, Japan). The pH was measured with a Crison Basic 20 (Crison Instruments SA, Barcelona, Spain) pH-meter and carefully adjusted with the suitable amount of NaOH and HCl or NaOD and DCl. NMR experiments were acquired using a Bruker AVANCE 2 800 MHz spectrometer equipped with a cryoprobe.
Saturation transfer difference (STD) NMR
1H-STD NMR spectra were acquired at 308 K. The ECD DC-SIGN:ligand samples were prepared in 25 mM Tris-d11, 150 mM NaCl, 4 mM CaCl2 in D2O at pH 8. The concentration of ECD DC-SIGN (tetramer) was set to 11 µM, and 500 µM that of the ligand, resulting in a ratio of protein-binding-sites:ligand of 1: 11. An in-house 1D-1H-STD NMR sequence using 90° PC9 pulses for protein saturation was employed for all STD experiments. 1024 scans were employed, with 2 s of total saturation time and a relaxation delay of 2.5 s. The on– and off-resonance NMR spectra were registered in the interleaved mode. On-resonance saturation was set at δ 7.3 ppm, while the off-resonance saturation was set at δ 100 ppm. The STD NMR spectra were obtained by subtracting the on-resonance spectrum from the off-resonance spectrum. The analysis of the spectra was carried out using the proton signal with the strongest STD effect as reference (100% of STD effect). On this basis, the relative STD intensities for the other protons of the molecule were calculated. A control STD NMR experiment was performed with a sample containing only the ligand (Man30), acquired exactly under the same conditions as in the presence of the protein.
2D-Noesy NMR
2D-NOESY experiments for the free ligands were performed with a ligand concentration of 1 mM in D2O. The experiments were performed at 298 K, using 50 and 400 ms mixing times. 2D-NOESY experiments in the presence of protein were also acquired using 25 mM Tris-d11, 150 mM NaCl, 4 mM CaCl2 in D2O at pH 8. A concentration of 11 µM of ECD DC-SIGN (tetramer) was used with a ligand concentration of 500 µM, resulting in a 1:45 protein (tetramer):ligand ratio. The experiments were acquired at 298 K using an NOESY sequence and 50, 200, and 600 ms mixing time.
The ISPA (Isolated Spin Pair Approximation) method was used to calculate proton-proton distances using 2D NOE cross-peaks. The integrals of the NOE cross-peaks between two protons “a” and “c” (Iac). was calculated using TopSpin 3.5.6 program. The NOE is inversely proportional to the sixth power of the distance between two nuclei corresponding to the NOE cross peak. The inter-proton distance between two protons “a” and “c” (rac) was then calculated from the integral of the volume of its NOE cross peak (Iac) using the relationship:
| (1) |
where, rab is the known distance between two protons, with Iab being the volume of its cross-peak. The distance H1-H2 = 2.52 Å was taken as reference for distance calculations.
For the proton-proton distance estimations in the free-state, the NOESY experiments with 400 ms mixing time was used, while in the bound-state, the 50 ms mixing time NOESY was employed.
Chemical shift perturbation (CSP) analysis
Samples containing 43 µM 15N labelled CRD DC-SIGN in 25 mM Tris, 4 mM CaCl2, 150 mM NaCl at pH 8.0 in 90 % H2O and 10 % D2O were prepared. 1H,15N-HSQC spectra were acquired at 310 K upon increasing additions of the ligands 29 and 30 (10–30-70–100-127 equivalents and 10–30-70–100-130 equivalents, respectively). The titrations were analyzed using CcpNmr Analysis software. The KD values were obtained by the fitting module in CcpNMR v2.4. The CSP of the protein-backbone NH groups were calculated using the formula: Δδ (ppm) = [(ΔδH2+ (0.14ΔδN)2)/2]1/2 and the results were graphically plotted.
Molecular dynamics
Molecular dynamics (MD) simulations (500 ns with a time step of 2 fs) were performed with the Amber 18 package program (Case et al., 2018, Lee et al., 2018) using the GLYCAM 06j-1 (Kirschner et al., 2008) force field for the glycan. The initial structure was set at the centre of a cubic TIP3P water (Jorgensen et al., 1983) box with the distance between solute and box set at 14 Å. A two-stage optimization approach was allowed: the first part minimizes only the positions of the water molecules and ions, and the second one was an unrestrained minimization of all the atoms in the simulation cell. The system was then heated by incrementing the temperature from 0 to 300 K under a constant pressure of 1 atm and periodic boundary conditions.
Model building for the complexes
DC-SIGN models for the O3-O4 and O4-O3 binding modes were constructed by superimposing the target Man residue in the ligand into the Ca2+-bound Man residue described in the crystallographic coordinates of two different DC-SIGN/Man complexes. In particular, PDB code: 1SL4 (Guo et al., 2004) was employed for the O3-O4 binding mode and PDB code: 2XR5 (Thépaut et al., 2013) for the O4-O3 binding mode (Fig. S7 and S8 in Supplementary data).
