Abstract
The RNA-dependent RNA polymerase of vesicular stomatitis virus (VSV), a nonsegmented negative-strand RNA virus, directs two discrete RNA synthetic processes, transcription and replication. Available evidence suggests that the two short extragenic regions at the genomic termini, the 3′ leader (Le) and the complement of the 5′ trailer (TrC), contain essential signals for these processes. We examined the roles in transcription and replication of sequences in Le and TrC by monitoring the effects of alterations to the termini of subgenomic replicons, or infectious viruses, on these RNA synthetic processes. Distinct elements in Le were found to be required for transcription that were not required for replication. The promoter for mRNA transcription was shown to include specific sequence elements within Le at positions 19 to 29 and 34 to 46, a separate element at nucleotides 47 to 50, the nontranscribed leader-N gene junction. The sequence requirements for transcription within the Le region could not be supplied by sequences found at the equivalent positions in TrC. In contrast, sequences from either Le or TrC functioned well to signal replication, indicating that within the confines of the VSV termini, the sequence requirements for replication were less stringent. Deletions engineered at the termini showed that the terminal 15 nucleotides of either Le or TrC allowed a minimal level of replication. Within these confines, levels of replication were affected by both the extent of complementarity between the genomic termini and the involvement of the template in transcription. In agreement with our previous observations, increasing the extent of complementarity between the natural termini increased levels of replication, and this effect was most operative at the extreme genome ends. In addition, abolishing the use of Le as a promoter for transcription enhanced replication. These analyses (i) identified signals at the termini required for transcription and replication and (ii) showed that Le functions as a less efficient promoter for replication than TrC at least in part because of its essential role in transcription. Consequently, these observations help explain the asymmetry of VSV replication which results in the synthesis of more negative- than positive-sense replication products in infected cells.
The 11,161-nucleotide (nt) nonsegmented negative-sense RNA genome of vesicular stomatitis virus (VSV), tightly encapsidated by its nucleocapsid protein (N), forms the active template for transcription and replication by the viral polymerase (18). The same core polymerase, a complex of the phosphoprotein (P) and large subunit (L), is required for both RNA synthetic processes, but other than the requirement for a constant supply of N protein during replication (2, 34), the mechanism by which these two discrete activities are regulated is unknown.
The viral genome comprises a 47-nt leader region (Le), five transcriptional units for the N, P, M (matrix), G (glycoprotein), and L mRNAs, and a 59-nucleotide trailer region (Tr), arranged in the order 3′ Le-N-P-M-G-L-Tr 5′. The 3′ Le is thought to act as a promoter for transcription of the viral mRNAs and also as a promoter for replication to yield the positive-sense antigenome. The 3′ end of the positive-strand antigenome, which is the complement of the 59-nt genomic 5′ Tr (TrC), is thought to act as the promoter for replication of progeny negative-sense genomes. Thus, Le is used as a promoter for two discrete RNA synthetic processes, transcription and replication, whereas TrC acts exclusively as a promoter of replication. Consistent with the use of Le and TrC as promoters of replication, they share common sequence elements and are identical at 25 of 46 positions for VSV (Indiana). The areas of identity are localized at nt 1 to 8, 13 to 18, 30 to 33, whereas significant sequence divergence occurs at nt 9 to 12, 19 to 29, and 34 to 46 (Fig. 1A). While it is generally accepted that the short terminal regions of all the Mononegavirales contain cis-acting signals essential for encapsidation, replication, transcription, and assembly (7, 9, 12, 14, 16, 31–33, 53), the requirements at the termini for transcription and replication are poorly defined; this in part is due to the multifunctional nature of the termini, making dissection of their roles complex.
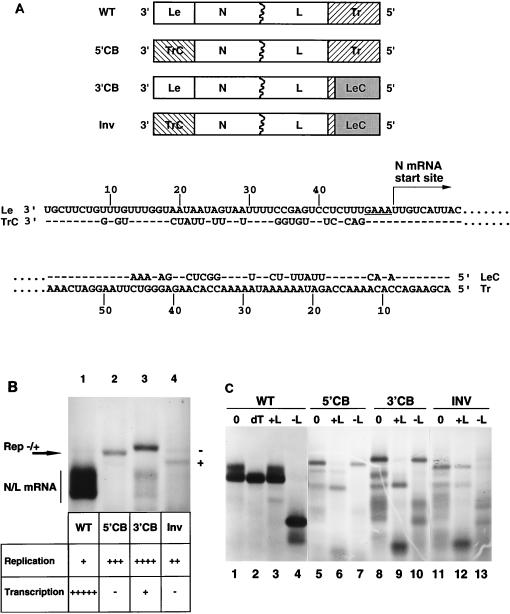
FIG. 1.
Examination of the roles of the VSV genomic termini in transcription and in replication. (A) Structures of VSV genomic analog and mutant VSV replicons showing the four possible permutations of the genomic termini. Negative-sense genomic analogs of the VSV genome were constructed to contain the wild-type genomic (WT), 5′ copyback (5′CB), 3′ copyback (3′CB), and inverted (Inv) termini. Sequences of the 3′ Le and TrC and of the 5′ Tr and LeC are shown with dashes to indicate positions of sequence identity. Exchanges of the 3′ Le with TrC were performed so that the nontranscribed tetranucleotide GAAA (underlined) and the N gene start site, both required for transcription initiation, remained intact. (B) RNA synthesis from VSV genomic analogs. Cells were infected with vTF7-3, transfected with cDNAs for the VSV genomic analogs WT, 5′CB, 3′CB, and Inv (lanes 1 to 4, respectively) along with plasmids encoding the VSV N, P, and L proteins, and exposed to [3H]uridine (33 μCi/ml) in the presence of actinomycin D (10 μg/ml) as described previously (49). Cytoplasmic extracts were prepared, and total RNA was analyzed by electrophoresis on agarose-urea gels and visualized by fluorography, as described previously (49). The products were identified as described in the text and as shown in panel C. Semiquantitative values are given for the levels of replication and transcription, as described in the Materials and Methods. (C) Characterization of RNA products by RNase H cleavage. Purified labeled RNAs were annealed with the indicated strand- and sequence-specific oligonucleotides prior to exposure to RNase H as described in Materials and Methods and analyzed by electrophoresis on agarose-urea gels. Lanes: 0, no oligonucleotide; dT, oligo(dT), −L, negative-sense L-specific oligonucleotide corresponding to nt 10925 to 10909 of the complete genome sequence of VSV; +L, positive-sense L-specific oligonucleotide corresponding to nt 10925 to 10908 of the complete VSV genome sequence. The positive-sense N/L mRNA (lanes 4 and 10) and positive-sense replication (Rep) products (lanes 7, 10, and 13) were cleaved by RNase H after annealing to −L. Negative-sense replication products were cleaved by RNase H when annealed with +L (lanes 6, 9, and 12).
