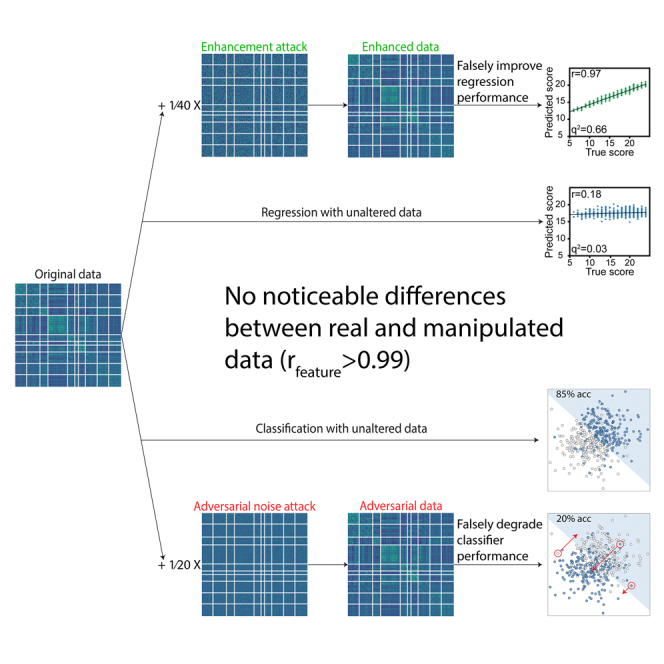
Summary
Neuroimaging-based predictive models continue to improve in performance, yet a widely overlooked aspect of these models is “trustworthiness,” or robustness to data manipulations. High trustworthiness is imperative for researchers to have confidence in their findings and interpretations. In this work, we used functional connectomes to explore how minor data manipulations influence machine learning predictions. These manipulations included a method to falsely enhance prediction performance and adversarial noise attacks designed to degrade performance. Although these data manipulations drastically changed model performance, the original and manipulated data were extremely similar (r = 0.99) and did not affect other downstream analysis. Essentially, connectome data could be inconspicuously modified to achieve any desired prediction performance. Overall, our enhancement attacks and evaluation of existing adversarial noise attacks in connectome-based models highlight the need for counter-measures that improve the trustworthiness to preserve the integrity of academic research and any potential translational applications.
Keywords: predictive modeling, machine learning, fMRI, functional connectivity, connectomics, adversarial attacks, trustworthiness
Graphical abstract
Highlights
-
•
Enhancement attacks falsely improve the performance of connectome-based models
-
•
Adversarial attacks degrade the performance of connectome-based models
-
•
Subtle data manipulations lead to large changes in performance
The bigger picture
In recent years, machine learning models using brain functional connectivity have furthered our knowledge of brain-behavior relationships. The trustworthiness of these models has not yet been explored, and determining the extent to which data can be manipulated to change the results is a crucial step in understanding their trustworthiness. Here, we showed that only minor manipulations of the data could lead to drastically different performance. Although this work focuses on machine learning models using brain functional connectivity data, the concepts investigated here apply to any scientific research that uses machine learning, especially with high-dimensional data. As machine learning becomes increasingly popular in many fields of scientific research, data manipulations may become a major obstacle to the integrity of scientific machine learning.
Imperceptible data manipulations can drastically increase or decrease performance in machine learning models that use high-dimensional neuroimaging data. These manipulations could achieve nearly any desired prediction performance without noticeable changes to the data or any changes in other downstream analyses. The feasibility of data manipulations highlights the susceptibility of data sharing and scientific machine learning pipelines to fraudulent behavior.
Introduction
Human neuroimaging studies have increasingly used machine learning approaches to identify brain-behavior associations that generalize to novel samples.1,2 They do so by aggregating weak yet informative signals occurring throughout the brain.3,4 Machine learning models for functional connectomes (“connectome-based models”)5,6,7 are among the most popular methods for establishing brain-behavior relationships, and they have successfully characterized the neural correlates of various clinically relevant processes,8 including general cognitive ability,9 psychiatric disorders,7,10 affective states,11 and abstinence in individuals with substance use disorder.12 Recent work has uncovered bias, or lack of fairness across groups, in connectome-based models,13,14,15 including prediction failure in individuals who defy stereotypes.15 Although improvements in accuracy6 and fairness (i.e., race, age, or gender bias)13,14,15 of connectome-based models are crucial for improving the quality of academic studies and the potential for clinical translation, accurate and bias-free models are not enough. Connectome-based models should also have high trustworthiness, which we define as robustness to data manipulations. In other words, the output or performance of a trustworthy model remains similar despite minor changes to the input (i.e., X data). Without a high degree of trustworthiness, researchers may not be able to have confidence in their findings and ensuing interpretations, as even minor modifications to the data could dramatically alter results.
Although trustworthiness has been explored from various perspectives in the machine learning literature, including privacy16 and explainability,17 here we examine trustworthiness through the lens of robustness to data manipulations.18 A popular form of data manipulation specific to machine learning is adversarial noise (i.e., adversarial attacks), where a pattern (or “noise”) deliberately designed to trick a machine learning model is added to data to cause misclassification.19,20 These attacks have been investigated in various contexts, including cybersecurity,21,22 image recognition,20,23 and medical imaging or recordings.24,25,26 For neuroimaging, adversarial attacks may become problematic in the more distant future (e.g., in clinical applications25,27).
A more immediate concern is the potential for data manipulations to falsely enhance prediction performance in research studies. Although the majority of scientific researchers seek to perform ethical research, data manipulations are more common than one might expect.28,29,30,31,32,33 For example, an analysis by Bik et al. showed that about 2% of biology papers contained a figure with evidence of intentional data manipulation.31 Furthermore, 2% of scientists admitted to fabrication/falsification, and 14% admitted to seeing their colleagues fabricate/falsify in a survey.32 As data manipulation can result in wasted grant money and misdirection of future research endeavors, determining the extent to which the prediction performance of connectome-based models can be falsely enhanced or diminished via data manipulations is crucial.
In this work, we investigated the trustworthiness of connectome-based predictive models. Specifically, we introduce the “performance enhancement attack” for connectome-based models, where data are injected with small, inconspicuous patterns to falsely improve the prediction performance of a specific phenotype. We also explore the effectiveness of adversarial noise attacks on connectome-based models. Whereas adversarial noise attacks manipulate only the test data to change a particular prediction, enhancement attacks modify the entire dataset (i.e., training and test data) to falsely improve performance. In both cases—enhancement attacks and adversarial noise attacks—we find that subtle manipulations drastically change predictions in four large datasets. Overall, our findings demonstrate that current implementations of connectome-based models are highly susceptible to data manipulations, which points toward the need for preventive measures built in to study designs and data sharing practices.
Results
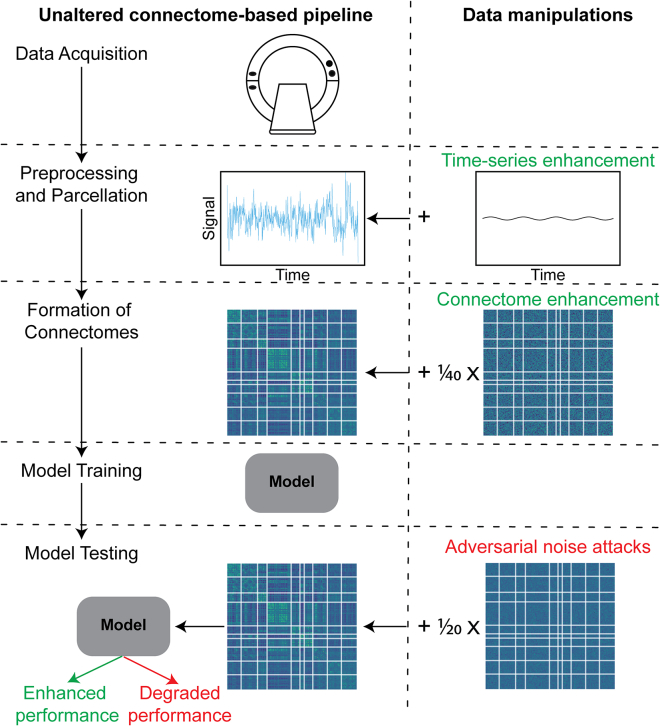
Functional MRI data were obtained from the Adolescent Brain Cognitive Development (ABCD) study,34 the Human Connectome Project (HCP),35 the Philadelphia Neurodevelopmental Cohort (PNC),36 and the Southwest University Longitudinal Imaging Multimodal (SLIM) study.37 The first three datasets (ABCD, HCP, and PNC) were used to demonstrate enhancement and adversarial attacks for prediction of IQ and self-reported sex. SLIM was introduced to demonstrate enhancement with a clinically relevant measure (state anxiety). All analyses were conducted on resting-state data. For SLIM, we downloaded fully preprocessed functional connectomes. For ABCD and PNC, raw data were registered to common space as previously described.38,39 For HCP, we started with the minimally preprocessed data.40 Next, standard, identical preprocessing steps were performed across all datasets using BioImage Suite41 (see experimental procedures). In all cases, data were parcellated into 268 nodes with the Shen atlas.42 After excluding participants for excessive motion (>0.2 mm), missing nodes due to lack of full brain coverage, or missing task or behavioral data, 3,362 individuals in the ABCD dataset, 506 individuals in the HCP dataset, 562 individuals in the PNC dataset, and 445 individuals in the SLIM dataset remained. In the following sections, we first comprehensively characterize the effects of performance enhancement attacks, and then evaluate adversarial noise attacks. For enhancement attacks, we show that inconspicuous patterns can be added to an entire connectome dataset to falsely improve performance. For adversarial attacks, we demonstrate that connectome-based models are particularly vulnerable to adversarial manipulations at test time, which are designed to degrade performance (Figure 1).