Results and discussion
ELISA assessment of DC-SIGN carbohydrate specificity
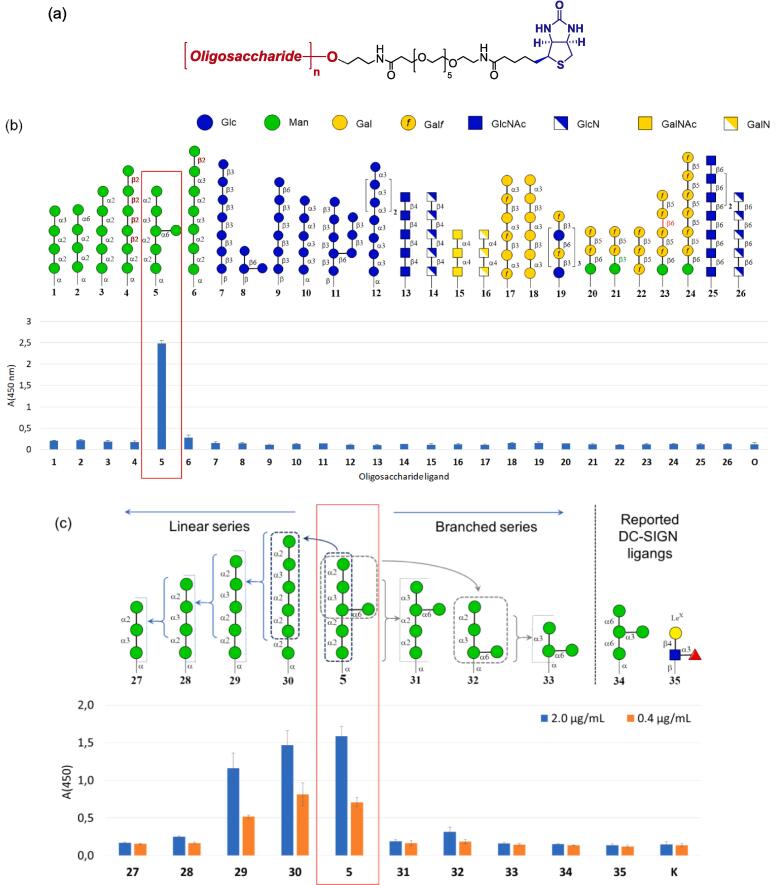
For initial assessment of carbohydrate specificity of DC-SIGN we created a series of synthetic biotinylated ligands 1–26 related to fragments of main polysaccharide types of Candida and Aspergillus cell-walls including α- and β-mannans, α- and β-glucans, galactomannan, galactosaminogalactan, chitin and chitosan together with β-(1 6)-D-glucosaminides 25 and 26 related to poly-(1 → 6)-N-acetyl-β-D-glucosamine (PNAG) and its N-deacetylated analogue dPNAG (Gening et al., 2007). All oligosaccharide ligands were equipped with a flexible hydrophilic hexaethylene glycol derived linker which permits the proper spatial presentation of glycans (Krylov and Nifantiev, 2020). These were loaded on streptavidin coated microtiter plates to produce a specific glycoarray used for the screening of DC-SIGN ligands (see Fig. 2b). Such glycoarrays have successfully been applied for screening of carbohydrate specific ligands in serum and monoclonal antibodies that recognise A. fumigatus galactomannan (Matveev et al., 2018, Schubert et al., 2019, Wong et al., 2020), α- and β-glucans (Komarova et al., 2018, Komarova et al., 2015, Matveev et al., 2019), galactosaminogalactan (Kazakova et al., 2020) and diverse bacterial polysaccharides (Argunov et al., 2019, Kurbatova et al., 2017).
Fig. 2.
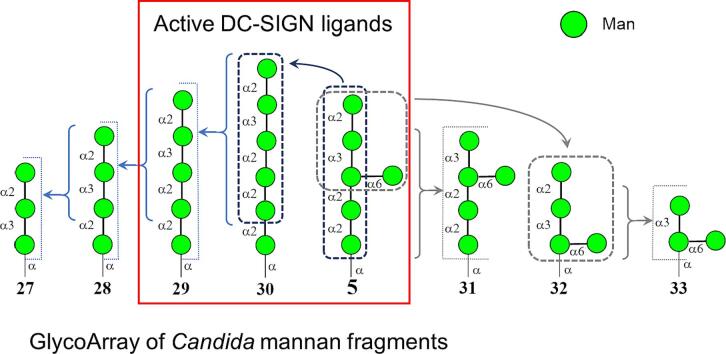
(a) Structure of biotinylated oligosaccharide ligands used in the creation of the glycoarray. (b) Results of screening of DC-SIGN carbohydrate specificity against a glycoarray assembled-up of glyco-ligands 1–29 (O – control) representing fragments of main polysaccharide types of Candida and Aspergillus cell-walls. (c) The results of the screening of DC-SIGN carbohydrate specificity on glycoarray built-up of glyco-ligands related to selected ligand 5 and additional ligands 34 and 35. The carbohydrate sequences are represented according to symbol carbohydrate nomenclature (Neelamegham et al. 2019).
The screens revealed DC-SIGN binding to the branched oligomannoside ligand 5 which is structurally related to the antigenic factor 4 of C. albicans mannan (Shibata et al., 1995). To specify which part of mannoside 5 determines the binding with DC-SIGN, a series of additional ligands 27–33 were generated, arrayed and screened (see Fig. 2c). This analysis revealed that the linear oligosaccharide ligands 29 and 30, which are structurally linked to the antigenic factor 13b of C. albicans mannan (Funayama et al., 1983) were also efficiently recognized by DC-SIGN. Ligands 5, 29 and 30 contain the common linear terminal fragment Man-α-(1 → 2)-Man-α-(1 → 3)-Man-α-(1 → 2)-Man-α-(1 → 2)-Man, which appears critical for recognition by DC-SIGN. The absence of a single (1 → 2)-linked Man unit from its “non-reducing” end (5 → 31; 29 → 28) drastically reduced the recognition of the mannooligosaccharide by DC-SIGN under these conditions. It was noticeable that the branched tetrasaccharide 32, an expected binder (Reina et al., 2008, Shahzad-ul-Hussan et al., 2017) was also barely recognised by DC-SIGN when presented in the array conditions.
We also compared DC-SIGN ligands 5 and 29 with the LeX trisaccharide 35, which is a known ligand for DC–SIGN (see for example refs.: Geissner et al., 2019, Guo et al., 2004, Pederson et al., 2014, Valverde et al., 2019a, van Liempt et al., 2006, Van Liempt et al., 2004). Interestingly, the LeX trisaccharide 35 was not as active as ligand for DC-SIGN as the ligands identified here. This fact was further evidenced by immobilization studies using biotin-streptavidin pairing that avoids possible problems associated with overloading of carbohydrate ligands. Our observations suggest that the presence of the simple tetrasaccharides 34 or 35 is not sufficient for strong binding to DC-SIGN under these experimental conditions. In fact, longer structures as those presented in glycoprotein N-linked mannan chains (see Fig. S1 in Supplementary data) are necessary for efficient binding, highlighting the important effect of epitope presentation in glycan recognition.
