Abstract
The adeno-associated virus type 2 (AAV-2) Rep78/Rep68 regulatory proteins are pleiotropic effectors of viral and cellular DNA replication, of cellular transformation by viral and cellular oncogenes, and of homologous and heterologous gene expression. To search for cellular proteins involved in mediating these functions, we used Rep68 as bait in the yeast two-hybrid system and identified the transcriptional coactivator PC4 as a Rep interaction partner. PC4 has been shown to mediate transcriptional activation by a variety of sequence-specific transcription factors in vitro. Rep amino acids 172 to 530 were sufficient and amino acids 172 to 224 were absolutely necessary for the interaction with PC4. The PC4 domains required for interaction were mapped to the C-terminal single-stranded DNA-binding domain of PC4. In glutathione S-transferase (GST) pull-down assays, in vitro-transcribed and -translated Rep78 or Rep68 proteins were bound specifically by GST-PC4 fusion proteins. Similarly, PC4 expressed in Escherichia coli was bound by GST-Rep fusion proteins, confirming the direct interaction between Rep and PC4 in vitro. Rep was found to have a higher affinity for the nonphosphorylated, transcriptionally active form of PC4 than for the phosphorylated, transcriptionally inactive form. The latter is predominant in nuclear extracts of HeLa or 293 cells. In the yeast system, but not in vitro, Rep-PC4 interaction was disrupted by a point mutation in the putative nucleotide-binding site of Rep68, suggesting that a stable interaction between Rep and PC4 in vivo is ATP dependent. This mutation has also been shown to impair Rep function in AAV-2 DNA replication and in inhibition of gene expression and inducible DNA amplification. Cytomegalovirus promoter-driven overexpression of PC4 led to transient accumulation of nonphosphorylated PC4 with concomitant downregulation of all three AAV-2 promoters in the absence of helper virus. In the presence of adenovirus, this effect was relieved. These results imply an involvement of the transcriptional coactivator PC4 in the regulation of AAV-2 gene expression in the absence of helper virus.
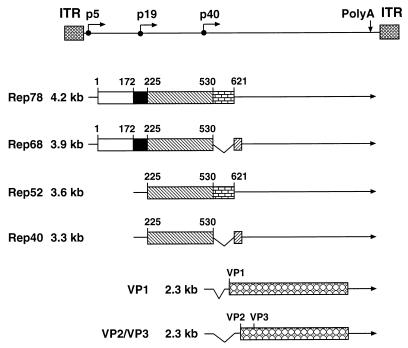
Adeno-associated virus type 2 (AAV-2) is a human parvovirus that requires coinfection with a helper virus, for example, adenovirus (3) or herpesvirus (10), for efficient DNA replication (8) and gene expression (11). However, a low level of helper-independent replication can also be detected in cells exposed to genotoxic agents (63–65). AAV-2 contains a linear single-stranded DNA genome of 4.7 kb (11, 55) with two open reading frames (ORFs) flanked by two 145-bp inverted terminal repeats (ITRs). The ORF in the right half of the AAV genome encodes the three structural proteins VP1, VP2, and VP3, while the ORF in the left half of the genome encodes four overlapping nonstructural proteins, termed Rep proteins. Rep78 and Rep68, a C-terminally spliced version of Rep78, regulate many steps in the AAV-2 life cycle, including DNA replication (23, 56), gene expression (41, 57), and site-specific integration (4, 53, 60). Rep78 and Rep68 possess the following activities: DNA-binding, site- and strand-specific endonuclease, helicase, and ATPase activities, all of which are required for AAV-2 DNA replication (32, 33, 62). The ITR DNA contains a Rep78 and a Rep68 binding site (ITR-RBS) in addition to the so-called terminal resolution site, which is nicked in a site- and strand-specific manner by Rep78 and/or Rep68 (32) in the course of AAV-2 DNA replication. Rep52 and Rep40 are N-terminally truncated forms of Rep78 and Rep68, respectively, that so far have only been implicated in the accumulation of single-stranded AAV-2 DNA (12). In analogy to Rep78 and Rep68, ATPase and helicase activities have recently also been demonstrated for Rep52 (54).
The role of Rep78 and Rep68 in the regulation of AAV-2 gene expression is strongly dependent on the presence or absence of a helper virus. In the absence of helper virus, AAV-2 efficiently integrates into the host chromosomal DNA to establish a latent infection. The low level of AAV-2 gene expression during latency (44) can be explained by the ability of Rep78 and/or Rep68 to negatively regulate the three AAV-2 promoters, p5, p19, and p40, in the absence of helper virus (7, 31, 39, 58). The downregulation of the p5 promoter is mediated by the Rep binding site (p5RBS) in the p5 promoter (40, 49). In the presence of helper virus, both the p19 and p40 promoters are activated by Rep78 and/or Rep68, a process which is dependent upon the ITR-RBS (49, 59), the p5RBS, and additional elements in the p19 and p40 promoters (46, 50, 51). Among these additional elements are binding sites for the cellular transcription factor Sp1 (50, 51), which has been shown to interact directly with Rep78 and Rep68 (29, 50). The p5 promoter is both activated by Rep78 and Rep68 in the presence of adenovirus mediated by the ITR-RBS and repressed by Rep78 and Rep68 mediated by the p5RBS (49).
The Rep proteins have a variety of activities outside the AAV-2 life cycle. The Rep78 and Rep68 proteins have been shown to repress viral and cellular promoters (2, 5, 25, 27, 28, 31, 42), including the H-ras, c-fos, c-myc oncogene promoters, the human immunodeficiency virus (HIV) type 1 long terminal repeat (LTR), and the human papillomavirus type 16 and 18 (HPV18) upstream regulatory regions (URRs). The Rep78 and Rep68 proteins inhibit cellular DNA replication (68), herpes simplex virus (HSV)-induced amplification of chromosomally integrated simian virus 40 (SV40) DNA (22), and bovine papillomavirus DNA amplification (26). Furthermore, the Rep78 and Rep68 proteins suppress cellular transformation by bovine papillomavirus DNA (24), adenovirus (17), or the E1A/EJras oncogene combination (35). An inhibition of cell proliferation leading to the accumulation of cells in the G1/S phase of the cell cycle has been reported upon infection with purified AAV-2 virions (61) and upon induction of Rep78 expression (67).
To date, relevant targets for these diverse activities of Rep78/Rep68 have been identified only partially. The inhibition of cellular transformation by E1A/EJras correlates with the inhibition of the corresponding oncogene promoters (20, 25). Specific cis-regulatory elements binding the AAV-2 Rep78 and Rep68 proteins have been described for the AAV-2 p5 promoter, the HIV LTR, and the H-ras promoter (5, 6, 40, 47). In addition, Rep78 interacts with transcription factor Sp1 to exert its inhibitory effect on the H-ras promoter (29). In contrast, mutational analysis of the HPV18 URR revealed that the cis elements involved in Rep-mediated inhibition are redundant, which suggests that Rep interacts with either some general mediator of sequence-specific transcription factors or a component of the basal transcription machinery (31).
In this report, we identify the transcriptional coactivator PC4 as an interaction partner of the large Rep proteins Rep78 and Rep68. PC4 was identified by virtue of its ability to activate transcription in the presence of a variety of sequence-specific transcription factors in a reconstituted transcription system (18, 37). The interaction of Rep with PC4 establishes a link between the transcriptional effects of Rep78/Rep68 and the general transcription machinery. Furthermore, concurrence of Rep domains needed for interaction with PC4 in vivo with those described to be important for Rep-mediated inhibition of HSV-induced SV40 DNA amplification and inhibition of cellular transformation by E1A/EJras suggests a direct involvement of Rep-PC4 interaction in these inhibitory processes.
MATERIALS AND METHODS
Yeast two-hybrid screen.