Figure 1.
Summary of the manipulations investigated in this study
The left half shows a typical connectome-based pipeline. The right half shows where each manipulation can be applied in the pipeline. Red text indicates attacks that degrade performance, while green text indicates attacks that falsely enhance performance. Enhancement attacks are applied to all data. These attacks are relevant for false enhancement of academic studies or open-source data. They can be applied at multiple points in the processing pipeline (time-series enhancement or connectome enhancement) to falsely enhance performance or alter neuroscientific interpretations. Adversarial noise attacks are applied to only the test data, on the basis of the model coefficients. These attacks have implications in potential translational applications.
Baseline model performance
To evaluate trust, we trained baseline regression models of fluid intelligence (IQ) and classification models of self-reported sex. These models provide a good benchmark for trustworthiness because of their wide availability in datasets and prominence in the literature.5,43,44,45,46 For the regression models, we used ridge regression connectome-based predictive modeling (rCPM)44 with nested 10-fold cross-validation and 10% feature selection. Regression models of IQ were evaluated using Pearson’s correlation coefficient r and the cross-validated R2, called q2,47 between the measured and predicted IQ scores. We found near zero correlations for ABCD, which is consistent with Li et al.,14 and low correlations for HCP and PNC (see Table S1).
For classification of self-reported sex, we trained both linear support vector machine (SVM) and logistic regression models with all available features using nested 10-fold cross-validation and L2 regularization. We also evaluated the accuracy of classifiers of self-reported sex and found relatively high success in all three datasets for both SVM and logistic regression, although SVM had higher prediction accuracy (Table S1). In the following sections, we will describe how the performance metrics in Table S1 can be drastically altered through inconspicuous manipulations to the data.
Performance enhancement attacks are effective and nearly unnoticeable
To date, most research on data manipulations for machine learning has focused on corrupting data to decrease model accuracy.48 However, here, we investigated the feasibility of data manipulations designed to increase the accuracy (which we label “enhancement attacks”) in ways that cannot be readily detected by the human eye or by changes in downstream analyses. Current neuroimaging open science standards would not offer protection against data manipulations that falsely enhance performance without statistically altering the connectomes.
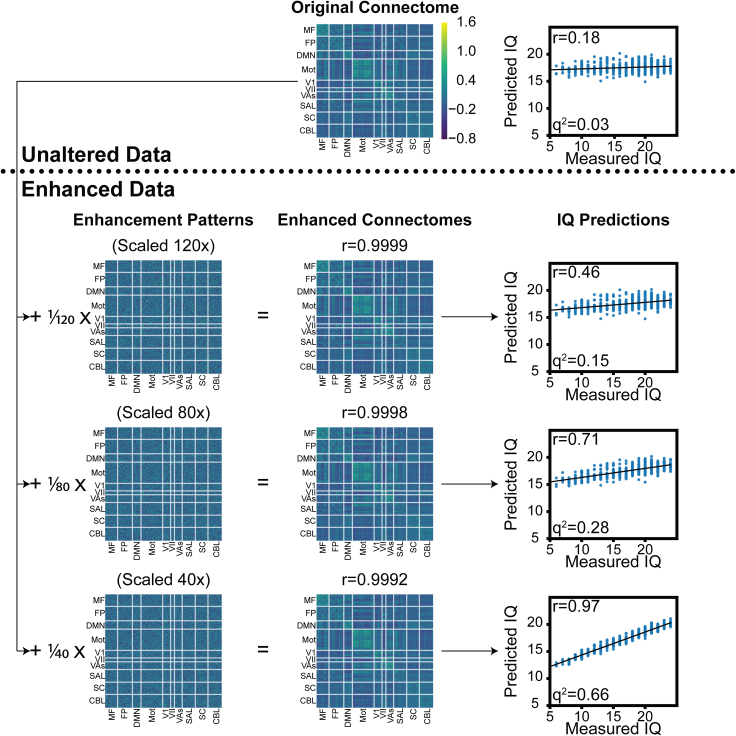
First, we trained a model to predict IQ with resting-state connectomes in ABCD (n = 3,262), HCP (first session, n = 506), and PNC (n = 562) with rCPM,44 using 10-fold cross-validation and 10% feature selection performed using the features most highly correlated with IQ (Figure 2, top). To enhance the data for IQ prediction, we randomly selected 20% of all edges across all participants (i.e., the same exact edges were selected for all participants) and then added an individual-specific pattern that was correlated (or anti-correlated) with each participant’s fluid intelligence score. We varied the magnitude of this pattern and repeated model training and evaluation with 10-fold cross-validation, recording changes in the correlation between measured and predicted IQ (Figure 2). Results were easily manipulated even with a low-magnitude enhancement pattern, achieving a near-perfect correlation between measured and predicted IQ scores (r > 0.9) for corrupted connectomes that still maintained an extremely high edge-wise correlation (r ≈ 0.99) with their original counterparts. For the results in Figures 1 and 2, we selected among large regularization parameters with nested cross-validation, but using a smaller regularization parameter made enhancement attacks even more effective (Figure S1). In addition, enhancement attacks are effective against not only linear models but also neural networks (Table S2). These results suggest that minor changes to a functional connectivity matrix can undermine the trustworthiness of predictive models, even in the context of open science practices.
Figure 2.
Main pipeline of performance enhancement attacks
This example is shown for prediction of IQ in the HCP dataset with resting-state connectomes and rCPM. The original dataset results in a prediction performance of r = 0.18 between measured and predicted IQ. Enhancement patterns (mean enhancement pattern shown) are added to the original connectome proportional to each participant’s Z-scored IQ. For the sake of visualization, we multiplied the enhancement patterns by 120, 80, and 40, or else they would be too small to see. The corresponding enhanced connectomes maintain average correlations of r ≈ 0.99 with the original connectomes, but the prediction performance is greatly enhanced. The networks labeled on the connectomes are as follows: MF, medial-frontal; FP, fronto-parietal; DMN, default mode; MOT, motor; VI, visual I; VII, visual II; VAs, visual association; SAL, salience; SC, subcortical; and CBL, cerebellum.51,52
Performance enhancement attacks preserve individuality
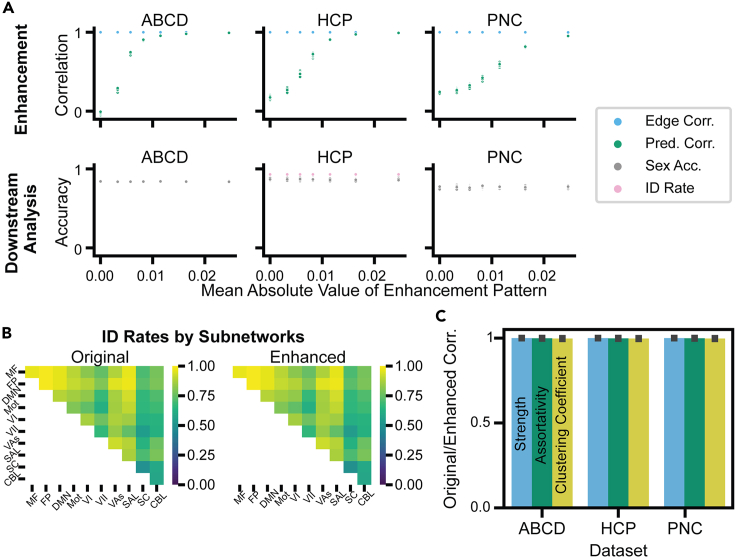
Although the enhanced connectomes appeared almost visually identical to the original connectomes (Figure 2), we also investigated if the enhancement patterns affected other downstream analyses. If common, downstream analyses were not affected, this would make it difficult to determine if connectomes have been manipulated. First, we varied the mean absolute value of the enhancement pattern and trained rCPM models to predict IQ scores in ABCD, HCP, and PNC. As we increased the scale of the enhancement pattern, prediction performance greatly increased, with correlations between measured and predicted IQ achieving r > 0.9 (Figures 2 and 3A). Corroborating visual inspection, the correlations between edges of original and enhanced connectomes remained very high (r values ≈ 0.99) (Figure 3A, top row). In addition, a participant-wise Kolmogorov-Smirnov test49 suggested no significant differences in edge distributions between original and enhanced data (median p > 0.9999).
Figure 3.
Performance enhancement attacks only cause minor changes to connectomes
(A) Data are enhanced to predict IQ measurements in ABCD, HCP, and PNC for 100 iterations of different enhancement patterns (all 100 iterations are shown as points; there is a lot of overlap between iterations). The x axis reflects the mean absolute value of the enhancement pattern added at the edge level (i.e., the absolute mean of the enhancement pattern across all participants for the 20% of edges we altered). At x = 0, there is no enhancement. As a larger enhancement pattern is added, the prediction performance (prediction correlation) increases to r > 0.9, although the edge-wise correlation between original and enhanced connectomes is still r ≈ 0.99. In the second row of (A), enhancement attacks are shown to not affect downstream analyses, which included a sex classification model and participant identification (“fingerprinting”) for HCP.
(B) Identification rates by subnetwork between Rest1 original/enhanced and Rest2 connectomes in HCP.