Binding in solution assessment by NMR
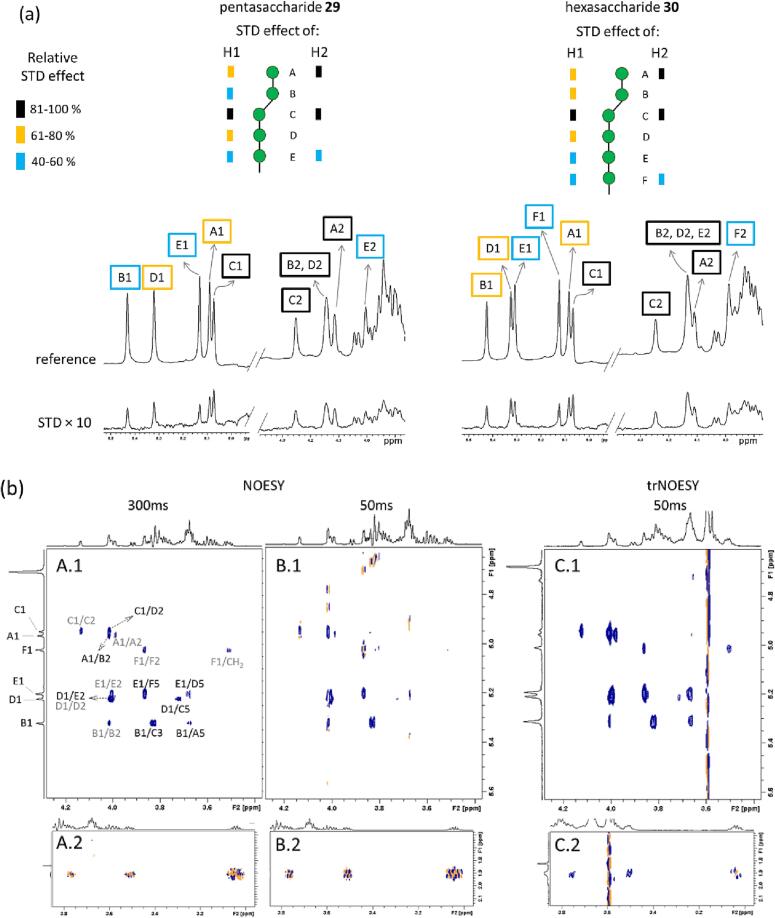
The interaction of DC-SIGN with the pentasaccharide 29 and hexasaccharide 30 was further studied in solution by NMR, using both ligand-based and receptor-based NMR complementary approaches. Ligand-based 1H-STD-NMR experiments showed very similar results for both ligands, with clear saturation transfer for most ligand protons (Fig. 3a and Fig. S2-S6 in Supplementary data), suggesting that they are all close to the lectin surface upon binding. This fact can be the result of two different scenarios: either there is in one binding mode involving the whole ligand structure, or there are multiple binding modes in equilibrium. Due to significant overlapping in the δ 3.4–4.0 ppm region, the STD NMR analysis was based mainly on Man H1 and H2 signals, which could be unambiguously identified for each Man residue (see Fig. S2 and S4 in Supplementary data). For both ligands, the residues at the reducing-end (E in 29 and E and F in 30) showed the weakest STD effect, while residue C showed the strongest STD effect. For the pentasaccharide 29, residue A showed a relative STD effect similar to C, although overall, very small differences were observed among residues A to D.
Fig. 3.
(a) 1H-STD-NMR analysis of the interaction of pentasaccharide 29 and hexasaccharide 30 with DC-SIGN based on H1 and H2 protons of the Man residues. Quantification is provided in the Supplementary data (Fig. S3 and S5). Expansion of the 1H-STD spectra at the Man H1 and H2 regions is shown (ratio lectin (tetramer): ligand = 1:45). Residues are named from A to F starting from the non-reducing-end. (b) NOESY (left, A and B with mixing times 300 ms and 50 ms respectively) and trNOESY (right, C with mixing time 50 ms) spectra of hexasaccharide 30 in the free and DC-SIGN bound states, respectively. Two different regions are shown for every experiment: on top (A.1, B.1 and C.1) the anomeric correlations, and below (A.2, B.2 and C.2) the correlations of the aminopropyl moiety. The NOE cross peaks are labeled: inter-residue (black) and intra-residue (grey). NOE correlation of the free hexasaccharide 30 (left, A.1 and B.1) are negative (blue contours) under the conditions used (see experimental section), except for those of the aminopropyl moiety at the reducing-end (A.2 and B.2, mixed orange/blue contours). NOE correlations are very weak at low mixing times (50 ms, B.1). In the presence of DC-SIGN (ratio lectin-binding-sites: 30 = 1:11) NOE correlations at 50 ms were negative and strong (C.1). Correlations of the –OCH2CH2- moiety of the aminopropyl moiety (3.7/1.9 and 3.5/1.9 ppm) were negative (C.2). The carbohydrate sequences are represented according to symbol carbohydrate nomenclature (Neelamegham et al. 2019). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Next, NOESY spectra for the free glycan were acquired, as well as further trNOESY, in the presence of DC-SIGN, to unravel the conformational features of these molecules in the free and the DC-SIGN bound states (Asensio et al., 1995, Valverde et al., 2019a). For the free ligand the cross peaks in the NOESY spectra at short mixing times (50 ms) were weak and negative for those protons belonging to the sugar moiety and close to zero (dominated by zero-quantum effect) for the protons at the reducing-end and aminopropyl part (Fig. 3b, B.1 and B.2 respectively). This indicates the existence of faster motions at the reducing-end. At longer mixing times (Fig. 3b, A), all NOE correlations became more intense. In the presence of DC-SIGN all the cross peaks were strong and negative at 50 ms, including those at the aminopropyl moiety (Fig. 3b, C.2). Although the cross-peaks of the sugar moiety did not change NOE sign in the presence or in the absence of the protein, their different NOE evolution with the mixing time, as well as the behavior of the protons at the aminopropyl moiety, proved that, under these conditions, the ligand is under the transferred NOE regime (Valverde et al., 2019b). The comparison of the glycan NOE cross peaks for the free and bound states (Fig. 3b) revealed a very similar NOE pattern, strongly suggesting that no major changes around the glycosidic linkages take place for any of the ligands upon DC-SIGN binding.
