Abstract
The interferon-regulated 2-5A/RNase L pathway plays a major role in the antiviral and antiproliferative activities of these cytokines. Several viruses, however, have evolved strategies to escape the antiviral activity of the 2-5A/RNase L pathway. In this context, we have cloned a cDNA coding for the RNase L inhibitor (RLI), a protein that specifically inhibits RNase L and whose regulated expression in picornavirus-infected cells down regulates the activity of the 2-5A/RNase L pathway. We show here that RLI increases during the course of human immunodeficiency virus type 1 (HIV-1) infection, which may be related to the downregulation of RNase L activity that has been described to occur in HIV-infected cells. In order to establish a possible causal relationship between these observations, we have stably transfected H9 cells with RLI sense or antisense cDNA-expressing vectors. The overexpression of RLI causes a decrease in RNase L activity and a twofold enhancement of HIV production. This increase in HIV replication correlates with an increase in HIV RNA and proteins. In contrast, reduction of RLI levels in RLI antisense cDNA-expressing clones reverses the inhibition of RNase L activity associated with HIV multiplication and leads to a threefold decrease in the viral load. This anti-HIV activity correlated with a decrease in HIV RNA and proteins. These findings demonstrate that the level of RLI, via its modulation of RNase L activity, can severely impair HIV replication and suggest the involvement of RLI in the inhibition of the 2-5A/RNase L system observed during HIV infection.
Interferons (IFN) control various cellular functions and participate in host defense against viral and microbial agents through multiple induced pathways (1). The 2-5A/RNase L pathway is one of the major pathways induced by IFN. It is implicated in some of the antiviral mechanisms of IFN and might play a role in the regulation of RNA turnover and stability (12). IFN induces four different forms of human 2-5A-synthetase which, upon activation by double-stranded RNA (dsRNA), convert ATP into an unusual series of oligomers known as 2-5A. 2-5A then activates RNase L, a latent endoribonuclease, which inhibits protein synthesis by cleavage of mRNA at the 3′ side of UpNp sequences (11, 13, 40). During viral infection this antiviral pathway can be activated, since several viruses produce dsRNA structures that can activate 2-5A-synthetase. The presence of 2-5A has been demonstrated in cells infected with encephalomyocarditis (EMC) virus (38), vaccinia virus (22), or reovirus (19).
Although 2-5A has long been considered to be the unique regulator of the 2-5A/RNase L pathway, we have cloned and characterized a polypeptide inhibitor of the 2-5A pathway (referred to as RNase L inhibitor [RLI]). RLI cDNA codes for a 68-kDa protein whose mRNA is not regulated by IFN. When expressed in a reticulocyte lysate, RLI induces neither 2-5A degradation nor irreversible modification of RNase L (3); however, it antagonizes the binding of 2-5A by the latter and thus its nuclease activity, since 2-5A binding is a prerequisite to RNase L dimerization and activation (10, 31).
Despite the presence of double-stranded viral RNA structures capable of activating the 2-5A/RNase L pathway and the presence of high concentrations of 2-5A, in several cases no RNase L activity could be detected. Several viruses appear to have developed strategies to counteract the antiviral activity of the 2-5A/RNase L pathway. For example, during herpes simplex virus type 1 and 2 (HSV-1 and HSV-2) infection, 2-5A derivatives are synthesized that behave as 2-5A antagonists (7). Similarly, infection by vaccinia virus leads to an inhibition of 2-5A-synthetase activity and to the degradation of 2-5A (20). Recently, Rivas et al. have shown that vaccinia virus E3L protein is an inhibitor of 2-5A-synthetase (23). Finally, EMC virus downregulates RNase L activity through the increased expression of RLI (18).
Along the same lines, an inhibition of RNase L activity has been observed during the course of human immunodeficiency virus (HIV) infection. RNase L is inactive in peripheral blood mononuclear cell extracts from AIDS patients, despite the presence of its 2-5A activator (5). Likewise, the 2-5A binding activity of RNase L in lymphocytes isolated from AIDS and pre-AIDS patients was approximately 65% lower than that found in controls (39). In experimental infection of H9 cells with HIV type 1 (HIV-1), a strong enhancement of 2-5A-synthetase activity and a small increase of RNase L activity were observed. Both enzymes reached maximal levels at day 3 after the onset of HIV-1 infection and declined sharply thereafter. Interestingly, RNase L can degrade HIV-1 transcripts during the early steps of infection, and HIV-1 transcript accumulation coincides with the decrease of RNase L activity (29, 35). These studies suggest that there is an accumulation of an inhibitor of the 2-5A/RNase L pathway that interferes with 2-5A binding through the formation of an inhibitor-RNase L complex. On the other hand, different studies have suggested that direct expression or activation of the 2-5A/RNase L pathway enzymes can lead to suppression of HIV replication. For example, the stable expression of transfected 2-5A-synthetase leads to the suppression of HIV replication (27, 28). Direct activation of RNase L with phosphorothioate-phosphodiester 2-5A derivatives inhibits HIV-1 reverse transcriptase and HIV-induced syncytium formation (33). Recently, Maitra and Silverman have shown that overexpression of RNase L can suppress HIV-1 replication (17). These results show that the 2-5A/ RNase L pathway is potentially able to regulate HIV replication but is rapidly inhibited after the early stages of HIV infection.