A yeast two-hybrid screening system was used to isolate cDNAs encoding proteins that are able to interact with the AAV-2 regulatory protein Rep68. Yeast strain HF7c (Clontech) was transformed sequentially by using the lithium acetate method with the bait plasmid pGBT-Rep68, which encodes the Gal4 DNA-binding domain fused in frame to Rep68 and a cDNA library from noninfected HeLa cells fused to the Gal4 transactivation domain in plasmid pGAD424 (Clontech). The cDNA had been generated by the method of Gubler and Hoffman (21) (Stratagene cDNA synthesis kit) and had been ligated to EcoRI/SalI-digested pGAD424 plasmid via EcoRI/XhoI linkers (Stratagene cDNA synthesis kit). Double transformants were selected on SD medium (described by Clontech in the manual for the Matchmaker two-hybrid system) lacking tryptophan, leucine, and histidine and assayed for β-galactosidase activity by filter assays (as described by Clontech). Positive colonies were restreaked on the same medium and assayed again for β-galactosidase activity. Colonies confirmed positive were grown in SD medium lacking leucine, and library plasmids were isolated and transformed into electrocompetent Escherichia coli HB101. Bacterial transformants were selected on M9 minimal medium (2 mM MgSO4, 0.1 mM CaCl2, 12.8 g of Na2HPO4 · 7H2O per liter, 3 g of KH2PO4 per liter, 0.5 g of NaCl per liter, 1.0 g of NH4Cl per liter) containing ampicillin (100 μg/ml), proline (40 μg/ml), 1 mM thiamine, 0.4% glucose, and an amino acid mixture medium (described by Clontech in the manual for the matchmaker two-hybrid system) lacking leucine. Library plasmids were repurified from bacterial transformants and retransformed into HF7c yeast cells together with pGBT9, pGBT9-Rep68, and pLAM5 encoding a human lamin C (Clontech). Yeast transformants were tested for growth on SD medium lacking histidine and for β-galactosidase activity to eliminate false positives.
Production and purification of GST fusion proteins.
Cultures of E. coli M15 transformed with the plasmids encoding glutathione S-transferase (GST) fusion proteins were grown at 30°C to an optical density at 600 nm of 0.6 to 0.8. Production of GST fusion proteins was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 30°C. Cells were harvested by centrifugation and lysed by sonication in lysis buffer (50 mM phosphate [pH 7.8], 300 mM NaCl, 1% [vol/vol] Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride). Cell debris was removed by centrifugation, and the supernatant was adsorbed to glutathione-Sepharose beads (Pharmacia). Glutathione-Sepharose beads with bound GST fusion proteins were collected by centrifugation and washed four times with lysis buffer.
GST pull-down assays.
35S-labeled Rep78 and Rep52 were obtained by coupled in vitro transcription-translation (Promega) of pBS-Rep78 and pBS-Rep52 in rabbit reticulocyte lysates with T7 RNA polymerase in the presence of [35S]methionine. Equal amounts of labeled proteins were incubated with 5 μg of either GST alone or GST-PC4 fusion proteins bound to 20 μl of glutathione-Sepharose beads (Pharmacia) for 1 h at 4°C in 1 ml of buffer A20 (20 mM HEPES NaOH [pH 7.9], 10% glycerol, 1 mM EDTA, 10 mM MgCl2, 4 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 20 mM KCl) containing 1% Triton X-100. The beads were washed five times in 1 ml of buffer A20 containing 1% Triton X-100, boiled in sodium dodecyl sulfate (SDS) sample buffer (43), and loaded on a 15% polyacrylamide gel (SDS-polyacrylamide gel electrophoresis [PAGE]). 35S-labeled Rep78 and Rep52 were visualized by autoradiography.
Recombinant PC4 proteins used in the GST pull-down assays were purified from the corresponding GST-PC4 fusion proteins. One hundred micrograms of GST-PC4 fusion proteins bound to glutathione-Sepharose beads were incubated for 16 h with 10 U of thrombin protease (Pharmacia) in 50 mM KCl–10 mM MgCl2–20 mM Tris-HCl (pH 7.5). The PC4 released into the supernatant was separated from the glutathione-bound GST moiety by centrifugation and bound to 10 mg of single-stranded DNA-cellulose (Sigma) for 2 h at 4°C. Single-stranded DNA cellulose-bound PC4 was recovered by centrifugation, washed three times with buffer A100 (buffer A containing 100 mM KCl), and eluted in 200 μl of buffer A1000 (buffer A containing 1,000 mM KCl). To generate the phosphorylated form of PC4, 15 μg of PC4 was incubated after thrombin cleavage with 500 U of recombinant casein kinase II (Calbiochem) prior to incubation with single-stranded DNA-cellulose.
For the GST pull-down assays, 1 μg of purified PC4 proteins was incubated for 1 h at 4°C with 10 μg of purified GST alone or GST-Rep fusion proteins bound to 40 μl of glutathione beads in a total of 1 ml of buffer A20. The beads were washed five times with buffer A20 and boiled in 60 μl of SDS sample buffer. Ten microliters of each binding assay mixture was then analyzed for PC4 content by SDS–15% PAGE and immunoblotting.
Generation of polyclonal PC4 antiserum.
His-tagged PC4 (pQE31-PC4) was expressed in E. coli and purified over an Ni-nitrilotriacetic acid column under denaturing conditions in the presence of 8 M urea as described elsewhere (Qiagen manual). Purified PC4 was precipitated with acetone and resuspended in phosphate-buffered saline at a concentration of 1 mg/ml. A rabbit was immunized with 200 μg of PC4 mixed 1:1 with complete Freund’s adjuvant followed by two boosts after 3 and 6 weeks with 200 μg of PC4 mixed 1:1 with incomplete Freund’s adjuvant.
Western blot analysis.
Nuclear extracts were essentially prepared as described previously (1). Protein samples were analyzed on SDS–15% PAGE gels (43). Proteins were electrophoretically transferred to nitrocellulose membranes (transfer buffer containing 20% methanol, 25 mM Tris-Cl, and 192 mM glycine). PC4 proteins were detected with the polyclonal rabbit anti-PC4 antiserum (1:500 dilution) and a peroxidase-coupled secondary antibody and by using enhanced chemoluminescence detection (Amersham) as described by the supplier.
Cell culture, virus infection, and transfection.
HeLa cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum, 100 μg of penicillin and streptomycin per ml, and 2 mM glutamine at 37°C under 5% CO2. Transfection of noninfected and adenovirus type 2-infected HeLa cells were performed by the protocol of Chen and Okayama (13, 14) in a slightly modified form. Briefly, the day before infection or transfection, 6 × 105 HeLa cells in 10 ml of medium were seeded into 10-cm-diameter culture dishes and incubated at 37°C under 5% CO2. For adenovirus type 2 infection, the medium was removed and the cells were incubated for 2 h with adenovirus type 2 (multiplicity of infection, 10) in a total volume of 1,000 μl. After the 2-h incubation period, 9 ml of DMEM was added and the cells were incubated for an additional 1 h prior to transfection. For transfection, 12 μg of DNA was mixed with 450 μl of 280 mM CaCl2 and 450 μl of 2× BES and incubated for 15 min at room temperature. The transfection mixture was added to the cells, and cells were incubated for 16 h at 37°C under 5% CO2. After removal of the supernatant, the cells were washed once with serum-free medium and then incubated in medium containing 10% fetal calf serum for 8 h at 37°C under 5% CO2.
Northern blot analysis.
RNA from 3 × 106 cells was isolated in accordance with published protocols (15) by guanidine isothiocyanate lysis. Equal amounts of RNA, based on the measurement of optical density at 260 nm, were electrophoresed on a 1% agarose-formaldehyde gel (50 mM HEPES [pH 7.8], 1 mM EDTA, 6% formaldehyde) and transferred to a nylon membrane (GeneScreen; Du Pont NEN) by capillary blotting in 25 mM phosphate buffer (pH 6.8) overnight. For detection of AAV-2 transcripts, a 1.6-kb HindII fragment of pTAV2-0 (22) was labeled by random priming. The filters were hybridized in hybridization solution (7% SDS [wt/vol], 0.125 M sodium phosphate buffer [pH 7.2], 0.25 M NaCl, 1 mM EDTA, 45% [vol/vol] formamide) at 42°C for 16 to 30 h. The filters were washed four times in 2× SSC–0.1% SDS at 42°C for 5 min and subsequently two times with 0.1× SSC–0.1% SDS at 65°C for 30 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Filters were air dried and autoradiographed at room temperature on Fuji RX films.
Plasmids.