Viral transcription, which is the predominant RNA synthetic activity in infected cells, occurs first. Available evidence suggests that the polymerase uses a single entry site at the 3′ end of the genome (17) and transcribes a 47-nt leader RNA which is neither capped or polyadenylated (10, 11, 26) and five capped and polyadenylated mRNAs whose relative abundance decreases with distance from the 3′end: N > P > M > G > L (1, 3, 47). At each of these gene junctions, a conserved sequence signals termination and polyadenylation of the upstream mRNA and transcription of the downstream mRNA (4, 5, 39, 43).
During replication, in a reaction that requires continuous N protein synthesis (34), the polymerase initiates at the exact 3′ end of the negative-sense genome and generates a complete genome-length positive strand, ignoring the transcriptive stop and start signals. In turn, the 3′ end of the positive strand (TrC) promotes synthesis of the genome-length negative strand. Replication is asymmetric, producing an approximately 5 to 10-fold excess of negative-sense genomes over positive-sense antigenomes in infected cells (24, 38, 40, 42, 48). This led to the suggestion that TrC is a more effective promoter of replication than Le. Support for this notion came from studies of defective interfering (DI) particle RNAs, which outcompete wild-type virus at the level of replication. The most efficiently replicating class of DI RNAs are generated by a copyback mechanism so that the 3′ terminus is complementary to the 5′ terminus, and thus the promoters for both positive- and negative-strand synthesis are TrC. However, such a copyback arrangement of the termini increases the extent of complementarity between the termini, which enhances levels of replication and could account for the replicative advantage of these DI RNAs (49).
To examine the cis-acting requirements at the termini of the VSV genome for transcription and replication, we made a series of alterations to the Le and Tr regions of subgenomic replicons and of infectious virus and determined the effects on RNA synthesis. The alterations included compensatory changes at both termini to distinguish the effects on RNA synthesis of altering either the sequence or the extent of complementarity between the termini. The results reported here define the 15 most terminal nucleotides at each end of the genome as essential for replication and show that both the primary sequence and the extent of complementarity between the natural termini regulate RNA synthesis. These studies identify distinct sequences at the termini essential for transcription but not replication, and they show that Le functions as a less efficient promoter of replication than TrC, at least in part because of its essential role in transcription.
MATERIALS AND METHODS
Plasmid construction and transfections.
Previously we described a specialized transcription plasmid, p8(−) (49), referred to here as plasmid pWT, (wild type) that was designed to generate a VSV subgenomic replicon that contained the wild-type genomic Le and Tr regions surrounding a single transcriptional unit that comprised of a fusion of the 5′ end of N mRNA with the 3′ end of the L mRNA (the WT replicon [Fig. 1A]). The genomic termini of pWT were altered by either restriction fragment exchange or site-directed mutagenesis using standard techniques (37). The primary structures of the altered terminal regions were determined by sequence analysis through the mutated regions, and the sequences of the altered termini are shown in the relevant figures. In addition, selected mutations were incorporated into the genomic termini of an infectious cDNA clone of VSV, pVSV1(+) (50). As described previously, plasmids expressing either VSV subgenomic replicons (33, 49) or the full-length VSV antigenome (50), together with plasmids encoding the VSV N, P, and L proteins, each under the control of T7 promoters, were transfected into baby hamster kidney (BHK-21) cells that were infected with a recombinant vaccinia virus, vTF7-3 (21), expressing T7 RNA polymerase.
Analysis of RNA synthesis from subgenomic replicons.
RNA synthesis was examined by direct metabolic labeling of cells 15 h posttransfection. Cells were exposed for 5 h to [3H]uridine (33 μCi/ml) in the presence of actinomycin D (10 μg/ml). Cells were harvested and either total RNA or N-protein-encapsidated RNA was analyzed as described elsewhere (33, 49). Autoradiographs of fluorogrammed gels were scanned with a PDI densitometer 320i, and a semiquantitative value was assigned to relative levels of replication and transcription. These semiquantitative analyses are representative of at least three independent experiments, between which no significant variation was observed. Exact quantitation of the genomic and antigenomic replication products was not possible for technical reasons. These RNA species typically comigrate. In addition, because of the large quantity of T7 transcripts synthesized as a source of the replicon RNA in transfected cells, hybridization or reverse transcriptase (RT)-mediated PCR (RT-PCR) could not be used to accurately distinguish and quantitate the products of replication.
Primer extension was used to map the positions of the 5′ ends of the mRNAs and positive-sense replication products, using Moloney murine leukemia virus (MoMLV) RT (GIBCO/BRL) and a primer that annealed to positive-sense RNA in the L gene at positions 10925 to 10908 of the complete VSV genome sequence, as described previously (49). Where indicated, RNA species were further characterized by annealing to sequence- and strand-specific oligonucleotides followed by incubation with RNase H as described elsewhere (5).
Characterization of recombinant viruses.
Recombinant VSV was recovered, purified, and amplified in BHK-21 cells as described elsewhere (50). To confirm that recovered viruses contained the indicated mutation in the genomic termini, viral RNA was purified from 108 PFU of amplified VSV and reverse transcribed with avian myeloblastosis virus RT and a primer that annealed to negative-sense RNA at nt 11026 to 11043 of the complete genome sequence. One-tenth of this reaction was used for DNA amplification by PCR using the primer described above and a second primer that annealed to the extreme 3′ end of positive-sense VSV RNA (nt 11161 to 11144). These PCR products were cloned and sequenced by standard methods. To analyze viral RNA synthesis, cells were infected at a multiplicity of infection of 3, exposed to [3H]uridine in the presence of actinomycin D (10 μg/ml) for 3 h, and analyzed as described previously (50).
RESULTS
Effects on RNA synthesis of switching the genomic termini.