RLI is a logical candidate to be the inhibitor induced by HIV infection. We have therefore studied the modulation of RNase L and RLI expression in H9 cells following HIV-1 infection. We have established that if RNase L activity decreased during the course of HIV-1 infection, the RNase L protein decreases only transiently and the amount of RLI protein increases. We have stably transfected H9 cells with RLI sense or antisense cDNA-expressing vectors and followed the course of de novo HIV-1 infection in two independent RLI sense clones and two independent RLI antisense clones. As expected, the overexpression of RLI decreases RNase L activity and enhances HIV-1 production while, on the contrary, the reduction of the RLI level correlates with an increase in RNase L activity and a decrease in virus production. Altogether, these data suggest that RLI could be a cellular inhibitor induced by HIV-1 to inhibit the 2-5A/RNase L pathway.
MATERIALS AND METHODS
Cells and virus.
H9 cells (human T lymphocyte cell line) were grown in RPMI Glutamax medium (Gibco BRL) supplemented with 10% (vol/vol) fetal calf serum. Strain IIIB of HIV-1 was grown in H9 cells and titrated by a reverse transcriptase (RT) assay (21).
For HIV-1 infection, H9 cells (3 × 108) were centrifuged and the cell pellet was resuspended in 150 ml with HIV-1 strain IIIB corresponding to 4.5 × 106 cpm of RT activity (15, 16). After 1 h of incubation at 37°C, the cells were seeded at a final concentration of 2 × 105 cells per ml in RPMI Glutamax containing 10% (vol/vol) fetal calf serum.
Expression vectors and transfections.
The human RLI cDNA (3) was subcloned in the sense or antisense orientation in pcDNA3 (Invitrogen) by standard procedures (26). Then, 5 × 105 H9 cells were transfected by electroporation (250 mV) with 10 μg of plasmid DNA. The empty pcDNA3 vector was used as a control. Stable transfectants were selected by culturing the cells in the presence of 1 mg of G418 (Gibco BRL) per ml. Individual clones were isolated by limit dilution and analyzed for the expression of the transfected cDNA. Clones expressing the transfected antisense cDNA were named VAS1 and VAS2; clones expressing the transfected sense cDNA were named VS1 and VS2, and the clone expressing the transfected empty vector was named VV.
Cell extracts.
After infection, cells were collected at the times indicated and were resuspended in 1 volume of hypotonic buffer (0.5% [vol/vol] Nonidet P-40, 20 mM HEPES [pH 7.5], 10 mM potassium acetate, 15 mM magnesium acetate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin per ml, 150 μg of leupeptin per ml), disrupted in a Dounce homogenizer, and centrifuged at 10,000 × g as described previously (25). The supernatant (S10) was pipetted off, and its protein concentration was determined by spectrophotometry (37).
Western blot analysis.
Cell extracts were analyzed by Western blot according to the method of Towbin et al. (34). Proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to a nitrocellulose membrane. For antibody revelation, the nitrocellulose membrane was soaked for 1 h in phosphate-buffered saline (PBS) (140 mM NaCl–2 mM KCl–8 mM Na2HPO4–1.5 mM KH2PO4 supplemented with 5% [wt/vol] skim milk) and then held overnight at 4°C with the polyclonal antibodies against RNase L (1/1,000) or RLI (1/500) that we have previously described and characterized (18) or with anti-gp120 (1/100) (a generous gift of D. Misse, ORSTOM, Montpellier, France) or anti-GAPDH (1/3,000) (a generous gift of G. Cathala, IGM, Montpellier, France) in the same buffer. The filter was washed with PBS supplemented with 0.05% (vol/vol) Tween 20 and incubated for 1 h at room temperature with donkey anti-rabbit immunoglobulin antibody conjugated to horseradish peroxidase (1/2,000; Amersham) in PBS supplemented with 5% (wt/vol) skim milk. Chemiluminescence was generated by the NEN kit. The gels were scanned, and the protein bands were quantified by image analysis with the Intelligent Quantifier program (BioImage Systems Corp.). Results are expressed as an average of three independent experiments. Standard deviations are indicated by vertical bars in the figures.
2-5A binding activity of RNase L during infection of cells with HIV-1.
H9 cells, as well as the VV, VS, and VAS transfectants (3 × 108 cells), were infected with HIV-1 (4.5 × 106 cpm of RT activity) (15, 16). Cells were harvested at various times after the onset of infection as indicated. Cell extracts (S10) were prepared as described above. The radiobinding assay (14) was performed with S10 cell extracts (600 μg of proteins) as a source of RNase L and 2-5A4-3′-[32P]pCp (2-5ApCp) (20,000 cpm, 3,000 Ci/mmol) as a probe. The radiobinding assay was utilized with the modifications previously described (2, 4). Results are expressed as an average of three independent experiments for each cell clone (H9, VV, VS1, VS2, VAS1, and VAS2). Standard deviations are indicated by vertical bars.