The Gal4 DNA-binding domain–Rep fusion plasmids were cloned as follows. Plasmid pGBT9 (Clontech) encoding the Gal4 DNA-binding domain was cut with SalI and PstI and ligated to a SalI/PstI fragment from pHIV-LTR-OVEC (31) containing an EcoRV site derived from Bluescript SK2 adjacent to the PstI site. The resulting plasmid was digested with SalI and partially digested with EcoRV to excise only the pHIV LTR-OVEC-derived insert. The resulting vector was ligated with a Rep78 encoding a XhoI/SmaI fragment from pKEXRep78 (31) to obtain pGBT9-Rep78. pGBT9-Rep78 was cut with NotI and partially cut with XbaI and ligated to the Rep encoding NotI/XbaI fragments from pKEXRep68, pKEXRep52, pKEXRep40, and pKEXStop531 (31) to generate the corresponding pGBT9-Rep fusion constructs. pGBT9-M172/530 was constructed by replacing the N-terminal NotI/SalI fragment from pGBT9-Stop531 with the corresponding fragment from pKEXM172 (36). pGBT9-M172/243 was generated by complete BamHI digestion of pGBT-M172/530 with BamHI and religation of the 250-bp insert encoding Rep amino acids 172 to 243 with the BamHI vector fragment. pGBT9-Rep68K340H was constructed by replacing the amino-terminal NotI/SalI Rep fragment of pGBT-Rep68 with the corresponding fragment from pKEXRep78K340H (31). pGBT9-M172/530K340H was constructed by replacing the internal BamHI/SalI Rep fragment from pGBT9-M172/530 with the corresponding fragment from pKEXRep78K340H.
The pGAD424-PC4 C-terminal deletion mutants were all derived from one of the original pGAD424-PC4 clones obtained in the initial two-hybrid screen with pGBT9-Rep68 as a bait plasmid. pGAD424-PC4 was digested with EcoRI/HindII, EcoRI/DraI, or EcoRI/BamHI, and the corresponding N-terminal PC4 fragments were ligated to an EcoRI/SmaI- or EcoRI/BamHI-digested pGAD424 vector to obtain pGBT9-PC4(1-22), pGBT9-PC4(1-77), and pGBT9-PC4(1-91), respectively. pGAD424-PC4(62-127) and pGAD424-PC4(82-127) were generated through PCR amplification of the corresponding sequences from pGAD424-PC4 with the N-terminal primer TAACATGTTTCAGATT or GCTAATTGATATTAGA, respectively, and the C-terminal primer CAACTAGAACAGTACA, which binds in the 3′ nontranslated region of PC4, and ligation of the PCR products to a SmaI-digested pGAD424 vector.
pGEX-PC4 was constructed by PCR amplification of the PC4 coding region from pGAD424-PC4 with the primer pair CATGCCTAAATCAAAG–CAACTAGAACAGTACA and blunt end ligation to a BamHI-blunted/SmaI pGEX-4T3 vector (Pharmacia). For pGEX-PC4(62-127) and pGEX-PC4(82-127), the corresponding pGAD424-PC4 EcoRI/PstI fragments were subcloned first into Bluescript SK2, excised from Bluescript with SalI/NotI, and subcloned into pGEX-4T3.
The N-terminal part of Rep68 in pGEX-Rep68 was amplified from pKEXRep68 with the primers GGCGGAATTCCATGCCGGGGTTTTAC, generating an EcoRI site at the N terminus of Rep, and AGTCGCGCTGCAGCTTCTC, binding at the first internal PstI site of Rep68. The PCR product was cloned into Bluescript SK2 through EcoRI/PstI digestion, excised from Bluescript with EcoRI/NotI, and subcloned into pGEX-4T3 to generate pGEX-RepN. pGEX-RepN was digested with XbaI, partially digested with PstI, and ligated to a PstI/XbaI fragment derived from pKEXRep68 containing the rest of the Rep68 coding frame. pGEX-M172/530 and pGEXRep40 were constructed from the corresponding pKEX plasmids by ligation of the NcoI-blunted/XbaI Rep fragments to BamHI-blunted/XbaI-digested pGEX-RepN. To generate pGEX-Rep68K340H and pGEX-M172/530K340H the NcoI/XbaI Rep sequences from pGBT9-Rep68K340H and pGBT9-M172/530K340H, respectively, were subcloned into NcoI/XbaI-digested pGEX-M172/530.
For construction of the bacterial His-tagged PC4 expression vector pQE31-PC4, PC4 sequences were amplified from pGAD424-PC4 with the primers GCGCGGATCCTAAATCAAAGGAACTT, generating a BamHI site, and CGCGCTGCAGGAATTTTACAGTTTTCTT, generating a PstI site. The PCR product was cut with PstI, partially cut with BamHI, and ligated to a BamHI/PstI-digested vector, pQE31 (Qiagen).
pBS-Rep78 and pBS-Rep52 were constructed by subcloning the corresponding XhoI/XbaI fragments of pKEXRep78 and pKEXRep52, respectively, into Bluescript SK2 (Stratagene).
For generation of pKEX-PC4, an EcoRI/PstI PC4 fragment of pGAD424-PC4 was subcloned into Bluescript SK2, excised with XhoI/XbaI, and subcloned into pKEX-XL (52).
RESULTS
Rep68 interacts with transcriptional coactivator PC4 in a yeast two-hybrid system.
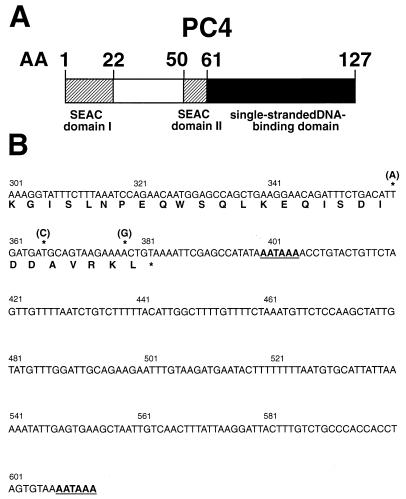
To select suitable Rep bait proteins for the yeast two-hybrid screening of an HeLa cDNA library for Rep-interacting proteins, we first determined which Rep domains (Fig. 1) activate transcription in yeast on their own when fused to the Gal4 DNA-binding domain. The authentic Rep proteins Rep78 and Rep52 and all mutant Rep proteins containing Rep amino acids 531 to 621 encoded by the major intron of AAV-2 (Fig. 1) showed intrinsic transactivation effects of reporter gene expression and hence were not suitable as bait proteins (data not shown). For this reason, and because Rep68 and Rep78 share most of their biological and enzymatic activities, initial two-hybrid screens were performed with a Gal4 DNA-binding domain–Rep68 fusion construct (pGBT9-Rep68). Yeast strain HF7c, which contains two Gal4-inducible reporter genes, HIS3 and lacZ, was sequentially cotransformed with pGBT9-Rep68 and an HeLa cDNA library (106 independent clones) fused to the Gal4 transactivation domain in plasmid pGAD424. In two independent screens with 2 × 106 and 1 × 107 transformants, respectively, a total of 24 clones capable of growing on plates lacking histidine and staining positive for β-galactosidase expression were obtained. The coding region of nine of these clones was found to correspond to the transcriptional coactivator PC4 (Fig. 2A) recently described independently by Ge and Roeder (18) and Kretzschmar et al. (37). PC4 enhances transcriptional activation by different sequence-specific transcription factors in a reconstituted system with purified general transcription factors (18, 37). Compared to the published sequence (18, 37), three nucleotide exchanges in the very 3′ part of PC4, neither of which affected the predicted amino acid sequence of PC4, were found in all clones (Fig. 2B). In the 3′ nontranslated region of PC4, two polyadenylation signals spaced 200 nucleotides apart were identified (Fig. 2B). As a specificity control, the pGAD424-PC4 clones were separately retransformed into yeast strain HF7c together with pGBT9, pGBT9-Rep68, or a pGBT9-lamin fusion plasmid. Only cotransformation of pGAD424-PC4 and pGBT9-Rep68 gave rise to colonies capable of growing on plates lacking histidine and staining positive for β-galactosidase expression, confirming PC4 as a true positive Rep interaction partner in the two-hybrid system.
FIG. 1.