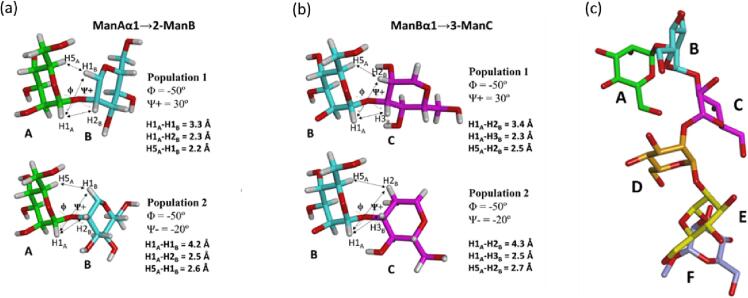
To assess the bound conformation of the ligands, a conformational analysis was carried out, paying attention to the conformation around the glycosidic torsion angles, following established NMR-based methodologies (Clavel et al., 2007). The torsion angles (Alonso et al., 2016) around the α(1 → 2)- and α(1 → 3)-linkages are defined as follows: φ as H1Man(i + 1)–C1Man(i + 1)–O–CXMan(i) and ψ as HXMan(i)–CXMan(i)–O–C1Man(i + 1) (Nóbrega and Vázquez, 2003). The major conformations around the different glycosidic linkages were deduced from the NOE analysis and compared to the conclusions of previous studies carried out for similar oligosaccharides (Clavel et al., 2007). Inter-residue proton-proton distances were calculated using the ISPA method (Gronenborn and Clore, 1985) as described in the experimental section. MD calculations were performed for the free 30. As described earlier (Clavel et al., 2007), for each of the α-Man-(1 → 2)-α-Man and α-Man-(1 → 3)-α-Man linkages, two populations are predicted, both with φ values in agreement with the exo-anomeric effect (-50°), and ψ values either in the negative (ca. −20°) or positive (ca. + 30) regions.
Focusing on the key NOE signals of the α-(1 → 2) linkage (Fig. 4a), the H1(i + 1)–H1(i) NOE is specific for the positive ψ-conformer, while medium-strong H5(i + 1)–H1(i) and H1(i + 1)–H2(i) inter-glycosidic NOEs may take place in either conformation (see also Table S1 in the Supplementary data). For the α-(1 → 3) linkage (Fig. 4b), the H1(i + 1)–H2(i) is also exclusive for the positive ψ-conformer, while medium-strong H5(i + 1)–H2(i) and strong H1(i + 1)–H3(i) NOEs are expected for both conformers. The presence of weak H1(i + 1)–H1(i) NOEs between all the Man residues involved in α-(1 → 2)-linkage (Table S1), along with the presence of additional inter-residue NOEs for the H1(i + 1)–H2(i) (strong) and H5(i + 1)–H1(i) (medium) proton pairs, indicates the presence of a major ψ conformer (ca. + 30°) and an exo-anomeric syn orientation around φ (-60°), although the possible presence of a minor population with a negative ψ (ca. −20°) geometry cannot be discounted. For the Man-α-(1 → 3)-Man linkage, the presence of the weak H1(i + 1)–H2(i) NOE suggests the predominance of the exo-syn (-60°/30°) conformation, also supported by the strong H1(i + 1)–H3(i), H5(i + 1)–H2(i) NOEs. Again, the presence of a sub-population of the negative ψ conformer cannot be ruled out.
Fig. 4.
Conformational analysis for hexasaccharide 30. The two possible populations around ψ torsion angle of (a) the α-(1 → 2)- and (b) the α-(1 → 3)-glycosidic linkages. The distances between the key inter-residue protons are highlighted. Population 1 is the major one for both a1 α-(1 → 2)- and α–(1 → 3)-linkages according to NOE data. (c) Perspective of the major conformation proposed for 30 based on the experimentally observed NOEs and the geometry models taken from MD simulations. A bent shape is present for the terminal Man-α-(1 → 2)-Man-α–(1 → 3)-Man moiety. The observed NOEs are basically identical in the presence of DC-SIGN, strongly suggesting that this is also the preferred bound conformation.
These interglycosidic conformations were then combined to generate the possible 3D shapes of the hexasaccharide. The predicted structure suggests a bent shape for the terminal Man-α-(1 → 2)-Man-α-(1 → 3)-Man fragment (Fig. 4c).
Thus, the combined STD NMR/trNOESY approach has shown that both ligands 29 and 30 are DC-SIGN binders and that several residues of the mannan are involved in the binding events. The mannose units at the “reducing end” (E and F) of the oligosaccharide chains are less involved in DC-SIGN recognition. The oligosaccharides adopt a bent-shape at the terminal end, and no major conformational changes occur upon DC-SIGN binding.
Receptor-based NMR
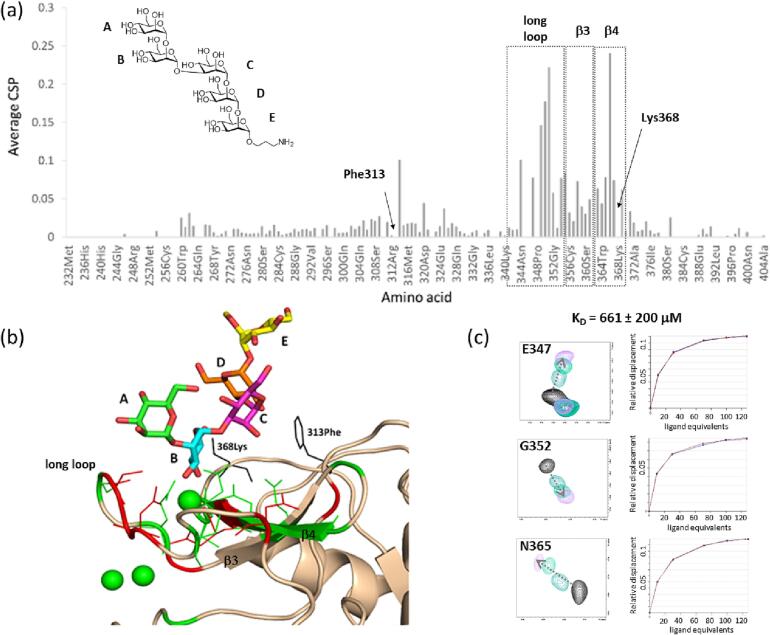
The carbohydrate recognition domain (CRD) of DC-SIGN, in its 15N-labeled form, was employed to deduce the lectins residues affected by glycan binding. Titration experiments were carried out as described in the experimental section to estimate the binding affinities. Clear chemical shift perturbations (CSP) of the lectin’s 1H,15N backbone cross-peaks were observed in the 1H,15N-HSQC NMR titrations, which supports the ligand binding by DC-SIGN. The most affected residues were those located around the calcium ion at the primary binding site of the immune receptor. In particular, residues at the long loop and β3 and β4 strands showed significant CSP, although distal residues F313 and T314 were also substantially affected (Fig. 5). These perturbations can be satisfactorily explained by the reported X-ray crystallographic structures of DC-SIGN with various oligomannosides, which showed an extended binding site towards F313 (PDBs: 1K9I, 1SL4 and 2XR6 (Feinberg et al., 2001, Guo et al., 2004, Sutkeviciute et al., 2014)). As observed for the binding of other oligosaccharides to the DC–SIGN CRD, the estimated dissociation constant (KD) were weak, ca. 0.6 mM and 1.1 mM for the penta- and hexasaccharide, respectively.