RNA analysis.
Total cellular RNA was prepared by using the guanidine thiocyanate-lithium chloride procedure (6). Northern blot hybridizations were performed according to standard techniques (26). Probes (HIV long terminal repeat [LTR] and GAPDH [glyceraldehyde-3-phosphate dehydrogenase]) were synthesized by the multiprime procedure (random primer DNA labeling system; Gibco-BRL). After autoradiography, mRNA was quantified by image analysis with the Intelligent Quantifier program (BioImage Systems Corp.). Each lane was normalized with the GAPDH probe.
RT assay.
At each indicated time after the onset of infection, 50 μl of cell culture supernatant was added to 10 μl of TNE buffer (20 mM Tris [pH 7.8], 1 mM EDTA, 100 mM NaCl, 0.1% [vol/vol] Triton X-100) and 20 μl of reagent buffer (250 mM Tris HCl [pH 7.8], 25 mM MgCl2, 500 mM KCl, 50 mM dithiothreitol, 0.5 mM EGTA, poly(rA)-oligo(dT10) [optical density = 0.025], 2.5 μl of [methyl-3H]dTTP [82 Ci/mmol, 1 mCi/ml]) and then incubated for 2 h at 37°C. For each reaction 50 μl was spotted onto a GF/C filter (Whatman) and incubated in 10% (vol/vol) trichloroacetic acid (TCA)–12 mM pyrophosphate. The filters were then washed three times in 5% (vol/vol) TCA–12 mM pyrophosphate, rinsed with 100% ethanol, dried, and counted. The results are expressed as an average of three independent experiments for each cell clone (H9, VV, VS1, VS2, VAS1, and VAS2). The standard deviations are indicated by vertical bars.
RESULTS
RNase L activity decreases in HIV-1-infected cells.
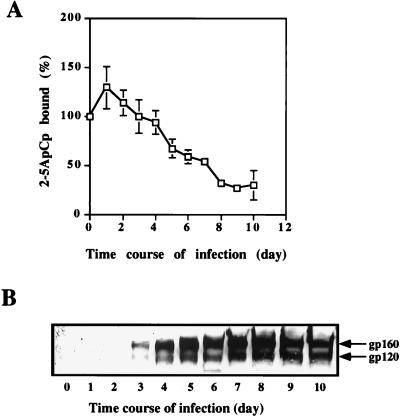
The nuclease activity of RNase L is strictly dependent on its activation by 2-5A (31), so we measured the RNase L activity in HIV-1-infected H9 cells with the 2-5A radiobinding assay (14). This assay quantifies the binding of a radioactive 2-5ApCp probe to RNase L in unfractionated cell extracts. In keeping with the data described in the introduction, we observed a decrease in the binding of 2-5ApCp by RNase L in extracts from HIV-1-infected H9 cells. 2-5A binding decreases by as early as 3 days after the onset of HIV-1 infection and reaches a minimum 10 days after the onset of HIV-1 infection (Fig. 1A). The virus production was monitored by Western blot analyses of the HIV-1 gp120 polypeptides in cell extracts. The polyclonal antibody against gp120 we have used also recognizes its gp160 precursor. gp120-gp160 polypeptides could be detected at day 3 postinfection and reached maximal levels at days 7 and 8 (Fig. 1B).
FIG. 1.
RNase L activity during HIV-1 infection. H9 cells were infected with HIV-1 and harvested at the indicated times. (A) Cell extracts (600 μg of proteins) were tested without further fractionation in a 2-5A radiobinding assay. Results are expressed as the percentage of 2-5ApCp bound. 100% is the binding level in uninfected cells at time zero. (B) H9 cells were infected with HIV-1 and harvested at the indicated times. For each time point, 200 μg of protein was analyzed by SDS-PAGE and Western blotting with polyclonal antibody against gp160/gp120. An autoradiograph of the gel is presented.
The decrease of RNase L activity monitored by the 2-5A radiobinding assay in unfractionated cell extracts could result from a decrease in RNase L itself or from the induction of an inhibitor. Polyclonal antibodies against RNase L and RLI have been used to quantify RNase L and RLI during the course of HIV-1 infection.
RLI increases during HIV-1 infection.