Genome organization, transcripts, and protein products of AAV-2. The viral genome is shown in the upper part of the figure. The ITRs are represented by hatched boxes; the three promoters at map units 5, 19, and 40 are indicated by right-angled arrows; and the common polyadenylation (polyA) site for all transcripts at map position 96 is indicated by a vertical arrow. The transcripts encoding the regulatory (Rep) proteins and the structural (VP) proteins of AAV-2 are shown in the lower part of the figure together with the corresponding protein products. Untranslated regions of the transcripts are indicated by solid lines, introns are shown as carats, while the coding regions are represented by boxes. Open, closed, and different shaded boxes indicate various domains of Rep: the N-terminal domain (amino acids 1 to 172) involved in sequence-specific DNA binding, the amino acids 172 to 225 important for Rep-mediated inhibition of cell transformation and DNA amplification, the central domain (amino acids 225 to 530) common to all four Rep proteins, the major intron of AAV-2 (amino acids 531 to 621), and the 7 C-terminal amino acids specific for Rep68/Rep40. Characteristic amino acid positions are given above the boxes.
FIG. 2.
(A) Schematic representation of the transcriptional coactivator PC4. The two SEAC domains are indicated by hatched boxes, while the C-terminal ssDBD is represented by a solid box. Amino acid (AA) positions are given above the boxes. (B) Nucleotide sequence of the 3′ translated and untranslated region of PC4 with the deduced C-terminal amino acid sequence. The numbering of nucleotides is in accordance with that described in reference 18. The three nucleotide exchanges found in comparison to the published sequence which have no effect on the amino acid sequence are marked by asterisks. In the region of the nucleotide exchanges, the published sequence (18, 37) contains the C-terminal PCR primers corresponding to the sequence of the mouse homolog of human PC4, which were used for the amplification and cloning of the human PC4 sequence. The two polyadenylation signals (sequence AATAAA) are underlined.
Rep-PC4 interaction in yeast requires Rep amino acids 172 to 224 and the region comprising the putative Rep nucleotide-binding site.
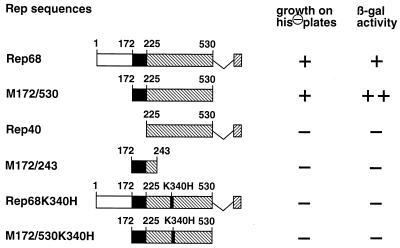
To identify Rep domains involved in Rep-PC4 interaction, a series of pGBT9-Rep fusion plasmids with deletions within the Rep68 coding region were tested in cotransformations with full-length pGAD424-PC4 in the yeast two-hybrid system. Simultaneous deletion of the 7 C-terminal and 171 N-terminal amino acids of Rep68 (M172/530) (Fig. 3) did not abolish interaction with PC4 in yeast (Fig. 3). In contrast, the β-galactosidase activity of M172/530 cotransformants was higher than that of Rep68 cotransformants. This result was confirmed in additional two-hybrid screens with pGBT9-M172/530 as a bait plasmid, where 196 of 200 selected positives were found to correspond to PC4 by colony hybridization (data not shown). Further deletion of 53 N-terminal amino acids in Rep40, however, totally abolished interaction with PC4 in the yeast two-hybrid system (Fig. 3). Apparently, Rep amino acids 172 to 224 are necessary for Rep-PC4 interaction but they are not sufficient for interaction if expressed as a Gal4 DNA-binding domain fusion protein (M172/243) (Fig. 3). This result can be explained either by the incorrect folding of this small polypeptide sequence in the context of the Gal4 DNA-binding domain or by the participation of additional Rep domains located within the Rep40 coding region in PC4 interaction. This region common to all four Rep proteins contains a putative nucleotide-binding site implicated in the enzymatic and biological activities of the large Rep proteins Rep78/Rep68. A well-characterized point mutation in this nucleotide-binding site changing lysine 340 to histidine abolished binding to PC4 both for Rep68 and M172/530 in the two-hybrid system (Fig. 3). This same point mutation abolishes the inhibitory effects of Rep on gene expression (31), on SV40 DNA amplification (36), and on cell transformation (35). Though suggestive, it has not yet been demonstrated whether this region actually binds ATP.
FIG. 3.
Interaction of different Rep constructs with full-length PC4 in the two-hybrid system. Different parts of the Rep coding region were fused in frame to the Gal4 DNA-binding domain in pGBT9 and cotransformed into yeast HF7c cells together with full-length PC4 fused to the Gal4 transactivation domain in pGAD424. The Rep sequences fused to the Gal4 DNA-binding domain are shown schematically on the left. On the right, growth of double transformants on SD plates lacking leucine, tryptophan, and histidine (his⊖ plates) is indicated in the first column and β-galactosidase (β-gal) activity assayed with transformants selected on SD plates lacking leucine and tryptophan is indicated in the second column. Symbols: ++, blue-green color observed within 0.5 to 1 h; +, blue-green color observed within 2 to 4 h; −, no evidence for color change after an overnight incubation at 30°C.
Rep interacts with the single-stranded DNA-binding domain of PC4.
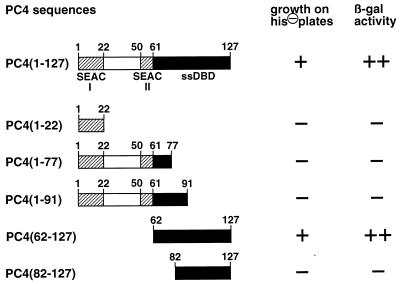
For the characterization of PC4 domains involved in Rep-PC4 interaction, a series of C-terminal and N-terminal deletion mutants of PC4 were fused in frame to the Gal4 transactivation domain in pGAD424 and examined for interaction with M172/530 in yeast. This Rep version was chosen because it showed the strongest reporter gene activation of all tested Rep constructs in cotransformations with full-length PC4 (Fig. 3). The protein sequence of PC4 displays two so-called SEAC (37) domains (Fig. 2A), stretches of consecutive serine residues followed by a stretch of acidic residues. These SEAC domains are located between amino acids 2 and 22 and between amino acids 50 and 61 (Fig. 2A and 4) and have homology to similar domains present in several transcriptional regulators of the alphaherpesvirus family, i.e., IE62 of varicella-zoster virus, ICP4 of HSV type 1, and IE180 of pseudorabies virus. However, neither a pGAD424-PC4 C-terminal deletion mutant retaining the first SEAC domain, PC4(1-22), nor a construct retaining both SEAC domains, PC4(1-77), was positive for interaction with M172/530 (Fig. 4). Furthermore, PC4(1-91), a PC4 deletion mutant in which only the 36 carboxy-terminal amino acids were deleted, was also completely negative. This mutant has been shown to retain almost full coactivator function in transcriptional activation (37), demonstrating that PC4 domains involved in Rep interaction do not correspond to those needed for transcriptional activation. In agreement with these results, the carboxy-terminal region of PC4, PC4(62-127), was positive for interaction with M172/530. This region contains a dimeric single-stranded DNA-binding domain (ssDBD) (9, 34). The further deletion of 20 N-terminal amino acids in PC4(82-127) abolished interaction. Taken together, these results demonstrate that amino acid residues in the N-terminal part of the ssDBD as well as the C-terminal part of the ssDBD are important for Rep-PC4 interaction.
FIG. 4.
Interaction of different PC4 constructs with M172/530 (Rep amino acids 172 to 530) in the two-hybrid system. Full-length PC4 and different truncated versions of PC4, shown schematically on the left, were fused in frame to the Gal4 transactivation domain in pGAD424 and cotransformed into yeast HF7c cells together with M172/530 fused to the Gal4 DNA-binding domain in pGBT9. On the right, growth of double transformants on SD plates lacking leucine, tryptophan, and histidine (his⊖) is shown in the first column, while β-galactosidase (β-gal) activity assayed with transformants selected on SD plates lacking leucine and tryptophane is shown in the second column. Symbols: ++, blue-green color observed within 0.5 to 1 h; +, blue-green color observed within 2 to 4 h; −, no evidence for color change after an overnight incubation at 30°C.
35S-labeled Rep78 interacts directly with GST-PC4 fusion proteins in vitro.
