Abstract
Bile acids are detergents derived from cholesterol that function to solubilize dietary lipids, remove cholesterol from the body, and act as nutrient signaling molecules in numerous tissues with functions in the liver and gut being the best understood. Studies in the early 20th century established the structures of bile acids, and by mid-century, the application of gnotobiology to bile acids allowed differentiation of host-derived “primary” bile acids from “secondary” bile acids generated by host-associated microbiota. In 1960, radiolabeling studies in rodent models led to determination of the stereochemistry of the bile acid 7-dehydration reaction. A two-step mechanism was proposed, which we have termed the Samuelsson-Bergström model, to explain the formation of deoxycholic acid. Subsequent studies with humans, rodents, and cell extracts of Clostridium scindens VPI 12708 led to the realization that bile acid 7-dehydroxylation is a result of a multi-step, bifurcating pathway that we have named the Hylemon-Björkhem pathway. Due to the importance of hydrophobic secondary bile acids and the increasing measurement of microbial bai genes encoding the enzymes that produce them in stool metagenome studies, it is important to understand their origin.
Supplementary key words: bile acids, intestinal lipid metabolism, gut microbiome, bile acid dehydroxylation, allo-bile acids, enterohepatic circulation
The Ancients (amongst whom is Aristotle) thought it [bile]
to be a mere excrement, and to be of no other use than by
its acrimony to promote the excretion of the guts.
-Thomas Gibson, The Anatomy of Humane Bodies Epitomized, (1682)
Man's liver is a brownish blob.
That does a most prodigious job.
It manufactures gall, or bile.
And normally keeps some on file.
Stored neatly in a pear-shaped sac.
From there the liver's yields attack.
The food man eats, to change its state.
By methods man can't duplicate,
Or even halfway understand.
He ought to treat this outsize gland,
With due respect and loving care.
To keep it in top-notch repair,
Because to get along at all.
Man needs an awful lot of gall.
-Irene Warsaw, JAMA, 1975, v. 231, p. 1260.
Despite many excellent recent reviews in the bile acid-microbiome field (1, 2, 3, 4, 5), so far, the historical developments leading up to our present mechanistic understanding of the microbial 7-dehydroxylation of bile acids have not been adequately covered (Fig. 1). Strikingly, derivatives of the hydrophobic secondary bile acid products of microbial 7-dehydroxylation, such as deoxycholic acid (DCA) and lithocholic acid (LCA), are, in general, preferred ligands for many host nuclear receptors (farnesoid X receptor (also known as NR1H4) (6, 7, 8), pregnane X receptor (or NR1I2) (9), vitamin D3 receptor (10), constitutive androstane receptor (or NR1I3) (11), retinoic acid receptor gamma T (12, 13), nuclear receptor 4A) (14) and G protein-coupled receptors (TGR5, (GPBAR1) (15, 16), muscarinic receptors (CHRM2, CHRM3) (17), sphingosine-1-phosphate receptor 2) (18) over the primary bile acids from which they derive. What has emerged in recent decades is that the ‘Western lifestyle’ of inactivity, diets low in fiber and high in processed carbohydrates and saturated fats, increases both the amount of bile entering the gastrointestinal (GI) tract and the hydrophobicity of the bile acid pool thereby affecting host metabolic function (3, 19) and increasing the risk of hepatobiliary and GI cancers in humans (2).
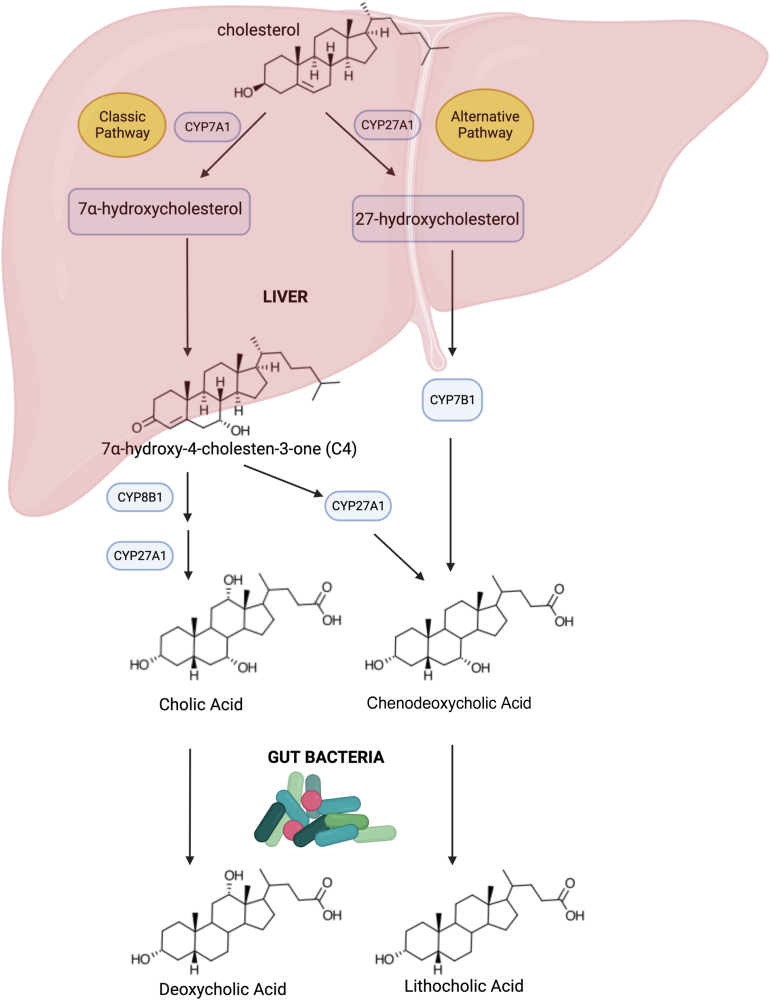
Fig. 1.
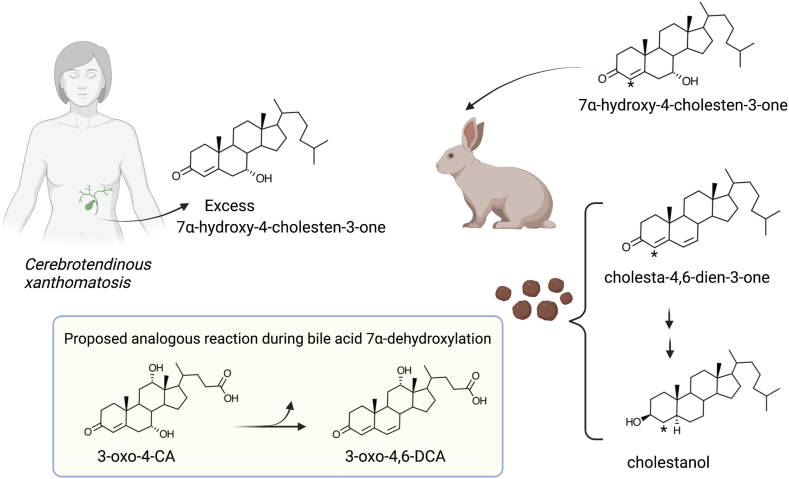
Bile salt biotransformations by intestinal bacteria. In the human liver, two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA), are synthesized from cholesterol via the classic (neutral) pathway in hepatocytes. CDCA is also synthesized to a lesser degree by the alternative pathway. Bile acids are conjugated to taurine or glycine (not shown) in hepatocytes before secretion into the gallbladder. The intermediate, 7⍺-hydroxy-4-cholesten-3-one, is an important serum marker for BA synthesis, and it will later become apparent that its metabolism by intestinal bacteria provided important clues for the analogous 7⍺-dehydroxylation of CA and CDCA to deoxycholic acid (DCA) and lithocholic acid (LCA), respectively.
Thus, due to the importance of hydrophobic secondary bile acids and the increasing measurement of microbial genes encoding the enzymes that produce them (termed “bile acid inducible” or “bai” genes) in stool metagenome studies, it is important to understand how they came to be discovered and characterized. Here, we critically revisit this history and attempt to objectively identify the key researchers involved, the important observations made and to define the models of bile acid 7-dehydroxylation proposed during the 20th century, and how our knowledge has expanded into the 21st century. As a disclaimer and as Goethe warned, "Writing history is always a questionable thing. Despite best intentions one runs the risk of being unbalanced; indeed, whoever undertakes such a venture declares in advance that (s)he will cast some in the light and some in the shadow.” So, with that said, we apologize for any omissions of scientists and discoveries that we have made in this brief review of the history, biochemistry, and microbiology of bile acid 7-dehydroxylation.
A brief history of bile acids
Notions of the biliary cycle, inspired by the feedback mechanism of blood circulation introduced by William Harvey, date back to the 17th century (20, 21). The introductory Gibson quote encapsulates the ancient view of bile prior to the paradigm shift of cycles and feedback that supplanted earlier notions of “flux and reflux” that date back to Aristotle and the idea of bile as a mere propellant of stool through the GI tract. Yet Western thinkers were starting to become skeptical of the wisdom of prior centuries. Indeed, Chinese materia medica was utilizing animal biles as early as the Zhou dynasty (c. 1046-256 BCE) (22). Johann Baptista van Helmont proclaimed bile as viscus nobile et vitale (23). It was Mauritius van Reverhorst who applied Harvey’s reasoning that only recirculation explains the large volume of daily blood flow to the biliary system in his description of a motus bilis circularis, an enterohepatic circulation of bile. In further departure from the Aristotelian influence, Reverhorst experimentally confirmed his hypothesis in 1691 by cannulating the bile duct of a dog and measuring with respect to time (a recent Galilean invention), the volume of collected bile in a living animal (24). From this experiment, it was clear that the amount of bile entering the small bowel far exceeded the amount excreted in feces. Reverhorst reasoned there were two fractions, an “earthy” sediment found in the feces and a “volatile” fraction that passes through “pores” in the intestine that enters the circulatory system and returns to the liver. We now identify these “pores” as high affinity transmembrane transport proteins expressed in the apical and basolateral surfaces of ileocytes (25).
The early attempts at chemical analysis of bile read more like alchemy (to us today) but, by contrast, were ultimately successful. Treatment of bile with lead acetate resulted in separation of two substances designated “bile resin” and “picromel” (a colorless, bittersweet fluid obtained from the lead acetate filtrate) or as Berzelius termed them “choleic acid” and “bilin” (26). It was Demarcay, an associate of Justus von Liebig, who first coined the term “bile acid” to denote the nitrogen-free acidic fraction of bile obtained by alkali treatment (27). Strecker identified two acidic components in ox bile, one containing nitrogen (glycocholic acid) and the other with nitrogen and sulfur (TCA) and deduced the correct chemical formula C24H40O5 for cholic acid (CA) (28). Subsequently, Mylius performed elemental analysis on another bile acid isolated from ox bile and termed it “DCA” due to it being one oxygen less than CA (29). LCA was later purified from gallbladder stones (30) and chenodeoxycholic acid (CDCA, Greek χήν (chen), "goose") from goose bile roughly a decade later (31). A wealth of information on the early chemistry of bile acids is reviewed in detail elsewhere (26, 32, 33, 34, 35).
Adolf Windaus and Heinrich Wieland were awarded the 1927 and 1928 Nobel Prize in Chemistry, respectively, for their laborious work on probable (although ultimately incorrect) structures shared by steroids, including cholesterol and bile acids (32). During the 1930s, the X-ray crystallographic work by Desmond Bernal explored the correct dimensions for ergosterol, cholesterol, and vitamin D (36). Bernal determined that sterols form double layers, with long, thin molecular dimensions more in line with elongated long-chain alcohols. This was inconsistent with the Wieland-Windaus formula that implied a compact molecular structure in which the ABC rings meeting at C9 (32). Subsequent experimental studies arrived at the now accepted cyclopentanoperhydrophenanthrene sterol nucleus with the five carbon side chain of bile acids that was subsequently confirmed by total chemical synthesis (21).
Collectively, this work fundamentally revolutionized our understanding of the constituents of one important fraction of bile and the structural relation of bile acids to cholesterol. What was yet unclear is the origin of each bile acid, their building blocks, and how their structures change during transit through the mammalian GI tract, a habitat densely populated by microbes.
The establishment of “secondary” bile acids
Starting in the 1930s, evidence was accumulating that bile acids were metabolized by intestinal bacteria. While proposed as early as 1896, it was Frankel in 1936 who first reported the in vitro hydrolysis of conjugated bile acids by animal and human intestinal content (37). Work by Schmidt and colleagues at Christ Hospital in Cincinnati with guinea pigs led to the identification of CA metabolites in the cecum, in cecal suspensions, and by a guinea pig cecal isolate of Alcaligenes faecalis which converted CA to tri-keto CA under aerobic conditions (38). During the 1950s and 1960s, Sune Bergstrӧm, Bengt Samuelsson, Jan Sjövall, Arne Norman, Sven Lindstedt, Anders Kallner, Henry Danielsson, Bengt Gustafsson, and Tore Midtvedt (Fig. 2), together with a team of talented coworkers, first at the University of Lund and later at the Karolinska Institutet in Sweden, would go on to make fundamental contributions to bile acid chemistry and biology (21, 39). Key to these discoveries was the nascent development of radiolabeling biochemicals, chromatographic methods to separate them, automatic liquid scintillation counters to quantify them, anaerobic microbial culturing to obtain pure cultures of bacteria responsible for metabolizing them, and gnotobiology to determine causation between microbe and bile acid metabolite (21, 39, 40).
Fig. 2.
A selection of prominent researchers in the field of bile acid biochemistry and microbiology. (See text for descriptions and acknowledgments for photo permissions).
Cholesterol was first labeled with deuterium in 1943 by the Nobel laureate, Konrad Bloch, who made a direct metabolic relationship between cholesterol and bile acid formation in an animal model (41). Soon after, [4-14C] and [26-14C]cholesterol began to be synthesized, allowing fundamental studies on the major pathways of cholesterol degradation in the liver of several mammalian species, including humans (39). In 1953, Bergstrӧm and Norman administered [4-14C]cholesterol to rats, observing formation of [4-14C]TCA in bile and multiple unconjugated derivatives of CA in the feces (42). That same year, Bergstrӧm and colleagues then developed a C-14 method to directly label bile acids at the C24 (carboxyl) position (43). In 1955, Lindstedt and Norman injected rats intraperitoneally with [24-14C]CA and [24-14C]CDCA and observed a complex pattern of bile acid metabolites, identical to those observed following administration of [4-14C]cholesterol (42, 44). Norman (1955) administered [24-14C]CA to conventional rats and rats treated with antibiotics and concluded that the [24-14C]CA was converted to [24-14C]TCA in the liver and deconjugated by bacteria in the intestine (45, 46).