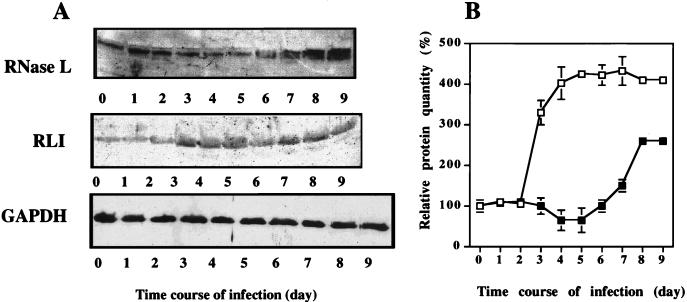
RNase L was quantified by a Western blot assay with the RNase L-specific polyclonal antibody described previously (18). RNase L was detected at all time points, decreasing transiently (by 40%) between days 4 and 6 (Fig. 2) and then increasing. The decrease of RNase L activity observed in cell extracts during the time course of HIV-1 infection (Fig. 1) coincides with the increased protein level, particularly between days 6 and 9, suggesting that there is the accumulation of an inhibitor. Since RLI inhibits the binding of 2-5ApCp to RNase L (3), it is a logical candidate for this role and, indeed, it increases during the time course of HIV-1 infection (Fig. 2). Variations in RNase L and RLI during HIV-1 infection are not linked to a nonspecific variation of protein synthesis, as shown by the behavior of the GAPDH protein used as a control (Fig. 2A).
FIG. 2.
Analysis of RNase L and RLI proteins during HIV-1 infection. H9 cells were infected with HIV-1 and harvested at the indicated times. (A) For each time point, 200 μg of proteins was analyzed by SDS-PAGE and Western blotting with polyclonal antibodies against RNase L, RLI, or GAPDH as indicated. (B) A densitometric analysis of the gels is presented (□, RLI; ■, RNase L). Vertical bars represent the standard deviations obtained with three independent experiments. 100% corresponds to the amount of RLI or RNase L protein in untreated cells at time zero.
The stable transfection of an RLI antisense or RLI sense construction modifies RNase L activity during HIV-1 infection.
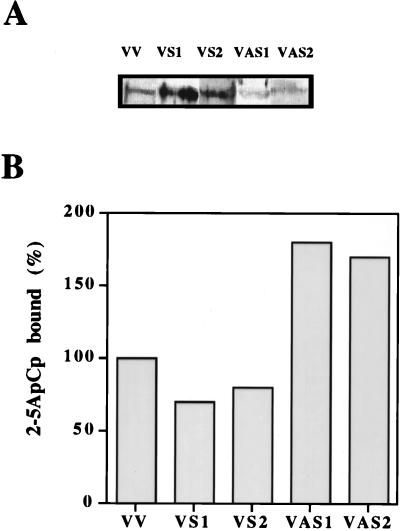
To determine the effects of regulating RLI levels on HIV-1 infection, H9 cells were stably transfected with RLI constructions in either the sense or the antisense orientation. H9 cell transfected clones were characterized by a Western blot assay with a polyclonal antibody against RLI protein (Fig. 3A) and with the 2-5ApCp binding activity of RNase L (Fig. 3B). Clones expressing the RLI antisense construction, the RLI sense construction, and the empty vector have been named VAS, VS, and VV, respectively. Two independent clones expressing higher levels of RLI (VS1 and VS2) demonstrated lower 2-5A binding activity by RNase L than cells transfected with the empty vector. On the contrary, in two independent clones expressing lower levels of RLI (VAS1 and VAS2), the 2-5A binding activity of RNase L is increased (Fig. 3B).
FIG. 3.
Expression of RLI and RNase L activity in H9 cells stably transfected with antisense and sense RLI cDNA. (A) Extracts (200 μg of proteins) from H9 cells stably transfected with the VV empty vector, with the sense RLI cDNA construction (VS1 and VS2), or with the antisense RLI cDNA construction (VAS1 and VAS2) were analyzed by SDS-PAGE and Western blotting with polyclonal antibody against RLI. (B) Cell extracts (600 μg of protein) from the different transfectants were tested without further fractionation in a 2-5A radiobinding assay. Results are expressed as the percentage of 2-5ApCp bound. 100% is the binding level in control VV cells.
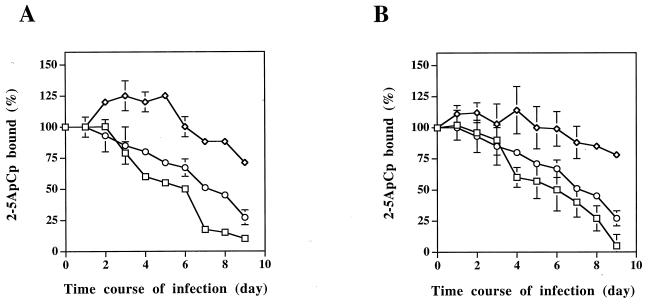
The 2-5A binding activity of RNase L during HIV-1 infection was analyzed in these clones expressing different levels of RLI (Fig. 4). HIV-1 infection of the VV clone gives rise to a 68% decrease in RNase L activity, a result in line with the data presented in Fig. 1 for the nontransfected H9 cells. A significantly larger decrease in RNase L activity is observed in the clones expressing the RLI sense construction: 90% for VS1 and 95% for VS2 (Fig. 4A and B). On the other hand, the clones expressing the RLI antisense construction lead to a much lower inhibition of RNase L activity: 29% for VAS1 and 22% for VAS2 (Fig. 4) upon HIV-1 infection. As expected, therefore, overexpression of RLI increases the HIV-1-associated down regulation of RNase L activity, and lower expression of RLI antagonizes the HIV-1-associated down regulation of RNase L activity.