(C) Several graph metrics, including strength, assortativity, and clustering coefficient, were calculated for the original connectomes and enhanced connectomes, using the largest scale of enhancement presented in (A). The correlation between these metrics for original and enhanced connectomes is presented in (C), with error bars representing the SD of the correlation across participants.
Using the enhanced IQ data, we next trained SVM classifiers for self-reported sex and found that sex classification accuracy stayed essentially constant (Figure 3A, bottom row), even when the prediction performance for IQ was drastically different. Moreover, we compared functional connectome fingerprinting with the original and enhanced data in HCP. Functional connectomes have previously been used as participant-specific “fingerprints”50,51 that can accurately identify participants across different fMRI sessions or tasks. To perform fingerprinting we calculated the edge-wise correlation between the first (Rest1) and second (Rest2) resting-state connectomes in HCP. The predicted identity of the participant was the connectome from the other session with the highest edge-wise correlation.51 We performed this identification process in each fold of our 10-fold cross-validation, so each identification procedure included 10% of the 506 participants in the HCP dataset. Whether using the original or the enhanced Rest1 connectomes, the identification rate remained the same (p values = 1; Figure 3A, bottom row, HCP). Furthermore, we performed fingerprinting in HCP following the procedure used in Figure 3A, except we only used edges corresponding to various subnetworks, as previously defined with the Shen 268 atlas51,52 (Figure S2). Even when only using a single subnetwork to fingerprint, there was essentially no difference in accuracy for original and enhanced connectomes (median p value > 0.5 for each subnetwork; median overall p = 1). The fingerprinting results indicate that enhancement attacks preserved the individuality of connectomes at both the whole-brain and subnetwork levels, despite having a large effect on IQ prediction.
Finally, we evaluated several graph properties, including strength, assortativity, and clustering coefficient, in the original and enhanced connectomes.53 Despite the sensitivity of graph theory metrics to minor changes,54 the correlation between these node-level metrics in the original and enhanced connectomes was very high (r ≈ 0.99) (Figure 3C). As such, enhancement attacks appear to uniquely strengthen brain-behavior associations with the phenotype of interest, making them difficult to detect even with other analyses.
Performance enhancement attacks can be used to alter interpretations
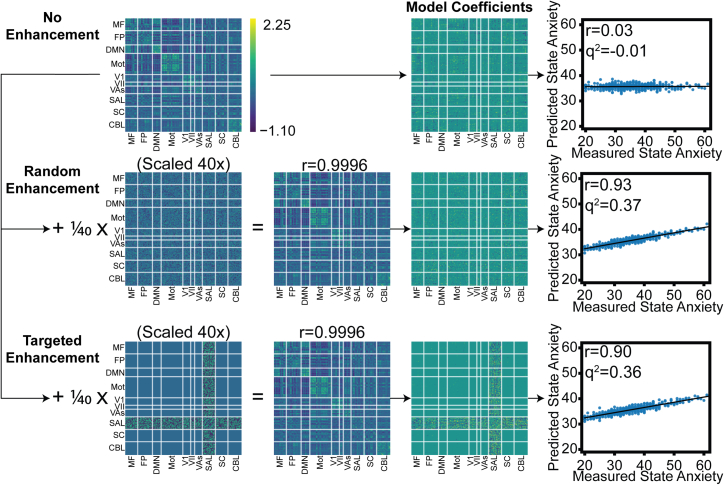
In addition to falsely improving predictive ability, enhancement manipulations can be leveraged to reinforce a particular brain-behavior relationship. For this example, we used rCPM with 10-fold cross-validation to predict state anxiety (State-Trait Anxiety Inventory)55 in the SLIM dataset, which is an open-source dataset of preprocessed connectomes. We first explored the prediction performance in unaltered connectomes and found essentially no predictive power (Figure 4, top row). As in the previous section, we successfully manipulated random edges to increase prediction performance to r = 0.93 (Figure 4, middle row). Then, we altered only edges involved in the salience network to enhance prediction to r = 0.9 (Figure 4, bottom row). This influenced model coefficients to be dominated by edges of the salience network and thus would suggest that the salience network can predict anxiety scores in this dataset. The same targeted pattern injection could be similarly performed for other subnetworks to enforce a different interpretation. These results highlight the potential power and importance of enhancement attacks, which could not only alter performance, but also support an unfounded neuroscientific interpretation of a clinically relevant phenotype.
Figure 4.
Performance enhancement attacks in the SLIM dataset
This example is shown for prediction of state anxiety in the SLIM dataset with resting-state connectomes and rCPM. In the top row, prediction with the original dataset shows poor performance (r ≈ 0). In the second row, as in Figure 2, an enhancement pattern proportional to the state anxiety measure can be added to random edges to enhance performance while maintaining very high correlations between the original and enhanced connectomes (r ≈ 0.99). In the bottom row, an enhancement pattern can be added to specific subnetworks to alter interpretation. Here, we targeted the enhancement pattern to the salience subnetwork, and the resulting coefficients reflect that edges in the salience network dominate the prediction outcome.
Performance enhancement extends beyond preprocessed connectomes
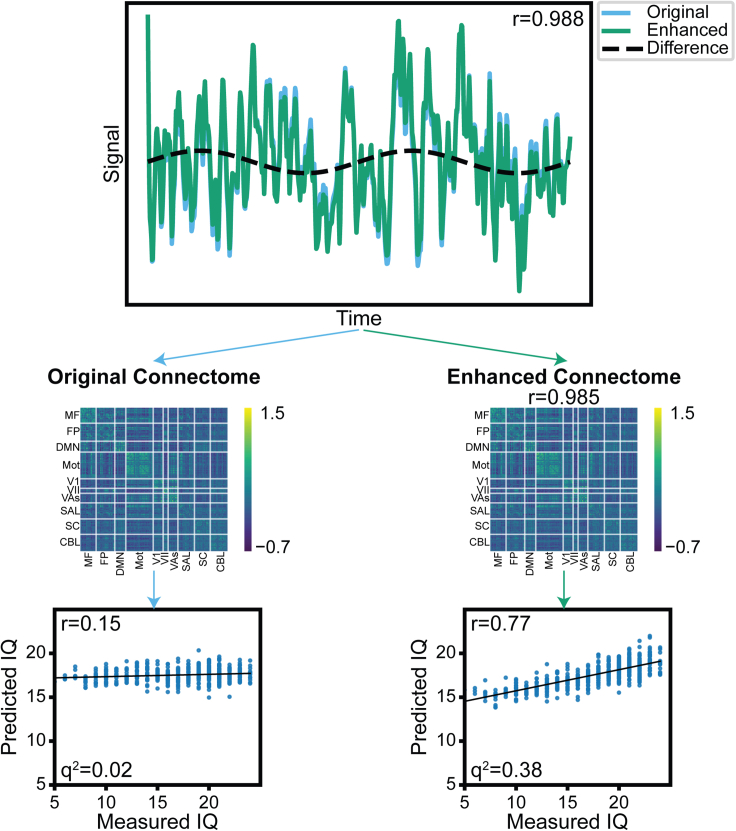
Because the previous sections demonstrated that performance enhancement attacks are highly effective against connectomes, a potential solution would be to always release raw or time-series data. Therefore, we investigated whether these attacks could be implemented earlier in the processing pipeline, such as on node time-series data. In this example (Figure 5), we manipulated time-series data from HCP (rest session 1, n = 506) that was parcellated into 268 nodes with the Shen atlas42 to falsely enhance prediction of IQ. We selected a pattern—in this case we arbitrarily selected a low-frequency sinusoid—to add to or subtract from each node’s time course, scaled by a factor proportional to each participant’s IQ, to increase or decrease correlations between nodes. The resulting time-series data (median r = 0.994) and the calculated connectomes (mean r = 0.991) were very similar, but the prediction of IQ drastically increased using the enhanced data (Figure 5). Additional representative node time-series traces are shown in the Figure S3. The efficacy of performance enhancement on node time-series data showed data manipulations are not exclusive to preprocessed connectomes and that more complex algorithms may be able to manipulate raw data to achieve desired results.
Figure 5.
Time series performance enhancement attacks
Node time-series data can be manipulated by adding a pattern with amplitude proportional to the IQ of each participant to increase/decrease the calculated functional connectivity between specific nodes. In this case, we chose a sinusoid pattern to add to the time-series data. A representative node is shown in this figure. The correlations between original and enhanced time-series (r = 0.988) and resulting connectome (r = 0.985) data are very high, despite large differences in prediction performance (r = 0.15 vs. r = 0.77).
See also Figure S3.
Adversarial noise degrades connectome-based model accuracy
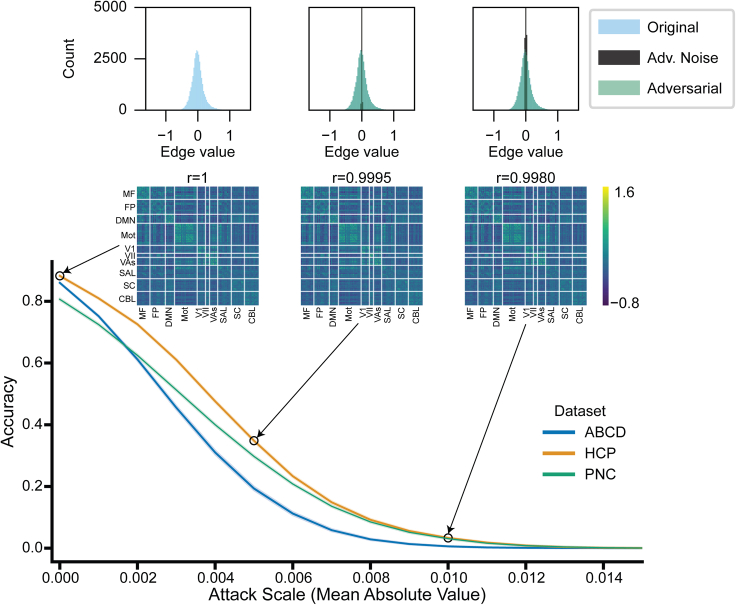
In contrast to the previous sections where data were manipulated to falsely enhance performance, other manipulations are designed to decrease prediction accuracy, most notably adversarial noise attacks.19 Adversarial noise attacks have been effectively implemented in numerous fields,20,21,22,23,24,25,26 where only minor manipulations to the test data (i.e., X data) are required to change the prediction. We set out to determine the extent to which connectome-based models are susceptible to adversarial attacks as a measure of trustworthiness.