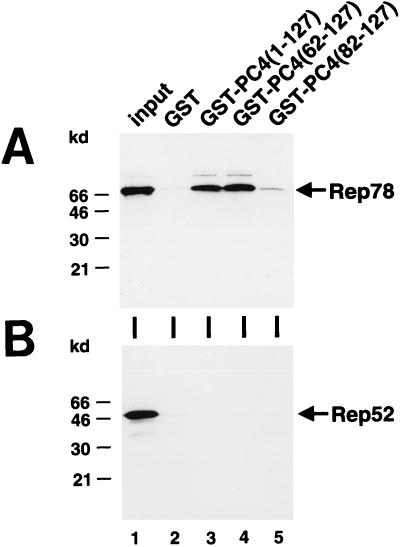
To confirm the data obtained by the yeast two-hybrid system and to demonstrate a direct interaction between Rep and PC4, GST-PC4 fusion proteins containing either full-length PC4 or truncated PC4 polypeptides were expressed in E. coli. The GST-PC4 fusion proteins, or GST alone, were bound to glutathione-Sepharose beads, purified, and subsequently incubated with in vitro-transcribed and -translated Rep78, Rep68, or Rep52. Both Rep78 (Fig. 5A) and Rep68 (data not shown) were specifically pulled down by GST fusions of full-length PC4 or the ssDBD of PC4 [Fig. 5A, GST-PC4(1-127) or GST-PC4(62-127), respectively], whereas Rep52 was not (Fig. 5B). Controls with GST alone showed only a very weak unspecific retention of Rep78 (Fig. 5). This unspecific retention was also seen with empty beads alone (data not shown) and is probably due to precipitation of Rep78 during the incubation. A weak signal only marginally higher than that with the GST-negative control was obtained with the GST-PC4 fusion in which the first 20 amino acids of the ssDBD had been deleted [Fig. 5A, GST-PC4(82-127)]. These experiments demonstrate a direct binding of PC4 to Rep and extend the yeast two-hybrid system results, which were obtained with Rep68, to Rep78. Due to its intrinsic transactivatory properties, Rep78 could not be tested separately in the two-hybrid system (see above).
FIG. 5.
Interaction of 35S-labeled Rep78 with GST-PC4 fusion proteins in vitro. Rep78 (A) and Rep52 (B) were transcribed and translated in vitro in the presence of [35S]methionine. Equal amounts of labeled proteins were incubated with glutathione-Sepharose-bound GST protein alone or the following GST-PC4 fusion proteins: PC4(1-127) expression full-length PC4, PC4(62-127) expressing the ssDBD of PC4, and PC4(82-127) expressing the 46 C-terminal amino acids of PC4. Bound proteins were collected by centrifugation, washed, boiled in SDS sample buffer, and analyzed by SDS-PAGE with subsequent autoradiography. The lane labeled “input” depicts the amount of labeled Rep proteins used in the pull-down assay.
Rep binds both the nonphosphorylated and the phosphorylated form of PC4 in vitro.
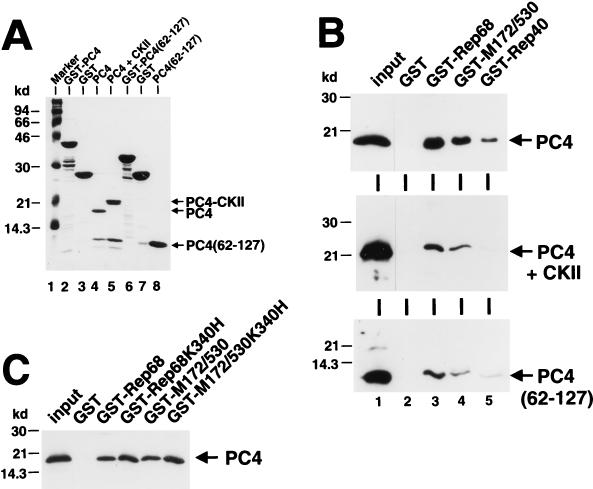
In HeLa nuclear extracts, most of PC4 is present in a phosphorylated form, most likely generated by casein kinase II through phosphorylation at serine residues within SEAC domain I (19). In the GST-PC4 fusion proteins used in the pull-down experiments, PC4 supposedly is present in the nonphosphorylated form. To examine whether Rep also interacts with phosphorylated PC4, pull-down assays with GST-Rep fusion proteins and purified bacterially expressed PC4, in both the nonphosphorylated and phosphorylated forms, were performed. PC4 was purified from GST-PC4 fusion protein by thrombin cleavage and further purification on single-stranded DNA cellulose (Fig. 6A, lane 4). Phosphorylated PC4 was obtained by treatment with casein kinase II. It can be clearly differentiated from nonphosphorylated PC4 by virtue of its reduced mobility in SDS-PAGE analysis (Fig. 6A, lanes 4 and 5). Additionally, the ssDBD of PC4 was similarly purified from the corresponding GST fusion protein (Fig. 6A, lanes 6 to 8). Purified PC4 proteins were incubated with GST-Rep fusion proteins or GST alone bound to glutathione-Sepharose beads, and after extensive washing, the bound fractions were subjected to immunoblot analysis with a polyclonal rabbit anti-PC4 serum. Both the nonphosphorylated and phosphorylated forms of PC4 (Fig. 6B, upper and middle panels) were retained specifically by GST-Rep68 and GST-M172/530 fusion proteins. It is important to note that in several independent experiments, the nonphosphorylated form of PC4 (Fig. 6B, upper panel) was bound with significantly higher affinity than the phosphorylated form of PC4 was (Fig. 6B, middle panel). In contrast to the results obtained with the two-hybrid system, a weak but specific binding was also observed with the GST-Rep40 fusion protein. The isolated ssDBD of PC4 was also bound by GST-Rep68 and GST-M172/530 fusion proteins (Fig. 6B, lower panel) with an affinity similar to that of the phosphorylated form of full-length PC4.
FIG. 6.
Interaction of different forms of bacterially expressed PC4 proteins with GST-Rep fusion proteins in vitro. (A) Purification of PC4 proteins. GST-PC4 (lane 2) and GST-PC4(62-127) (lane 6) were cleaved with thrombin protease while bound to glutathione-Sepharose beads. Beads were pelleted by centrifugation with the PC4 moieties of the fusion proteins released into the supernatant. The GST moiety bound to the glutathione-Sepharose beads is also shown (lanes 3 and 7). The PC4 proteins were further purified on single-stranded DNA-cellulose and eluted with high-salt buffer [lane 4, PC4; lane 8, PC4(62-127)]. For phosphorylation, PC4 was incubated with casein kinase II prior to purification on single-stranded DNA-cellulose (lane 5). The positions of the purified PC4 proteins are indicated by arrows. (B and C) GST pull-down assays of purified PC4 proteins. (B) One microgram of either PC4 (upper panel), PC4 treated with casein kinase II (middle panel), or PC4(62-127) (lower panel) was incubated with 10 μg of either the GST protein alone or different GST-Rep fusion proteins bound to glutathione-Sepharose beads. Bound proteins were collected by centrifugation, washed, boiled in SDS sample buffer, and analyzed by SDS-PAGE and Western blotting with a polyclonal rabbit anti-PC4 serum. The lane labeled “input” depicts the amount of purified proteins used in the pull-down assay. (C) One microgram of PC4 was incubated with 10 μg of either the GST protein alone or different GST-Rep fusion proteins and analyzed as described for panel B.
Binding of purified PC4 to GST-Rep fusion proteins is not affected by mutation of the Rep nucleotide-binding site.
Since a point mutation in the Rep nucleotide-binding site abolished Rep-PC4 interaction in the two-hybrid system, we also examined the influence of the same mutation on binding of purified PC4 to GST-Rep fusion proteins. Unexpectedly, mutated GST-Rep68 and GST-M172/530 bound nonphosphorylated PC4 with an affinity similar to that of the nonmutated proteins (Fig. 6C). Possibly, the point mutation in the nucleotide-binding site does not lead to a change in the overall conformation of the Rep proteins and the basic affinity for PC4 in vitro but rather affects some secondary ATP-dependent transition, which leads to the stabilization of a primary Rep-PC4 complex in vivo.
Influence of PC4 overexpression of AAV-2 gene expression.