In 1958, Norman and Sjövall administered [24-14C]CA and [24-14C]7-keto-DCA into rats and observed that bile acid hydrolysis occurs in the cecum, along with DCA formation (47). It was not clear whether 7-keto-DCA is an intermediate in the formation of DCA, or in equilibrium with CA, but this work established that both 7-keto-DCA and DCA are microbial products of CA. The results of similar experiments demonstrated that LCA is a microbial product of the 7-dehydroxylation of CDCA (48). In 1960, Norman demonstrated the conversion of CA to DCA after anaerobic culturing of mixed rat cecal microbiota (49). Two years later, Norman and Shorb reported the conversion of [24-14C]CA and [24-14C]CDCA by anaerobic suspensions of human stool bacteria (50). They suggested that the insolubility of LCA in aqueous culture may explain the relatively low levels of LCA observed in the bile relative to DCA, which must be absorbed into portal circulation more efficiently. Based on these collective observations, Bergström, Danielsson, and Samuelsson would suggest the use of the terms “primary” bile acid (principally C7-hydroxylated) as host derived and “secondary” bile acid as microbial conversion products (originally narrowly implied as C7-dehydroxylated) (39).
Another important technology was being developed in parallel to determine the function of host-associated microbes. Louis Pasteur believed commensals were absolutely required for multicellular life, while Marceli Nencki and Élie Mechnikov argued that vigor could be enhanced and life prolonged without contaminating and toxin-producing microorganisms (51, 52, 53). Over a ten-year period, starting in 1885, germ-free (GF) guinea pigs were delivered and maintained on sterile milk, settling the question that life was possible without commensal microbes (54, 55, 56).
It was only natural for additional questions to be raised, spurring improvements in technology to determine the role of host-associated microbiota in animal physiology and metabolism. A stainless steel, autoclavable gnotobiotic isolator for the rearing of GF rats was reported in 1946 by Bengt Gustafsson in Sweden (57) and the same year by Reyniers and coworkers in the USA (58). In 1957, Gustafsson, Bergström, Lindstedt, and Norman showed that [24-14C]CA administration to GF rats resulted in fecal excretion of [24-14C]TCA, although monocolonization with a strain of Clostridium perfringens resulted in deconjugation and accumulation of [24-14C]CA in feces. The bile acid pool size in GF rats was notably larger than that in conventional rats, despite lower daily fecal excretion (59), an observation only recently explained through the antagonistic effects of tauro-β-muricholic acid on farnesoid X receptor signaling (60).
The work of Gustafsson, Midtvedt, and Norman in the 1960s comparing bile acids in intestinal content between conventional and GF rats conclusively showed that microbes biotransform bile acids in vivo (61, 62). Monocolonization of GF rats with Escherichia coli resulted in the appearance of 7-oxoDCA, indicating the presence of bile acid 7ɑ-hydroxysteroid dehydrogenase (7ɑ-HSDH) activity (63); the gene encoding bile acid 7ɑ-HSDH and the structure was identified many decades later (64, 65). The strictly anaerobic bile acid 7ɑ-dehydroxylating bacteria had evaded isolation up to 1960 when Samuelsson and Bergström turned to clever radiolabeling strategies that led to a proposed mechanism by which the C7 hydroxyl group is removed by gut microbiota.
The Samuelsson-Bergström model of bile acid 7ɑ-dehydroxylation
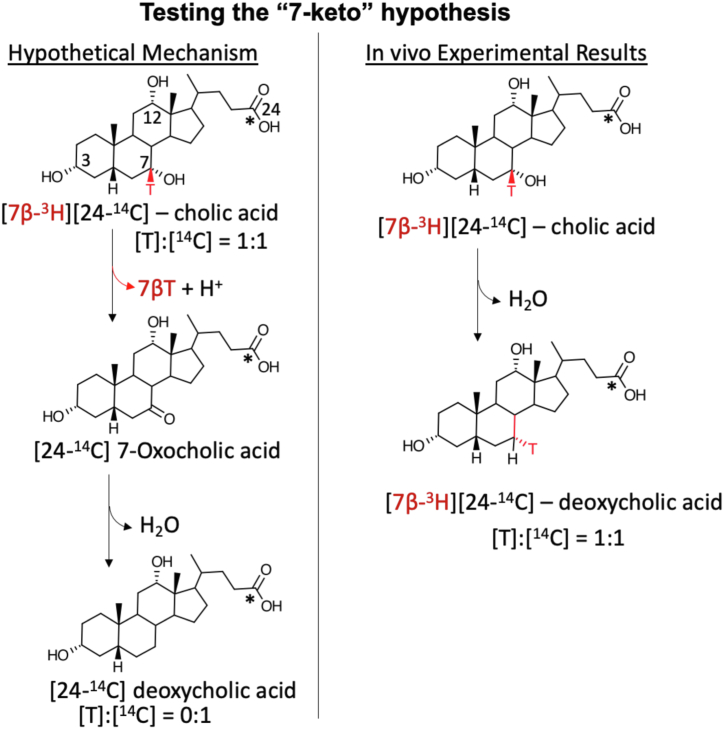
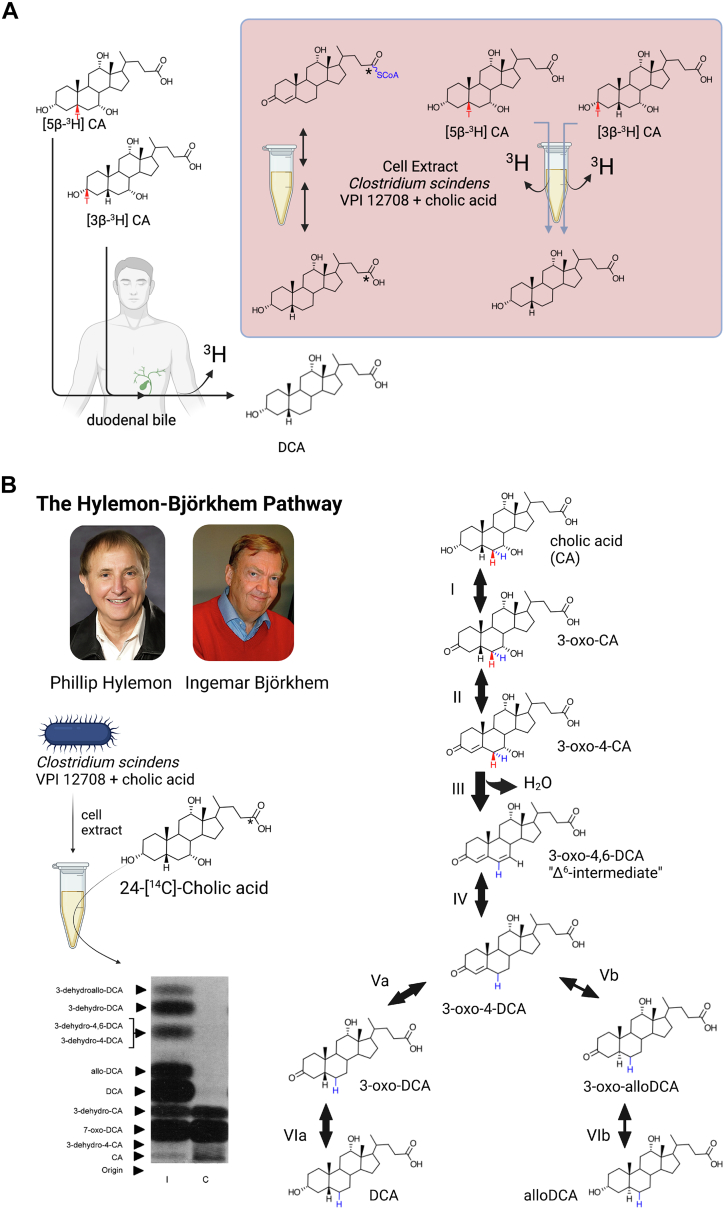
Early work had established that 7-oxoDCA is formed from CA in the intestine of rats (47) and rabbits (66). Administration of either [24-14C]CA or [24-14C]7-oxoDCA to rats resulted in the formation of [24-14C]DCA. Two possibilities were proposed, the first is that bile acid 7ɑ-dehydroxylation may proceed through a 7-keto-DCA intermediate. Alternatively, 7-keto-DCA was shown to be converted to CA by bacterial enzymes which would then be 7ɑ-dehydroxylated to DCA (66, 67). Formation of a 7-oxoDCA intermediate through dehydrogenation proceeds via removal of the 7ɑ-hydroxy proton and 7β-hydrogen. The “7-keto-intermediate” hypothesis was thus tested by observing the fate of the tritium label during metabolism of [24-14C][7β-3H]CA in vivo (Fig. 3). The “7-keto-intermediate” hypothesis was disproved after it was observed in experiments that no change occurred in the ratio of [3H]:[14C] in DCA (as [24-14C][7ɑ-3H]DCA) recovered from intestinal contents when compared to the ratio of [3H]:[14C] recovered in CA (as [24-14C][7β-3H]CA), indicating the retention of the tritium at C7 in the formation of DCA (66).
Fig. 3.
Testing the “7-keto-intermediate” hypothesis. Early experiments in rodent models observed the conversion of radiolabeled CA to both 7-oxoDCA and DCA. It was hypothesized that 7-oxoDCA may serve as an intermediate between CA and DCA. To test this hypothesis, 3H was added to [24-14C]CA at the C7 position. If 7-oxoDCA was an intermediate, it was expected that after in vivo metabolism of [7β-3H][24-14C]CA ([3H]:[14C] ratio = 1:1), fecal extraction of the end products should yield a [3H]:[14C] ratio of 0:1, indicating a loss of the [7β-3H]. However, the experimental results yielded DCA with a [3H]:[14C] ratio of 1:1, indicating that 7-oxoDCA is not an intermediate in the bile acid 7⍺-dehydroxylation pathway. DCA, deoxycholic acid.
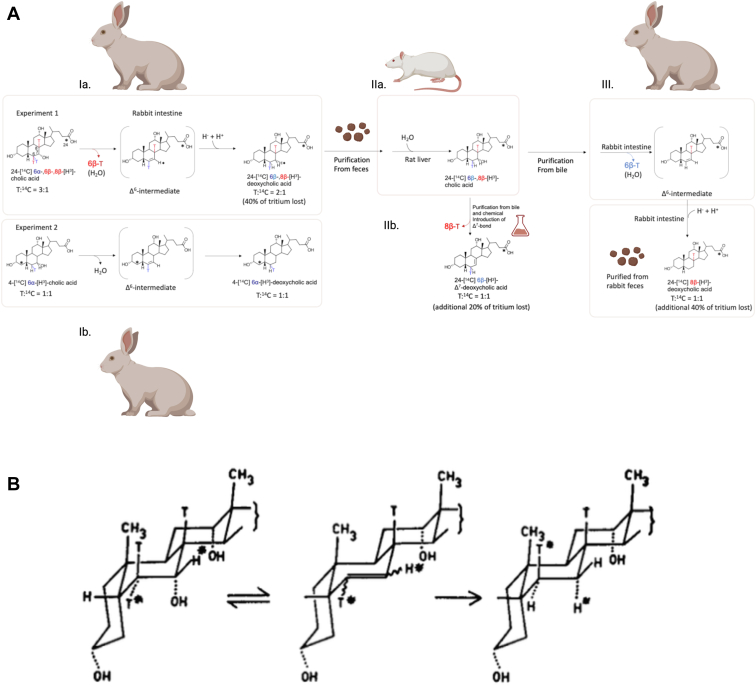
Following up on their observations, a subsequent model of bile acid 7-dehydroxylation was proposed in the 1960s by the Nobel laureates Bengt Samuelsson and Sune Bergström (66, 67, 68). The Samuelsson-Bergström model was based on elegant experiments determining the position and stereochemistry from which tritium loss occurred during the in vivo metabolism of [24-14C][6ɑ,6β,8β-3H]CA and [4-14C][6-3H]cholesterol (converted to [4-14C][6ɑ-3H]CA in the liver) in rats and rabbits (39, 69). The experimental results led to the proposal for a two-step mechanism (Fig. 4): a diaxial transelimination of the 7ɑ-hydroxyl group and 6β-hydrogen yielding 3ɑ,12ɑ-dihydroxy-5β-chol-6-enoic acid (Δ6-intermediate), followed by transhydrogenation of the Δ6-intermediate to DCA (39). Furthermore, the formation of a Δ6-intermediate is consistent with the observed retention of the C7-tritium in the “7-keto-intermediate” experiment (Fig. 3) (66, 67).
Fig. 4.
The Bergström-Samuelsson model of bile acid 7-dehydroxylation. A: A series of in vivo experiments administering [24-14C]6⍺-,6β-,8β-[3H]CA ([T]:[14C] ratio of 3:1 where T = tritium or 3H) (experiment 1) or [4-14C]6⍺-[3H]CA ([T]:[14C] ratio of 1:1) in rats and/or rabbits were performed in order to determine the stereochemistry for bile acid 7⍺-dehydroxylation. It was determined in previous studies that in [24-14C]6⍺-,6β-,8β-[3H]CA, 40% of the tritium was at 6⍺-position, 40% at 6β-position, and 20% at 8β-position (total 100%). In experiment 1, the radiolabeled CA was first administered to intact rabbits and DCA was recovered from feces, and the chromatographically purified compound was observed to have a [T]:[14C] ratio of 2:1 with a 40% loss of tritium (Ia.). From this experiment, it was clear that one of the C6-tritium labels was removed; however, it could not be determined which one from this experiment. Experiment 2 administered [4-14C]6⍺-[3H]CA to rabbits, and the chromatographically purified fecal DCA metabolite had a [T]:[14C] ratio of 1:1, indicating retention of the 6⍺-tritium (Ib.). Additional experiments were performed to further validate loss of the 6β-tritium and the fate of the 8β-tritium. The [24-14C]6β-,8β-[3H]DCA purified from rabbit feces in experiment 1 was then administered to bile duct cannulated rats (IIa.). Unlike the rabbit, the rat liver hydroxylates DCA, leading to removal of the 7⍺-[hydrogen] but retention of the 6β-tritium yielding [24-14C]6β-,8β-[3H]CA in bile. To determine the fate of the 8β-tritium, a Δ7-intermediate was chemically generated from the radiolabeled CA obtained from rat bile, resulting in a [T]:[14C] = 1:1 or another 20% loss of the tritium content (40% remaining), indicating elimination of the 8β-tritium (IIb.). To further confirm that the 6β-tritium is eliminated, the [24-14C]6β-,8β-[3H]CA obtained from rat bile was administered again to the rabbit and after fecal extraction, the [T]:[14C] ratio was reduced to 1:1 or another 40% loss of tritium (20% remaining) (III.). B: The original 1960 Samuelsson figure describing the proposed two-step mechanism of bile acid 7-dehydroxylation: (1) the 6β-tritium and 7⍺-OH is removed (diaxial trans-elimination); (2) the hydrogens added to C6 and C7 of the Δ6-intermediate are added trans to each other (inversion of the 7β-hydrogen to the 7⍺-position (denoted with star) and 6⍺-hydrogen is brought into the 6β-position), yielding DCA. Reprinted with permission (67). DCA, deoxycholic acid.