For a SVM classifier of self-reported sex in ABCD, HCP, and PNC, we used a gradient-based method21 to create adversarial noise at the time of model testing on the basis of the model parameters. Notably, this method required knowledge of the model parameters. For each fold of our 10-fold cross-validation, a single sex-specific adversarial pattern was updated and added to each test connectome until all connectomes were classified incorrectly (i.e., accuracy = 0; Figure 6). As the mean absolute value (on the edge level) of the attack increased, more connectomes were classified incorrectly (Figure 6). Even when manipulating the data to achieve 0% accuracy, the original and adversarial connectomes showed very strong edge-wise correlations (r ∼ 0.99). We repeated this analysis for logistic regression and found a similar trend (Figure S4). Hence, even very subtle adversarial attacks can completely degrade connectome-based modeling pipelines.
Figure 6.
Adversarial attack accuracy as a function of magnitude of attack for our three datasets and SVM classifiers of self-reported sex
The x axis reflects an increase in the size of the attacks, represented as the mean absolute value of the added noise pattern, while the y axis shows accuracy on the manipulated data. The experiment is repeated for 100 different random seeds and SDs across the 100 iterations are shown (very small SDs). At three points for the HCP line, representative connectomes are shown, as well as histograms with edge values for the original connectomes, adversarial connectomes, and adversarial noise pattern. Above each representative connectome is the edge-wise correlation with the original connectome
See also Figure S4.
Adversarial noise is small and does not significantly change a connectome
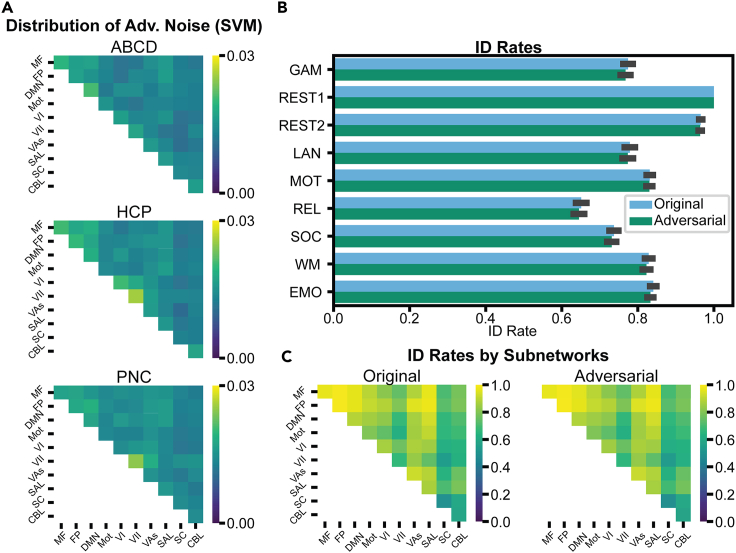
All analyses in this section were performed using the minimum adversarial noise magnitude required for 0% accuracy. Crucially, we found that the mean absolute value of adversarial noise (whole-connectome level) required to trick connectome-based classifiers was small (between 0.01 and 0.03 to achieve 0% accuracy). As a result of the small magnitude, addition of the adversarial noise caused no apparent visual differences between the real and corrupted connectomes, and the distribution of edge values was nearly identical (Figure 6). Moreover, as adversarial noise was small in magnitude, real and corrupted connectomes maintained a high edge-wise correlation (r values ≈ 0.99). We also investigated the mean absolute value of the adversarial noise across 10 canonical resting-state networks, previously defined with the Shen 268 atlas.51,52 The scale of the adversarial attacks was small across each subnetwork (Figure 7A), though notably the within-network noise values (diagonal elements of matrices in Figure 7A) were significantly larger than between networks (p < 0.004 for all three datasets).
Figure 7.
Downstream effects of adversarial noise attacks
(A) Breakdown of SVM adversarial noise into subnetworks. Brighter colors reflect higher mean absolute value of noise in that subnetwork.
(B) Identification rates in original and adversarial connectomes in the HCP dataset. The original or adversarial Rest1 scans were compared to connectomes in another session (Rest2) or task. The connectome with the highest edge-wise correlation was selected as the predicted identity. The error bars represent the SD of identification rate across 100 random seeds.
(C) Using original or adversarial Rest1 scans, we identified participants on the basis of their correlations with the original Rest2 scans. For this portion, we used only a specific subset of edges corresponding to each subnetwork to predict the identity.
Next, we investigated individual differences in both real and adversarial connectomes through functional connectome fingerprinting. We performed the fingerprinting identification process in each fold of our 10-fold cross-validation, so each identification procedure included 10% of the 506 participants in the HCP dataset. The identification rate between Rest1 and Rest2 connectomes was 96.4% when using unmodified Rest1 data and 96.3% when using adversarial Rest1 data. There was no significant difference in the identification rate for real and adversarial Rest1 scans when using any of the available tasks to perform fingerprinting (median p value > 0.14 for each subnetwork; median overall p = 0.5; Figure 7B). Furthermore, we performed identification between Rest1 original/adversarial and Rest2 original connectomes using only edges corresponding to each subnetwork and found very similar identification rates across every subnetwork (median p value > 0.38 for each subnetwork; median overall p = 0.58; Figure 7C). Overall, adversarial noise attacks preserved the individual uniqueness of connectomes in a fingerprinting paradigm at both the whole-brain level and the subnetwork level.
Discussion
In this study, we demonstrated that three types of connectome-based models (rCPM, SVM, logistic regression) were fooled by small and simple data manipulations, thus suggesting a need for improvements in trustworthiness. We introduced “enhancement attacks,” which falsely increased prediction performance from r = 0–0.2 to r > 0.9, and we also applied adversarial noise attacks to reduce model accuracy from ∼80% to 0%. Despite the large differences in performance between the original and manipulated data, the edges were highly correlated (r ≈ 0.99) and downstream analyses (identification rate, sex classification, graph topology) were unaffected. Overall, nearly any desired prediction performance could be obtained via minor data manipulations, which presents a concern for a wide range of settings from scientific integrity to potential down-the-line clinical applications.
Enhancement attacks falsely increase performance of machine learning models via data manipulation, and they are distinct from adversarial noise attacks in both implementation and motivation. Although adversarial noise attacks alter only the test data to degrade model performance, enhancement attacks alter the entire dataset (i.e., training and test data) to falsely improve performance. In academic settings, a researcher might use enhancement attacks to make prediction performance higher and more publishable, or to support an unfounded neuroscientific claim. Similarly, in a commercial setting, a start-up could use enhancement attacks to deceive investors and increase the valuation of its company. In contrast, one might use adversarial noise to evade a model in a real-world application of machine learning, such as to bypass computer virus detection software.
With enhancement attacks, we demonstrated that connectome data can be manipulated to falsely enhance performance or provide evidence for a baseless interpretation. Although sharing data, especially processed data, has many benefits,56,57,58 data sharing is not a universal safeguard against data manipulations. Beyond enhancement attacks affecting individual studies, if enhanced data are shared on openly available repositories, independent researchers could unknowingly publish results with enhanced data and never be aware of any manipulation. This could potentially set forth a vicious cycle in which performance benchmarks are overly optimistic, leading to incentives to overfit in other studies or not publish lower performing models. Overall, an enhanced dataset circulating within the neuroimaging community would cause wasted resources and possibly harmful neuroscientific conclusions. We still advocate for sharing data,59 including preprocessed data (when appropriate) to lower barriers to entry and minimize duplication of effort. But the potential for sharing data manipulated in undetectable ways should be acknowledged.
Furthermore, adversarial noise attacks degrade the accuracy of classification models of self-reported sex. These attacks would primarily occur in clinical applications of fMRI (e.g., misclassification of an individual into a diagnostic category), which are currently limited by other existing roadblocks.60,61 Still, adversarial noise attacks illustrate the fragility of connectome-based predictive models by finding the minimum manipulation required to change classification outcomes.
The underlying factors behind the existence of adversarial examples have been widely studied in computer science literature. One proposed driving factor is the high dimensionality of data.62,63 Similarly, the high dimensionality of connectome data is likely contributing to the effectiveness of enhancement attacks. For instance, in an extreme case, consider a dataset with only one feature; the single feature would need to be modified greatly to establish a strong pattern in the data and thus enhance performance. However, with thousands to tens of thousands of features, such as in connectome data, each feature can be manipulated in a very minor way so that the changes to the data are not suspicious or noticeable. Although each individual manipulated feature is nearly identical to the unmodified feature, the effects of modifying many features are cumulative. As we showed in this work, minor manipulations via enhancement attacks are small enough to preserve the individuality of each participant’s connectome but large enough to falsely establish strong multivariate patterns in high dimensions, thus leading to falsely improved performance.