Fig. 5.
(a) CSP analysis for DC-SIGN close to saturation with pentasaccharide 29 (127 equivalents). Besides the observed CSPs for amino acids in the long loop (Asn349, Asn350 and Val351), residues in strands β3 and β4 (Glu354, Glu358 and Asn365, Asp366 respectively) were also highly perturbed, and some signals from residues such as Phe313 and Lys389 disappeared upon addition of the ligand and did not recover their intensity. (b) A 3D model for one of the proposed binding modes, O3-O4 binding mode through residue B (see Fig. S7 and S8 in Supplementary data) showing the amino acid residues described above. The residues that show the highest experimental CSPs are highlighted in red (perturbed > 2σ) and green (perturbed > σ). Residues whose HSQC signals disappeared, and their intensities are not recovered are shown in black. (c) Snippets of the 1H,15N-HSQC spectra showing the shift of selected amino acid cross-peaks during the titration, and corresponding fitting curve for KD estimation (as obtained from CcpNMR analysis software). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Thus, the ligand- and receptor-based NMR data demonstrate that the pentasaccharide 29 and hexasaccharide 30 are DC-SIGN binders in solution. The combined STD NMR/trNOESY/CSP experimental evidence strongly suggest that, in solution, multiple binding modes coexist in equilibrium for the ligand. On average, all the Man residues of the oligosaccharides are close to the protein surface during a given period of time. Indeed, it is not possible to explain all the experimental STD profiles with a single binding complex. This fact is not surprising, since it has been reported that Man residues can bind DC-SIGN in different binding modes at the monosaccharide or the disaccharide level (Daniel Martínez et al., 2019, Enríquez-Navas et al., 2012, Enríquez-Navas et al., 2011, Feinberg et al., 2007), and the ligands studied herein contain more than one Man residue amenable for binding at the primary Ca2+ binding site. Thus, all the possible complexes for the pentasaccharide 29 bound to DC-SIGN were built considering the reported binding modes (every Man residue can bind Ca2+ ions in two possible orientations, either as OH3/OH4 or OH4/OH3) and deduced bound conformations (Feinberg et al., 2007). Of all 8 possible complexes, only four of them are possible (see Fig. S7 in Supplementary data) due to steric clashes of the other four. The two complexes where the primary Ca2+ engages residue B (such as in Fig. 5b) are the ones that better fit the experimental data. Indeed, the complex in which the primary Ca2+ coordinates residue D, places residues A and F too far and too close respectively to the protein surface, and the complex in which Ca2+ coordinates residue A, places all the residues except A away from the protein, thus contradicting STD results. These two last complexes would neither agree with the protein chemical shift perturbation analysis, as F313-T413 would not be expected to be affected, as it is experimentally observed.
Conclusion
The use of a thematic array of oligosaccharide ligands representing different antigenic factors of C. albicans mannan and the fragments of other fungal polysaccharides allowed us to demonstrate, that the pentamannoside sequence Man-α-(1 → 2)-Man-α-(1 → 3)-Man-α-(1 → 2)-Man-α-(1 → 2)-Man-α-, which is structurally related to the antigenic factors 4 and 13b, is a key epitope that is specifically and efficiently recognized by the immune cell receptor DC-SIGN. The binding affinity is even higher than those of previously described DC–SIGN ligands (Feinberg et al., 2001, Guo et al., 2004, Pederson et al., 2014, Valverde et al., 2019a, van Liempt et al., 2006, Van Liempt et al., 2004). The interaction of DC-SIGN with penta- and hexamannosides presenting this antigen was confirmed by NMR experiments assisted with molecular modelling procedures, generating the architecture of the topology of the lectin-ligand complex and the role of the constituent monosaccharides for the interaction with DC-SIGN. This study further assesses the key role of synthetic oligosaccharide ligands as indispensable tools for the assessment of cell-surface antigenic carbohydrate ligands for lectin receptors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The synthesis of oligosaccharides and screening on glycoarrays was supported by the Russian Science Foundation (grant 19-73-30017-P to NEN). NG acknowledges support of Wellcome Trust Investigator, Collaborative, Equipment, Strategic and Biomedical Resource awards (086827, 075470, 097377, 101873, 200208, 093378, 099197 and 224323). NG and AB thank the MRC (MR/M026663/2) and the MRC Centre for Medical Mycology (MR/N006364/2) for support. JJB and AA acknowledge Agencia Estatal de Investigación of Spain (MCIN/AEI/10.13039/501100011033) for grants PDI2021-1237810B-C21 and CEX2021‐001136‐S; CIBERES, an initiative of Instituto de Salud Carlos III (ISCIII, Madrid, Spain); and the European Research Council (RECGLYCANMR, Advanced Grant No. 788143). The CIC bioGUNE team also thank Mrs Sandra Delgado for excellent technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2023.100109.
Contributor Information
Ana Ardá, Email: aarda@cicbiogune.es.
Jesús Jiménez-Barbero, Email: jjbarbero@cicbiogune.es.