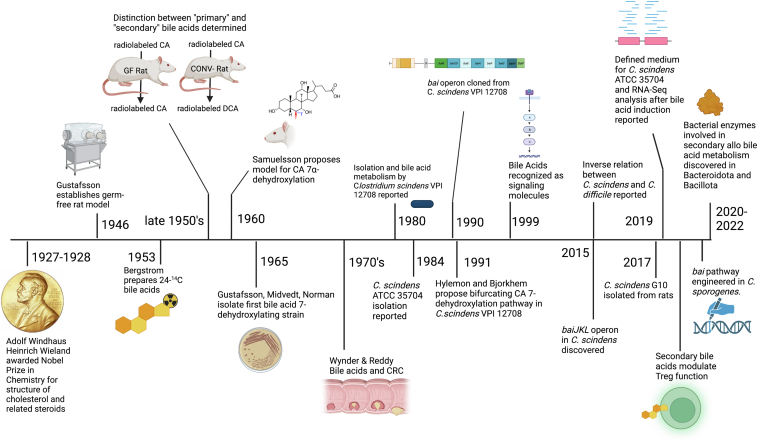
The proposed Δ6-intermediate could not be identified in these early studies, due to the lack of bile acid 7-dehydroxylating bacterial isolates from which in vitro studies could be performed (39). However, support for this model came from chemical synthesis of the Δ6-intermediate, which when added to cultures of certain clostridia was converted to DCA (70, 71, 72, 73). Ironically, it was subsequent work in clostridia that revealed that the removal of the C7 hydroxyl group was far more complicated than initially thought (See Fig. 5 for Timeline).
Fig. 5.
A timeline of key events in bile acid microbiology. (See text for descriptions).
Isolation of bile acid 7-dehydroxylating bacteria
The first reported isolation of a bacterium capable of converting CA to DCA was at Harvard University in 1962, but the isolate was lost after a series of transfers in anaerobic medium (74). Successful isolation and maintenance of several bile acid 7-dehydroxylating bacterial isolates was reported in 1966 by Bengt Gustafsson, Tore Midtvedt, and Arne Norman at the Karolinska Institutet (75). The classification of their isolates, including “strain II”, were referred to as “lactobacillus” (75, 76), but the authors suggested these isolates “most nearly resemble the members of the genus Clostridium”. The inconclusive taxonomic assignment was due to uncertainty as to whether spores were observed, consistent with Clostridium or “sporelike” pleomorphisms characteristic of other genera (75).
Shortly thereafter, an isolate from rabbit feces suspected to be causal in the formation of gallstones composed largely of glycoallodeoxycholic acid was reported a few years later by Erwin Mosbach (Fig. 2) and associates at St. Luke’s Hospital in New York (77). A difficulty inherent in much of the history of steroid microbiology of the gut is that the fate of these strains is not clear; there is no record of deposition of these early strains in a culture collection. Moreover, it is always a task going back into earlier literature to determine the taxonomic methods used for identification since many of these have now changed. Early reports of the isolation of bile acid 7-dehydroxylating bacteria placed these organisms in the genera Bacteroides or Lactobacillus (68, 77, 78). Subsequent studies found no evidence that intestinal Bacteroides spp. convert CA to DCA (79), although recent work has revived the possibility of C7-dehydroxylation by Bacteroides strains (80). These characteristics, including lack of observable sporulation, are consistent with what has been learned about bile acid 7-dehydroxylating clostridia that have in recent decades been classified based on molecular phylogeny (81, 82, 83).
Most known bile acid 7-dehydroxylating isolates are in the Clostridium clusters and go by an increasingly expanding list of reclassified genera. Indeed, in 1969, Hattori and Hayakawa reported isolation of “Bacteroides strain 28S'' with bile acid 7ɑ-dehydroxylating activity (68). During the late 1970s, this strain was later reclassified as Clostridium leptum VPI 10900 by Holdeman and Moore at the “VPI Anaerobe Lab” at Virginia Tech (79, 84). Phillip Hylemon and coworkers demonstrated conversion of [24-14C]CA to [24-14C]DCA and [24-14C]CDCA to [24-14C]LCA by C. leptum VPI 10900 (79). Unfortunately, preparation of cell-free extracts from which to observe CA dehydroxylation intermediates could not be done with C. leptum, as activity was lost upon cell lysis even under strictly anaerobic conditions (79). It was around this time that Eubacterium sp. VPI 12708, later reclassified as Clostridium scindens VPI 12708 (81), was isolated from the stool of a colon cancer patient by Rainer Hammann in Bonn, Germany (85). Additional isolates were reported by Fusae Takamine (Fig. 2) (86), Seiju Hirano, Noriyuki Masuda and colleagues in Japan (87, 88), Thomas Clavel and colleagues in Germany (89, 90), and James E. Wells, Phillip B. Hylemon, and colleagues in the USA (91).
A landmark paper described the separation of bile acid 7-dehydroxylating activity into strains with “high” activity (C. scindens and Peptacetobacter hiranonis) and those with “low” activity (Paeniclostridium sordelli, C. leptum, and Clostridium hylemonae) based on ∼100-fold differences in conversion of CA to DCA (86). Starting with its initial characterization in 1980 at the VPI Anaerobe Lab (84), C. scindens VPI 12708 has served as a model organism (despite lack of genetic tractability) for resolution of the complex biochemistry and enzymology of bile acid 7-dehydroxylation (92). More recently, C. scindens ATCC 35074, isolated in the early 1980s by Victor Bokkenheuser (Fig. 2) and coworkers in New York, has been instrumental to the “omics” era in the analysis of bile acid 7-dehydroxylating bacteria in the human gut microbiome (93, 94, 95, 96).
The Hylemon-Björkhem pathway of cholic acid 7-dehydroxylation
Another model for bile acid 7-dehydroxylation was developed decades after the work of Samuelsson and Bergström based on collaborations between Ingemar Björkhem and fellow steroid chemists at the Karolinska Institutet and Phillip Hylemon, a microbiologist at the Medical College of Virginia (now VCU School of Medicine) in the USA during the 1980s and 1990s.
The Björkhem lab focused studies on the formation of cholestanol (5α-cholestan-3β-ol) in patients with the rare inborn error in bile acid metabolism called cerebrotendinous xanthomatosis (CTX) (97). Aside from identifying 7α-hydroxy-4-cholesten-3-one (C4) as a biological marker for bile acid synthesis in the liver (98), the Björkhem lab determined that patients with CTX had a marked increase in biliary excretion of C4 (97). It was hypothesized that C4 was a key intermediate in the formation of cholestanol, which accumulates in xanthomas in CTX patients. Skrede and Björkhem (1982) observed that [4-14C]C4 was converted by the intestinal microbiota of rabbits to [4-14C]cholesta-4,6-dien-3-one and 4-cholesten-3-one (99). In this work, the authors drew the analogy between a possible mechanism of bile acid 7α-dehydroxylation by C. scindens VPI 12708 and the 7α-dehydroxylation of C4 by intestinal bacteria (Fig. 6) (99) and later in liver microsomes (100). This novel hypothesis suggested the formation of a 3-oxo-Δ4-bile acid intermediate and C7-dehydration, yielding a 3-oxo-Δ4,6-intermediate not predicted by the Samuelsson-Bergström model nor from the then contemporary work in C. scindens VPI 12708.
Fig. 6.
The metabolism of C4 in cerebrotendinous xanthomatosis patients and rabbits provided a potential mechanism for bile acid 7-dehydroxylation by intestinal bacteria. Studies by Ingemar Björkhem in CTX patients with radiolabeled 7⍺-hydroxy-4-cholesten-3-one (C4) indicated that the C7⍺-hydroxyl group was highly labile and converted to cholesta-4,6-dien-3-one and then to cholestanol by intestinal bacteria. An analogy was made between 7⍺-dehydroxylation of C4 and the 7⍺-dehydroxylation of CA and CDCA by intestinal bacteria, in a mechanism distinct from that proposed in the Bergström-Samuelsson model, which did not include a 3-oxo-Δ4-steroid intermediate. CDCA, chenodeoxycholic acid; CTX, cerebrotendinous xanthomatosis.
The formation of 3-oxo-Δ4-CA proceeds through oxidation of the 3ɑ-hydroxy group (removal of the 3ɑ-hydroxy proton and 3β-hydride) followed by introduction of a double bond in the A ring (C4-C5 dehydrogenation). The administration of CA tritiated at C3 or C5 as well as coadministration of [24-14C]CA to healthy human volunteers followed by collection of duodenal bile provided a key in vivo test of both the Samuelsson-Bergström model and the mechanism proposed by Björkhem (101, 102). The same experiments were performed with whole cells or cell extracts of C. scindens VPI 12708. In both in vivo and in vitro experiments, the tritium label was removed during CA conversion to DCA giving strong support for the “3-oxo- Δ4-CA intermediate” hypothesis which was analogous to C4 predicted by Skrede and Björkhem. In this scheme, C7-dehydroxylation would then yield a “Δ6-intermediate” (3-oxo-Δ4,6-DCA) distinct from the “Δ6-intermediate” (3ɑ,12ɑ-dihydro-5β-chol-6-enoic acid) predicted by Samuelsson and Bergström (Fig. 4).
The above results are also consistent with the formation of 3-oxo-Δ4-DCA, one of the reductive products on the path from 3-oxo-Δ4,6-DCA to DCA, observed in the cell extracts of C. scindens VPI 12708 (102). Indeed, the Hylemon lab subsequently observed the formation of hydrophilic, potentially conjugated CA intermediates during the incubation of radiolabeled unconjugated bile acids with cell-free extracts of C. scindens VPI 12708 (99). After addition of [24-14C]DCA to cell extracts, they identified conjugation to a nucleotide analog that would later be identified as coenzyme A (99). In collaboration with Jan Svöjall, the bile acid linked to CoA in the cell extracts of C. scindens VPI 12708 was determined to be 3-oxo-Δ4-DCA. This indicated that cell extracts were catalyzing in the oxidative direction from DCA → 3-oxo-DCA → 3-oxo-Δ4-DCA (Fig. 7A). In earlier studies by Kallner, radiolabeled 3-oxo-Δ4-DCA was injected into the reductive environment of the rat cecum where it was converted primarily to radiolabeled DCA (103, 104). While tempting to interpret the in vivo studies of Kallner as early support for 3-oxo-Δ4-DCA as an intermediate in bile acid 7-dehydroxylation, other bacteria, such as Clostridium paraputrificum and Bacteroides sp., lack the ability to convert CA to DCA but express enzymes that reduce 3-oxo-Δ4-steroids (105, 106, 107).
Fig. 7.
The Hylemon-Björkhem pathway of cholic acid 7-dehydroxylation. A: To determine whether a 3-oxo-Δ4 intermediate was involved in bacterial 7⍺-dehydroxylation of bile acids, Hylemon and Björkhem determined the fate of [3β-3H]CA + [24-14C]CA and [5β-3H]CA + [24-14C]CA in human volunteers and cell extracts of Clostridium scindens VPI 12708, a model bile acid 7⍺-dehydroxylating human gut bacterium. These experiments confirmed the loss of [3β-3H] and [5β-3H] labels during conversion of CA to DCA indicating formation of a 3-oxo-Δ4 intermediate inconsistent with the Bergström-Samuelsson model. B: Further studies by Phillip B. Hylemon at VCU Medical School and Ingemar Björkhem at the Karolinska Institutet (pictured) with [24-14C]CA in cell extracts of C. scindens VPI 12708 yielded a complex pattern of CA intermediates when cells were induced by CA (“I”) relative to cells grown without bile acids (“C”). Each bile acid intermediate was scraped from the TLC plate, and structures were determined by MS/MS analysis. Each bile acid intermediate was chemically synthesized and shown to be converted to DCA in cell extracts of C. scindens VPI 12708, verifying these as intermediates in the pathway (except for 7-oxoDCA). Unexpectedly, derivatives of allodeoxycholic acid (alloDCA) were also detected. Identification of these CA intermediates led to the elucidation of the complex, bifurcating Hylemon-Björkhem pathway of cholic acid 7-dehydroxylation. We have highlighted the hydrogens (red & blue) that Samuelsson had tritiated at carbon-6 for reference. DCA, deoxycholic acid.
Analyzing the multiple products formed after incubation of radiolabeled CA with cell extracts of C. scindens VPI 12708 led Hylemon and Björkhem to propose a detailed biochemical pathway for CA conversion to DCA by intestinal clostridia (Fig. 7B). Addition of each chemically synthesized, radiolabeled CA intermediate to cell extracts of C. scindens VPI 12708 resulted in DCA formation, consistent with the hypothesized mechanism (108). We therefore propose that the pathway deduced from their studies be named the Hylemon-Björkhem pathway (Fig. 7B).
Bai genes
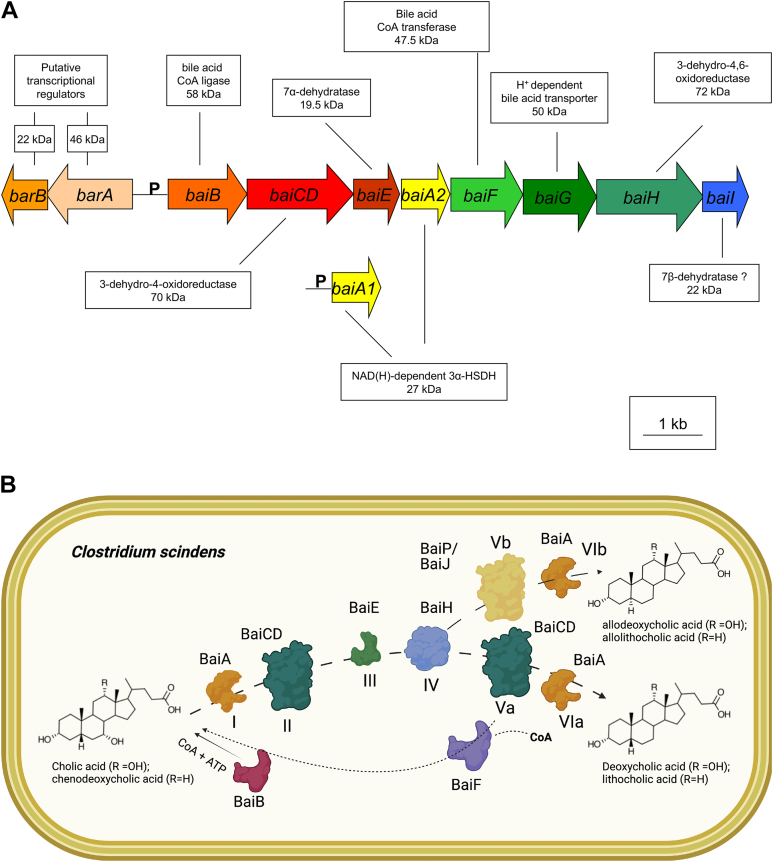
The identification and characterization of Bai enzymes over the ensuing three decades, mainly in the Hylemon lab, further support the Hylemon-Björkhem pathway of CA 7-dehydroxylation. Early studies determined that the expression of Bai proteins was inducible by the addition of CA or CDCA, but not the 7β-hydroxy epimer of CDCA, ursodeoxycholic acid (UDCA), or the secondary bile acid, DCA (85, 109). Comparison of two-dimensional gels from control and CA-induced whole cells of C. scindens VPI 12708 led to identification of several inducible polypeptides (71). N-terminal sequencing of CA-inducible polypeptides led to a reverse genetic approach to clone the ∼12-kb bai operon in its entirety a decade later by the Hylemon lab (110). The order of bai genes is largely conserved among intestinal clostridia, and in C. scindens VPI 12708 strain, it is baiBCDEAFGHI (Fig. 8A) (111).