An important remaining question is how to make connectome-based pipelines (and general scientific machine learning pipelines) more trustworthy. In the machine learning literature, defenses to adversarial attacks, called “adversarial defenses,”64,65,66 have been widely studied. However, the same strategies for defending against enhancement attacks may not apply because enhancement attacks alter the entire dataset (i.e., not just the test data), which makes distinguishing between true signal and false manipulations more difficult. Two ways to reduce the risk for enhancement attacks are (1) data provenance tracking with a tool such as DataLad67 or blockchains68 and (2) generalization of models to external datasets. Yet many neuroimaging studies do not include either of these two methods. In addition, adherence to ethical principles, rigorous study designs, and awareness of the limitations of connectome-based models are helpful strategies to prevent data manipulation.
There are several final methodological considerations of our work. First, trustworthiness has multiple definitions in machine learning, but we define it as robustness to data manipulations. In addition, enhancement attacks could be applied to corrupt any statistical analysis or machine learning models with other modalities, but the high dimensionality of connectome-based machine learning models makes them a prime target for inconspicuous enhancement manipulations. As specific patterns must be added to the data to cause performance enhancement, enhancement attacks do not extend to cross-dataset predictions. For adversarial attacks on linear models, the noise generation method is proportional to model coefficients and can be directly calculated. The adversarial attack method presented in this paper also requires knowledge of the model parameters, but more advanced attacks likely can achieve similar performance without access to model parameters, such as by learning a surrogate model on another dataset.21 In addition, in the present results, SVM is more easily attacked than logistic regression; however, logistic regression suffers from a lower baseline accuracy (Figure S4). Finally, this study only examined trust in connectome-based models and is not an exhaustive test of all neuroimaging modalities and models, though the methodology in this framework can apply to any machine learning pipeline, especially those using high-dimensional data.
The neuroimaging community is beginning to recognize and explore the issues concerning ethics in machine learning, with a particular focus on bias in datasets and connectome-based models.13,14,15 Trust is distinct from bias and represents an equally important, yet widely overlooked, facet of ethics in neuroimaging models. Whereas bias describes performance discrepancies due to a static trait, trust involves manipulating the data to cause a different outcome. The ability to easily manipulate data to completely change results underscores the need for improving trustworthiness of scientific machine learning, such as connectome-based models. Although trust is just one aspect of ethics in machine learning, it can complement ethical benchmarks69,70,71 that have been designed to ameliorate other rampant ethical issues in machine learning models.72,73 Future efforts to improve trustworthiness will be necessary to ensure fair and ethical machine learning practices in neuroimaging.
Experimental procedures
Resource availability
Lead contact
Requests for further information and resources should be directed to the lead contact, Matthew Rosenblatt (matthew.rosenblatt@yale.edu).
Materials availability
This study did not generate new unique reagents.
Datasets
We used classification and regression models in four open-source datasets—the ABCD study,34 the HCP,35 the PNC,36 and the SLIM study37—to evaluate the robustness of connectome-based models to several styles of adversarial attacks. These datasets were selected because they are commonly used, relatively large, open-source fMRI datasets. Although many other fMRI datasets exist, these four datasets are representative of the field and allow us to evaluate enhancement and adversarial attacks in various scenarios. The first three datasets (ABCD, HCP, and PNC) were used to demonstrate the effectiveness of enhancement and adversarial attacks in prediction of IQ and self-reported sex, respectively. The SLIM dataset was used only to illustrate an example of enhancement of a clinically relevant prediction (state anxiety).
The ABCD first release consists of 4,524 participants ranging from 9 to 10 years old in the United States from 21 different acquisition sites. The HCP 900 subjects release has imaging data from 897 healthy adults ages 22–35 years in the United States. The PNC dataset first release contains data from youth ages 8–21 years in the greater Philadelphia area, with multimodal neuroimaging data collected in 1,000 participants. The SLIM dataset has 595 healthy young adults ages 17–27 years in China.
Processing
For the ABCD and PNC datasets, fMRI data were motion corrected and the Shen atlas was warped to single participant space as previously described.38,39 For the HCP dataset, we started with the minimally preprocessed HCP data.40 Further preprocessing steps were performed using BioImage Suite41 and are the same across studies. Several covariates of no interest were regressed from participants’ functional data including linear and quadratic drifts, mean cerebrospinal fluid signal, mean white matter signal, and mean global signal. For additional control of possible motion-related confounds, a 24-parameter motion model (including six rigid body motion parameters, six temporal derivatives, and these terms squared) was regressed from the data. The data were temporally smoothed with a Gaussian filter (approximate cutoff frequency = 0.12 Hz). We then applied a canonical gray matter mask defined in common space, so only voxels in the gray matter were used in further calculations. Denoised data were parcellated into 268 nodes using the Shen atlas.42 Next, the mean time courses of each node pair were correlated, and correlation coefficients were Fisher transformed, generating a connectome for each participant. For the HCP dataset, connectomes for each phase encoding (i.e., RL and LR) were calculated independently and then averaged together. For the SLIM dataset, preprocessed connectomes were downloaded, with preprocessing steps described in.37 After excluding participants for excessive motion (>0.2 mm) and missing nodes due to lack of full brain coverage, 3,362 individuals in the ABCD dataset, 506 in the HCP dataset, 561 in the PNC dataset, and 445 in the SLIM dataset were retained.
Baseline regression models
For all baseline regression models, we trained ridge-regression connectome-based predictive models (rCPM)44 in MATLAB (The MathWorks) with 10-fold cross-validation and a nested 10-fold cross-validation to select the L2 regularization parameter, λ. For feature selection, we correlated each edge with the phenotype of interest and picked the top 10% of edges with the lowest p values. In the nested folds, we performed a grid search for λ. We used MATLAB’s default settings for a grid search over λ (detailed description: https://www.mathworks.com/help/stats/lasso.html). In sum, we searched over a geometric sequence with the maximum being the largest λ that gives a nonnull model and the minimum being λmax∗10−4. λ is then selected as the largest λ for which the mean squared error (MSE) is within 1 standard error of the minimum MSE. With this method, the λ is generally very high, often λ ≈ 100–150 (log[λ] ≈ 4.6–5). To explore the effectiveness of enhancement attacks across a wide variety of λ, we also included a parameter sensitivity analysis (Figure S1).
For Table S2, we compared ridge regression models and neural networks in HCP resting-state data using 100% of available features in both cases and enhancing 100% of edges, as opposed to 20% in the analyses in the main text. We used ridge regression with a regularization parameter of 1,000 and all default parameters for the neural network (MLPRegressor function in scikit-learn75). These results demonstrate that enhancement attacks at various scales are still effective in neural networks. Furthermore, in theory, neural networks should be able to learn non-linear enhancement patterns, whereas ridge regression models cannot.
For ABCD, HCP, and PNC, the phenotype of interest was a fluid intelligence (IQ) measurement. For ABCD, Raven’s progressive matrices76 were used, scaled by age (mean 6.31, SD 2.56, range 1–17, median 6), and the same measure, though not scaled by age, was used for HCP (mean 17.54, SD 4.45, range 5–24, median 19). For PNC, IQ was assessed using the Penn Matrix Reasoning test (mean 12.28, SD 4.04, range 0–23, median 12).77 For SLIM, the phenotype of interest was the state anxiety score, as assessed by the State-Trait Anxiety Inventory55 (mean 35.66, SD 8.28, range 20–65, median 35).
The main metric we used to determine prediction performance was Pearson’s r between original and predicted phenotypes. We also reported the cross-validation R2, called q2,47 which is defined as
Baseline classification models
We trained both SVM (linear kernel) and logistic regression models in MATLAB to predict self-reported sex in ABCD, HCP, and PNC. The self-reported sex of participants by dataset was: ABCD (1,653 female, 1,609 male), HCP (270 female, 236 male), and PNC (318 female, 244 male). Models were trained with 10-fold cross-validation, with nested 5-fold cross-validation to select an L2 regularization parameter. For the L2 regularization hyperparameter search, we used MATLAB’s default search for the “fitclinear” function (https://www.mathworks.com/help/stats/fitclinear.html), which is Bayesian optimization, as described in (https://www.mathworks.com/help/stats/bayesianoptimization.html). Essentially, it uses Bayesian optimization to search within the range of [1e-5/number of training samples, 1e5/number of training samples]. We used accuracy as our primary evaluation metric, though we also reported sensitivity and specificity in Table S1.
Connectome enhancement
We enhanced IQ prediction in ABCD, HCP, and PNC. To enhance connectome data, we first Z-scored the phenotypic measurements.
Then, we randomly selected 20% of all edges e to manipulate. For each of the selected edges, we added a value with magnitude proportional to each participant’s Z-scored IQ.
where ei,j represents edge i for participant j, and yz,j is the Z-scored phenotype for participant j.
Whether we added or subtracted each value was randomly determined for each edge. The results presented in Figure 3 used k = {0, 0.004, 0.007, 0.01, 0.014, 0.02, 0.03}. k = 0 was used as a reference and means that no enhancement was performed.
After injecting the enhancement pattern into each connectome, rCPM models were re-trained, as described above, to predict each phenotype. Because we injected patterns proportional to the phenotype of interest, we would expect performance to be falsely enhanced. We repeated enhancement for each value of k for 100 different random seeds and recorded r and q2 at each iteration.
Enhancement downstream analyses
In addition to r and q2, we evaluated enhanced connectomes in several other analyses—self-reported sex classification, functional connectome fingerprinting, and investigation of graph properties—to determine their similarity with original connectomes.