FIG. 4.
2-5A binding activity in cells stably transfected with RLI sense and antisense constructions during HIV-1 infection. H9 cells transfected with the empty vector (○, VV), with the antisense RLI cDNA construction (◊, VAS), or with the sense RLI cDNA construction (□, VS) were infected with HIV-1 and harvested at the indicated times. Extracts (600 μg of protein) were tested without further fractionation in a 2-5A radiobinding assay. Results are expressed as the percentage of 2-5ApCp bound. 100% is the binding obtained at time zero in uninfected cells. Vertical bars represent the standard deviations obtained with three independent experiments. Clones VV, VAS1, and VS1 are shown in panel A, and clones VV, VAS2, and VS2 are shown in panel B.
The production of HIV is proportional to the RLI level.
The RLI protein level modulates RNase L activity. We therefore analyzed whether the replication of HIV-1 is modified in the different transfectants. Virus production was monitored by different techniques: the measurement of RT activity in the cell culture supernatant, Northern blot analysis of HIV mRNA transcripts, and Western blot analysis of the HIV-1 gp160/gp120 polypeptides in cell extracts.
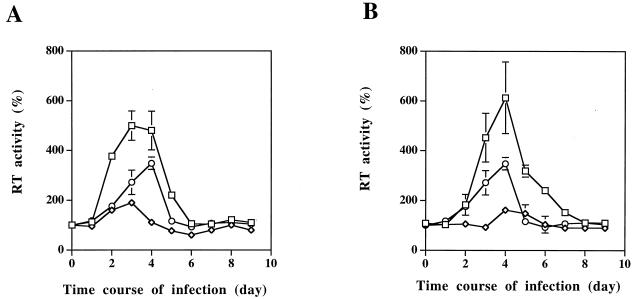
As clearly shown in Fig. 5, the clones expressing the RLI sense constructions (VS1 and VS2) produced significantly larger amounts of virus than did the control cells, as monitored by the RT activity peak. On the contrary, the RT activity was highly repressed (by 75%) in the clones expressing the RLI antisense construction (VAS1 and VAS2) (Fig. 5).
FIG. 5.
RT activity in clones expressing sense or antisense RLI constructions. H9 cells transfected with the empty vector (○, VV), with the antisense RLI cDNA construction (◊, VAS), or with the sense RLI cDNA construction (□, VS) were infected with HIV-1. Virus production was monitored by supernatant RT assay every day. 100% is the RT activity observed in uninfected cells. Vertical bars refer to the standard deviations obtained with three independent experiments. Clones VV, VAS1, and VS1 are shown in panel A, and clones VV, VAS2, and VS2 are shown in panel B.
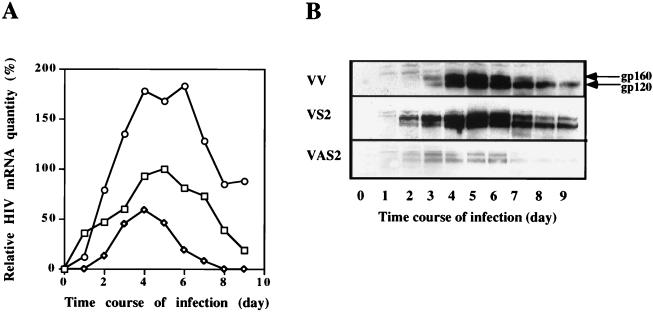
The mechanism of the pro-HIV effect of overexpressing RLI was then investigated by detection of the total levels of the HIV-1 mRNA in Northern blots. The amounts of HIV RNA are doubled in cells transfected with the RLI sense vector (VS2) compared to cells transfected with the empty vector (VV). By contrast, the anti-HIV effect of lower expression of RLI was confirmed, since viral RNA levels are halved in cells transfected with the RLI antisense vector (VAS2) (Fig. 6A). This difference in HIV-1 RT activity and mRNA was also observed at the protein level.
FIG. 6.
HIV-1 mRNA level and expression of gp160/gp120 during HIV-1 infection in cells transfected with RLI sense and antisense cDNA constructions. H9 cells transfected with the antisense RLI cDNA construction (◊, VAS2), with the sense RLI cDNA construction (○, VS2), or with the empty vector (□, VV) were infected with HIV-1 and harvested at the indicated times. (A) For each time point, 20 μg of total RNA was analyzed by Northern blot. A quantification of total HIV mRNA by the Intelligent Quantifier program is represented. (B) For each time point, 200 μg of protein was analyzed by SDS-PAGE and Western blotting with a polyclonal antibody against gp160/gp120.