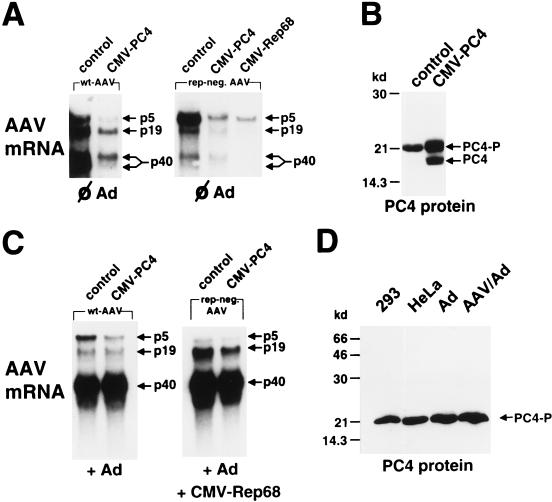
PC4, for which we could demonstrate an interaction with the large Rep proteins Rep78 or Rep68 in vivo and in vitro, was originally described as a coactivator needed for transcriptional activation by different sequence-specific transcription factors in vitro. We therefore reasoned that PC4 alone or together with Rep78 and Rep68 might be involved in the regulation of AAV-2 gene expression from its three promoters, p5, p19, and p40. PC4 was overexpressed by transfection of a plasmid expressing PC4 under the control of the cytomegalovirus (CMV) promoter (CMV-PC4). Plasmids spanning the full-length genome of AAV-2 (pTAV2-0) or a rep-negative plasmid (pTAV2-3) were cotransfected into noninfected or adenovirus-infected HeLa cells. As a control, the empty expression vector was transfected instead of CMV-PC4. AAV-2 mRNA steady-state levels were monitored 24 h posttransfection. The PC4 expression pattern was examined in parallel by Western blotting whole-cell extracts. In the absence of the CMV-PC4 expression construct, we were not able to detect the nonphosphorylated form of PC4 (Fig. 7B, control) in whole-cell extracts. Neither nuclear extracts of noninfected HeLa or 293 cells nor nuclear extracts of adenovirus- or AAV-2–adenovirus-infected cells showed a shift to the nonphosphorylated form of PC4. The latter showed only a rise in the absolute level of the phosphorylated form of PC4 in comparison to noninfected HeLa cells (Fig. 7D). In contrast, transfection of CMV-PC4 led to the accumulation of nonphosphorylated PC4 with a parallel rise in the amount of phosphorylated PC4 (Fig. 7B). Based on consideration of the transfection efficiency and further transfection experiments with tagged PC4 expression constructs, which differ from the authentic PC4 in their migration behavior (data not shown), we estimate that expression of PC4 under control of the CMV promoter led to an approximately 5- to 10-fold overexpression of phosphorylated PC4 per transfected cell. In the absence of helper virus, overexpression of PC4 and/or accumulation of nonphosphorylated PC4 downregulated all three AAV-2 promoters, p5, p19, and p40, both in the presence (Fig. 7A, wt-AAV) and in the absence (Fig. 7A, rep-neg. AAV) of Rep. In the presence of Rep, the inhibition was most pronounced for the 2.3-kb spliced mRNA(s) encoding the AAV-2 structural proteins (Fig. 7A, p40, lower arrow). With the rep-negative AAV-2 genome, downregulation of the AAV-2 promoters was similar to that observed after overexpression of Rep68, which is also under control of the CMV promoter. In the presence of adenovirus, inhibition of the AAV promoters by overexpression of PC4 was relieved (Fig. 7C). Only some minor inhibition of the p5 promoter could be observed.
FIG. 7.
Effect of PC4 overexpression on AAV-2 gene expression. (A and C) Noninfected (A) or adenovirus type 2 (multiplicity of infection, 10)-infected (C) HeLa cells were cotransfected with equal amounts (6 μg) of pTAV2-0 (wt-AAV; left panel) or pTAV2-3 (rep-neg.; right panel) and either the empty CMV expression vector (control) or the vector expressing PC4 or Rep68 as indicated. In the rep-neg. AAV lanes of panel C, 3 μg of CMV-Rep68 was added for cotransfection. At 24 h posttransfection, the cells were assayed for AAV-2 mRNA levels by Northern blot analysis. AAV-2 mRNAs transcribed from the p5, p19, and p40 promoters are indicated by arrows. In the experiment illustrated in panel A, about 10- to 20-fold-longer exposure times were used relative to those in panel C to account for the reduced AAV gene expression in the absence of helper virus. (B) Parallel PC4 Western blot analysis of whole-cell extracts of cells cotransfected with pTAV2-0 and either the empty vector or CMV-PC4. Arrows indicate the positions of nonphosphorylated PC4 (PC4) and phosphorylated PC4 (PC4-P). (D) PC4 Western blot analysis of nuclear extracts from noninfected 293 cells (293), noninfected HeLa cells (HeLa), and HeLa cells infected for 24 h with adenovirus type 2 (Ad) or coinfected with AAV-2 and adenovirus type 2 (AAV/Ad). The arrow indicates the position of phosphorylated PC4 (PC4-P).
DISCUSSION
The AAV-2 Rep78/Rep68 proteins are essential regulators of AAV-2 DNA replication and gene expression (8, 11). They are also pleiotropic effectors of viral and cellular DNA replication (22, 26, 68), of heterologous gene expression (2, 25, 27, 31, 42), and of cell transformation by oncogenic viruses (17) or by transfected oncogenes (35). An inhibition of cell proliferation upon induction of Rep78 expression has been demonstrated (67), and the growth-inhibitory effects of AAV-2 on primary cells in the absence of apparent viral gene expression (61) might be due to functional Rep78 associated with the incoming AAV-2 virion (38). The aim of this study was to identify cellular proteins involved in mediating the pleiotropic effects of the Rep proteins by direct protein-protein interactions. With a yeast-based two-hybrid screen, one predominant Rep68-interacting protein was detected and identified to be the recently described transcriptional coactivator PC4 (18, 37). PC4 has been studied mostly in vitro, where it enhances activation of the basal transcription complex by a variety of sequence-specific transcription factors. Transcriptional activation by PC4 encompasses different types of activation domains, like the acidic domains of Gal4-AH or VP16, the proline-rich activation domain of CTF, and the glutamine-rich activation domain of Sp1 (18). A direct physical interaction between PC4 and these different activation domains was demonstrated by electrophoretic mobility shift assays (18) or by GST pull-down assays. PC4 also interacts with components of the general transcription machinery, namely, with TFIIA or with the TATA box-binding protein (TBP) in a manner dependent upon the presence of TFIIA (18). It is assumed that PC4 acts initially during TFIIA-TFIID-promoter (DA) complex formation (34) and its function is dependent both on TBP-associated factors and on TFIIH (45).
Expression of the reporter genes in the yeast two-hybrid system was not due to the transactivatory properties of PC4 since (i) interaction with Rep68 was not detected with a C-terminal deletion mutant of PC4 [PC4(1-91)] that has been shown to retain full transactivation in the in vitro system (37) and (ii) cotransformation of PC4 with other proteins fused to the Gal4 DNA-binding domain did not stimulate reporter gene expression. The significance of the detected Rep-PC4 interaction is further confirmed by the fact that PC4 was almost exclusively pulled out of an HeLa cDNA library with the central part of the Rep coding region (M172/530) as bait; of 200 transformants that screened positive, 196 corresponded to PC4 or at least a very closely related protein.
We have demonstrated that the large Rep proteins Rep78 and Rep68 but not the small Rep proteins Rep52 and Rep40 are capable of interacting with PC4 in vivo. While the amino-terminal domain (amino acids 1 to 171) of Rep78/Rep68 was dispensable for interaction with PC4, interaction in the yeast two-hybrid system was completely lost upon further deletion of Rep amino acids 172 to 224 (corresponds to Rep52/Rep40). Some weak residual binding of purified PC4, however, was observed with the GST-Rep40 fusion protein in the pull-down assays in vitro, suggesting an involvement of Rep amino acids 225 to 530 in the Rep-PC4 interaction. Further evidence that these amino acids contribute to the binding of PC4 was obtained from the lack of interaction seen with the Rep fragment comprising amino acids 172 to 234 and from the abrogating effect of a point mutation at amino acid position 340 within the putative Rep nucleotide-binding site on the Rep-PC4 interaction in vivo. Thus, amino acids 172 to 224 are essential but not sufficient for the Rep-PC4 interaction. We observed that M172/530 consistently led to a higher reporter gene expression in the two-hybrid system, in comparison to Rep68, which contains 171 additional amino-terminal amino acids. Possible explanations for this observation are the following: (i) the conformation of the Rep moiety of the fusion proteins may differ between the in vivo and the in vitro system or (ii) the Gal4-Rep68 fusion protein might downregulate its own expression or the expression of yeast genes indirectly involved in reporter gene expression. Since we were unable to obtain any yeast transformants when we expressed the Gal4 DNA-binding domain Rep68 fusion protien under a stronger promoter as opposed to what was found for the Gal4 DNA-binding domain M172/530 fusion protein (data not shown), we would favor the latter explanation. A point mutation in the Rep nucleotide-binding site changing amino acid 340 from lysine to histidine completely abolished the interaction of both Rep68 and M172/530 with PC4 in the two-hybrid system but had no effect on PC4 binding in GST pull-down assays. Binding data obtained in GST pull-down assays clearly reflect specific binding of Rep and PC4. However, this may not be sufficient for formation of a functional complex in vivo, where a secondary ATP-dependent transition may be required. Both Rep and PC4 have DNA-binding activity, so stabilization of the Rep-PC4 complex may be achieved by DNA binding in vivo. This assumption is in line with our inability to detect an association between Rep and PC4 in coimmunoprecipitation experiments, where nucleic acids are lost during extract preparation.