For self-reported sex classification, we used the data that was enhanced for IQ prediction to train a SVM classifier as described in baseline classification models. At each different scale of the enhancement k, we compared the sex classification performance to that of the unaltered dataset, using accuracy as the main metric.
We also performed functional connectome fingerprinting50,51 following the method of Finn et al.51 We performed fingerprinting only in the HCP dataset because it has two resting-state scans. A total of 50 or 51 participants were used at a time for fingerprinting evaluation, corresponding to all the connectomes in a single fold of the 10-fold cross-validation enhancement pipeline (10% of 506 HCP participants). With 50 participants, the identification rate of random guessing is 2%. To predict identity, we first calculated the edge-wise correlation of each participant’s Rest1 connectome with their Rest2 connectome, and the predicted identity of each participant corresponded to the Rest2 connectome with the highest edge-wise correlation with their Rest1 connectome. We repeated the fingerprinting process using both original Rest1 connectomes and Rest1 connectomes that had been enhanced for IQ prediction.
In a further implementation of fingerprinting, we identified participants using only edges that are part of selected subnetworks, as previously defined with the Shen 268 atlas.51,52 10 networks were defined on the basis of nodes (Figure S2): medial-frontal (MF), fronto-parietal (FP), default mode (DMN), motor (MOT), visual I (VI), visual II (VII), visual association (VAs), salience (SAL), subcortical (SC), cerebellum (CBL). On the basis of these 10 networks, we defined 55 subnetworks, where each subnetwork was defined as edges belonging to each pair of networks. For example, 10 subnetworks involved the MF network: MF-MF, MF-FP, MF-DMN, MF-MOT, MF-VI, MF-VII, MF-VAs, MF-SAL, MF-SC, MF-CBL. The fingerprinting procedure followed the same process as above, except edge-wise correlations were calculated only with edges belonging to one of the 55 subnetworks.
Moreover, we evaluated several graph properties for the positive edges only, including strength, assortativity, and clustering coefficient, in the original and enhanced connectomes.53 All three of these properties are node-level metrics, and we evaluated the similarity between original and enhanced connectomes by correlating the node-level metrics for each of these measures. Strength is the sum of edge weights for each node. Assortativity measures how similar the strengths are between connected nodes. The clustering coefficient is the mean weight of triangles for each node.53
Targeted enhancement
In an extension of the above attack, we constrained the injected enhancement pattern to a specific resting-state network instead of a random subset of edges. We performed this enhancement in the SLIM37 dataset (n = 445) to predict state anxiety scores from the State-Trait Anxiety Inventory.55 After manipulating edges of one network, we repeated model training with rCPM. We also evaluated the model coefficients (averaged over the 10 folds of cross-validation) to assess how changing a specific network altered the distribution of coefficients across networks.
Time-series enhancement
We used HCP node time-series data (268-node Shen atlas; 1,200 time points) and prediction of IQ as an example of time-series enhancement. We arbitrarily selected an enhancement pattern as a sinusoid with four periods across the 1,200 time points. For each participant, the enhancement pattern (sinusoid) was scaled by a factor proportional to their IQ score and the original correlation of that edge with the IQ scores. Then, for each edge, we found the two nodes corresponding to that particular edge. If the edge was positively correlated with IQ, we added the participant-specific enhancement pattern to both node time-series. If the edge was negatively correlated with IQ, we added the enhancement pattern to one node and subtracted it from the other node.
After enhancing the time-series data, we computed the connectomes by taking the correlation between each pair of nodes and applying the Fisher transform. Then, rCPM models were trained with 10-fold cross-validation to predict IQ. We recorded the similarity (Pearson’s r) between the original and enhanced time-series data along with similarity between the resulting original and enhanced connectomes. Although there may be other more effective strategies to enhance time-series data, we performed this simple attack as a proof of concept.
Adversarial noise attacks
In both classifiers (SVM, logistic regression), we used a gradient-based attack following the method of Biggio et al.21 The attacks occurred on the test data at the time of model testing and are “white-box” attacks, meaning that they required access to the model parameters. Let our decision function be represented by g(x) with input features x. A participant was classified as female if g(x) < 0 and male if g(x) > 0. The goal of the adversarial noise attack was to manipulate all the true female (male) connectomes such that the adversarial g(x) > 0 (<0). The noise for female and male connectomes were optimized separately.
The loss functions were
and
Adversarial noise was initialized to zeros and iteratively updated on the basis of the gradient:
where is the step size.
For linear SVM and logistic regression, the derivative term was given by:
for coefficients β. For these linear models, this process just simplifies to adding adversarial noise proportional to the coefficients.
We also explored how the adversarial noise was distributed across subnetworks by taking the mean absolute value of the adversarial noise in each subnetwork.
Adversarial noise downstream analyses
We compared functional connectome fingerprinting rates between original and adversarial Rest1 connectomes, as previously described. Notably, we performed fingerprinting with the Rest1 connectomes and each of the eight other tasks in HCP (Rest2, gambling, language, motor, relational, social, working memory, emotion). We also performed subnetwork-specific fingerprinting.
Statistics
As previously described, performance of regression models was quantified with Pearson’s r or cross-validation R2, called q2, between measured and predicted phenotypes. Classifiers were assessed by prediction accuracy, sensitivity, and specificity. For comparisons of real and manipulated connectomes, we computed Pearson’s r between original and manipulated edges, to which we refer to as “edge-wise correlation” throughout this work. To assess the significance of the differences in ID rate and accuracy between original and manipulated data, we used McNemar’s test78 and reported the median p value across the 100 iterations.
Acknowledgments
This study was supported by National Insititute of Mental Health grant R01MH121095 (obtained by R.T.C. and D.S.). M.R. was supported by the National Science Foundation Graduate Research Fellowship under grant DGE-2139841. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. R.X.R. was supported by the National Research Service Award (award 5T32GM100884-09) from the National Institute of General Medicine. M.L.W. was supported by the National Institute on Drug Abuse (T32DA022975). S.N. was supported by the National Institute of Mental Health (K00MH122372). Data were provided in part by the HCP, WU-Minn Consortium (principal investigators David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 National Institutes of Health institutes and centers that support the National Institutes of Health Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. Additional data were provided by the PNC (principal investigators Hakon Hakonarson and Raquel Gur; phs000607.v1.p1). Support for the collection of these datasets was provided by grant RC2MH089983 awarded to Raquel Gur and RC2MH089924 awarded to Hakon Hakonarson. Data obtained from the SLIM study were funded by The National Natural Science Foundation of China (31271087, 31470981, 31571137, 31500885); National Outstanding young people plan, the Program for the Top Young Talents by Chongqing, the Fundamental Research Funds for the Central Universities (SWU1509383, SWU1509451); Natural Science Foundation of Chongqing (cstc2015jcyjA10106); Fok Ying Tung Education Foundation (151023); the General Financial Grant from the China Postdoctoral Science Foundation (2015M572423, 2015M580767); Special Funds from the Chongqing Postdoctoral Science Foundation (Xm2015037); and Key Research for Humanities and Social Sciences of Ministry of Education (14JJD880009). Furthermore, some data used in the preparation of this article were obtained from the ABCD Study (abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children ages 9–10 years and follow them over 10 years into early adulthood. The ABCD study is supported by the National Institutes of Health and additional federal partners under awards U01DA041022, U01DA041028, U01DA041048, U01DA041089, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, and U24DA041147. A full list of supporters is available at abcdstudy.org. A listing of participating sites and a complete listing of the study investigators can be found at abcdstudy.org/principal-investigators.html. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD data repository grows and changes over time. The ABCD data used in this report came from NIMH Data Archive digital object identifier 10.15154/1504041.
Author contributions
Conceptualization, M.R., R.X.R., M.L.W., and D.S.; Methodology, M.R., M.L.W., and D.S.; Software, M.R. and R.X.R.; Formal Analysis, M.R. and D.S.; Investigation, M.R.; Data Curation, A.S.G., C.H., W.D., R.T.C, and D.S.; Writing—Original Draft, M.R. and D.S.; Writing—Review and Editing, M.R., R.X.R., M.L.W., W.D., C.H., A.S.G., S.N., and D.S.; Visualization, M.R., R.X.R., M.L.W., S.N., and D.S.; Supervision, M.L.W., S.N., and D.S.; Funding Acquisition, R.T.C. and D.S.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: May 15, 2023
Contributor Information
Matthew Rosenblatt, Email: matthew.rosenblatt@yale.edu.
Dustin Scheinost, Email: dustin.scheinost@yale.edu.
Supplemental information
Data and code availability
-
•
All four datasets used in this study are open-source: ABCD (NIMH Data Archive, https://nda.nih.gov/abcd),34 HCP (ConnectomeDB database, https://db.humanconnectome.org),35 PNC (dbGaP Study, accession code: phs000607.v3.p2, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2),36 and SLIM (INDI, http://fcon_1000.projects.nitrc.org/indi/retro/southwestuni_qiu_index.html).37 Data collection was approved by the relevant ethics review board for each of the four datasets.
-
•
BioImage Suite tools used for processing can be accessed at (https://bioimagesuiteweb.github.io/alphaapp/). MATLAB scripts for trust analyses are available on GitHub (https://github.com/mattrosenblatt7/trust_connectomes) and Zenodo74 (https://doi.org/10.5281/zenodo.7750583).