Fig. 8.
The bile acid inducible operon and bile acid enzymes catalyzing each step of the Hylemon-Björkhem pathway. A: The bile acid inducible (bai) regulon in Clostridium scindens VPI 12708 has been identified (Modified from Ridlon et al. (92)). B: Enzymes corresponding to each reaction step of the Hylemon-Björkhem pathway (I-VIa/VIb), as well as enzymes involved in bile acid-coenzyme A metabolism.
Deconjugation of bile acids is a prerequisite for bile acid 7-dehydroxylation (79, 109, 112, 113). The inhibition of bile salt hydrolase (BSH) activity should thus also decrease the rate of bile acid 7-dehydroxylation, as has recently been demonstrated (114, 115). Studies reported in the 1980s on cofactor requirements for DCA formation with dialyzed cell extracts of C. scindens VPI 12708 indicated that the NAD+/NADH ratio and addition of flavins are important for activity (71, 72, 85). Molecular cloning and much of the enzymology of the Hylemon-Björkhem pathway were worked out in the Hylemon lab (92, 111). Unconjugated primary bile acids are imported into the cell by a 50-kDa proton-dependent transporter encoded by the baiG gene (116). Consistent with the identification of coenzyme A conjugates in the cell extracts of C. scindens VPI 12708, free bile acids were shown to be ligated to coenzyme A in an ATP-dependent manner through the bile acid CoA ligase (BaiB) or through coenzyme A transferase activity (BaiF, BaiK) (117, 118, 119). The conversion of CA to DCA in cell extracts of C. scindens VPI 12708 is enhanced by the addition of NAD+, a cofactor important in oxidoreduction reactions (72). Bile acids with a 3ɑ-hydroxy group (CA, CDCA, and UDCA) or 3-oxo-group (3-oxo-CA, 3-oxo-CDCA, 3-oxo-UDCA) are substrates for bile acid 7-dehydroxylation, while iso-bile acids (3β-hydroxy) are not substrates (120). This is because the first oxidation step is catalyzed by NAD+-dependent 3ɑ-HSDH encoded by the baiA1 and baiA2 genes (121, 122). The BaiA1 and BaiA2 enzymes are unique among HSDH enzymes in being specific for coenzyme A conjugates (121, 122). The rational formation of iso-CA and iso-CDCA may represent a strategy to decrease the rate of formation of secondary bile acids such as DCA and LCA. After formation of 3-oxo-cholyl∼SCoA, the 72-kDa flavoprotein, BaiCD, catalyzes a stereospecific NADH-dependent oxidation yielding 3-oxo-Δ4-cholenyl∼SCoA (123). Taken together, CoA addition followed by oxidations catalyzed by BaiA and BaiCD yields the “3-oxo-Δ4-intermediate” identified by Hylemon and Björkhem.
The Hylemon lab originally chromatographically separated “7ɑ-dehydroxylase” and “NADH:flavin oxidoreductase” activities from cell extracts of C. scindens VPI 12708. They later determined that the rate-limiting 7ɑ-dehydration catalyzed by the 19.5-kDa BaiE reaction results in the formation of chemically stable conjugated double-bond system in the 3-oxo-Δ4,6-DCA∼CoA intermediate (124). Work with analogous structures (i.e., C4) indicates that dehydration proceeds by both an enzymatic mechanism, as well as spontaneous dehydration through raising the pH into the basic range (99, 100). There is some uncertainty as to whether this reaction involves a trans elimination or cis elimination of water, with the former suggested by radiolabel studies, while the latter is suggested by X-ray crystallography (39, 125). It is also unclear at what point the majority of CoA is hydrolyzed or transferred. The BaiE recognizes both 3-oxo-Δ4-cholenyl∼SCoA and 3-oxo-Δ4-cholenoic acid (125). A 3-oxo-Δ4-deoxycholenyl∼CoA intermediate has also been observed in cell extracts, suggesting CoA transfer may occur after dehydration (102).
The “NADH:FOR” protein was determined to be the flavin-dependent BaiH (126, 127), which catalyzes the formation of 3-oxo-Δ4-UDCA from 3-oxoUDCA (123). Recent work has identified the BaiH as also catalyzing bile acid Δ6-reductase activity, converting 3-oxo-Δ4,6-DCA to 3-oxo-Δ4-DCA (128). It is thus highly probable that the conversion of Samuelsson’s Δ6-intermediate (3ɑ,12ɑ-dihydroxy-5β-chol-6-enoic acid) to DCA in cell extracts of C. scindens VPI 12708 was catalyzed by the BaiH or BaiN (129), although this has not yet been demonstrated. The BaiCD was shown to reduce 3-oxo-Δ4-DCA to 3-oxo-DCA (128). Finally, the BaiA1 and BaiA2 catalyze the final reductive step, yielding DCA (121). Candidate efflux transporters involved in the export of secondary bile acids have recently been identified through transcriptomic analysis after CA induction of C. scindens ATCC 35704 (94). Engineering of the baiBACDEFH genes into Clostridium sporogenes in the Fischbach lab has been shown to be sufficient to confer biochemical conversion of CA to DCA to this nonbile acid 7-dehydroxylating species (128). In vitro incubation of BaiB, BaiA, BaiCD, BaiE, BaiF, and BaiH with ATP, CoA, and NAD+ was both necessary and sufficient to convert CA to DCA (128). Recent work also demonstrates that the baiN gene product functions in an analogous two-step reaction catalyzed by BaiH and BaiCD (129). Taken together, Bai enzymes catalyzing each step of the Hylemon-Björkhem pathway from CA to DCA have been identified (Fig. 8B).
A short digression on early hints of the mechanism of CA degradation
The microbiologist Stanley Falkow famously stated that, “The world is covered in a fine patina of feces” (130). Our agricultural efforts bear this out. The manuring of soils creates rich sources of carbon, including bile salts. A gram of animal manure contains roughly 1–8 mg of bile salts, and manured soil concentrations of bile acids are estimated in the millimolar range. It is thus not surprising that soil microbiota have evolved complex biochemical pathways to mineralize bile salts to CO2 (131).
Evidence for a multistep C7 dehydroxylation of CA by the aerobic soil bacterium Arthrobacter simplex was reported by Hayakawa and Samuelsson (1964) at the Karolinska Institutet (132). Rigorous identification of bile acid intermediates included 3-oxoCA, 3-oxo-Δ4-CA, 3-oxo-Δ1,4-DCA, 3-oxo-Δ4,6-DCA, and 3-oxoDCA (132). In the introduction of the Hayakawa and Samuelsson paper, the authors lamented the barriers to studying the analogous pathway in anaerobic bacteria in human stool samples: “The predominant microbiological reaction involves elimination of the 7ɑ-hydroxyl group of cholic acid and chenodeoxycholic acid, leading to formation of deoxycholic acid and lithocholic acid. This reaction has been demonstrated in vitro with microorganisms obtained from rat and man. However, despite considerable efforts directed at the isolation of the microorganism(s) responsible for this transformation, it has so far not been possible to identify the microorganism(s) involved. For this reason, the mechanism of the reaction has only been studied in vivo with bile acids specifically labeled with tritium. However, nothing has been learned about the enzymatic details.”(132).
Two years later, in 1966, pure cultures of seven human fecal bacteria capable of bile acid 7-dehydroxylation were isolated at the Karolinska Institutet by Gustafsson, Midtvedt, and Norman. Thus, Samuelsson possessed the methodology for identifying the key 3-oxo-Δ4-DCA and 3-oxo-Δ4,6-DCA intermediates in aerobic bile acid degradation in soils and for confirming the reduction of the C6-C7 double bond during enterohepatic circulation of radiolabeled CA in humans, and shortly thereafter, in C7-dehydroxylating stool bacteria that had been isolated by collaborators at the same University (75). In hindsight, the ingredients were there for bile acid pathway intermediates to have been outlined decades earlier. However, the research focus of Bergström and Samuelsson had shifted towards prostaglandins during the early 1960s once their group moved from Lund University to the Karolinska Institutet, work for which they would later share the Nobel Prize in Physiology and Medicine (133).
Bacterial shape-shifting of bile acids
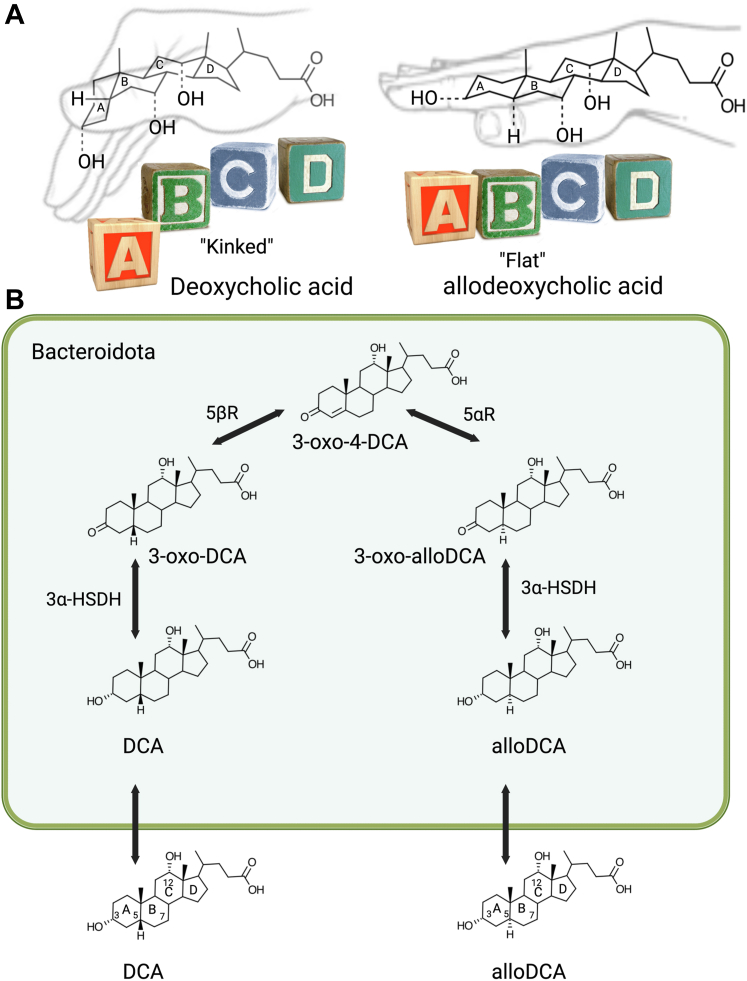
The usefulness of a scientific model is determined by its ability to incorporate and explain new phenomena. The “3-oxo-Δ4-intermediate” introduced by the Hylemon-Björkhem pathway raises the possibility for the formation of secondary bile acid isomers with distinct steroid ring shapes not predicted by the Samuelsson-Bergström model. So far, we have described the conversion of “kinked” primary bile acids produced by the human liver to “kinked” secondary products such as DCA and LCA. However, microbes can generate “flat” stereoisomers known as secondary “allo” bile acids which include allodeoxycholic acid (alloDCA) and allolithocholic acid (alloLCA) (Fig. 9A) (103, 104). AlloDCA was first isolated from rabbit bile by Kishi in 1936, who misidentified it as “lagodeoxycholic acid” (the 12β-hydroxy epimer of DCA) but was properly identified decades later after purification from rabbit feces and bile by Danielsson and colleagues (1963) (134) and from rabbit gallstones by Alan Hofmann and Erwin Mosbach (both pictured in Fig. 2) (135). Indeed, early studies reported the formation of gallstones enriched with glycoallodeoxycholic acid in rabbits fed 5ɑ-cholestan-3β-ol (135, 136). Gallstone formation in this model was prevented through neomycin treatment (137), and a neomycin-sensitive fecal isolate “FA 1/146” that converted CA to DCA and alloCA to alloDCA isolated by Bokkenheuser, Mosbach, and colleagues indicated a causal role for the gut microbiome in generating alloDCA, which is absorbed and accumulates in bile in this model (77).
Fig. 9.
An indirect pathway for the microbial formation of allo-secondary bile acids. A: Allo-bile acids are “flat” owing to the A/B-trans ring structures, while host bile acids and secondary bile acids are “kinked” due to A/B-cis ring structures. B: The pathway from CA to alloDCA can be thought of as the “direct pathway”, but an “indirect pathway” to alloDCA catalyzed by enzymes encoded by the Bacteroidota results in an equilibrium between DCA and alloDCA through a 3-oxo-Δ4-DCA intermediate. So far, the Bacteroidota have not been shown to encode a complete set of bai-like genes, instead, expression of 3⍺-HSDH, 3-ketosteroid-5β-reductase (5βR), and 3-ketosteroid-5⍺-reductase (5⍺R) allow isomerization of the A/B-rings. alloDCA, allodeoxycholic acid; Bai, bile acid-inducible; DCA, deoxycholic acid; HSDH, hydroxysteroid dehydrogenase.