VSV genomic RNA contains at the 3′ terminus the 47-nt extragenic Le and at the 5′ terminus the 59-nt extragenic Tr. The genomic 3′ Le and antigenomic 3′ TrC are thought to act as the promoters for RNA synthesis. Consequently, any of the following four combinations of the termini should yield replicable RNA: (i) the wild-type termini, 3′ Le-Tr 5′; (ii) a 5′ copyback arrangement of the termini, 3′ TrC-Tr 5′, as found for many DI particles; (iii) a 3′ copyback arrangement of the termini, 3′ Le-LeC 5′; and (iv) the arrangement found on the positive-strand, 3′ TrC-LeC 5′. As a first step to determine whether distinct sequences within the termini were required for either transcription or replication, the RNA synthetic activity of the subgenomic WT replicon (Fig. 1A) was compared to that of each of the three other possible terminal permutations by engineering these changes into pWT to provide replicons 5′CB, 3′CB, and Inv (Fig. 1A). The terminal regions were exchanged so that the sequences and locations of the leader-N gene junction and of the N gene start site, important requirements for transcription (see below), remained intact. Because Le and TrC differ slightly in size, when TrC was exchanged with Le in 3′CB and Inv, 9 nt of TrC sequence remained at the antigenomic 3′ terminus (Fig. 1A).
RNA synthesis was examined by transfection of pWT into vTF7-3-infected BHK-21 cells together with plasmids expressing the trans-acting N, P, and L proteins required to support the recovery of replicable RNA transcribed from the transfected cDNA, as described previously (49). The major product of RNA synthesis obtained from pWT (Fig. 1B, lane 1) was previously identified by oligo(dT) chromatography, Northern blotting, and primer extension analysis as the N/L mRNA product of transcription (49). Consistent with the levels of transcription and replication observed in VSV-infected cells, replication of the wild-type genomic RNA was the minor synthetic event, and a band that corresponds to both the positive- and negative-sense replication products was just visible above the N/L mRNA (Fig. 1B, lane 1). Replication products were previously identified by the fact that they were encapsidated with N protein and were immunoprecipitated by anti-N antiserum. Primer extension analysis confirmed that both positive- and negative-sense products of genome replication were synthesized (49).
In this analysis, the RNA products were further characterized by RNase H cleavage following incubation with sequence- and strand-specific oligonucleotides (Fig. 1C). For example, the positive-sense mRNA was cleaved by RNase H after annealing with a negative-sense L-specific oligonucleotide, −L, (Fig. 1C, lane 4), but not when incubated with a positive-sense L-specific oligonucleotide, +L (Fig. 1C, lane 3). This mRNA migrated as a doublet that resolved to a single discrete product when exposed to RNase H after annealing to oligo(dT) (Fig. 1C, lane 2), suggesting that it was present in two forms that differed in the extent of polyadenylation. This is most likely a consequence of the presence of the recombinant vaccinia virus vTF7-3, which encodes its own poly(A) polymerase (6).
Replication was the exclusive RNA synthetic activity following transfection of 5′CB (Fig. 1B, lane 2), and it was more abundant than observed for the WT replicon (Fig. 1B, lane 1). The gels used here resolve RNA on the basis of both size and base composition, which results in the separation of the genomic and antigenomic replication products of 5′CB (Fig. 1B, lane 2). Using oligonucleotide-directed RNase H cleavage, we identified the slower-migrating product as negative-sense genomic RNA, and it was more abundant than the faster-migrating antigenomic positive-sense RNA (Fig. 1C, lanes 5 to 7).
3′CB replicated to higher levels than 5′CB and transcribed at approximately 1/10 of the level observed for WT (Fig. 1B, Lane 3). Again, replication products were predominantly negative sense, but in this case the positive- and negative-sense replication products comigrated (Fig. 1C, lanes 8 to 10).
Inv replicated at higher levels than WT, but it did not direct transcription (Fig. 1B, lane 4). Oligonucleotide-directed RNase H cleavage showed that more of the positive-sense than the negative-sense replication product of Inv was produced (Fig. 1B, lane 4; Fig. 1C, lanes 11 to 13).
Since neither Inv nor 5′CB was a template for mRNA synthesis (Fig. 1B, lanes 2 and 4), these exchanges showed that the Le contained signals essential for transcription that were not provided by TrC despite sequence identity at 25 of 46 positions. In addition, these data supported our previous observation that increasing the extent of complementarity between the natural genomic termini enhanced replication, as evidenced by comparing 3′CB and 5′CB, in which the termini are perfectly complementary for 49 nt, with WT and Inv, in which 21 positions are mismatched (Fig. 1B). These data also showed that replication was enhanced as the use of Le as a promoter of transcription was decreased, as shown by comparing replication levels of WT and Inv, which have the same degree of terminal complementarity but in which Inv is not a template for transcription.
Effects of deletions in Le on RNA synthesis.
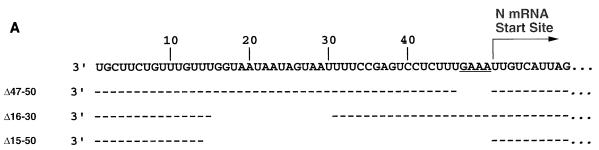
The promoter exchanges described above demonstrated that Le contained essential signals for transcription and replication. To narrow down the specific signals at the genomic 3′ terminus required for either transcription or replication, we engineered deletions into the 3′ terminus of the WT replicon (Fig. 2A) and examined the effects on RNA synthesis (Fig. 2B). Deletion of the nontranscribed leader-N gene junction, Δ47–50, abolished transcription but enhanced replication (Fig. 2B, lane 2), indicating that it was essential for transcription. Deletion of nt 15 to 50 resulted in a template that was inactive for transcription but allowed a low level of replication (Fig. 2B, lane 4), indicating that nt 15 to 50 are not essential for replication. While the inability of Δ15–50 to transcribe could be attributed to deletion of the leader-N gene junction, each of the promoter exchanges described above maintained the leader-N gene junction intact, indicating that a separate specific sequence element within Le is essential for transcription. Therefore, in all subsequent changes to the Le, the leader-N gene junction, the N transcriptional start site, and the terminal 15 nt of the genome were kept intact.
FIG. 2.