References
- 1.Whelan R., Garavan H. When optimism hurts: inflated predictions in psychiatric neuroimaging. Biol. Psychiatry. 2014;75:746–748. doi: 10.1016/j.biopsych.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Gabrieli J.D.E., Ghosh S.S., Whitfield-Gabrieli S. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015;85:11–26. doi: 10.1016/j.neuron.2014.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cremers H.R., Wager T.D., Yarkoni T. The relation between statistical power and inference in fMRI. PLoS One. 2017;12:e0184923. doi: 10.1371/journal.pone.0184923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble S., Mejia A.F., Zalesky A., Scheinost D. Improving power in functional magnetic resonance imaging by moving beyond cluster-level inference. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2203020119. e2203020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen X., Finn E.S., Scheinost D., Rosenberg M.D., Chun M.M., Papademetris X., Constable R.T. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc. 2017;12:506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui Z., Gong G. The effect of machine learning regression algorithms and sample size on individualized behavioral prediction with functional connectivity features. Neuroimage. 2018;178:622–637. doi: 10.1016/j.neuroimage.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Du Y., Fu Z., Calhoun V.D. Classification and prediction of brain disorders using functional connectivity: promising but challenging. Front. Neurosci. 2018;12:525. doi: 10.3389/fnins.2018.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg M.D., Casey B.J., Holmes A.J. Prediction complements explanation in understanding the developing brain. Nat. Commun. 2018;9:589. doi: 10.1038/s41467-018-02887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H., Finn E.S., Rosenberg M.D. Neural signatures of attentional engagement during narratives and its consequences for event memory. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021905118. e2021905118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen A.N., Barch D.M., Petersen S.E., Schlaggar B.L., Greene D.J. Machine learning with neuroimaging: evaluating its applications in psychiatry. Biol. Psychiatry. Cogn. Neurosci. Neuroimaging. 2020;5:791–798. doi: 10.1016/j.bpsc.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldfarb E.V., Rosenberg M.D., Seo D., Constable R.T., Sinha R. Hippocampal seed connectome-based modeling predicts the feeling of stress. Nat. Commun. 2020;11:2650. doi: 10.1038/s41467-020-16492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yip S.W., Scheinost D., Potenza M.N., Carroll K.M. Connectome-based prediction of cocaine abstinence. Am. J. Psychiatry. 2019;176:156–164. doi: 10.1176/appi.ajp.2018.17101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benkarim O., Paquola C., Park B.-Y., Kebets V., Hong S.-J., de Wael R.V., Zhang S., Thomas Yeo B.T., Eickenberg M., Ge T., et al. The cost of untracked diversity in brain-imaging prediction. bioRxiv. 2021 doi: 10.1101/2021.06.16.448764. Preprint at. [DOI] [Google Scholar]
- 14.Li J., Bzdok D., Chen J., Tam A., Ooi L.Q.R., Holmes A.J., Ge T., Patil K.R., Jabbi M., Eickhoff S.B., et al. Cross-ethnicity/race generalization failure of behavioral prediction from resting-state functional connectivity. Sci. Adv. 2022;8:eabj1812. doi: 10.1126/sciadv.abj1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene A.S., Shen X., Noble S., Horien C., Hahn C.A., Arora J., Tokoglu F., Spann M.N., Carrión C.I., Barron D.S., et al. Brain–phenotype models fail for individuals who defy sample stereotypes. Nature. 2022;609:109–118. doi: 10.1038/s41586-022-05118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brundage M., Avin S., Wang J., Belfield H., Krueger G., Hadfield G., Khlaaf H., Yang J., Toner H., Fong R., et al. Toward trustworthy AI development: mechanisms for supporting verifiable claims. arXiv. 2020 doi: 10.48550/arXiv.2004.07213. Preprint at. [DOI] [Google Scholar]
- 17.Rawal M., Rawat S., Amant Recent advances in trustworthy explainable artificial intelligence: status, challenges and perspectives. IEEE Transactions on Artificial Intelligence. 2021;1:1. doi: 10.1109/TAI.2021.3133846. [DOI] [Google Scholar]
- 18.Eshete B. Making machine learning trustworthy. Science. 2021;373:743–744. doi: 10.1126/science.abi5052. [DOI] [PubMed] [Google Scholar]
- 19.Szegedy C., Zaremba W., Sutskever I., Bruna J., Erhan D., Goodfellow I., Fergus R. Intriguing properties of neural networks. arXiv. 2013 doi: 10.48550/arXiv.1312.6199. Preprint at. [DOI] [Google Scholar]
- 20.Goodfellow I.J., Shlens J., Szegedy C. Explaining and harnessing adversarial examples. arXiv. 2014 doi: 10.48550/arXiv.1412.6572. Preprint at. [DOI] [Google Scholar]
- 21.Biggio B., Corona I., Maiorca D., Nelson B., Šrndić N., Laskov P., Giacinto G., Roli F. Machine Learning and Knowledge Discovery in Databases. Springer Berlin Heidelberg; 2013. Evasion attacks against machine learning at test time; pp. 387–402. [DOI] [Google Scholar]
- 22.Demontis A., Melis M., Biggio B., Maiorca D., Arp D., Rieck K., Corona I., Giacinto G., Roli F. Yes, machine learning can be more secure! a case study on android malware detection. IEEE Trans. Dependable Secure Comput. 2019;16:711–724. doi: 10.1109/TDSC.2017.2700270. [DOI] [Google Scholar]
- 23.Hendrycks D., Zhao K., Basart S., Steinhardt J., Song D. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021. Natural adversarial examples; pp. 15262–15271. [DOI] [Google Scholar]
- 24.Paschali M., Conjeti S., Navarro F., Navab N. Medical Image Computing and Computer Assisted Intervention. 2018. Generalizability vs. Robustness: investigating medical imaging networks using adversarial examples; pp. 493–501. [DOI] [Google Scholar]
- 25.Finlayson S.G., Bowers J.D., Ito J., Zittrain J.L., Beam A.L., Kohane I.S. Adversarial attacks on medical machine learning. Science. 2019;363:1287–1289. doi: 10.1126/science.aaw4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han X., Hu Y., Foschini L., Chinitz L., Jankelson L., Ranganath R. Deep learning models for electrocardiograms are susceptible to adversarial attack. Nat. Med. 2020;26:360–363. doi: 10.1038/s41591-020-0791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finlayson S.G., Chung H.W., Kohane I.S., Beam A.L. Adversarial attacks against medical deep learning Systems. arXiv. 2018 doi: 10.48550/arXiv.1804.05296. Preprint at. [DOI] [Google Scholar]
- 28.Acuna D.E., Brookes P.S., Kording K.P. Bioscience-scale automated detection of figure element reuse. bioRxiv. 2018 doi: 10.1101/269415. Preprint at. [DOI] [Google Scholar]
- 29.Bucci E.M. Automatic detection of image manipulations in the biomedical literature. Cell Death Dis. 2018;9:400. doi: 10.1038/s41419-018-0430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cicconet M., Elliott H., Richmond D.L., Wainstock D., Walsh M. Image Forensics: detecting duplication of scientific images with manipulation-invariant image similarity. arXiv. 2018 doi: 10.48550/arXiv.1802.06515. Preprint at. [DOI] [Google Scholar]
- 31.Bik E.M., Casadevall A., Fang F.C. The prevalence of inappropriate image duplication in biomedical research publications. mBio. 2016;7:e00809-16. doi: 10.1128/mBio.00809-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanelli D. How many scientists fabricate and falsify research? A systematic review and meta-analysis of survey data. PLoS One. 2009;4:e5738. doi: 10.1371/journal.pone.0005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Marzouki S., Evans S., Marshall T., Roberts I. Are these data real? Statistical methods for the detection of data fabrication in clinical trials. BMJ. 2005;331:267–270. doi: 10.1136/bmj.331.7511.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casey B.J., Cannonier T., Conley M.I., Cohen A.O., Barch D.M., Heitzeg M.M., Soules M.E., Teslovich T., Dellarco D.V., Garavan H., et al. The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Essen D.C., Smith S.M., Barch D.M., Behrens T.E.J., Yacoub E., Ugurbil K., WU-Minn HCP ConsortiumMinn HCP Consortium The Wu-Minn human connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satterthwaite T.D., Connolly J.J., Ruparel K., Calkins M.E., Jackson C., Elliott M.A., Roalf D.R., Hopson R., Prabhakaran K., Behr M., et al. The Philadelphia Neurodevelopmental Cohort: a publicly available resource for the study of normal and abnormal brain development in youth. Neuroimage. 2016;124:1115–1119. doi: 10.1016/j.neuroimage.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W., Wei D., Chen Q., Yang W., Meng J., Wu G., Bi T., Zhang Q., Zuo X.-N., Qiu J. Longitudinal test-retest neuroimaging data from healthy young adults in southwest China. Sci. Data. 2017;4:170017. doi: 10.1038/sdata.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greene A.S., Gao S., Scheinost D., Constable R.T. Task-induced brain state manipulation improves prediction of individual traits. Nat. Commun. 2018;9:2807. doi: 10.1038/s41467-018-04920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapuano K.M., Rosenberg M.D., Maza M.T., Dennis N.J., Dorji M., Greene A.S., Horien C., Scheinost D., Todd Constable R., Casey B.J. Behavioral and brain signatures of substance use vulnerability in childhood. Dev. Cogn. Neurosci. 