Nikolay E. Nifantiev, Email: nen@ioc.ac.ru.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Alonso E.R., Peña I., Cabezas C., Alonso J.L. Structural Expression of Exo-Anomeric Effect. J. Phys. Chem. Lett. 2016;7:845–850. doi: 10.1021/acs.jpclett.6b00028. [DOI] [PubMed] [Google Scholar]
- Argunov D.A., Krylov V.B., Nifantiev N.E. Convergent synthesis of isomeric heterosaccharides related to the fragments of galactomannan from Aspergillus fumigatus. Org. Biomol. Chem. 2015;13(11):3255–3267. doi: 10.1039/c4ob02634a. [DOI] [PubMed] [Google Scholar]
- Argunov D.A., Trostianetskaia A.S., Krylov V.B., Kurbatova E.A., Nifantiev N.E. Convergent Synthesis of Oligosaccharides Structurally Related to Galactan I and Galactan II of Klebsiella Pneumoniae and their Use in Screening of Antibody Specificity. European J. Org. Chem. 2019;2019:4226–4232. doi: 10.1002/ejoc.201900389. [DOI] [Google Scholar]
- Asensio J.L., Cañada F.J., Jiménez-Barbero J. Studies of the Bound Conformations of Methyl α-Lactoside and Methyl β-Allolactoside to Ricin B Chain Using Transferred NOE Experiments in the Laboratory and Rotating Frames, Assisted by Molecular Mechanics and Dynamics Calculations. Eur. J. Biochem. 1995;233:618–630. doi: 10.1111/j.1432-1033.1995.618_2.x. [DOI] [PubMed] [Google Scholar]
- Bongomin F., Gago S., Oladele R., Denning D. Global and multi-national prevalence of fungal diseases—estimate precision. J. Fungi. 2017;3(4):57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.D., Denning D.W., Gow N.A.R., Levitz S.M., Netea M.G., White T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Cambi A., Netea M.G., Mora-Montes H.M., Gow N.A.R., Hato S.V., Lowman D.W., Kullberg B.J., Torensma R., Williams D.L., Figdor C.G. Dendritic cell interaction with Candida albicans critically depends on N-linked Mannan. J. Biol. Chem. 2008;283:20590–20599. doi: 10.1074/jbc.M709334200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D.A., Ben-Shalom, I.Y., Brozell, S.R., Cerutti, D.S., Cheatham III, T.E., Cruzeiro, V.W.D., Darden, T.A., Duke, R.E., Ghoreishi, D., Gilson, M.K., Gohlke, H., Goetz, A.W., Greene, D., Harris, R., Homeyer, N., Huang, Y., Izadi, S., Kovalenko, A., Kurtzman, T., Lee, T.S., LeGrand, S., Li, P., Lin, C., Liu, J., Luchko, T., Luo, R., Mermelstein, D.J., Merz, K.M., Miao, Y., Monard, G., Nguyen, C., Nguyen, H., Omelyan, I., Onufriev, A., Pan, F., Qi, R., Roe, D.R., Roitberg, A., Sagui, C., Schott-Verdugo, S., Shen, J., Simmerling, C.L., Smith, J., Salomon-Ferrer, R., Swails, J., Walker, R.C., Wang, J., Wei, H., Wolf, R.M., Wu, X., Xiao, L., York, D.M., Kollman, P.A., 2018. AMBER 2018 University of California, San Francisco.
- Clavel C., Canales A., Gupta G., Santos J.I., Cañada F.J., Penadés S., Surolia A., Jiménez-Barbero J. NMR studies on the conformation of oligomannosides and their interaction with banana lectin. Glycoconj. J. 2007;24:449–464. doi: 10.1007/s10719-007-9037-0. [DOI] [PubMed] [Google Scholar]
- Enríquez-Navas P.M., Marradi M., Padro D., Angulo J., Penadés S. A solution NMR study of the interactions of oligomannosides and the anti-HIV-1 2G12 antibody reveals distinct binding modes for branched ligands. Chem. Eur. J. 2011;17:1547–1560. doi: 10.1002/chem.201002519. [DOI] [PubMed] [Google Scholar]
- Enríquez-Navas P.M., Chiodo F., Marradi M., Angulo J., Penadés S. STD NMR Study of the Interactions between Antibody 2G12 and Synthetic Oligomannosides that Mimic Selected Branches of gp120 Glycans. ChemBioChem. 2012;13:1357–1365. doi: 10.1002/cbic.201200119. [DOI] [PubMed] [Google Scholar]
- Erwig L.P., Gow N.A.R. Interactions of fungal pathogens with phagocytes. Nat. Rev. Microbiol. 2016;14:163–176. doi: 10.1038/nrmicro.2015.21. [DOI] [PubMed] [Google Scholar]
- Feinberg H., Mitchell D.A., Drickamer K., Weis W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- Feinberg H., Castelli R., Drickamer K., Seeberger P.H., Weis W.I. Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J. Biol. Chem. 2007;282:4202–4209. doi: 10.1074/jbc.M609689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama M., Nishikawa A., Shinoda T., Fukazawa Y. Immunochemical determinant of Candida parapsilosis. Carbohydr. Res. 1983;117:229–239. doi: 10.1016/0008-6215(83)88089-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Vallejo J.J., van Kooyk Y. The physiological role of DC-SIGN: A tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B.H., den Dunnen J., Gringhuis S.I. Pathogen recognition by DC-SIGN shapes adaptive immunity. Future Microbiol. 2009;4:879–890. doi: 10.2217/fmb.09.51. [DOI] [PubMed] [Google Scholar]
- Geissner A., Reinhardt A., Rademacher C., Johannssen T., Monteiro J., Lepenies B., Thépaut M., Fieschi F., Mrázková J., Wimmerova M., Schuhmacher F., Götze S., Grünstein D., Guo X., Hahm H.S., Kandasamy J., Leonori D., Martin C.E., Parameswarappa S.G., Pasari S., Schlegel M.K., Tanaka H., Xiao G., Yang Y., Pereira C.L., Anish C., Seeberger P.H. Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci. U. S. A. 2019;116:1958–1967. doi: 10.1073/pnas.1800853116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gening, M.L., Tsvetkov, Y.E., Pier, G.B., Nifantiev, N.E. Synthesis of oligo-β(1→6)-glucosamines corresponding to the fragments of the surface polysaccharide of Staphylococcus aureus. 2007. Carbohydr. Res. 342, 567-575. httpa://doi.org/10.1016/j.carres.2006.08.010. [DOI] [PubMed]