Effects of RNA synthesis of deletions at the genomic 3′ terminus. (A) Sequences of the deletions. Sequences of the negative-sense genomic Le of the WT replicon and each of the deletions is sown 3′ to 5′, highlighting the N mRNA start site. Dashes indicate positions of sequence identity; blanks indicate deleted nucleotides (B) Agarose-urea gel analysis of actinomycin D-resistant RNAs synthesized in cells transfected with the indicated VSV replicon and the VSV N, P, and L support plasmids. RNAs were labeled, harvested, and analyzed as described in Materials and Methods. Rep, replication product. (C) Characterization of the products of RNA synthesis of Δ16–30 by RNase H analysis. Cells were transfected with Δ16–30, and the products of RNA synthesis were labeled and harvested as described in Materials and Methods. Purified RNAs were annealed with sequence- and strand-specific oligonucleotides prior to exposure to RNase H, and the products were analyzed by electrophoresis on agarose-urea gels as described in the text. Lanes 1 and 2, WT RNAs incubated with the indicated oligonucleotides. Lane 1 (0), no primer; lane 2 (dT), oligo(dT), shown to indicate the different mobility of the WT and Δ16–30 RNAs; lanes 3 to 8, Δ16–30 RNAs incubated with the indicated oligonucleotides. −Le (lane 7) is −LeΔ16–30, a negative-sense oligonucleotide designed to anneal to the deleted positive-sense leader of Δ16–30. (D) Primer extension analysis on the positive-sense products of RNA synthesis. Total cellular RNA was examined by primer extension with MoMLV RT and a primer (indicated by the arrow) which annealed to positive-sense RNA at positions 10925 to 10909 of the complete VSV genome sequence. As a control to demonstrate that the products were dependent on the VSV polymerase, parallel primer extension reactions were performed with RNAs extracted from cells in which the plasmid encoding the large polymerase subunit (pL) was omitted from the transfections (−). Positions of the products of transcription (192 nt) and replication (242 nt for WT; 227 nt for Δ16–30) are indicated. A dideoxynucleotide sequence ladder of the WT replicon generated with the same primer is shown for comparison.
Methylation protection studies previously indicated that nt 16 to 30 of Le region were contacted by the P component of the viral polymerase (23). To determine whether this region was required for transcription, nt 16 to 30 of Le were deleted, to yield pΔ16–30. The products of RNA synthesis obtained from Δ16–30 were of unexpected mobility (Fig. 2B, lane 3). However, the majority were positive sense, as demonstrated by their cleavage with RNase H after incubation with oligonucleotide −L (Fig. 2C, lane 5) but not with oligonucleotide +L (Fig. 2C, lane 6). Some of the products were polyadenylated, as shown by RNase H cleavage after incubation with oligo(dT) (Fig. 2C, lane 4). Thus, Δ16–30 produced polyadenylated positive-sense RNAs that were both larger and smaller than the N/L mRNA obtained from pWT (Fg. 2C; compare lanes 1 and 2 with lanes 3 and 4). To examine whether these RNAs contained leader or trailer sequence at the 5′ or 3′ termini, purified RNAs were incubated with either a negative-sense Δ16–30 leader-specific oligonucleotide (−LeΔ16–30) or a negative-sense trailer-specific oligonucleotide (−Tr) prior to exposure to RNase H. These analyses showed that some RNAs contained the positive-sense leader linked to the mRNA (Fig. 2C, lane 7) and some contained a positive-sense trailer region (Fig. 2C, lane 8).
Because of the unusual size of the positive-sense transcription products observed for Δ16–30, we examined whether the 5′ terminus of any transcripts mapped to the authentic N gene start site. Primer extension analysis was performed with an oligonucleotide that annealed to positive-sense VSV RNA at residues 10925 to 10909 of the complete VSV genome sequence as depicted in Fig. 2D. As predicted, extension of this primer by MoMLV RT on the positive-sense replication product of WT yielded a product of 242 nt, and a product of 192 nt was obtained from extension on the mRNA (Fig. 2D, lane 1); both products were absent when the plasmid expressing the L protein was omitted from the transfection (Fig. 2D, lane 2). For technical reasons, Δ16–30 was generated from a positive-sense transcript; thus, products of replication could not be distinguished from the primary T7 transcript or the Le-mRNA readthrough (Fig. 2D, lane 3, 227 nt). However, as shown by the presence of a polymerase-dependent product at 192 nt the 5′ termini of some transcripts mapped to the authentic N gene start site (Fig. 2D, lane 3). These data show that deletion of nt 16 to 30 of Le did not prevent polymerase recognition or mRNA synthesis, but the major products were of unexpected mobility and contained leader and or trailer sequences (Fig. 2C, lanes 3 to 8). These data suggest that the polymerase recruited by Δ16–30 was unable to efficiently obey the stop and start signals found at the Le-N and L-Tr junctions. For this reason, further analysis of Le by deletion mutagenesis was not carried out. As an alternative approach to map the requirements in Le for transcription, we exchanged the distinct regions of Le and TrC; these analyses are described later.
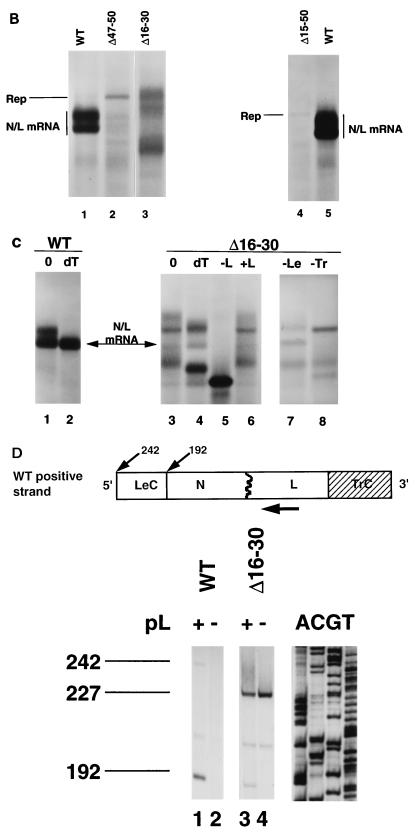
Effects of deletions in trailer on RNA synthesis.