In 1991, Hylemon et al. (108) reported the formation of [24-14C]alloDCA as a product of [24-14C]CA metabolism in cell extracts of C. scindens VPI 12708. This indicates that the Hylemon-Björkhem model is a multistep, bifurcating pathway that can only be explained through formation of 3-oxo-4 intermediates (Fig. 7B) (108). In the path from CA (5β-H) to DCA (5β-H) via a 3-oxo-Δ4 intermediate, the A/B-ring stereochemistry is retained. The BaiCD is a 3-keto-Δ4-5β-oxidoreductase enzyme responsible for removal and reintroduction of the 5β-H during 7-dehydroxylation of CA (Figs. 7B and 8B) (128). However, an alternative path from CA (5β-H) to alloDCA (5ɑ-H), which changes the A/B ring stereochemistry, leads to the prediction that C. scindens encodes a bile acid 3-keto-Δ4-5ɑ-oxidoreductase enzyme that utilizes 3-oxo-Δ4-DCA and 3-oxo-Δ4-LCA as substrates.
Enzymatic basis for allo-secondary bile acid formation in the Hylemon-Björkhem pathway
In 2019, the complete genome and global transcriptomic responses of C. scindens ATCC 35704 to bile acids was reported (94). Among genes significantly upregulated by CA induction was a predicted pyridine nucleotide-dependent urocanate reductase that we hypothesized to encode a bile acid 5ɑ-reductase involved in the formation of alloDCA (94). Our subsequent work confirmed that this gene, named baiP, encodes a bile acid 5ɑ-reductase which acts in concert with BaiA1 to convert 3-oxo-Δ4-DCA to alloDCA and 3-oxo-Δ4-LCA to alloLCA (138). Prior work by Ridlon and Hylemon identified a bai gene polycistron that was named baiJKL and was located adjacent to baiA1 in C. scindens VPI 12708 and C. hylemonae TN271 (117). The BaiK is a homolog of the BaiF which is involved in bile acid CoA transfer and shares this function (117). The baiJ was predicted to encode a flavin-dependent 3-ketosteroid dehydrogenase hypothesized to be involved in sterol A/B ring oxidoreduction. Phylogenetic analysis of BaiP from C. scindens ATCC 35704 revealed BaiJ as a homolog. Functional characterization of recombinant BaiJ from C. scindens VPI 12708 expressed in E. coli confirmed the conversion of 3-oxo-Δ4-DCA and 3-oxo-Δ4-LCA to allo-secondary bile acids (138). Taken together, the identification of baiJ and baiP genes supports the observed bifurcation proposed by the Hylemon-Björkhem pathway, yielding both DCA and alloDCA, through a 3-oxo-Δ4-DCA intermediate.
Phylogenetic analysis and Hidden Markov model searches of BaiJ, BaiP, and BaiE, and BaiCD in human gut metagenomes revealed enrichment in colorectal cancer patients relative to adenoma and healthy controls (138). The Oscillospiraceae was recently identified as a novel group of uncultured bile acid 7-dehydroxylating bacteria (139), and these taxa were also identified as having the genetic potential for generating secondary allo-bile acids (138). We have referred to the formation of secondary allo-bile acids from primary bile acids as the “direct pathway” since primary bile acids are converted to secondary allo-bile acids by a single bacterial strain (138). There are other mechanisms by which secondary allo-bile acids can be generated by gut microbiota through the metabolism of DCA and LCA.
An “indirect pathway” to secondary allo-bile acids
The indirect pathway describes the equilibrium between DCA and alloDCA or LCA and alloLCA through a 3-oxo-Δ4-intermediate (Fig. 9B). This “indirect pathway” first requires conversion of CA to DCA or CDCA/UDCA to LCA by one bacterial strain, followed by oxidation and ring epimerization by another strain. The in vivo conversion of radiolabeled DCA to radiolabeled alloDCA after intracecal injection in rabbits was first reported in 1963 by Danielsson (134). A biochemical pathway was proposed in 1967 by Anders Kallner, who studied the metabolic fate of tritiated DCA, 3-oxo-DCA, 3-oxo-Δ4-DCA, 3-oxo-alloDCA, and alloDCA by the gut microbiota in rats (103, 104). In all cases, a mixture of radiolabeled DCA and alloDCA was obtained, confirming an equilibrium between these compounds in the GI tract (Fig. 9B). Loss of the 3β-tritium from [3β-3H][24-14C]DCA during in vivo conversion to [24-14C]alloDCA confirmed the formation of a 3-oxo intermediate (103, 104).
A later study by Edenharder (1988) reported the conversion of 5β-bile acids to 5ɑ-bile acids by fecal Bacteroides isolates (78). Work in the chicken microbiome identified a gene annotated as “steroid 5ɑ-reductase” (5AR) similar to mammalian SRD5A1 widespread among the Bacteroidota (140). A recent study (107) by Kenya Honda and colleagues reported that strains of Parabacteroides merdae, Odoribacter laneus, and Bacteriodes dorei incubated with 3-oxo-Δ4-LCA resulted in conversion to isoalloLCA. This confirms that the Bacteroidota (formerly Bacteroidetes) express 5AR that recognize bile acids and indicate the expression of bile acid 3β-HSDH (described in the next section) (107). Twenty other Bacteroidota strains were shown to convert 3-oxo-Δ4-LCA to 3-oxo-alloLCA, and disruption of the 5AR gene in P. merdae prevented the formation of alloLCA derivatives (107). The 5AR gene was clustered with predicted 3β-HSDH and NADH:flavin oxidoreductase (similar to BaiCD and BaiH), hypothesized to function as bile acid 5β-reductase. Several strains were found to convert secondary bile acids such as 3-oxoLCA or isoLCA (5β-reduced) to isoalloLCA. However, the conversion of CDCA to isoalloLCA required the combined metabolism of C. scindens (capable of C7-dehydroxylation) with Bacteroidota strains (107). So far, the Bacteroidota have not been shown to encode a complete set of bai-like genes. The contribution of C. scindens to allo-secondary bile acid production (through the direct pathway) in coculture with Bacteroidota strains is not clear. Collectively, these data demonstrate that Bacteroidota strains possess an “indirect pathway” allowing isomerization of the bile acid A/B-rings.
While isoalloLCA derivatives in stool are measured at relatively low micromolar concentrations, these concentrations (1–3 μM isoalloLCA) are sufficient to inhibit the growth of gram-positive pathogens, including Clostridioides difficile (107). Thus, while “kinked” secondary bile acids DCA and LCA are inhibitory against many gram-negatives (141), the formation of “flat”, 3β-derivatives such as isoalloLCA appear to be inhibitory for many gram-positive bacteria (86). The relative contributions of the direct and indirect pathways to the formation of secondary allo-bile acids in the GI tract are unexplored.
The special case of bile acids with an epimerized C7 hydroxyl group
Navigators and watchers of the night sky, both professional and amateur, look to the handle of the constellation Ursa Minor (“little bear”) to find the North Star. For bile acid researchers, “urso” bile acids hold a special affection. UDCA is a major primary biliary bile acid of bears (genus Ursus) (22, 142) and has been utilized in traditional medicine, along with many other animal biles, for centuries (22). There is a “little bear” in all of us, as judged by the measurement of minor quantities of UDCA in feces, blood, and bile (21). Unlike bears (and mice), whose “urso” biliary component is produced primarily by enzymes encoded in their genome (thus a primary bile acid), it is the epimerization action of the human gut microbiota responsible for the formation of UDCA as a secondary bile acids in humans (21). UDCA is synthesized on the order of 1,000 metric tons for therapeutic uses each year, primarily for the treatment of primary biliary cholangitis (21).
The Hylemon-Björkhem model in humans is further complicated in the case of 7β-hydroxy bile acids such as the therapeutic bile acid, UDCA. Two mechanisms have been determined for the removal of the 7β-hydroxy group from UDCA: 1) epimerization of UDCA to CDCA followed by 7ɑ-dehydroxylation by the concerted action of intestinal bacteria and 2) direct 7β-dehydroxylation by species such as C. scindens (143).
Regarding the second mechanism, it is presumed, although not yet demonstrated, that the BaiG imports UDCA which is then ligated to CoA by the BaiB, BaiF, or BaiK, followed by oxidation by BaiA (143). Prior work provided qualitative evidence that the BaiH is a stereospecific enzyme involved in the second oxidative step involved in the formation of 3-oxo-Δ4-UDCA(∼SCoA), while the BaiCD catalyzes the same step with 7ɑ-hydroxy bile acid epimers (123). The BaiE is stereospecific and does not recognize 3-oxo-Δ4-UDCA∼SCoA as a substrate. Indeed, bile acid 7ɑ-dehydratase (BaiE) and 7β-dehydratase activity in C. scindens can be detected and separated by FPLC (124). The baiI gene product is a homolog of BaiE in the SnoAL_4 family of proteins, the only gene in the bai operon for which a function has yet to be ascribed and is hypothesized to have bile acid 7β-dehydratase activity (92).
After the rate-limiting 7β-dehydratase step, a 3-oxo-Δ4,6-LCA derivative is formed as is also the case with the 7ɑ-dehydration of CDCA. Thus, reduction of this intermediate is expected to follow the aforementioned BaiH → BaiCD→ BaiA path towards LCA or BaiH→BaiP/BaiJ→BaiA path towards alloLCA. Thus, 7ɑ-dehydroxylation and 7β-dehydroxylation are predicted to proceed by the same series of steps, with the substitution of stereospecific enzymes at the step leading up to and including 7β-dehydration.
Recent developments and prospects in bile acid 7-dehydroxylation
An inverse relationship has been reported between C. scindens and DCA levels and C. difficile colonization (144). Several studies have incorporated C. scindens ATCC 35704 into small, defined consortia, providing insights into colonization, metabolism, and competition (145, 146, 147). In a murine model of C. difficile infection, addition of C. scindens ATCC 35704 was shown to have minor effects on the composition of the “Oligo-MM12” consortium but significantly decreased early colonization and pathogenesis of C. difficile, which was able to overcome colonization resistance by 72 h postinfection with concomitant decline in DCA levels (145). The inhibition of C. difficile strains by DCA-producing clostridia in vitro is reported to be strain-dependent, and C. difficile is ultimately able to outcompete these bacteria in defined communities (148). Secondary bile acids common in mice (eg, ω-MCA, HDCA, and MDCA) have also shown to inhibit C. difficile germination (149); however, human strains of C. scindens and other bile acid 7-dehydroxylating are not capable of biotransforming mouse bile acids (150). Eyssen reported isolation of a strain from rat feces that could 7-dehydroxylate murine bile acids; however, the phylogenetic position of this strain was not determined, and, to our knowledge, this strain was not preserved (150).
Recently, C. scindens G10 was reported to have been isolated from rat feces; however, only conversion of CA to DCA was tested (151). Future studies should determine whether strain G10, if indigenous to rats and mice, is capable of bile acid 7-dehydroxylation of muricholic acids. To further complicate this relationship, C. scindens ATCC 35704 was shown to generate tryptophan-based antibiotics that target C. difficile vegetative growth in synergy with the inhibitory effects of secondary bile acids (152). So far genetic manipulation of C. scindens ATCC 35704 has remained elusive, and determining the causal relationship between bai genes, antimicrobial peptide formation, and C. difficile colonization and pathogenesis remains to be determined (145, 153).
C. scindens DSM 100975 was recently isolated from pig feces and the complete genome sequenced (89). Comparative genomics between the human isolate ATCC 35704 and the pig isolate DSM 100975 share 2966 of 3655 genes, with the ∼675 genes unique to both strains composed largely of mobile genetic elements and phage genes. There is strict conservation of the bai gene cluster between the two strains, and in both cases, global transcriptomics revealed that the bai genes are significantly induced by CA, but not DCA (89, 94). It is not currently known whether this strain is able to convert hyocholic acid (3α,6α,7α-trihydroxy-5β-cholan-24-oic acid) to hyodeoxycholic acid (3α,6α-dihydroxy-5β-cholan-24-oic acid). The gnotobiotic piglet is a useful translational model, and the acquisition of C. scindens strains from different vertebrate species will allow future exploration into strain-specific adaptations to the gut environment.
Hayakawa and Hattori (1970) showed that P. sordellii NCIB 6929 produce DCA and 7-oxo-DCA from CA in vitro (154). Indeed, the first strains that were preserved in culture collections were Paraclostridium bifermentans ATCC 9714 and P. sordellii NCIB 6929 (154). A study in 1997 by Doerner et al. (86) compared the rate of bile acid 7-dehydroxylation activity from available isolates and separated the strains into two groups differing >100 fold in conversion rate of CA to DCA (86). P. sordellii is a “low” activity strain; however, it is unclear how this strain differs from “high” activity strains such as C. scindens or P. hiranonis. Possibilities include lack of a complete bai operon, changes in enzymatic or transporter activity, differences in promoter sequence or lack of inducibility, or perhaps other mechanisms exist beyond known bai genes. Indeed, we previously located two putative bai gene clusters in the P. sordellii ATCC 9714 genome consisting of baiA_baiCD and baiH_baiE, as well as a putative NADP-dependent bile acid 7α-HSDH (111). Predicted upstream regulatory proteins differ between P. sordellii (IclR) and canonical bai operons such as that found in C. scindens VPI 12708 (AraC). Could the baiACDHE genes (HSDH, 3-oxo- Δ4-reductase(s), and dehydratase) constitute an absolute minimal set of structural genes necessary for the conversion of CA to DCA?
Only a single study has reported the lack of 7-dehydroxylation in gnotobiotic gerbils colonized with defined consortium with or without P. sordellii ATCC 9714 (155). Another study noted a lack of DCA formation when bi-associating gnotobiotic mice with BSH-expressing Bacteroides distasonis and the 7-dehydroyxlating strains P. hiranonis TO931 or Eubacterium sp. strain 36S; the absence of DCA formation was explained by a lack of TCA conversion to CA (156). Successful deconjugation and 7-dehydroxylation of CA in this mouse model was reported after additional Bacteroides strains and Bilophila wadsworthia were added (157). In the P. sordelli colonization study, it was reported that 90% of conjugated CA and CDCA were deconjugated to free bile acids suggesting substrate limitation was not the issue (155). Further studies should examine the in vivo role of “low activity” 7-dehydroxylating bacteria to secondary bile acid formation.
A more perplexing observation from our genomic queries is the case of strains of C. leptum, which has been shown to convert CA and CDCA, but not UDCA, to secondary bile acids (79, 158). Conversion by this strain is known to be slow and inefficient (86). The rate-limiting bile acid 7α-dehydratase (BaiE) is in the SnoaL_4 family, and a search of BaiE from C. scindens VPI 12708 against the genome of C. leptum VPI 10900 revealed a single candidate with low sequence identity genome sequence of P. sordellii VPI 9048 genome that shares 48% AA sequence identity with the baiE gene product of C. scindens. Upstream of the putative baiE gene, we located an NADH-dependent flavin oxidoreductase with 47% AA sequence identity with the baiCD gene product and 64% AA sequence similarity with the baiH gene product from C. scindens. However, other bai gene candidates were not clearly observed. Perhaps this suggests a distinct mechanism by which this species converts CA to DCA. Could this be the 7α-dehydratase and Δ6-reductase of Bergström and Samuelsson?