The PC4 domains involved in Rep-PC4 interaction do not correspond to those required for transcriptional activation in vitro. Transcriptional activity of PC4 mostly involves an amino-terminal domain of 61 amino acids containing two so-called SEAC domains (consecutive serine residues followed by an acidic stretch) which display homology to several transcriptional regulators of the alphaherpesvirus family such as IE62, ICP4, and IE180 of varicella-zoster virus, HSV type 1, and pseudorabies virus, respectively (37). A C-terminal deletion mutant of PC4 comprising roughly two-thirds (amino acids 1 to 91) of the coding region, including both SEAC domains, retains full transactivation activity but was completely negative for interaction with Rep. The interaction with Rep was mapped to the carboxy-terminal ssDBD (34) of PC4 (amino acids 62 to 127). Amino-terminal (amino acids 62 to 81) as well as carboxy-terminal (amino acids 92 to 127) residues of the ssDBD were absolutely required for Rep binding. Phosphorylation of serine residues mainly at SEAC domain I negatively regulates the transcriptional activity of PC4 (19, 37), and in nuclear extracts of HeLa cells, PC4 was found predominantly in this phosphorylated transcriptionally inactive form, which in vivo is most probably generated by casein kinase II (19). In vitro, we found a stronger affinity of Rep for nonphosphorylated PC4, the form which we could barely detect in nuclear extracts of HeLa or 293 cells. In vivo, however, Rep-PC4 interaction does not seem to be limited exclusively to this nonphosphorylated form of PC4, since Rep also binds to the isolated ssDBD of PC4, which more closely mimics the phosphorylated form of PC4 (34).
Several lines of evidence support an involvement of the Rep-PC4 interaction in Rep-mediated regulation of cellular and viral gene expression, DNA amplification, and cell transformation. The Rep domains involved in Rep-PC4 interaction correspond to those that are needed for Rep-mediated inhibition of HSV-induced SV40 DNA amplification (36) and overlap those involved in Rep-mediated inhibition of cellular transformation by E1A/EJras (35, 66). A well-characterized point mutation in the putative nucleotide-binding site of Rep, which does not affect the DNA-binding properties of Rep78/Rep68, abolishes both the interaction with PC4 in vivo and the inhibition of DNA amplification (36). This point mutation also abolishes Rep78-mediated inhibition of the HPV18 URR and the HIV LTR (31). An involvement of direct Rep-PC4 interaction in the downregulation of heterologous promoters is further suggested by the redundancy of cis elements in the HPV18 URR required for inhibition (31), since PC4 mediates transcriptional activation by a variety of unrelated activation domains of sequence-specific transcription factors. In contrast to other Rep-responsive promoters like the AAV-2 p5 and p19 promoters and the H-ras promoter (5), the HPV18 URR does not contain a consensus Rep binding site (31). Therefore, at least three mechanisms of Rep-mediated inhibition of homologous and heterologous promoters, which are not mutually exclusive, may exist. The first mechanism involves direct binding of Rep to sequence elements within the promoter, depends on the amino-terminal DNA-binding domain of Rep78 and Rep68 but not on a functional nucleotide-binding site, and could account for inhibition of the AAV-2 p5 promoter (31, 40) and the H-ras promoter (20), which exhibit strong binding by Rep78/Rep68. The second mechanism does not involve sequence-specific DNA binding of Rep, but rather protein-protein interactions of Rep with coactivators like PC4, depends upon a functional nucleotide-binding site, and largely accounts for inhibition of promoters like the HPV18 URR and the HIV LTR. PC4 has been suggested to play a similar role in the formation of the transcriptional preinitiation complex as HMG1 (45), which has been demonstrated to also bind directly to Rep78 and Rep68 and enhance Rep-mediated inhibition of the p5 promoter (16). Very recently, the interaction of Rep78 with a central component of the basal transcription apparatus, the TBP, has also been reported (30). A third mechanism involved in Rep-mediated transcriptional regulation would be protein-protein interactions with sequence-specific transcription factors like Sp1 (29, 50) which have been implicated both in the transactivation of the AAV-2 p19 promoter (50) and inhibition of the H-ras promoter (29). Since both Sp1 and Rep are capable of binding to PC4, there may also exist trimeric complexes of Sp1, Rep, and PC4 with various functions in transcriptional activation and inhibition.
Rep-mediated inhibition of the AAV-2 p5 promoter appears to involve sequence-specific binding of Rep to its recognition site (p5RBS) as well as mechanisms dependent on a functional Rep nucleotide-binding site (40). We could demonstrate that overexpression of PC4 also leads to an inhibition of the AAV-2 promoters with concomitant accumulation of the nonphosphorylated, transcriptionally active form of PC4. This may suggest that the nonphosphorylated active form of PC4 leads to downregulation of AAV gene expression. This repression was mostly relieved in the presence of helper virus. However, interpretation remains speculative in view of the high background of phosphorylated PC4 in HeLa cells and the lack of data regarding PC4 function in vivo.
As stated above, the mutational analysis points to a possible role of Rep-PC4 interactions in the inhibition of HSV-induced SV40 DNA amplification. A function for PC4 in SV40 DNA replication was suggested due to the existence of common complexes of PC4 with RPA (replication protein A) on single-stranded DNA (48). Although not similar at the level of primary amino acid sequence, the dimeric single-stranded binding sites (ssDBDs) of PC4 and RPA show a clear structural homology (9). PC4 stimulates as well as inhibits SV40 DNA replication in vitro, depending upon the relative concentrations of PC4 and RPA (48). By binding to the ssDBD of PC4, Rep might alter the single-stranded DNA-binding properties of PC4 and thus inhibit SV40 DNA replication. When we addressed the issue of a functional role of PC4 in AAV DNA replication in the presence of adenovirus, we obtained controversial data (data not shown). Both stimulatory and inhibitory effects of PC4 overexpression on AAV replication were observed, depending on the concentration of the large Rep proteins Rep78 and Rep68. A detailed elucidation of the role of PC4 in Rep-mediated regulation of DNA replication, gene expression, and cell transformation will require a more profound knowledge of the function of PC4, especially the phosphorylated form of PC4, in vivo.