The above analysis demonstrated that the Le-N gene junction was essential for transcription but not replication (Fig. 2B, lane 2) and that TrC was unable to function as a promoter of transcription, acting exclusively as a promoter of replication (Fig. 1B, lane 4). In addition, analysis of Δ15–50 demonstrated that 15 nt of the genomic 3′ Le functioned as a promoter for replication, albeit inefficiently. To map the requirements for replication provided by TrC, we made a series of deletions in the 5′ Tr region of WT. These deletions were engineered from the naturally occurring AflII site located 51 nt from the 5′ end of the WT replicon, using either exonuclease III or site-directed mutagenesis, to yield plasmids designed to generate RNAs in which the genomic 5′ termini were incrementally reduced from 51 to 14 nt (Fig. 3A). These replicons were named to reflect the number of authentic nucleotides remaining at their 5′ termini; their sequence are shown in Fig. 3A. Because replication is only a minor event for replicons with wild-type termini, the direct effect of these deletions on replication was not readily visualized by agarose-urea gel analysis. However, these deletions decreased transcription incrementally (Fig. 3B, lanes 1 to 6), so that transcription was 1/20 of the wild-type level for WT/22 and only just above background for WT/14. The decrease in transcription products was presumably a consequence of the reduction in levels of RNA replication, providing less negative-sense template for the polymerase, as there were no changes to the promoter for transcription, namely, the genomic 3′ Le. While levels of transcription from these replicons were only an indirect measure of their ability to replicate, primer extension analyses confirmed that WT/14 synthesized the positive-sense replication product, but at greatly reduced levels (data not shown). Coupled with results of analyses of the deletions at the genomic 3′ terminus, these data suggest that the terminal 15 nt of either end of the genome allowed a low level of replication. A direct assessment of the effects of these deletions on RNA replication was provided by introducing these same alterations into a cDNA clone of DI-T, a template that exclusively replicated. In this study, deletions that progressively reduced TrC from 59 to 14 nt were shown to cause an incremental decrease in levels of replication and also to affect the balance of positive- and negative-sense replication products. These results are described in detail in the accompanying report (52).
FIG. 3.
Progressive deletions in the genomic 5′ Tr of the WT replicon, generated as described in Materials and Methods. Sequences of the altered regions are shown as genomic, negative sense, 3′ to 5′. These replicons were named to reflect the number of authentic nucleotides remaining at the 5′ terminus. Positions of identity are indicated by dashes; numbers indicate positions of boundaries of the deletions. (B) Effects on RNA synthesis of deletions in the genomic 5′ Tr region. Each of the replicons was transfected into BHK-21 cells; RNAs were labeled in the presence of actinomycin D, purified, and examined by electrophoresis on agarose-urea gels as described in the text. Rep, replication product. Note that the altered mobility of the products of transcription of WT/29 reflects the deletion of the polyadenylation site (underlined in panel A). These transcripts thus contain a positive-sense copy of 29 nt of the genomic Tr at their 3′ termini.
Effect on polymerase behavior of exchanges between the termini.
The previous exchanges and deletions showed that sequences between nt 15 and 46 of the 3′ Le region, and a separate element, nt 47 to 50, were required for transcription and that these requirements could not be provided by TrC. However, as exemplified by Δ16–30, further mapping by deletions was complicated by the synthesis of RNA species of unexpected mobility.
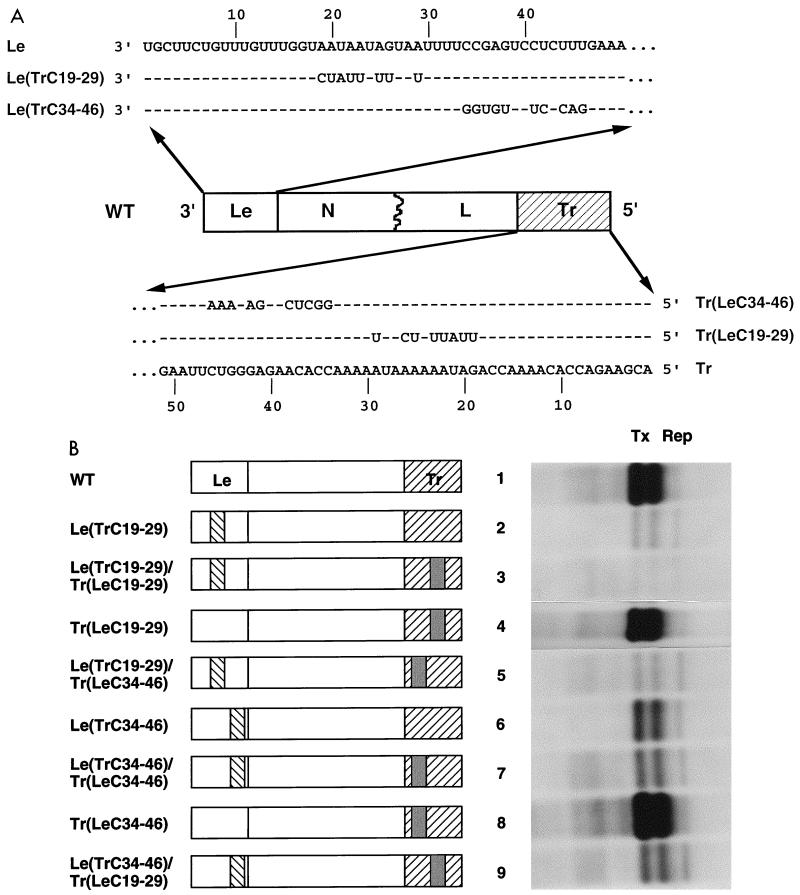
The sequences of the 3′ termini of replicon WT, which was an active template for transcription, and replicons 5′CB and Inv, which were not, are distinct at 21 of 46 positions shown in Fig. 1A. As mentioned above, the regions of sequence difference fell into three groups, nt 9 to 12, nt 19 to 29, and nt 34 to 46. As these were the only sequence changes between the 3′ termini of the WT replicon and those of either 5′CB or Inv, we concluded that these sequences were responsible for the failure of 5′CB and Inv to transcribe. As an alternative approach to the use of deletions in Le to examine the requirements for transcription, regions of Le were exchanged for the equivalent positions of TrC. Since it was not practical to generate all possible permutations of these 21 nt we exchanged groups of them as shown in Fig. 4A. Since we demonstrated previously that exchange of nt 9, 11, and 12 of Le for TrC resulted in only a minor alteration in RNA synthesis (49), these changes were not reanalyzed here.
FIG. 4.