The amount of the HIV-1 gp160/gp120 proteins was evaluated by Western blot assays with a polyclonal antibody. In the VV clone, gp160/gp120 polypeptides could be detected as early as 3 days postinfection and reached maximal levels at days 5 and 6 (Fig. 6B). The high RT activity in the VS2 clone was correlated with the presence of gp160/gp120 as early as 2 days after the onset of infection; the amount of both proteins increased to maximum levels at days 4, 5, and 6. On the other hand, gp160/gp120 polypeptides were poorly expressed in the VAS2 clone and were absent from day 7 (Fig. 6B).
DISCUSSION
HIV replication decreases when RLI is underexpressed in human cells.
We show here that RLI can be a suppressor of HIV replication when it is underexpressed in human H9 cells by transfection with an RLI antisense cDNA construction. On the contrary, when RLI is overexpressed in human H9 cells transfected with an RLI sense construction, HIV replication is greatly increased.
The strategy we used here bypasses the requirement for IFN treatment to obtain inhibition of viral growth. This strategy has been used with success in different systems to study the role of several IFN-inducible proteins, and it has revealed the importance of these proteins in the antiviral action of IFN. Moreover, this strategy has overcome problems associated with viruses that have developed powerful tools to block specific IFN pathways. Indeed, as outlined in the introduction, during the course of a number of viral infections RNase L activity is not detectable, despite high levels of 2-5A-synthetase, 2-5A, or even RNase L (see reference 32 for a review). These observations provided strong evidence that viruses can inactivate the 2-5A/RNase L pathway. For example, the expression of 2-5A synthetase and/or RNase L in mouse cells restores apoptosis inhibited by the vaccinia virus E3L protein (23) and inhibits replication of EMC virus (8, 9, 24, 32, 36). Similarly, we have previously demonstrated that when RLI expression is decreased in HeLa cells by transfected antisense RLI cDNA constructions, RNase L activity is increased during EMC virus infection and EMC virus production is lower (18).
HIV inhibits the 2-5A/RNase L pathway during the time course of infection (29, 30); similarly, expression of 2-5A-synthetase cDNA from an HIV-1 LTR reduced HIV replication in HeLa T4+ cells (27, 28). The overexpression of RNase L in Jurkat cells and in peripheral blood lymphocytes severely impairs HIV replication (17). It is worth noting that the expression of RNase L in the different systems cited above seems to produce a much greater antiviral effect than did the expression of 2-5A-synthetase (17, 23). Levels of RNase L represent a central limiting component to the antiviral activity of the 2-5A/RNase L pathway.
RNase L activity is correlated with RLI levels in HIV-1-infected human cells.
In the present report evidence is presented that modifying the RLI levels consequently gives rise to different levels of RNase L activity in HIV-1-infected cells and also affects viral production (Fig. 3 and 4).
Western blot assays with antibodies against RNase L and RLI reveal that the amount of RNase L is only transiently decreased upon HIV-1 infection and that RLI is simultaneously induced (Fig. 2). We provide evidence here that the loss of RNase L activity during HIV-1 infection is due not only to a decrease in the level of the RNase L protein itself but also to the accumulation of the RNase L inhibitor, RLI. We have already established that the ratio between RNase L and RLI in cell extracts governs the overall activity of the 2-5A/RNase L system, even when 2-5A is present at doses largely sufficient to activate RNase L (3). When RLI is decreased by the expression of an RLI antisense cDNA construction, the antiviral activity of RNase L against EMC virus is increased (18). These experiments demonstrate that slight changes in the ratio between these two proteins could profoundly modify the 2-5A binding activity of RNase L. The increased RLI level observed here is sufficient to account for the inhibition of RNase L activity observed during HIV-1 infection.
Moreover, the experiments described so far indicate a correlation among HIV-1 infection, the reversible inactivation of RNase L, and RLI induction. Modifying the level of expression of RLI through the expression of antisense or sense constructions represents an attractive possibility to substantiate this hypothesis. The inhibition of RNase L activity by HIV-1 varies in function of the amount of RLI (Fig. 4). The overexpression of RLI giving rise to greater inhibition of RNase L activity correlated with a higher viral production. On the contrary, blocking of RLI by antisense construction reverses the inhibition of RNase L activity observed during HIV-1 infection and, consequently, inhibits viral production. These results are clearly illustrated in Fig. 5 and 6. We could observe a great difference between the VAS and VS clones in the production of HIV-1 at both the mRNA and protein levels.
The 2-5A/RNase L pathway is a major component of the antiviral activity of IFN. Viruses have developed numerous strategies to circumvent this pathway. Many of these viral inhibitors are now being identified, though their presence was first described several years ago (see reference 32 for a review). RLI, the RNase L inhibitor that we have cloned and identified, is implicated in the inhibition of the 2-5A/RNase L pathway by EMC virus (3, 18). The present study describes a new strategy that HIV-1 has evolved to inhibit cellular antiviral defenses. The identification of the antagonistic pathways developed by viruses is important in understanding the physiopathology of viral infection and in the implementation of new and more efficient antiviral strategies. In particular, it would be of interest to understand how HIV-1 is able to induce RLI and to inhibit the cellular antiviral defenses.