ACKNOWLEDGMENT
S. Weger was supported by a grant from the FORGEN program of the Bayerische Forschungsstiftung.
REFERENCES
- 1.Andrews N A, Fuller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:249. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoni B A, Rabson A B, Miller I L, Trempe J P, Chejanovsky N, Carter B J. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J Virol. 1991;65:396–404. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchison R W, Casto B C, Hammond W M. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 4.Balague C, Kalla M, Zhang W W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchu R B, Kotin R M, Hermonat P L. The regulatory rep protein of adeno-associated virus binds to sequences within the c-H-ras promoter. Cancer Lett. 1994;86:23–31. doi: 10.1016/0304-3835(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 6.Batchu R B, Hermonat P L. The trans-inhibitory Rep78 protein of adeno-associated virus binds to TAR region DNA of the human immunodeficiencey virus type 1 long terminal repeat. FEBS Lett. 1995;367:267–271. doi: 10.1016/0014-5793(95)00584-v. [DOI] [PubMed] [Google Scholar]
- 7.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berns K I. Parvoviridae and their replication. In: Fields B N, Knipe D M, editors. Virology. 2nd ed. Vol. 2. New York, N.Y: Raven Press; 1990. pp. 1743–1763. [Google Scholar]
- 9.Brandsen J, Werten S, van der Vliet P C, Meisterernst M, Kroon J, Gros P. C-terminal domain of transcription factor PC4 reveals dimeric ssDNA-binding site. Nat Struct Biol. 1997;4:900–903. doi: 10.1038/nsb1197-900. [DOI] [PubMed] [Google Scholar]
- 10.Buller R M, Janik J E, Sebring E D, Rose J A. Herpes simplex virus types 1 and 2 completely help adeno-associated virus replication. J Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter B J, Trempe J P, Mendelson E. Adeno-associated virus gene expression and regulation. In: Tijssen P, editor. Handbook of parvoviruses. I. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 227–254. [Google Scholar]
- 12.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 13.Chen A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 14.Chen A, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomzynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De la Maza L M, Carter B J. Inhibition of adenovirus oncogenicity in hamsters by adeno-associated virus DNA. J Natl Cancer Inst. 1981;67:1323–1326. [PubMed] [Google Scholar]
- 18.Ge H, Roeder R G. Purification, cloning and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 19.Ge H, Zhao Y, Chait B T, Roeder R G. Phosphorylation negatively regulates the function of coactivator PC4. Proc Natl Acad Sci USA. 1994;91:12691–12695. doi: 10.1073/pnas.91.26.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiger, C., and R. Heilbronn. Unpublished data.
- 21.Gubler U, Hoffman B J. A simple and very efficient method for generating cDNA libraries. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 22.Heilbronn R, Bürkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–3018. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermonat P L. The adeno-associated virus Rep78 gene inhibits cellular transformation induced by bovine papillomavirus. Virology. 1989;172:253–261. doi: 10.1016/0042-6822(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 25.Hermonat P L. Inhibition of H-ras expression by the adeno-associated virus Rep78 transformation suppressor gene product. Cancer Res. 1991;51:3373–3377. [PubMed] [Google Scholar]
- 26.Hermonat P L. Inhibition of bovine papillomavirus plasmid DNA replication by adeno-associated virus. Virology. 1992;189:329–333. doi: 10.1016/0042-6822(92)90710-7. [DOI] [PubMed] [Google Scholar]
- 27.Hermonat P L. Downregulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 1994;81:129–136. doi: 10.1016/0304-3835(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 28.Hermonat P L. Adeno-associated virus inhibits human papillomavirus type 16: a viral interaction implicated in cervical cancer. Cancer Res. 1994;54:2278–2281. [PubMed] [Google Scholar]
- 29.Hermonat P L, Santin A D, Batchu R B. The adeno-associated virus Rep78 major regulatory/transformation suppressor protein binds cellular Sp1 in vitro and evidence of a biological effect. Cancer Res. 1996;56:5299–5304. [PubMed] [Google Scholar]
- 30.Hermonat P L, Santin A D, Batchu R B, Zhan D. The adeno-associated virus Rep78 major regulatory protein binds the cellular TATA-binding protein in vitro and in vivo. Virology. 1998;245:120–127. doi: 10.1006/viro.1998.9144. [DOI] [PubMed] [Google Scholar]
- 31.Hörer H, Weger S, Butz K, Hoppe-Seyler F, Geisen C, Kleinschmidt J A. Mutational analysis of adeno-associated virus Rep protein-mediated inhibition of heterologous and homologous promoters. J Virol. 1995;69:5485–5496. doi: 10.1128/jvi.69.9.5485-5496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 33.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaiser K, Stelzer G, Meisterernst M. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 1995;14:3520–3527. doi: 10.1002/j.1460-2075.1995.tb07358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khleif S N, Myers T, Carter B J, Trempe J P. Inhibition of cellular transformation by the adeno-associated virus rep gene. Virology. 1991;181:738–741. doi: 10.1016/0042-6822(91)90909-u. [DOI] [PubMed] [Google Scholar]
- 36.Kleinschmidt J A, Möhler M, Weindler F W, Heilbronn R. Sequence elements of the adeno-associated virus rep gene required for suppression of herpes-simplex-virus induced DNA amplification. Virology. 1995;206:254–262. doi: 10.1016/s0042-6822(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 37.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 38.Kube D M, Ponnazhagan S, Srivastava A. Encapsidation of adeno-associated virus type 2 Rep proteins in wild-type and recombinant progeny virions: Rep-mediated growth inhibition of primary human cells. J Virol. 1997;71:7361–7371. doi: 10.1128/jvi.71.10.7361-7371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kyöstiö S R M, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyöstiö S R M, Wonderling R S, Owens R A. Negative regulation of the adeno-associated virus (AAV) p5 promoter involves both the p5 rep binding site and the consensus ATP-binding motif of the AAV Rep68 protein. J Virol. 1995;69:6787–6796. doi: 10.1128/jvi.69.11.6787-6796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation of the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Labow M A, Graf L H, Jr, Berns K I. Adeno-associated virus gene expression inhibits cellular transformation by heterologous genes. Mol Cell Biol. 1987;7:1320–1325. doi: 10.1128/mcb.7.4.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Laughlin C A, Cardellichio C B, Coon H C. Latent infection of KB cells with adeno-associated virus type 2. J Virol. 1986;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik S, Guermah M, Roeder R G. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarty D M, Christensen M, Muzyczka N. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oelze I, Rittner K, Sczakiel G. Adeno-associated virus type 2 rep gene-mediated inhibition of basal gene expression of human immunodeficiency virus type 1 involves its negative regulatory functions. J Virol. 1994;68:1229–1233. doi: 10.1128/jvi.68.2.1229-1233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Z-Q, Ge H, Amin A A, Hurwitz J. Transcrition-positive cofactor PC4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J Biol Chem. 1996;271:22111–22116. doi: 10.1074/jbc.271.36.22111. [DOI] [PubMed] [Google Scholar]
- 49.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira D J, Muzyczka N. The cellular transcription factor SP1 and an unknown cellular protein are required to mediate Rep protein activation of the adeno-associated virus p19 promoter. J Virol. 1997;71:1747–1756. doi: 10.1128/jvi.71.3.1747-1756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira D J, Muzyczka N. The adeno-associated virus type 2 p40 promoter requires a proximal SP1 interaction and a p19 CArG-like element to facilate Rep transactivation. J Virol. 1997;71:4300–4309. doi: 10.1128/jvi.71.6.4300-4309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rittner K, Stöppler H, Pawlita M, Sczakiel G. Versatile eucaryotic vectors for strong and constitutive transient and stable gene expression. Methods Mol Cell Biol. 1991;2:176–181. [Google Scholar]
- 53.Samulski R J. Adeno-associated virus: integration at a specific chromosomal locus. Curr Opin Genet Dev. 1993;3:74–80. doi: 10.1016/s0959-437x(05)80344-2. [DOI] [PubMed] [Google Scholar]
- 54.Smith R H, Kotin R M. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J Virol. 1998;72:4874–4881. doi: 10.1128/jvi.72.6.4874-4881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tratschin J-D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tratschin J-D, Tal J, Carter B J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral rep gene product. Mol Cell Biol. 1986;6:2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trempe J P, Carter B J. Regulation of adeno-associated virus gene expression in 293 cells: control of mRNA abundance and translation. J Virol. 1988;62:68–74. doi: 10.1128/jvi.62.1.68-74.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weger S, Wistuba A, Grimm D, Kleinschmidt J A. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winocour E, Callahan M F, Huberman E. Perturbation of the cell cycle by adeno-associated virus. Virology. 1988;167:393–399. [PubMed] [Google Scholar]
- 62.Wonderling R S, Kyöstiö S R M, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yakobson B, Hrynko T A, Peak M J, Winocour E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J Virol. 1989;63:1023–1030. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yalkinoglu A, Heilbronn R, Bürkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 66.Yang Q, Kadam A, Trempe J P. Mutational analysis of the adeno-associated virus rep gene. J Virol. 1992;66:6058–6069. doi: 10.1128/jvi.66.10.6058-6069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Q, Chen F, Ross J, Trempe J P. Inhibition of cellular and SV40 DNA replication by the adeno-associated virus Rep proteins. Virology. 1995;207:246–250. doi: 10.1006/viro.1995.1072. [DOI] [PubMed] [Google Scholar]