2020;46:100878. doi: 10.1016/j.dcn.2020.100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glasser M.F., Sotiropoulos S.N., Wilson J.A., Coalson T.S., Fischl B., Andersson J.L., Xu J., Jbabdi S., Webster M., Polimeni J.R., et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi A., Scheinost D., Okuda H., Belhachemi D., Murphy I., Staib L.H., Papademetris X. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics. 2011;9:69–84. doi: 10.1007/s12021-010-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen X., Tokoglu F., Papademetris X., Constable R.T. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage. 2013;82:403–415. doi: 10.1016/j.neuroimage.2013.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biswal B.B., Mennes M., Zuo X.-N., Gohel S., Kelly C., Smith S.M., Beckmann C.F., Adelstein J.S., Buckner R.L., Colcombe S., et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. USA. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao S., Greene A.S., Constable R.T., Scheinost D. Combining multiple connectomes improves predictive modeling of phenotypic measures. Neuroimage. 2019;201:116038. doi: 10.1016/j.neuroimage.2019.116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weis S., Patil K.R., Hoffstaedter F., Nostro A., Yeo B.T.T., Eickhoff S.B. Sex classification by resting state brain connectivity. Cereb. Cortex. 2020;30:824–835. doi: 10.1093/cercor/bhz129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eliot L., Ahmed A., Khan H., Patel J. Dump the “dimorphism”: comprehensive synthesis of human brain studies reveals few male-female differences beyond size. Neurosci. Biobehav. Rev. 2021;125:667–697. doi: 10.1016/j.neubiorev.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 47.Scheinost D., Noble S., Horien C., Greene A.S., Lake E.M., Salehi M., Gao S., Shen X., O’Connor D., Barron D.S., et al. Ten simple rules for predictive modeling of individual differences in neuroimaging. Neuroimage. 2019;193:35–45. doi: 10.1016/j.neuroimage.2019.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biggio B., Nelson B., Laskov P. Poisoning attacks against support vector machines. arXiv. 2012 doi: 10.48550/arXiv.1206.6389. Preprint at. [DOI] [Google Scholar]
- 49.Massey F.J. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;46:68–78. doi: 10.1080/01621459.1951.10500769. [DOI] [Google Scholar]
- 50.Miranda-Dominguez O., Mills B.D., Carpenter S.D., Grant K.A., Kroenke C.D., Nigg J.T., Fair D.A. Connectotyping: model based fingerprinting of the functional connectome. PLoS One. 2014;9:e111048. doi: 10.1371/journal.pone.0111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn E.S., Shen X., Scheinost D., Rosenberg M.D., Huang J., Chun M.M., Papademetris X., Constable R.T. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noble S., Spann M.N., Tokoglu F., Shen X., Constable R.T., Scheinost D. Influences on the test–retest reliability of functional connectivity MRI and its relationship with behavioral utility. Cereb. Cortex. 2017;27:5415–5429. doi: 10.1093/cercor/bhx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Luo W., Greene A.S., Constable R.T. Within node connectivity changes, not simply edge changes, influence graph theory measures in functional connectivity studies of the brain. Neuroimage. 2021;240:118332. doi: 10.1016/j.neuroimage.2021.118332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spielberger C.D. Consulting Psychologists Press; 1983. Manual for the State-Trait Anxiety Inventory (Form Y) (“self-Evaluation Questionnaire”) [Google Scholar]
- 56.Cameron C., Yassine B., Carlton C., Francois C., Alan E., András J., Budhachandra K., John L., Qingyang L., Michael M., et al. The neuro bureau preprocessing initiative: open sharing of preprocessed neuroimaging data and derivatives. Front. Neuroinform. 2013;7 doi: 10.3389/conf.fninf.2013.09.00041. [DOI] [Google Scholar]
- 57.Mennes M., Biswal B.B., Castellanos F.X., Milham M.P. Making data sharing work: the FCP/INDI experience. Neuroimage. 2013;82:683–691. doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markiewicz C.J., Gorgolewski K.J., Feingold F., Blair R., Halchenko Y.O., Miller E., Hardcastle N., Wexler J., Esteban O., Goncalves M., et al. OpenNeuro: an open resource for sharing of neuroimaging data. bioRxiv. 2021 doi: 10.1101/2021.06.28.450168. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horien C., Noble S., Greene A.S., Lee K., Barron D.S., Gao S., O’Connor D., Salehi M., Dadashkarimi J., Shen X., et al. A hitchhiker’s guide to working with large, open-source neuroimaging datasets. Nat. Hum. Behav. 2021;5:185–193. doi: 10.1038/s41562-020-01005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dadi K., Rahim M., Abraham A., Chyzhyk D., Milham M., Thirion B., Varoquaux G., Alzheimer’s Disease Neuroimaging Initiative Benchmarking functional connectome-based predictive models for resting-state fMRI. Neuroimage. 2019;192:115–134. doi: 10.1016/j.neuroimage.2019.02.062. [DOI] [PubMed] [Google Scholar]
- 61.Specht K. Current challenges in translational and clinical fMRI and future directions. Front. Psychiatry. 2019;10:924. doi: 10.3389/fpsyt.2019.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilmer J., Metz L., Faghri F., Schoenholz S.S., Raghu M., Wattenberg M., Goodfellow I. The relationship between high-dimensional geometry and adversarial examples. arXiv. 2018 doi: 10.48550/arXiv.1801.02774. Preprint at. [DOI] [Google Scholar]
- 63.Chattopadhyay N., Chattopadhyay A., Gupta S.S., Kasper M. 2019 International Joint Conference on Neural Networks (IJCNN) 2019. Curse of dimensionality in adversarial examples; pp. 1–8. [DOI] [Google Scholar]
- 64.Meng D., Chen H. Proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security CCS ’17. Association for Computing Machinery; 2017. MagNet: a two-pronged defense against adversarial examples; pp. 135–147. [DOI] [Google Scholar]
- 65.Qiu S., Liu Q., Zhou S., Wu C. Review of artificial intelligence adversarial attack and defense technologies. 2019;9:909. doi: 10.3390/app9050909. [DOI] [Google Scholar]
- 66.Zhang Y., Liang P. In: Proceedings of the Twenty-Second International Conference on Artificial Intelligence and Statistics Proceedings of Machine Learning Research. Chaudhuri K., Sugiyama M., editors. PMLR; 2019. Defending against whitebox adversarial attacks via randomized discretization; pp. 684–693. [Google Scholar]
- 67.Halchenko Y., Meyer K., Poldrack B., Solanky D., Wagner A., Gors J., MacFarlane D., Pustina D., Sochat V., Ghosh S., et al. DataLad: distributed system for joint management of code, data, and their relationship. J. Open Source Softw. 2021;6:3262. doi: 10.21105/joss.03262. [DOI] [Google Scholar]
- 68.Bell J., LaToza T.D., Baldmitsi F., Stavrou A. 2017 IEEE/ACM 12th International Workshop on Software Engineering for Science (SE4Science) 2017. Advancing Open Science with Version Control and Blockchains; pp. 13–14. [DOI] [Google Scholar]
- 69.Mitchell M., Wu S., Zaldivar A., Barnes P., Vasserman L., Hutchinson B., Spitzer E., Raji I.D., Gebru T. Proceedings of the Conference on Fairness, Accountability, and Transparency FAT∗ ’19. Association for Computing Machinery; 2019. Model cards for model reporting; pp. 220–229. [DOI] [Google Scholar]
- 70.Raji I.D., Yang J. About ML: annotation and benchmarking on understanding and transparency of machine learning lifecycles. arXiv. 2019 doi: 10.48550/arXiv.1912.06166. Preprint at. [DOI] [Google Scholar]
- 71.Jiang H., Kim B., Guan M., Gupta M. Advances in Neural Information Processing Systems. 2018. To trust or not to trust A classifier; pp. 5541–5552. [Google Scholar]
- 72.Buolamwini J., Gebru T. Proceedings of the 1st Conference on Fairness, Accountability and Transparency Proceedings of Machine Learning Research (PMLR) 2018. Gender shades: intersectional accuracy disparities in commercial gender classification; pp. 77–91. [Google Scholar]
- 73.Turner Lee N. Detecting racial bias in algorithms and machine learning. J. Inf. Commun. Ethics Soc. 2018;16:252–260. doi: 10.1108/JICES-06-2018-0056. [DOI] [Google Scholar]
- 74.Rosenblatt M. 2023. Connectome-based machine learning models are vulnerable to subtle data manipulations. v1.0.0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 76.Bilker W.B., Hansen J.A., Brensinger C.M., Richard J., Gur R.E., Gur R.C. Development of abbreviated nine-item forms of the Raven’s standard progressive matrices test. Assessment. 2012;19:354–369. doi: 10.1177/1073191112446655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moore T.M., Reise S.P., Gur R.E., Hakonarson H., Gur R.C. Psychometric properties of the Penn computerized neurocognitive battery. Neuropsychology. 2015;29:235–246. doi: 10.1037/neu0000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All four datasets used in this study are open-source: ABCD (NIMH Data Archive, https://nda.nih.gov/abcd),34 HCP (ConnectomeDB database, https://db.humanconnectome.org),35 PNC (dbGaP Study, accession code: phs000607.v3.p2, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2),36 and SLIM (INDI, http://fcon_1000.projects.nitrc.org/indi/retro/southwestuni_qiu_index.html).37 Data collection was approved by the relevant ethics review board for each of the four datasets.
-
•
BioImage Suite tools used for processing can be accessed at (https://bioimagesuiteweb.github.io/alphaapp/). MATLAB scripts for trust analyses are available on GitHub (https://github.com/mattrosenblatt7/trust_connectomes) and Zenodo74 (https://doi.org/10.5281/zenodo.7750583).