Effects on RNA synthesis of exchanges between the genomic termini of the subgenomic WT replicon. (A) Sequence alterations to the genomic termini. Nucleotides 19 to 29 or 34 to 46 of the 3′ Le of WT were replaced with the equivalent nucleotides from TrC, yielding two altered leader regions, Le(TrC19–29) and Le(TrC34–46), whose sequences are shown. In addition, nt 19 to 29 or 34 to 46 of the genomic 5′ Tr of WT were substituted with the equivalent positions from LeC, yielding two altered trailer regions, Tr(LeC19–29) and Tr(LeC34–46), whose sequences are shown. For comparison, the sequences of the 3′ Le and 5′ Tr of WT are shown; positions of sequence identity are represented by dashes. The possible permutations of these two altered regions of Le and Tr were generated to yield a panel of eight additional replicons (Fig. 4B). LeC, solid fill; TrC, leftward diagonal lines. (B) Agarose-urea gel analysis of VSV-specific RNAs generated from genomic analogs with altered Le and Tr sequences. Following transfection of each replicon into BHK-21 cells, RNAs were labeled with [3H]uridine in the presence of actinomycin D and analyzed as described in Materials and Methods. The products of replication and transcription by the VSV polymerase are indicated by Rep and Tx, respectively.
The two other groups of sequence divergence, nt 19 to 29 and 34 to 46 of Le, were substituted by the corresponding regions of TrC in WT, yielding Le(TrC19–29) and Le(TrC34–46), respectively (Fig. 4A). These exchanges increased replication and decreased transcription, suggesting that both regions (nt 19 to 29 and 34 to 46) were essential components of the transcriptional promoter (Fig. 4B, lanes 2 and 6). However these exchanges increased the extent of complementarity between the termini from 25 of 46 nt for WT to 33 of 46 nt for Le(TrC19–29) and 35 of 46 nt for Le(TrC34–46). To determine whether the observed effects on RNA synthesis were due to either increases in the extent of complementarity, causing an increase in replication and decrease in transcription, or alteration of the primary sequence of Le, we engineered compensatory mutations into the 5′ Tr, resulting in two further replicons, Le(TrC19–29)/Tr(LeC19–29) and Le(TrC34–46)/Tr(LeC34–46). These mutations restored the wild-type terminal complementarity (25 of 46 positions) but did not restore wild-type RNA synthesis (Fig. 4B, lanes 3 and 7), thus demonstrating that nt 19 to 29 and 34 to 46 of Le influence the efficiency of transcription.
As a control, the effects of these alterations on the Tr region alone were examined. Exchange of nt 19 to 29 or 34 to 46 of Tr with LeC in WT yielded Tr(LeC19–29) and Tr(LeC34–46), respectively (Fig. 4A). These exchanges slightly increased levels of RNA replication (Fig. 4B, lanes 4 and 8). Levels of transcription were not inhibited but rather were enhanced, presumably as a consequence of the increased replication providing more negative-strand templates for the polymerase.
The significance of the extent of complementarity between the termini in regulating RNA synthesis was examined further, using two replicons with almost completely complementary termini (43 of 46 nt), Le(TrC19–29)/Tr(LeC34–46), and Le(TrC34–46)/Tr(LeC19–29) (Fig. 4A). Both were inefficient templates for mRNA synthesis (Fig. 4B, lanes 5 and 9), because highly complementary termini and alterations to nt 19 to 29 and 34 nt 46 of Le are associated with down-regulation of mRNA synthesis. As expected from the increase in terminal complementarity and decrease in transcription, both Le(TrC19–29)/Tr(LeC34–46) and Le(TrC34–46)/Tr(LeC19–29) replicated to higher levels than observed for WT. However, compared to 3′CB and 5′CB, in which the termini are perfectly complementary for 49 nt, Le(TrC19–29)/Tr(LeC34–46) and Le(TrC34–46)/Tr(LeC19–29) were less efficient templates for replication. We previously showed that the major enhancement of replication occurred when perfect complementarity between the natural termini was increased from 8 nt as found for the wild type up to 22 nt (49), suggesting that the greatest influence of complementarity between the termini on RNA synthesis occurs at the extreme genome ends. In comparison to 5′CB and 3′CB, replicons Le(TrC19–29)/Tr(LeC34–46) and Le(TrC34–46)/Le(TrC19–29) have mismatches at positions 9, 11, and 12, and this may account for their reduced ability to replicate.
Recombinant viruses containing altered termini.
The above analyses showed that nt 19 to 29 and 34 to 46 of Le contained essential signals for transcription and that these regions of Le could replace TrC sequences for efficient replication. These latter observations contrast with recent findings for the DI particle DI-T, in which the specific sequence of nt 31 to 45 of TrC were identified as a replication enhancer element that increased replication 4 to 15-fold (27). To examine further the importance of the altered termini in RNA synthesis and confirm our findings for subgenomic replicons, we engineered selected mutations in a full-length cDNA clone of infectious VSV, pVSV1(+), assayed the ability to rescue infectious virus, and examined the replication characteristics of the rescued virus (Table 1).
TABLE 1.
Recombinant viruses with altered termini
| Recombinant | Recoverya | Burst sizeb (PFU/cell) | Plaque sizec (mm) |
|---|---|---|---|
| pVSV1(+) | + | 1,650 | 2.3 |
| pV1(+)3′CB | − | ||
| pV1(+)Le(TrC19–28) | − | ||
| pV1(+)Tr(LeC19–28) | + | 1,850 | 1.9 |
| pV1(+)Tr(LeC34–46) | + | 1,150 | 1.7 |
Mutations were incorporated into an infectious cDNA clone of VSV, pVSV1(+), using standard techniques, and attempts were made to recover infectious virus as described in Materials and Methods. +, infectious virus recovered; −, virus not recovered from at least six independent transfections.
Determined for growth in BHK-21 cells at 37°C.
Determined on BSC40 cells incubated at 37°C for 28 h.