ACKNOWLEDGMENTS
We are very grateful to D. Misse (ORSTOM, Montpellier, France) for the gift of antibodies against HIV gp120, to M. Benkirane (IGH, UPR 1142, Montpellier, France) for the gift of HIV LTR plasmid, to G. Cathala (IGMM, Montpellier, France) for the gift of a polyclonal antibody against GAPDH, and to I. Robbins (IGMM, Montpellier, France) for revising the manuscript.
This work was supported by grants from Institut National de la Santé et de la Recherche Médicale to C.B. and from the Agence Nationale de la Recherche contre le SIDA to B.L. C.M. holds a predoctoral fellowship from the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Baron S, Coppenhaver D, Dianzani F, Fleischmann W, Hughes T, Klimpel G, Niesel D, Stanton G, Tyring S. Cell effects of IFNs and animal studies. In: Baron S, editor. Interferon: principles and medical applications. Galveston, Tex: The University of Texas; 1992. pp. 271–287. [Google Scholar]
- 2.Bayard B, Bisbal C, Lebleu B. Activation of ribonuclease L by (2′-5′)(A)4-poly(l-lysine) conjugates in intact cells. Biochemistry. 1986;25:3730–3736. doi: 10.1021/bi00360a038. [DOI] [PubMed] [Google Scholar]
- 3.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNase L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 4.Bisbal C, Salehzada T, Lebleu B, Bayard B. Characterization of two murine (2′-5′)(A)n-dependent endonucleases of different molecular mass. Eur J Biochem. 1989;179:595–602. doi: 10.1111/j.1432-1033.1989.tb14588.x. [DOI] [PubMed] [Google Scholar]
- 5.Carter W A, Strayer D R, Brodsky I, Lewin M, Pellegrino M G, Einck L, Henriques H F, Simon G L, Parenti D M, Scheib R G, et al. Clinical, immunological, and virological effects of ampligen, a mismatched double-stranded RNA, in patients with AIDS or AIDS-related complex. Lancet. 1987;i:1286–1292. doi: 10.1016/s0140-6736(87)90543-5. [DOI] [PubMed] [Google Scholar]
- 6.Cathala G, Savouret J F, Mendez B, West B L, Karin M, Martial J A, Baxter J D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2:329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- 7.Cayley P J, Davies J A, McCullagh K G, Kerr I M. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur J Biochem. 1984;143:165–174. doi: 10.1111/j.1432-1033.1984.tb08355.x. [DOI] [PubMed] [Google Scholar]
- 8.Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- 9.Coccia E M, Romeo G, Nissim A, Marziali G, Albertini R, Affabris E, Battistini A, Fiorucci G, Orsatti R, Rossi G B. A full-length murine 2-5A synthetase cDNA transfected in NIH-3T3 cells impairs EMCV but not VSV replication. Virology. 1990;179:228–233. doi: 10.1016/0042-6822(90)90292-y. [DOI] [PubMed] [Google Scholar]
- 10.Dong B, Silverman R H. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J Biol Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 11.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 12.Hassel B A, Zhou A, Sotomayor C, Maran A, Silverman R H. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 1993;12:3297–3304. doi: 10.1002/j.1460-2075.1993.tb05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr I M, Brown R E. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knight M, Cayley P J, Silverman R H, Wreschner D H, Gilbert C S, Brown R E, Kerr I M. Radioimmune, radiobinding and HPLC analysis of 2-5A and related oligonucleotides from intact cells. Nature. 1980;288:189–192. doi: 10.1038/288189a0. [DOI] [PubMed] [Google Scholar]
- 15.Lemaitre M, Guetard D, Henin Y, Montagnier L, Zerial A. Protective activity of tetracycline analogs against the cytopathic effect of the human immunodeficiency viruses in CEM cells. Res Virol. 1990;141:5–16. doi: 10.1016/0923-2516(90)90052-k. [DOI] [PubMed] [Google Scholar]
- 16.Locardi C, Petrini C, Boccoli G, Testa U, Dieffenbach C, Butto S, Belardelli F. Increased human immunodeficiency virus (HIV) expression in chronically infected U937 cells upon in vitro differentiation by hydroxyvitamin D3: roles of interferon and tumor necrosis factor in regulation of HIV production. J Virol. 1990;64:5874–5882. doi: 10.1128/jvi.64.12.5874-5882.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maitra R K, Silverman R H. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J Virol. 1998;72:1146–1152. doi: 10.1128/jvi.72.2.1146-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinand C, Salehzada T, Silhol M, Lebleu B, Bisbal C. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis virus (EMCV)-infected cells. Eur J Biochem. 1998;254:238–247. doi: 10.1046/j.1432-1327.1998.2540248.x. [DOI] [PubMed] [Google Scholar]