We think more likely that additional bile acid enzymes developed through convergent evolution from genes encoding members of the same or distinct protein families that serve analogous functions to those of canonical Bai enzymes. Indeed, Fe-S flavoproteins similar to BaiH and BaiCD are quite common, although their functions remain largely uncharacterized (159). Proteins in the short chain dehydrogenase/reductase, such as BaiA, are also widely distributed, and there are characterized 3ɑ-HSDHs (160, 161) distantly related to BaiA, indicating that additional examples will be found, perhaps in the genome of C. leptum. Further experiments will be required to resolve this issue, and C. leptum represents an interesting organism for future study in bile acid 7α-dehydroxylation.
Overcoming barriers
A significant limitation in mechanistic gut microbiome research at present is the lack of genetically tractable model organisms, particularly in the Firmicutes (Bacillota). However, recent breakthroughs in Firmicutes (Bacillota) genetic tools are starting to make possible determination of the causal role of microbial genes in biochemical pathways, including bile acid pathways (128, 162) and the role genes/pathways in complex host phenotypes (163). Faecalicatena contorta S122 was recently identified as a low abundance (0.016% relative abundance) organism widely distributed in human metagenomes which harbors bai genes and actively converts CA to DCA (163). A genetic manipulation pipeline for nonmodel clostridia was developed in this study and knockout of the baiH gene in F. contorta S122 was achieved rendering the mutant incapable of in vitro conversion of CA to DCA. Stable colonization in both complex and defined communities in gnotobiotic mice treated with dextran sodium sulfate resulted in significant mucosal inflammation associated with baiH-dependent expansion of Erysipelotrichaceae in the gut (163).
Conclusions
We hope that our decidedly and necessarily myopic historical focus will inspire others to obsess over the historical developments and natural history of similar biomolecules and most importantly, their relations to human health and disease. And we trust that such efforts will unearth as dedicated a cast of characters as we have attempted to outline in this story. Science is, after all, a human activity. With that being said, it is with great respect and admiration that we recognize Professors Phillip B. Hylemon and Ingemar Björkhem, with what, in our opinion, rightfully should be named after their dedicated career focus. This conclusion appeared to us obvious from our examination of the historical literature in bile acid 7-dehydroxylation and the foundation that preceded it.
There is a long precedence of naming metabolic schemes as such in the history of anaerobic bacteriology. Examples that are most famous include the Wood–Ljungdahl pathway of reductive acetogenesis attributed to the advances made by Harland G. Wood and Lars G. Ljungdahl (164) and the related Wolfe Cycle after the methanogen expert, Ralph S. Wolfe (165). In similar fashion, such scholars have had organisms named after them, such as Acetobacterium woodii (166), Clostridium ljungdahlii (167), Methanothermobacter wolfeii (168, 169), and in the present case, Clostridium hylemonae (81) and Hylemonella (170). The (unfathomably) ancient acetogenic and methanogenic pathways that utilize early Earth gases long predate vertebrates, and both pathways adapted to utilize the copious release of CO2 and H2 produced principally from carbohydrate fermentation in the gut of newcomers to this planet. But they have also adapted and evolved in the elbow-room-only environment of the GI tract in which, relatively speaking, methanogens, acetogens, and bile acid 7-dehydroxylating bacteria are low abundance bricks in the wall, albeit, in many cases, keystone species in the arch. Indeed, Phil Hylemon and one of the authors (JMR) have proposed a potential link between these pathways (120, 171). In light of the perspective of the past and the present, we look forward to the bright future of bile acid-microbiome-gastrointestinal-hepatobiliary discoveries.
Conflict of interest
J. M. R. trained in the laboratory of Phillip B. Hylemon. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors express their sincere appreciation to the following: 1) Ingemar Björkhem at the Karolinska Institutet in Stockholm, Sweden for the picture of himself and for providing helpful comments on the manuscript; 2) Phillip Hylemon at the Virginia Commonwealth University in Richmond for the picture of himself and for providing helpful comments on the manuscript; 3) the Lasker Awards Archives from the History of Medicine (IHM) at the National Library of Medicine for the picture of Sune Bergström; 4) the Karolinska Institutet and photographer Gunnar Ask for the picture of Bengt Samuelsson; 5) Elisabeth Norin at the Karolinska Institutet for the pictures of Tore Midtvedt and Bengt Gustafsson; 6) the late Alan Hofmann at the University of California, San Diego for the pictures of Erwin Mosbach, Jan Sjövall, Sven Lindstedt, and Henry Danielsson; 7) Elsevier for permitting us to reuse the picture of Alan Hofmann; 8) University of the Ryukyus in Okinawa, Japan for the picture of Fusae Takamine; 9) Anders Kallner at the Karolinska Institutet for the picture of himself; and 10) Carl Bokkenheuser for the picture of Victor Bokkenheuser. Figures were created with the software BioRender (https://www.biorender.com/).
Author contributions
J. M. R. conceptualization; J. M. R. investigation; J. M. R., S. L. D., and H. R. G. writing–original draft; J. M. R. visualization; J. M. R., S. L. D., and H. R. G. writing–review and editing.
Funding and additional information
We would like to acknowledge financial support from National Institutes of Health grants (R01 CA204808-01 [J. M. R., H. R. G.], R01 GM134423-01A1 [J. M. R.], GM145920-01 [J. M. R], R03 AI147127-01A1 [J. M. R.]) as well as UIUC Department of Animal Sciences Matchstick grant and Hatch ILLU-538-916 (J. M. R.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.O'Keefe S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia W., Xie G., Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs C.D., Trauner M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat. Rev. Gastroenterol. Hepatol. 2022;19:432–450. doi: 10.1038/s41575-021-00566-7. [DOI] [PubMed] [Google Scholar]
- 4.Collins S.L., Stine J.G., Bisanz J.E., Okafor C.D., Patterson A.D. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2022;21:236–247. doi: 10.1038/s41579-022-00805-x. [DOI] [PubMed] [Google Scholar]
- 5.Wise J.L., Cummings B.P. The 7-α-dehydroxylation pathway: an integral component of gut bacterial bile acid metabolism and potential therapeutic target. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1093420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makishima M., Okamoto A.Y., Repa J.J., Tu H., Learned R.M., Luk A., et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 7.Parks D.J., Blanchard S.G., Bledsoe R.K., Chandra G., Consler T.G., Kliewer S.A., et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Chen J., Hollister K., Sowers L.C., Forman B.M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 9.Staudinger J.L., Goodwin B., Jones S.A., Hawkins-Brown D., MacKenzie K.I., LaTour A., et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makishima M., Lu T.T., Xie W., Whitfield G.K., Domoto H., Evans R.M., et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 11.Guo G.L., Lambert G., Negishi M., Ward J.M., Brewer H.B., Jr., Kliewer S.A., et al. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J. Biol. Chem. 2003;278:45062–45071. doi: 10.1074/jbc.M307145200. [DOI] [PubMed] [Google Scholar]
- 12.Hang S., Paik D., Yao L., Kim E., Trinath J., Lu J., et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576:143–148. doi: 10.1038/s41586-019-1785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik D., Yao L., Zhang Y., Bae S., D'Agostino G.D., Zhang M., et al. Human gut bacteria produce Tau(Eta)17-modulating bile acid metabolites. Nature. 2022;603:907–912. doi: 10.1038/s41586-022-04480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W., Hang S., Fang Y., Bae S., Zhang Y., Zhang M., et al. A bacterial bile acid metabolite modulates T(reg) activity through the nuclear hormone receptor NR4A1. Cell Host Microbe. 2021;29:1366–1377.e9. doi: 10.1016/j.chom.2021.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama T., Miyamoto Y., Nakamura T., Tamai Y., Okada H., Sugiyama E., et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem. Biophys. Res. Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 16.Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 17.Raufman J.P., Cheng K., Zimniak P. Activation of muscarinic receptor signaling by bile acids: physiological and medical implications. Dig. Dis. Sci. 2003;48:1431–1444. doi: 10.1023/a:1024733500950. [DOI] [PubMed] [Google Scholar]
- 18.Nagahashi M., Yuza K., Hirose Y., Nakajima M., Ramanathan R., Hait N.C., et al. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 2016;57:1636–1643. doi: 10.1194/jlr.R069286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahlstrom A., Sayin S.I., Marschall H.U., Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 20.Reuben A. The biliary cycle of Moritz Schiff. Hepatology. 2005;42:500–505. doi: 10.1002/hep.20823. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann A.F., Hagey L.R. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J. Lipid Res. 2014;55:1553–1595. doi: 10.1194/jlr.R049437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D.Q., Carey M.C. Therapeutic uses of animal biles in traditional Chinese medicine: an ethnopharmacological, biophysical chemical and medicinal review. World J. Gastroenterol. 2014;20:9952–9975. doi: 10.3748/wjg.v20.i29.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Helmont J. Franz Mercuriusreference; Amsterdam: 1668. Onus medicinue, fe opera et opuscula omnia, Franz Mercurius. [Google Scholar]
- 24.van Reverhorst M. Jordanus Luchtmans; Leiden: 1696. Dissertatio anatomica-medica de motu bilis circulari eiusque morbis, Jordanus Luchtmans. [Google Scholar]
- 25.Dawson P.A., Karpen S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015;56:1085–1099. doi: 10.1194/jlr.R054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair P., Kritchevsky D. In: The Bile Acids: Chemistry, Physiology, and Metabolism. Nair P., Kritchevsky D., editors. Plenum Press; New York, NY: 1971. Chemistry of bile acids; pp. 1–9. [Google Scholar]
- 27.Demarçay H. Die natur der galle. Ann. Pharm. 1838;27:270–291. [Google Scholar]
- 28.Strecker A. Untersuchung der Ochsengalle. J. Prakt. Chem. 1849;46:137–146. [Google Scholar]
- 29.Mylius F. Ueber die cholsäure. Ber. Dtsch. Chem. Ges. 1886;19:369–379. [Google Scholar]
- 30.Fischer H. Zur kenntnis der gallenfarbstoffe. I. mitteilung. Hoppe Seyler's Z. Physiol. Chem. 1911:204–239. [Google Scholar]
- 31.Wieland H., Reverey G. Untersuchungen über die Gallensäuren. XXI. Mitteilung. Zur Kenntnis der menschlichen Galle. 1. Hoppe seyler's Z. Physiol. Chem. 1924;140:186–202. [Google Scholar]
- 32.Fieser L.F., Fieser M. Reinhold; New York: 1959. Steroids. [Google Scholar]
- 33.Haslewood G.A.D. Elsevier/North-Holland Biomedical Press; Amsterdam: 1978. The Biological Importance of Bile Salts. [Google Scholar]
- 34.Radt F. Elsevier/North-Holland Publishing Co.; Amsterdam: 1962. Elsevier’s Encyclopedia of Organic Chemistry. [Google Scholar]
- 35.Danielsson H., Sjövall J. Elsevier; Amsterdam: 1985. Sterols and Bile Acids. [Google Scholar]
- 36.Bernal J.D. Crystal structures of vitamin D and related compounds. Nature. 1932;129:277–278. [Google Scholar]
- 37.Frankel M. The biological splitting of conjugated bile acids. Biochem. J. 1936;30:2111–2116. doi: 10.1042/bj0302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt L.H., Hughes H.B., Green M.H., Cooper E. Studies on bile acid metabolism: II. The action of Alcaligenes faecalis on cholic acid. J. Biol. Chem. 1942;145:229–236. [Google Scholar]
- 39.Bergström S., Danielsson H., Samuelsson B. In: Lipide Metabolism. Bloch K., editor. John Wiley & Sons; New York, NY: 1960. Formation and metabolism of bile acids; pp. 291–336. [Google Scholar]
- 40.Sjövall J. Fifty years with bile acids and steroids in health and disease. Lipids. 2004;39:703–722. doi: 10.1007/s11745-004-1288-1. [DOI] [PubMed] [Google Scholar]
- 41.Bloch K., Berg B.N., Rittenberg D. The biological conversion of cholesterol to cholic acid. J. Biol. Chem. 1943;149:511–517. [Google Scholar]
- 42.Bergström S., Norman A. Metabolic products of cholesterol in bile and feces of rat. Steroids and bile acids. Proc. Soc. Exp. Biol. Med. 1953;83:71–74. doi: 10.3181/00379727-83-20269. [DOI] [PubMed] [Google Scholar]
- 43.Bergström S., Rottenberg M., Voltz J. The preparation of some carboxylabelled bile acids. Bile acids and steroids 2. Acta Chem. Scand. 1953;7:481–484. [Google Scholar]
- 44.Lindstedt S., Norman A. On the excretion of bile acid derivatives in feces of rats fed cholic acid-24-14C and chenodesoxycholic acid-24-14C. Bile acids and steroids 19. Acta Physiol. Scand. 1955;34:1–10. doi: 10.1111/j.1748-1716.1955.tb01219.x. [DOI] [PubMed] [Google Scholar]
- 45.Norman A., Grubb R. Hydrolysis of conjugated bile acids by clostridia and enterococci. Bile acids and steroids 25. Acta Pathol. Microbiol. Scand. 1955;36:537–547. doi: 10.1111/j.1699-0463.1955.tb04651.x. [DOI] [PubMed] [Google Scholar]
- 46.Norman A. Influence of chemotherapeutics on the metabolism of bile acids in the intestine of rats. Steroids and bile acids 17. Acta Physiol. Scand. 1955;33:99–107. doi: 10.1111/j.1748-1716.1955.tb01196.x. [DOI] [PubMed] [Google Scholar]
- 47.Norman A., Sjövall J. On the transformation and enterohepatic circulation of cholic acid in the rat. Bile acids and steroids 68. J. Biol. Chem. 1958;233:872–885. [PubMed] [Google Scholar]