A 3′CB version of pVSV1(+) was generated; however, despite numerous attempts, infectious virus was not recovered. This is consistent with the inefficient transcription observed for 3′CB (Fig. 1B, lane 3) and the inability of 3′CB RNAs to assemble into infectious particles (52). When nt 19 to 29 of Le were substituted with the corresponding sequences of TrC, generating pV1(+)Le(TrC19–29), infectious virus was also not recovered, consistent with the down-regulation of transcription observed for replicon Le(TrC19–29) (Fig. 4B, lane 2). In contrast, when nt 19 to 29 or 34 to 46 of Tr were replaced by the corresponding sequences of LeC to generate pV1(+)Tr(LeC19–29) and pV1(+)Tr(LeC34–46), infectious viruses were readily recovered. Viruses were purified and amplified in BHK-21 cells, the sequences of the altered Tr regions were confirmed by RT-PCR as described previously (50).
Viral RNA replication and transcription obtained for rV1-Tr(LeC19–29) and rV1-Tr(LeC34–46) were qualitatively similar to those of the WT replicon (Fig. 5, lanes 1 to 3), and quantitation of the abundance of these products revealed only minor variations in the relative amounts of transcription versus replication. Thus, these data demonstrate that nt 19 to 29 or 34 to 46 of Le efficiently replaced the equivalent positions of TrC for viral RNA synthesis. When the growth properties of these two recombinants were compared to those of the wild-type virus in cell culture, each showed similar burst size, but plaque size was slightly smaller (Table 1). These data thus showed that the replication enhancer sequence identified for DI-T (27) can be replaced by the equivalent sequences from Le for efficient viral growth in cell culture.
FIG. 5.
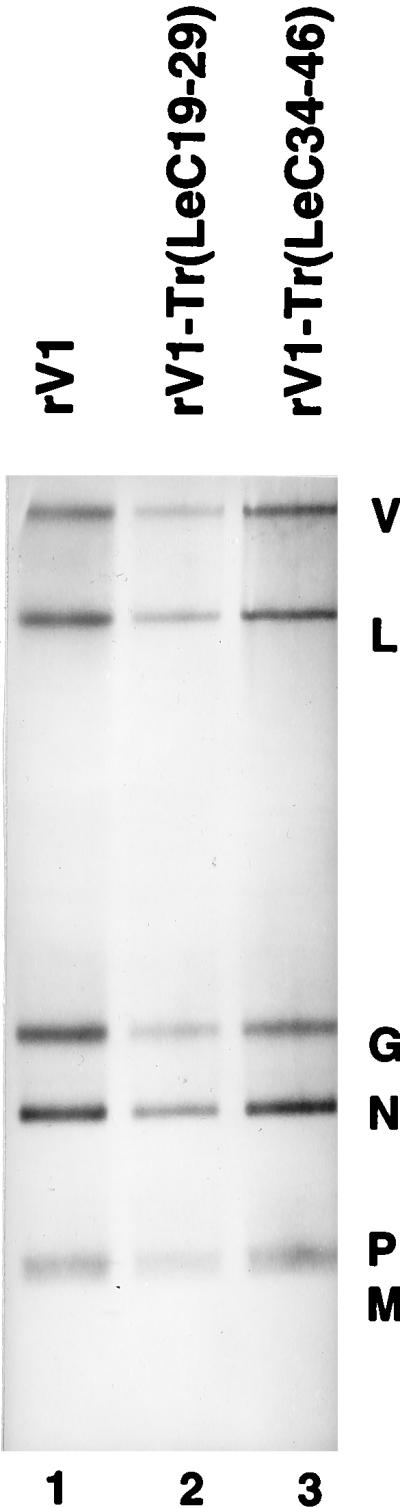
Agarose-urea gel analysis of RNA synthesis of recombinant viruses rV1-Tr(LeC19–29) and rV1-Tr(LeC34–46), with alterations to the genomic 5′ Tr that increased terminal complementarity, compared to the unmodified virus rV1 in BHK-21 cells as described in Materials and Methods. The genomic and antigenomic full-length replication products comigrate and are indicated by V; the viral mRNAs are identified; note that the P and M mRNAs comigrate.
DISCUSSION
In the work described here we analyzed the role of primary sequence and extent of complementarity at the VSV termini in regulating transcription and replication. These studies define sequences at the genomic 3′ terminus required for transcription of VSV mRNAs, identify regions at either terminus which are required or dispensable for replication, and emphasize that mutations engineered into the genomic termini affect multiple processes.
Transcription.
Exchange of Le for TrC and vice versa demonstrated that specific sequences within Le were essential for transcription but dispensable for efficient replication (Fig. 1A and B). Comparison of replication and transcription of replicons WT, 3′CB, and Inv showed that the unique sequences within Le that are not present in TrC were essential for transcription. Two approaches were used to further analyze the requirements for transcription and replication: (i) deletions and (ii) exchanges of sequences in Le for the corresponding nonidentical regions of TrC.
Deletions at the genomic 3′ terminus identified the nontranscribed Le-N gene junction as an essential requirement for mRNA synthesis. This was similar to findings for the nontranscribed dinucleotide present between the N-P, P-M, M-G, and G-L genes (5, 43), suggesting that the Le-N gene junction is necessary for transcription of the downstream gene. Also, results for deletions at the 3′ terminus showed that in addition to the Le-N gene junction, sequences between nt 15 and 50 were important for transcription but that the terminal 15 nt still allowed replication. Evaluation of the effects of further deletions at the genomic 3′ terminus on transcription proved complex, owing to the synthesis of products of an unusual size (discussed below). This led us to use the alternative approach of exchanges between the termini to map the requirements in Le for transcription.
Reciprocal exchanges between Le and TrC showed that sequences in Le between nt 19 and 46, which are distinct from those in TrC, influenced the efficiency of transcription. Specifically, exchange of nt 19 to 29 and 34 to 46 of Le for the equivalent nonidentical regions of TrC reduced transcription to approximately one-fifth and one-third respectively, of WT replicon levels (Fig. 4B) and prevented recovery of infectious virus from cDNA (Table 1). This effect on transcription was shown to be a consequence of alterations to the primary sequence of Le, rather than the extent of complementarity between the termini, by placing compensatory mutations in the genomic Tr (Fig. 4B).
Previous analyses of the requirements for transcription at an internal gene junction (43), and our unpublished observations for the N gene start signal (51), demonstrated that the conserved sequence 3′ UUGUCnnUAC 5′, although not analyzed here, is also essential for transcription. Thus, the 3′-terminal promoter for transcription comprises three separate and distinct elements: (i) the 3′ Le (particularly nt 19 to 29 and 34 to 46); (ii) the Le-N gene junction; and (iii) the N gene start signal.