- 19.Nilsen T W, Maroney P A, Baglioni C. Synthesis of (2′-5′)oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. J Virol. 1982;42:1039–1045. doi: 10.1128/jvi.42.3.1039-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paez E, Esteban M. Resistance of vaccinia virus to interferon is related to an interference phenomenon between the virus and the interferon system. Virology. 1984;134:12–28. doi: 10.1016/0042-6822(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 21.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rice A P, Roberts W K, Kerr I M. 2-5A accumulates to high levels in interferon-treated, vaccinia virus-infected cells in the absence of any inhibition of virus replication. J Virol. 1984;50:220–228. doi: 10.1128/jvi.50.1.220-228.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivas C, Gil J, Melkova Z, Esteban M, Diaz G M. Vaccinia virus E3L protein is an inhibitor of the interferon (IFN)-induced 2-5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 24.Rysiecki G, Gewert D R, Williams B R. Constitutive expression of a 2′,5′-oligoadenylate synthetase cDNA results in increased antiviral activity and growth suppression. J Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- 25.Salehzada T, Silhol M, Steff A M, Lebleu B, Bisbal C. 2′,5′-Oligoadenylate-dependent RNase L is a dimer of regulatory and catalytic subunits. J Biol Chem. 1993;268:7733–7740. [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schroder H C, Suhadolnik R J, Pfleiderer W, Charubala R, Muller W E. (2′-5′)Oligoadenylate and intracellular immunity against retrovirus infection. Int J Biochem. 1992;24:55–63. doi: 10.1016/0020-711x(92)90229-t. [DOI] [PubMed] [Google Scholar]
- 28.Schroder H C, Ugarkovic D, Merz H, Kuchino Y, Okamoto T, Muller W E. Protection of HeLa-T4+ cells against human immunodeficiency virus (HIV) infection after stable transfection with HIV LTR-2′,5′-oligoadenylate synthetase hybrid gene. FASEB J. 1990;4:3124–3130. doi: 10.1096/fasebj.4.13.1698680. [DOI] [PubMed] [Google Scholar]
- 29.Schroder H C, Wenger R, Kuchino Y, Muller W E. Modulation of nuclear matrix-associated 2′,5′-oligoadenylate metabolism and ribonuclease L activity in H9 cells by human immunodeficiency virus. J Biol Chem. 1989;264:5669–5673. [PubMed] [Google Scholar]
- 30.Schroder H C, Wenger R, Rottmann M, Muller W E. Alteration of nuclear (2′-5′)oligoriboadenylate synthetase and nuclease activities preceding replication of human immunodeficiency virus in H9 cells. Biol Chem Hoppe-Seyler. 1988;369:985–995. doi: 10.1515/bchm3.1988.369.2.985. [DOI] [PubMed] [Google Scholar]
- 31.Silverman R H. 2-5A-dependent RNase L: a regulated endoribonuclease in the interferon system. In: D’Alessio G, Riordan J F, editors. Ribonucleases: structure and functions. New York, N.Y: Academic Press, Inc.; 1997. pp. 515–551. [Google Scholar]
- 32.Silverman R H, Cirino N M. RNA decay by the interferon-regulated 2-5A system as a host defense against viruses. In: Hartford J B, Morris D R, editors. mRNA metabolism and post-transcriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 295–309. [Google Scholar]
- 33.Sobol R W, Fisher W L, Reichenbach N L, Kumar A, Beard W A, Wilson S H, Charubala R, Pfleiderer W, Suhadolnik R J. HIV-1 reverse transcriptase: inhibition by 2′,5′-oligoadenylates. Biochemistry. 1993;32:12112–12118. doi: 10.1021/bi00096a023. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ushijima H, Rytik P G, Schacke H, Scheffer U, Muller W E, Schroder H C. Mode of action of the anti-AIDS compound poly(I) · poly(C12U) (Ampligen): activator of 2′,5′-oligoadenylate synthetase and double-stranded RNA-dependent kinase. J Interferon Res. 1993;13:161–171. doi: 10.1089/jir.1993.13.161. [DOI] [PubMed] [Google Scholar]
- 36.Watling D, Serafinowska H T, Reese C B, Kerr I M. Analogue inhibitor of 2-5A action: effect on the interferon-mediated inhibition of encephalomyocarditis virus replication. EMBO J. 1985;4:431–436. doi: 10.1002/j.1460-2075.1985.tb03647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitaker J R, Granum P E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- 38.Williams B R, Golgher R R, Brown R E, Gilbert C S, Kerr I M. Natural occurrence of 2-5A in interferon-treated EMC virus-infected L cells. Nature. 1979;282:582–586. doi: 10.1038/282582a0. [DOI] [PubMed] [Google Scholar]
- 39.Wu J M, Chiao J W, Maayan S. Diagnostic value of the determination of an interferon-induced enzyme activity: decreased 2′,5′-oligoadenylate dependent binding protein activity in AIDS patient lymphocytes. AIDS Res. 1986;2:127–131. doi: 10.1089/aid.1.1986.2.127. [DOI] [PubMed] [Google Scholar]
- 40.Zhou A, Hassel B A, Silverman R H. Expression cloning of 2-5A-dependent RNase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753–765. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]