- 48.Norman A., Sjövall J. Formation of lithocholic acid from chenodeoxycholic acid in the rat. Bile acids and steroids 103. Acta Chem. Scand. 1960;14:1815–1818. [Google Scholar]
- 49.Norman A., Bergman S. The action of intestinal microorganisms on bile acids. Bile acids and steroids 101. Acta Chem. Scand. 1960;14:1781–1789. [Google Scholar]
- 50.Norman A., Shorb M.S. In vitro formation of deoxycholic and lithocholic acid by human intestinal microorganisms. Proc. Soc. Exp. Biol. Med. 1962;110:552–555. doi: 10.3181/00379727-110-27577. [DOI] [PubMed] [Google Scholar]
- 51.Basic M., Bleich A. Gnotobiotics: past, present and future. Lab. Anim. 2019;53:232–243. doi: 10.1177/0023677219836715. [DOI] [PubMed] [Google Scholar]
- 52.Nencki M. Bemerkungen zu einer Bemerkung Pasteur's. Arch. Exp. Pathol. Pharmakol. 1886:385–388. [Google Scholar]
- 53.Pasteur L. Observations relatives à la note précédente de M. Duclaux. Compt. Rend. Acad. Sci. 1885;100:68. [Google Scholar]
- 54.Nuttall G.H.F., Thierfelder H. Thierisches Leben ohne Bakterien im Verdauungskanal. Hoppe Seyler's Z. Physiol. Chem. 1896;22:109–121. [Google Scholar]
- 55.Nuttall G.H.F., Thierfelder H. Thierisches Leben ohne Bakterien im Verdauungskanal. (II. Mittheilung) Hoppe Seyler's Z. Physiol. Chem. 1897;22:62–73. [Google Scholar]
- 56.Luckey T. Academic Press; New York: 1963. Germfree Life and Gnotobiology. [Google Scholar]
- 57.Gustafsson B. Germ-free rearing of rats. Acta Anat. (Basel) 1946;2:376–391. doi: 10.1159/000140222. [DOI] [PubMed] [Google Scholar]
- 58.Reyniers J.A., Trexler P.C., Ervin R.F. Rearing germ-free albino rats. Lobund Rep. 1946:1–84. [PubMed] [Google Scholar]
- 59.Gustafsson B.E., Bergström S., Lindstedt S., Norman A. Turnover and nature of fecal bile acids in germfree and infected rats fed cholic acid-24-14C. Bile acids and steroids 41. Proc. Soc. Exp. Biol. Med. 1957;94:467–471. doi: 10.3181/00379727-94-22981. [DOI] [PubMed] [Google Scholar]
- 60.Sayin S.I., Wahlstrom A., Felin J., Jantti S., Marschall H.U., Bamberg K., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Gustafsson B.E., Norman A. Comparison of bile acids in intestinal contents of germfree and conventional rats. Proc. Soc. Exp. Biol. Med. 1962;110:387–389. doi: 10.3181/00379727-110-27526. [DOI] [PubMed] [Google Scholar]
- 62.Gustafsson B.E., Midtvedt T., Norman A. Metabolism of cholic acid in germfree animals after the establishment in the intestinal tract of deconjugating and 7α-dehydroxylating bacteria. Acta Pathol. Microbiol. Scand. 1968;72:433–443. doi: 10.1111/j.1699-0463.1968.tb00457.x. [DOI] [PubMed] [Google Scholar]
- 63.Gustafsson B.E., Norman A., Sjövall J. Influence of E. coli infection on turnover and metabolism of cholic acid in germ-free rats. Arch. Biochem. Biophys. 1960;91:93–100. doi: 10.1016/0003-9861(60)90460-4. [DOI] [PubMed] [Google Scholar]
- 64.Yoshimoto T., Higashi H., Kanatani A., Lin X.S., Nagai H., Oyama H., et al. Cloning and sequencing of the 7α-hydroxysteroid dehydrogenase gene from Escherichia coli HB101 and characterization of the expressed enzyme. J. Bacteriol. 1991;173:2173–2179. doi: 10.1128/jb.173.7.2173-2179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka N., Nonaka T., Tanabe T., Yoshimoto T., Tsuru D., Mitsui Y. Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry. 1996;35:7715–7730. doi: 10.1021/bi951904d. [DOI] [PubMed] [Google Scholar]
- 66.Bergström S., Lindstedt S., Samuelsson B. Bile acids and steroids. LXXXII. On the mechanism of deoxycholic acid formation in the rabbit. J. Biol. Chem. 1959;234:2022–2025. [PubMed] [Google Scholar]
- 67.Lindstedt S., Samuelsson B. Bile acids and steroids. LXXXIII. On the inter-conversion of cholic and deoxycholic acid in the rat. J. Biol. Chem. 1959;234:2026–2030. [PubMed] [Google Scholar]
- 68.Hayakawa S. Microbiological transformation of bile acids. Adv. Lipid Res. 1973;11:143–192. doi: 10.1016/b978-0-12-024911-4.50011-8. [DOI] [PubMed] [Google Scholar]
- 69.Samuelsson B. Bile Acids and steroids. 96. On the mechanism of the biological formation of deoxycholic acid from cholic acid. J. Biol. Chem. 1960;235:361–366. [Google Scholar]
- 70.Ferrari A., Beretta L. Activity on bile acids of a Clostridium bifermentans cell-free extract. FEBS Lett. 1977;75:163–165. doi: 10.1016/0014-5793(77)80076-8. [DOI] [PubMed] [Google Scholar]
- 71.White B.A., Cacciapuoti A.F., Fricke R.J., Whitehead T.R., Mosbach E.H., Hylemon P.B. Cofactor requiremets for 7α-dehydroxylation of cholic and chenodeoxycholic acid in cell extracts of the intestinal anaerobic bacterium, Eubacterium species V.P.I. 12708. J. Lipid Res. 1981;22:891–898. [PubMed] [Google Scholar]
- 72.White B.A., Paone D.A., Cacciapuoti A.F., Fricke R.J., Mosbach E.H., Hylemon P.B. Regulation of bile acid 7-dehydroxylase activity by NAD+ and NADH in cell extracts of Eubacterium species V.P.I. 12708. J. Lipid Res. 1983;24:20–27. [PubMed] [Google Scholar]
- 73.Ferrari A., Scolatico C., Beretta L. On the mechanism of cholic acid 7α-dehydroxylation by a Clostridium bifermentans cell-free extract. FEBS Lett. 1977;75:166–168. doi: 10.1016/0014-5793(77)80077-x. [DOI] [PubMed] [Google Scholar]
- 74.Portman O.W., Shah S., Antonis A., Jorgensen B. Alteration of bile salts by bacteria. Proc. Soc. Exp. Biol. Med. 1962;109:959–965. doi: 10.3181/00379727-109-27391. [DOI] [PubMed] [Google Scholar]
- 75.Gustafsson B.E., Midtvedt T., Norman A. Isolated fecal microorganisms capable of 7α-dehydroxylating bile acids. J. Exp. Med. 1966;123:413–432. doi: 10.1084/jem.123.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Midtvedt T. Microbial bile acid transformation. Am. J. Clin. Nutr. 1974;27:1341–1347. doi: 10.1093/ajcn/27.11.1341. [DOI] [PubMed] [Google Scholar]
- 77.Bokkenheuser V., Hoshita T., Mosbach E.H. Bacterial 7-dehydroxylation of cholic acid and allocholic acid. J. Lipid Res. 1969;10:421–426. [PubMed] [Google Scholar]
- 78.Edenharder R. Dehydroxylation of cholic acid at C12 and epimerization at C5 and C7 by Bacteroides species. J. Steroid Biochem. 1984;21:413–420. doi: 10.1016/0022-4731(84)90304-2. [DOI] [PubMed] [Google Scholar]
- 79.Stellwag E.J., Hylemon P.B. 7α-dehydroxylation of cholic acid and chenodeoxycholic acid by Clostridium leptum. J. Lipid Res. 1979;20:325–333. [PubMed] [Google Scholar]
- 80.Lucas L.N., Barrett K., Kerby R.L., Zhang Q., Cattaneo L.E., Stevenson D., et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems. 2021 doi: 10.1128/mSystems.00805-21. [DOI] [PubMed] [Google Scholar]
- 81.Kitahara M., Takamine F., Imamura T., Benno Y. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7α-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2000;50:971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 82.Kitahara M., Takamine F., Imamura T., Benno Y. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7α-dehydroxylating activity. Int. J. Syst. Evol. Microbiol. 2001;51:39–44. doi: 10.1099/00207713-51-1-39. [DOI] [PubMed] [Google Scholar]
- 83.Winter J., Morris G.N., O'Rourke-Locascio S., Bokkenheuser V.D., Mosbach E.H., Cohen B.I., et al. Mode of action of steroid desmolase and reductases synthesized by Clostridium "scindens" (formerly Clostridium strain 19) J. Lipid Res. 1984;25:1124–1131. [PubMed] [Google Scholar]
- 84.Holdeman L., Cato E., Moore W. Virginia Polytechnic Institute and State University, Blacksburg; Blacksburg: 1977. Anaerobe Laboratory Manual. [Google Scholar]
- 85.Hylemon P.B., Cacciapuoti A.F., White B.A., Whitehead T.R., Fricke R.J. 7α-Dehydroxylation of cholic acid by cell extracts of Eubacterium species V.P.I. 12708. Am. J. Clin. Nutr. 1980;33:2507–2510. doi: 10.1093/ajcn/33.11.2507. [DOI] [PubMed] [Google Scholar]
- 86.Doerner K.C., Takamine F., LaVoie C.P., Mallonee D.H., Hylemon P.B. Assessment of fecal bacteria with bile acid 7α-dehydroxylating activity for the presence of bai-like genes. Appl. Environ. Microbiol. 1997;63:1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hirano S., Nakama R., Tamaki M., Masuda N., Oda H. Isolation and characterization of thirteen intestinal microorganisms capable of 7α-dehydroxylating bile acids. Appl. Environ. Microbiol. 1981;41:737–745. doi: 10.1128/aem.41.3.737-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirano S., Masuda N., Oda H., Imamura T. Transformation of bile acids by mixed microbial cultures from human feces and bile acid transforming activities of isolated bacterial strains. Microbiol. Immunol. 1981;25:271–282. doi: 10.1111/j.1348-0421.1981.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 89.Wylensek D., Hitch T.C.A., Riedel T., Afrizal A., Kumar N., Wortmann E., et al. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat. Commun. 2020;11:6389. doi: 10.1038/s41467-020-19929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Streidl T., Karkossa I., Segura Munoz R.R., Eberl C., Zaufel A., Plagge J., et al. The gut bacterium Extibacter muris produces secondary bile acids and influences liver physiology in gnotobiotic mice. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2020.1854008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wells J.E., Berr F., Thomas L.A., Dowling R.H., Hylemon P.B. Isolation and characterization of cholic acid 7α-dehydroxylating fecal bacteria from cholesterol gallstone patients. J. Hepatol. 2000;32:4–10. doi: 10.1016/s0168-8278(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 92.Ridlon J.M., Kang D.J., Hylemon P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 93.Ridlon J.M., Ikegawa S., Alves J.M., Zhou B., Kobayashi A., Iida T., et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 2013;54:2437–2449. doi: 10.1194/jlr.M038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Devendran S., Shrestha R., Alves J.M.P., Wolf P.G., Ly L., Hernandez A.G., et al. Clostridium scindens ATCC 35704: integration of nutritional requirements, the complete genome sequence, and global transcriptional responses to bile acids. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.00052-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morris G., Winter J., Cato E., Ritchie A., Bokkenheuser V. Clostridium scindens sp. nov., a human intestinal bacterium with desmolytic activity on corticoids. Int. J. Syst. Evol. Microbiol. 1985;35:478–481. [Google Scholar]
- 96.Bokkenheuser V.D., Morris G.N., Ritchie A.E., Holdeman L.V., Winter J. Biosynthesis of androgen from cortisol by a species of Clostridium recovered from human fecal flora. J. Infect. Dis. 1984;149:489–494. doi: 10.1093/infdis/149.4.489. [DOI] [PubMed] [Google Scholar]
- 97.Björkhem I. Five decades with oxysterols. Biochimie. 2013;95:448–454. doi: 10.1016/j.biochi.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 98.Axelson M., Björkhem I., Reihnér E., Einarsson K. The plasma level of 7α-hydroxy-4-cholesten-3-one reflects the activity of hepatic cholesterol 7α-hydroxylase in man. FEBS Lett. 1991;284:216–218. doi: 10.1016/0014-5793(91)80688-y. [DOI] [PubMed] [Google Scholar]
- 99.Skrede S., Björkhem I. Biosynthesis of cholestanol from intestinal 7α−hydroxy-4-cholesten-3-one. J. Biol. Chem. 1982;257:8363–8367. [PubMed] [Google Scholar]
- 100.Skrede S., Buchmann M.S., Björkhem I. Hepatic 7α-dehydroxylation of bile acid intermediates, and its significance for the pathogenesis of cerebrotendinous xanthomatosis. J. Lipid Res. 1988;29:157–164. [PubMed] [Google Scholar]
- 101.Björkhem I., Einarsson K., Melone P., Hylemon P. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: evidence for a 3-oxo-Δ4-steroid intermediate. J. Lipid Res. 1989;30:1033–1039. [PubMed] [Google Scholar]
- 102.Coleman J.P., White W.B., Egestad B., Sjövall J., Hylemon P.B. Biosynthesis of a novel bile acid nucleotide and mechanism of 7α−dehydroxylation by an intestinal Eubacterium species. J. Biol. Chem. 1987;262:4701–4707. [PubMed] [Google Scholar]
- 103.Kallner A. On the biosynthesis and metabolism of allodeoxycholic acid in the rat. Bile acids and steroids 175. Acta Chem. Scand. 1967;21:315–321. doi: 10.3891/acta.chem.scand.21-0315. [DOI] [PubMed] [Google Scholar]
- 104.Kallner A. The transformation of deoxycholic acid into allodeoxycholic acid in the rat. Bile acids and steroids.174. Acta Chem. Scand. 1967;21:87–92. doi: 10.3891/acta.chem.scand.21-0087. [DOI] [PubMed] [Google Scholar]
- 105.Stokes N.A., Hylemon P.B. Characterization of Δ4-3-ketosteroid-5β-reductase and 3β-hydroxysteroid dehydrogenase in cell extracts of Clostridium innocuum. Biochim. Biophys. Acta. 1985;836:255–261. doi: 10.1016/0005-2760(85)90073-6. [DOI] [PubMed] [Google Scholar]
- 106.Bokkenheuser V.D., Winter J., Cohen B.I., O'Rourke S., Mosbach E.H. Inactivation of contraceptive steroid hormones by human intestinal clostridia. J. Clin. Microbiol. 1983;18:500–504. doi: 10.1128/jcm.18.3.500-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sato Y., Atarashi K., Plichta D.R., Arai Y., Sasajima S., Kearney S.M., et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. 2021;599:458–464. doi: 10.1038/s41586-021-03832-5. [DOI] [PubMed] [Google Scholar]
- 108.Hylemon P.B., Melone P.D., Franklund C.V., Lund E., Björkhem I. Mechanism of intestinal 7α-dehydroxylation of cholic acid: evidence that allo-deoxycholic acid is an inducible side-product. J. Lipid Res. 1991;32:89–96. [PubMed] [Google Scholar]
- 109.White B.A., Lipsky R.L., Fricke R.J., Hylemon P.B. Bile acid induction specificity of 7α-dehydroxylase activity in an intestinal Eubacterium species. Steroids. 1980;35:103–109. doi: 10.1016/0039-128x(80)90115-4. [DOI] [PubMed] [Google Scholar]
- 110.Mallonee D.H., White W.B., Hylemon P.B. Cloning and sequencing of a bile acid-inducible operon from Eubacterium sp. strain VPI 12708. J. Bacteriol. 1990;172:7011–7019. doi: 10.1128/jb.172.12.7011-7019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ridlon J.M., Harris S.C., Bhowmik S., Kang D.J., Hylemon P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes. 2016;7:22–39. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Batta A.K., Salen G., Arora R., Shefer S., Batta M., Person A. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 1990;265:10925–10928. [PubMed] [Google Scholar]