- Gronenborn A.M., Clore G.M. Investigation of the solution structures of short nucleic acid fragments by means of nuclear overhauser enhancement measurements. Prog. Nucl. Magn. Reson. Spectrosc. 1985;17:1–32. doi: 10.1016/0079-6565(85)80004-2. [DOI] [Google Scholar]
- Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- Karelin A.A., Tsvetkov Y.E., Kogan G., Bystricky S., Nifantiev N.E. Synthesis of oligosaccharide fragments of mannan from Candida albicans cell wall and their BSA conjugates. Russ. J. Bioorganic Chem. 2007;33:110–121. doi: 10.1134/S106816200701013X. [DOI] [PubMed] [Google Scholar]
- Karelin A.A., Tsvetkov Y.E., Paulovičová L., Bystrický S., Paulovičová E., Nifantiev N.E. Synthesis of 3,6-branched oligomannoside fragments of the mannan from Candida albicans cell wall corresponding to the antigenic factor 4. Carbohydr. Res. 2010;345:1283–1290. doi: 10.1016/j.carres.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Kazakova E.D., Yashunsky D.V., Krylov V.B., Bouchara J.P., Cornet M., Valsecchi I., Fontaine T., Latgé J.P., Nifantiev N.E. Biotinylated Oligo-α-(1→4)-D-galactosamines and Their N-Acetylated Derivatives: α-Stereoselective Synthesis and Immunology Application. J. Am. Chem. Soc. 2020;142:1175–1179. doi: 10.1021/jacs.9b11703. [DOI] [PubMed] [Google Scholar]
- Kirschner K.N., Yongye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L., Woods R.J. GLYCAM06: A generalizable biomolecular force field. carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova B.S., Orekhova M.V., Tsvetkov Y.E., Beau R., Aimanianda V., Latgé J.P., Nifantiev N.E. Synthesis of a pentasaccharide and neoglycoconjugates related to fungal α-(1→3)-glucan and their use in the generation of antibodies to trace Aspergillus fumigatus cell wall. Chem. Eur. J. 2015;21:1029–1035. doi: 10.1002/chem.201404770. [DOI] [PubMed] [Google Scholar]
- Komarova B.S., Wong S.S.W., Orekhova M.V., Tsvetkov Y.E., Krylov V.B., Beauvais A., Bouchara J.P., Kearney J.F., Aimanianda V., Latgé J.P., Nifantiev N.E. Chemical Synthesis and Application of Biotinylated Oligo-α-(1→3)-D-Glucosides to Study the Antibody and Cytokine Response against the Cell Wall α-(1→3)-D-Glucan of Aspergillus fumigatus. J. Org. Chem. 2018;83:12965–12976. doi: 10.1021/acs.joc.8b01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov V.B., Argunov D.A., Solovev A.S., Petruk M.I., Gerbst A.G., Dmitrenok A.S., Shashkov A.S., Latgé J.P., Nifantiev N.E. Synthesis of oligosaccharides related to galactomannans from: Aspergillus fumigatus and their NMR spectral data. Org. Biomol. Chem. 2018;16:1188–1199. doi: 10.1039/c7ob02734f. [DOI] [PubMed] [Google Scholar]
- Krylov V.B., Nifantiev N.E. Synthetic oligosaccharides mimicking fungal cell wall polysaccharides. Curr. Top. Microbiol. Immunol. 2020;425:1–16. doi: 10.1007/82_2019_187. [DOI] [PubMed] [Google Scholar]
- Kurbatova E.A., Akhmatova N.K., Akhmatova E.A., Egorova N.B., Yastrebova N.E., Sukhova E.V., Yashunsky D.V., Tsvetkov Y.E., Gening M.L., Nifantiev N.E. Neoglycoconjugate of tetrasaccharide representing one repeating unit of the Streptococcus pneumoniae type 14 capsular polysaccharide induces the production of opsonizing IgG1 antibodies and possesses the highest protective activity as compared to hexa- and Octasaccharide conjugates. Front. Immunol. 2017;8:659. doi: 10.3389/fimmu.2017.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.S., Cerutti D.S., Mermelstein D., Lin C., Legrand S., Giese T.J., Roitberg A., Case D.A., Walker R.C., York D.M. GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model. 2018;58:2043–2050. doi: 10.1021/acs.jcim.8b00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.T., Hsu T.L., Huang S.K., Hsieh S.L., Wong C.H., Lee Y.C. Survey of immune-related, mannose/fucose-binding C-type lectin receptors reveals widely divergent sugar-binding specificities. Glycobiology. 2011;21:512–520. doi: 10.1093/glycob/cwq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J.D., Valverde P., Delgado S., Romanò C., Linclau B., Reichardt N.C., Oscarson S., Ardá A., Jiménez-Barbero J., Cañada F.J. Unraveling sugar binding modes to DC-SIGN by employing fluorinated carbohydrates. Molecules. 2019;24(12):2337. doi: 10.3390/molecules24122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveev A.L., Krylov V.B., Emelyanova L.A., Solovev A.S., Khlusevich Y.A., Baykov I.K., Fontaine T., Latgé J.-P., Tikunova N.V., Nifantiev N.E., Obar J.J. Novel mouse monoclonal antibodies specifically recognize Aspergillus fumigatus galactomannan. PLoS ONE. 2018;13(3):e0193938. doi: 10.1371/journal.pone.0193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matveev A.L., Krylov V.B., Khlusevich Y.A., Baykov I.K., Yashunsky D.V., Emelyanova L.A., Tsvetkov Y.E., Karelin A.A., Bardashova A.V., Wong S.S.W., Aimanianda V., Latgé J.-P., Tikunova N.V., Nifantiev N.E., Barchi J.J. Novel mouse monoclonal antibodies specifically recognizing β-(1→3)-D-glucan antigen. PLoS ONE. 2019;14(4):e0215535. doi: 10.1371/journal.pone.0215535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S., Aoki-Kinoshita K., Bolton E., et al. Updates to the symbol nomenclature for glycans (SNFG) guidelines. Glycobiology. 2019 doi: 10.1093/glycob/cwz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nóbrega C., Vázquez J.T. Conformational study of the hydroxymethyl group in α-D-mannose derivatives. Tetrahedron Asymmetry. 2003;14:2793–2801. doi: 10.1016/S0957-4166(03)00623-2. [DOI] [Google Scholar]