- 113.Schmassmann A., Fehr H.F., Locher J., Lillienau J., Schteingart C.D., Rossi S.S., et al. Cholylsarcosine, a new bile acid analogue: metabolism and effect on biliary secretion in humans. Gastroenterology. 1993;104:1171–1181. doi: 10.1016/0016-5085(93)90289-o. [DOI] [PubMed] [Google Scholar]
- 114.Adhikari A.A., Ramachandran D., Chaudhari S.N., Powell C.E., Li W., McCurry M.D., et al. A gut-restricted lithocholic acid analog as an inhibitor of gut bacterial bile salt hydrolases. ACS Chem. Biol. 2021;16:1401–1412. doi: 10.1021/acschembio.1c00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Adhikari A.A., Seegar T.C.M., Ficarro S.B., McCurry M.D., Ramachandran D., Yao L., et al. Development of a covalent inhibitor of gut bacterial bile salt hydrolases. Nat. Chem. Biol. 2020;16:318–326. doi: 10.1038/s41589-020-0467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mallonee D.H., Hylemon P.B. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J. Bacteriol. 1996;178:7053–7058. doi: 10.1128/jb.178.24.7053-7058.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ridlon J.M., Hylemon P.B. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J. Lipid Res. 2012;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ye H.Q., Mallonee D.H., Wells J.E., Björkhem I., Hylemon P.B. The bile acid-inducible baiF gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A hydrolase. J. Lipid Res. 1999;40:17–23. [PubMed] [Google Scholar]
- 119.Mallonee D.H., Adams J.L., Hylemon P.B. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J. Bacteriol. 1992;174:2065–2071. doi: 10.1128/jb.174.7.2065-2071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harris S.C., Devendran S., Méndez-García C., Mythen S.M., Wright C.L., Fields C.J., et al. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243T. Gut Microbes. 2018;9:523–539. doi: 10.1080/19490976.2018.1458180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bhowmik S., Jones D.H., Chiu H.P., Park I.H., Chiu H.J., Axelrod H.L., et al. Structural and functional characterization of BaiA, an enzyme involved in secondary bile acid synthesis in human gut microbe. Proteins. 2014;82:216–229. doi: 10.1002/prot.24353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mallonee D.H., Lijewski M.A., Hylemon P.B. Expression in Escherichia coli and characterization of a bile acid-inducible 3α-hydroxysteroid dehydrogenase from Eubacterium sp. strain VPI 12708. Curr. Microbiol. 1995;30:259–263. doi: 10.1007/BF00295498. [DOI] [PubMed] [Google Scholar]
- 123.Kang D.J., Ridlon J.M., Moore D.R., II, Barnes S., Hylemon P.B. Clostridium scindens baiCD and baiH genes encode stereo-specific 7α/7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta. 2008;1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dawson J.A., Mallonee D.H., Björkhem I., Hylemon P.B. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 1996;37:1258–1267. [PubMed] [Google Scholar]
- 125.Bhowmik S., Chiu H.P., Jones D.H., Chiu H.J., Miller M.D., Xu Q., et al. Structure and functional characterization of a bile acid 7α−dehydratase BaiE in secondary bile acid synthesis. Proteins. 2016;84:316–331. doi: 10.1002/prot.24971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baron S.F., Hylemon P.B. Expression of the bile acid-inducible NADH:flavin oxidoreductase gene of Eubacterium sp. VPI 12708 in Escherichia coli. Biochim. Biophys. Acta. 1995;1249:145–154. doi: 10.1016/0167-4838(95)00034-r. [DOI] [PubMed] [Google Scholar]
- 127.Franklund C.V., Baron S.F., Hylemon P.B. Characterization of the baiH gene encoding a bile acid-inducible NADH:flavin oxidoreductase from Eubacterium sp. strain VPI 12708. J. Bacteriol. 1993;175:3002–3012. doi: 10.1128/jb.175.10.3002-3012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Funabashi M., Grove T.L., Wang M., Varma Y., McFadden M.E., Brown L.C., et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. 2020;582:566–570. doi: 10.1038/s41586-020-2396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harris S.C., Devendran S., Alves J.M.P., Mythen S.M., Hylemon P.B., Ridlon J.M. Identification of a gene encoding a flavoprotein involved in bile acid metabolism by the human gut bacterium Clostridium scindens ATCC 35704. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:276–283. doi: 10.1016/j.bbalip.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 130.Pollan M. Some of my best friends are germs. N. Y. Times Mag. 2013;15 [Google Scholar]
- 131.Feller F.M., Holert J., Yücel O., Philipp B. Degradation of bile acids by soil and water bacteria. Microorganisms. 2021;9:1759. doi: 10.3390/microorganisms9081759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hayakawa S., Samuelsson B. Transformation of cholic acid in vitro by Corynebacterium simplex. Bile acids and steroids. 132. J. Biol. Chem. 1964;239:94–97. [PubMed] [Google Scholar]
- 133.Bergström S., Samuelsson B. Isolation of prostaglandin E1 from human seminal plasma. Prostaglandins and related factors. 11. J. Biol. Chem. 1962;237:3005–3006. [PubMed] [Google Scholar]
- 134.Danielsson H., Kallner A., Sjövall J. On the composition of the bile acid fraction of rabbit feces and the isolation of a new bile acid: 3α, 12α-dihydroxy-5α-cholanic acid. Bile acids and steroids. 136. J. Biol. Chem. 1963;238:3840–3852. [PubMed] [Google Scholar]
- 135.Hofmann A.F., Mosbach E.H. Identification of allodeoxycholic acid as the major component of gallstones induced in the rabbit by 5α-cholestan-3β-ol. J. Biol. Chem. 1964;239:2813–2821. [PubMed] [Google Scholar]
- 136.Mosbach E.H., Bevans M. Formation of gall stones in rabbits fed 3β-cholestanol. Arch. Biochem. Biophys. 1956;63:258–259. doi: 10.1016/0003-9861(56)90030-3. [DOI] [PubMed] [Google Scholar]
- 137.Hofmann A.F., Bokkenheuser V., Hirsch R.L., Mosbach E.H. Experimental cholelithiasis in the rabbit induced by cholestanol feeding: effect of neomycin treatment on bile composition and gallstone formation. J. Lipid Res. 1968;9:244–253. [PubMed] [Google Scholar]
- 138.Lee J.W., Cowley E.S., Wolf P.G., Doden H.L., Murai T., Caicedo K.Y.O., et al. Formation of secondary allo-bile acids by novel enzymes from gut Firmicutes. Gut Microbes. 2022;14 doi: 10.1080/19490976.2022.2132903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim K.H., Park D., Jia B., Baek J.H., Hahn Y., Jeon C.O. Identification and characterization of major bile acid 7α-dehydroxylating bacteria in the human gut. mSystems. 2022;7 doi: 10.1128/msystems.00455-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Medvecky M., Cejkova D., Polansky O., Karasova D., Kubasova T., Cizek A., et al. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genomics. 2018;19:561. doi: 10.1186/s12864-018-4959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Islam K.B., Fukiya S., Hagio M., Fujii N., Ishizuka S., Ooka T., et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 142.Hofmann A.F., Hagey L.R., Krasowski M.D. Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ridlon J.M., Bajaj J.S. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm. Sin. B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A., et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Studer N., Desharnais L., Beutler M., Brugiroux S., Terrazos M.A., Menin L., et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell. Infect. Microbiol. 2016;6:191. doi: 10.3389/fcimb.2016.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marion S., Desharnais L., Studer N., Dong Y., Notter M.D., Poudel S., et al. Biogeography of microbial bile acid transformations along the murine gut. J. Lipid Res. 2020;61:1450–1463. doi: 10.1194/jlr.RA120001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mark Welch J.L., Hasegawa Y., McNulty N.P., Gordon J.I., Borisy G.G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reed A.D., Nethery M.A., Stewart A., Barrangou R., Theriot C.M. Strain-dependent inhibition of Clostridioides difficile by commensal clostridia carrying the bile acid-inducible (bai) operon. J. Bacteriol. 2020;202 doi: 10.1128/JB.00039-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Francis M.B., Allen C.A., Sorg J.A. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Eyssen H.J., De Pauw G., Van Eldere J. Formation of hyodeoxycholic acid from muricholic acid and hyocholic acid by an unidentified gram-positive rod termed HDCA-1 isolated from rat intestinal microflora. Appl. Environ. Microbiol. 1999;65:3158–3163. doi: 10.1128/aem.65.7.3158-3163.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tawthep S., Fukiya S., Lee J.Y., Hagio M., Ogura Y., Hayashi T., et al. Isolation of six novel 7-oxo- or urso-type secondary bile acid-producing bacteria from rat cecal contents. J. Biosci. Bioeng. 2017;124:514–522. doi: 10.1016/j.jbiosc.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 152.Kang J.D., Myers C.J., Harris S.C., Kakiyama G., Lee I.K., Yun B.S., et al. Bile acid 7α-dehydroxylating gut bacteria secrete antibiotics that inhibit Clostridium difficile: role of secondary bile acids. Cell Chem. Biol. 2019;26:27–34.e4. doi: 10.1016/j.chembiol.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Savidge T., Sorg J.A. Role of bile in infectious disease: the gall of 7α-dehydroxylating gut bacteria. Cell Chem. Biol. 2019;26:1–3. doi: 10.1016/j.chembiol.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hayakawa S., Hattori T. 7α-dehydroxylation of cholic acid by Clostridium bifermentans strain ATCC 9714 and Clostridium sordellii strain NCIB 6929. FEBS Lett. 1970;6:131–133. doi: 10.1016/0014-5793(70)80020-5. [DOI] [PubMed] [Google Scholar]
- 155.Wostmann B.S., Beaver-Johnson M., Wagner M. Lack of 7α-dehydroxylation in gnotobiotic gerbils associated with an octaflora including Clostridium sordellii. Prog. Clin. Biol. Res. 1985;181:107–110. [PubMed] [Google Scholar]
- 156.Narushima S., Itoh K., Takamine F., Uchida K. Absence of cecal secondary bile acids in gnotobiotic mice associated with two human intestinal bacteria with the ability to dehydroxylate bile acids in vitro. Microbiol. Immunol. 1999;43:893–897. doi: 10.1111/j.1348-0421.1999.tb01224.x. [DOI] [PubMed] [Google Scholar]
- 157.Narushima S., Itoha K., Miyamoto Y., Park S.H., Nagata K., Kuruma K., et al. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41:835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- 158.Stellwag E.J., Hylemon P.B. Characterization of 7α-dehydroxylase in Clostridium leptum. Am. J. Clin. Nutr. 1978;31:S243–S247. doi: 10.1093/ajcn/31.10.S243. [DOI] [PubMed] [Google Scholar]
- 159.Pascal Andreu V., Fischbach M.A., Medema M.H. Computational genomic discovery of diverse gene clusters harbouring Fe-S flavoenzymes in anaerobic gut microbiota. Microb. Genom. 2020;6 doi: 10.1099/mgen.0.000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Devlin A.S., Fischbach M.A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 2015;11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mythen S.M., Devendran S., Méndez-García C., Cann I., Ridlon J.M. Targeted synthesis and characterization of a gene cluster encoding NAD(P)H-dependent 3α-, 3β-, and 12α-hydroxysteroid dehydrogenases from Eggerthella CAG:298, a gut metagenomic sequence. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02475-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guo C.J., Allen B.M., Hiam K.J., Dodd D., Van Treuren W., Higginbottom S., et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science. 2019;366 doi: 10.1126/science.aav1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jin W.B., Li T.T., Huo D., Qu S., Li X.V., Arifuzzaman M., et al. Genetic manipulation of gut microbes enables single-gene interrogation in a complex microbiome. Cell. 2022;185:547–562.e22. doi: 10.1016/j.cell.2021.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Drake H.L. In: Acetogenesis. Drake H.L., editor. Springer US; Boston, MA: 1994. Acetogenesis, acetogenic bacteria, and the acetyl-CoA “Wood/Ljungdahl” pathway: past and current perspectives; pp. 3–60. [Google Scholar]
- 165.Thauer R.K. The Wolfe cycle comes full circle. Proc. Natl. Acad. Sci. U. S. A. 2012;109:15084–15085. doi: 10.1073/pnas.1213193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Balch W.E., Schoberth S., Tanner R.S., Wolfe R.S. Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int. J. Syst. Evol. Microbiol. 1977;27:355–361. [Google Scholar]
- 167.Tanner R.S., Miller L.M., Yang D. Clostridium ljungdahlii sp. nov., an acetogenic species in clostridial rRNA homology group I. Int. J. Syst. Evol. Microbiol. 1993;43:232–236. doi: 10.1099/00207713-43-2-232. [DOI] [PubMed] [Google Scholar]
- 168.Wasserfallen A., Nölling J., Pfister P., Reeve J., De Macario E.C. Phylogenetic analysis of 18 thermophilic Methanobacterium isolates supports the proposals to create a new genus, Methanothermobacter gen. nov., and to reclassify several isolates in three species, Methanothermobacter thermautotrophicus comb. nov., Methanothermobacter wolfeii comb. nov., and Methanothermobacter marburgensis sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:43–53. doi: 10.1099/00207713-50-1-43. [DOI] [PubMed] [Google Scholar]
- 169.Winter J., Lerp C., Zabel H.-P., Wildenauer F., König H., Schindler F. Methanobacterium wolfei, sp. nov., a new tungsten-requiring, thermophilic, autotrophic methanogen. Syst. Appl. Microbiol. 1984;5:457–466. [Google Scholar]
- 170.Spring S., Jackel U., Wagner M., Kampfer P. Ottowia thiooxydans gen. nov., sp. nov., a novel facultatively anaerobic, N2O-producing bacterium isolated from activated sludge, and transfer of Aquaspirillum gracile to Hylemonella gracilis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2004;54:99–106. doi: 10.1099/ijs.0.02727-0. [DOI] [PubMed] [Google Scholar]
- 171.Hylemon P.B., Harris S.C., Ridlon J.M. Metabolism of hydrogen gases and bile acids in the gut microbiome. FEBS Lett. 2018;592:2070–2082. doi: 10.1002/1873-3468.13064. [DOI] [PubMed] [Google Scholar]