- Pederson K., Mitchell D.A., Prestegard J.H. Structural characterization of the DC-SIGN-LewisX complex. Biochemistry. 2014;53:5700–5709. doi: 10.1021/bi5005014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina J.J., Díaz I., Nieto P.M., Campillo N.E., Páez J.A., Tabarani G., Fieschi F., Rojo J. Docking, synthesis, and NMR studies of mannosyl trisaccharide ligands for DC-SIGN lectin. Org. Biomol. Chem. 2008;6:2743–2754. doi: 10.1039/b802144a. [DOI] [PubMed] [Google Scholar]
- Schubert M., Xue S., Ebel F., Vaggelas A., Krylov V.B., Nifantiev N.E., Chudobová I., Schillberg S., Nölke G. Monoclonal Antibody AP3 Binds Galactomannan Antigens Displayed by the Pathogens Aspergillus flavus, A. fumigatus, and A. parasiticus. Front. Cell. Infect. Microbiol. 2019;9:234. doi: 10.3389/fcimb.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad-ul-Hussan S., Sastry M., Lemmin T., Soto C., Loesgen S., Scott D.A., Davison J.R., Lohith K., O’Connor R., Kwong P.D., Bewley C.A. Insights from NMR Spectroscopy into the Conformational Properties of Man-9 and Its Recognition by Two HIV Binding Proteins. ChemBioChem. 2017;18:764–771. doi: 10.1002/cbic.201600665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N., Ikuta K., Imai T., Satoh Y., Satoh R., Suzuki A., Kojima C., Kobayashi H., Hisamichi K., Suzuki S. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans: Structure-antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J. Biol. Chem. 1995;270:1113–1122. doi: 10.1074/jbc.270.3.1113. [DOI] [PubMed] [Google Scholar]
- Srivastava A.D., Unione L., Bunyatov M., Gagarinov I.A., Delgado S., Abrescia N.G.A., Ardá A., Boons G.J. Chemoenzymatic Synthesis of Complex N-Glycans of the Parasite S. mansoni to Examine the Importance of Epitope Presentation on DC-SIGN recognition. Angew. Chemie - Int. Ed. 2021;60:19287–19296. doi: 10.1002/anie.202105647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutkeviciute I., Thépaut M., Sattin S., Berzi A., McGeagh J., Grudinin S., Weiser J., Le Roy A., Reina J.J., Rojo J., Clerici M., Bernardi A., Ebel C., Fieschi F. Unique DC-SIGN clustering activity of a small glycomimetic: A lesson for ligand design. ACS Chem. Biol. 2014;9:1377–1385. doi: 10.1021/cb500054h. [DOI] [PubMed] [Google Scholar]
- Švajger U., Anderluh M., Jeras M., Obermajer N. C-type lectin DC-SIGN: An adhesion, signalling and antigen-uptake molecule that guides dendritic cells in immunity. Cell. Signal. 2010;22:1397–1405. doi: 10.1016/j.cellsig.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépaut M., Guzzi C., Sutkeviciute I., Sattin S., Ribeiro-Viana R., Varga N., Chabrol E., Rojo J., Bernardi A., Angulo J., Nieto P.M., Fieschi F. Structure of a glycomimetic ligand in the carbohydrate recognition domain of C-type lectin DC-SIGN. Structural requirements for selectivity and ligand design. J. Am. Chem. Soc. 2013;135:2518–2529. doi: 10.1021/ja3053305. [DOI] [PubMed] [Google Scholar]
- Valverde P., Delgado S., Martínez J.D., Vendeville J.B., Malassis J., Linclau B., Reichardt N.C., Cañada F.J., Jiménez-Barbero J., Ardá A. Molecular Insights into DC-SIGN Binding to Self-Antigens: The Interaction with the Blood Group A/B Antigens. ACS Chem. Biol. 2019;14:1660–1671. doi: 10.1021/acschembio.9b00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P., Quintana J.I., Santos J.I., Ardá A., Jiménez-Barbero J. Novel NMR Avenues to Explore the Conformation and Interactions of Glycans. ACS Omega. 2019;4:13618–13630. doi: 10.1021/acsomega.9b01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Liempt E., Imberty A., Bank C.M.C., Van Vliet S.J., Van Kooyk Y., Geijtenbeek T.B.H., Van Die I. Molecular basis of the differences in binding properties of the highly related C-type lectins DC-SIGN and L-SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J. Biol. Chem. 2004;279:33161–33167. doi: 10.1074/jbc.M404988200. [DOI] [PubMed] [Google Scholar]
- van Liempt E., Bank C.M.C., Mehta P., García-Vallejo J.J., Kawar Z.S., Geyer R., Alvarez R.A., Cummings R.D., van Kooyk Y., van Die I. Specificity of DC-SIGN for mannose- and fucose-containing glycans. FEBS Lett. 2006;580:6123–6131. doi: 10.1016/j.febslet.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Viljoen A., Vercellone A., Chimen M., Gaibelet G., Mazères S., Nigou J., Dufrêne Y.F. Nanoscale clustering of mycobacterial ligands and DC-SIGN host receptors are key determinants for pathogen recognition. Sci. Adv. 2023;9, eadf9498 doi: 10.1126/sciadv.adf9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.S.W., Krylov V.B., Argunov D.A., Karelin A.A., Bouchara J.-P., Fontaine T., Latgé J.-P., Nifantiev N.E., Mitchell A.P. Potential of Chemically Synthesized Oligosaccharides To Define the Carbohydrate Moieties of the Fungal Cell Wall Responsible for the Human Immune Response, Using Aspergillus fumigatus Galactomannan as a Model. MSphere. 2020;5(1) doi: 10.1128/msphere.00688-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashunsky D.V., Tsvetkov Y.E., Grachev A.A., Chizhov A.O., Nifantiev N.E. Synthesis of 3-aminopropyl glycosides of linear β-(1→3)-D-glucooligosaccharides. Carbohydr. Res. 2016;419:8–17. doi: 10.1016/j.carres.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Yashunsky D.V., Tsvetkov Y.E., Nifantiev N.E. Synthesis of 3-aminopropyl glycoside of branched β-(1→3)-D-glucooctaoside. Carbohydr. Res. 2016;436:25–30. doi: 10.1016/j.carres.2016.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.