Deletion of nt 16 to 30 of Le, a region previously mapped as a contact site for the P component of the polymerase (23), resulted in the synthesis of positive-sense RNAs of unexpected mobility (Fig. 2B, lane 3). Among the products observed was a positive-sense readthrough transcript of leader covalently linked to the N/L mRNA. In a separate analysis of the requirements for transcription, we have shown that an mRNA must be of a minimum size for termination of transcription to occur (51). Thus, it seems likely that deletion of nt 16 to 30, which reduced Le from 47 to 33 nt may prevent efficient termination of Le synthesis prior to the N gene start signal, resulting in the synthesis of a Le-mRNA readthrough transcript. In addition to these RNAs, replicon Δ16–30 synthesized products that contained Tr sequences (Fig. 2C, lane 8). These data show that the polymerase does not efficiently obey the stop and start signals present at the Le-N and L-Tr junctions of replicon Δ16–30. This is consistent with observations from a reconstituted in vitro system in which nt 16 to 30 of the VSV (New Jersey) Le region were shown to be dispensable for transcription, although the stop and start signals at the Le-N gene junction were ignored (41). Previously it was shown that binding of the L component of the polymerase to the nucleocapsid template requires prior binding of P (29) and that nt 16 to 30 of Le contact P (23). This finding raises the intriguing possibility that the polymerase complex is either incorrectly assembled or disoriented by deletion of nt 16 to 30 and hence fails to recognize the transcriptive stop and start signals. Further experiments will be required to examine these possibilities.
Replication.
In the present analysis, we examined requirements at the genomic termini for replication, showed that sequences derived from either Le or TrC functioned well in replication, and demonstrated that the levels of replication were affected in a complex manner that reflected the extent of complementarity between the termini and the involvement of a template in transcription.
In agreement with our previous observations (49), we typically observed that as the extent of complementarity between the termini was increased, levels of replication were enhanced. This was shown by comparing replicons WT and Inv, in which the termini are complementary for 25 of 46 positions, with 5′CB and 3′CB, in which the termini are perfectly complementary for 49 nt (Fig. 1A and B). Further evidence that the extent of complementarity between the termini influenced levels of replication was provided by deletions at the 5′ terminus of the transcriptionally inactive DI-T: deletions which incrementally reduced the extent of complementarity below 45 nt progressively reduced overall levels of replication (52).
While the role of complementarity between the genomic termni of the nonsegmented negative-strand RNA viruses remains controversial (27, 45), our previous analysis (49), combined with the observations reported here and in the accompanying report (52), show that the ends of the genome influence one another. This is most strikingly demonstrated by 3′CB, where changes at the 5′ end of the negative-strand dramatically alter the behavior of the polymerase at the WT 3′ end, switching the template activity from predominantly transcription to almost exclusively replication (Fig. 1B). The combination of all these data suggest that the greatest influence of complementarity between the termini on RNA synthesis was exerted by increasing perfect complementarity at the extreme genome ends within the natural lengths of Le and Tr and not by extending the length of complementarity beyond the termini. In contrast to the Mononegavirales, in the segmented negative-strand RNA viruses, interaction of the termini is widely accepted and appears to be required for RNA synthesis (8, 15, 19, 20, 22, 25, 28, 35, 36, 46).
Comparison of the levels of replication for replicons WT and Inv, which have the same degree of terminal complementarity, demonstrates that replication is also enhanced as use of Le as a promoter of transcription is decreased (Fig. 1B, lanes 1 and 4). Further evidence for this is provided by Δ47–50, which has wild-type genomic termini but, lacking the Le-N gene junction, was transcriptionally silent and replicated to higher levels than WT (Fig. 2B, lanes 1 and 2). These data suggest that Le functions as a less efficient promoter of replication than TrC, at least in part because of this additional role in promoting transcription, and suggest that transcription and replication are competing events promoted by Le. These observations provide a partial explanation for the asymmetry of replication observed for VSV.
That the asymmetry of replication is not simply a consequence of the dual promoter function of Le is evident from comparing the replication products of 3′CB, 5′CB, and Inv (Fig. 1B and C). For example, 5′CB, which has 49 nt of TrC as promoter for positive-strand synthesis and the entire 59 nt of TrC as promoter for negative-strand synthesis, synthesizes an excess of negative-strand replication products (Fig. 1C, lanes 5 to 7). This finding suggests that the promoter for negative-strand synthesis extends beyond the 49 nt of TrC that are present at the 3′ end of the negative strand in 5′CB. This is consistent with the observation that replication of DI-T, which has 45 nt of TrC as the promoter for positive-strand synthesis and the same TrC promoter for negative-strand synthesis as the wild-type virus, is asymmetric (approximately 60% negative strand). Precedent for an extended replication promoter is provided by the paramyxoviruses measles virus (13), Sendai virus (44), and simian virus 5 (30), where essential components of the promoter are located up to 90 nt away from the genome ends. In the case of 3′CB, an additional factor likely contributes to the observed asymmetry of replication (Fig. 1C, lanes 8 to 10). Here, the 3′ negative-strand Le acts as a promoter of transcription, which would decrease its use as a promoter of replication relative to the 3′ end of the positive strand, which, lacking a gene start signal, acts exclusively as a promoter of replication. Replication of Inv was also asymmetric, though here the positive-strand product was synthesized in excess of the negative strand. This observation is intriguing because Inv is transcriptionally inactive; thus, the Le at the 3′ end of the positive-strand promotes only replication, and to a lesser extent than the 49 nt of TrC found at the 3′ end of the negative strand. A possible explanation for these observations is that the promoters compete with one another for replicase and that TrC is intrinsically a stronger promoter of replication than Le. However, the observation that 3′CB replicates to the highest level observed for any combination of the termini would seem to be incompatible with this explanation. Thus, in conclusion, the data presented here indicate that the involvement of Le in transcription acts to decrease its use as a promoter of replication, and this may contribute to the asymmetry of replication observed in infected cells. However, as demonstrated by Inv, other factors also contribute to the regulation of positive- and negative-strand synthesis.
In the present analysis, we also delineated the sequence requirements at the termini for replication. Alternations to both termini that included deletions and exchanges identified sequences dispensable for replication, including nt 16 to 30, 34 to 46, and 47 to 50 of Le (Fig. 2B and 4B). Deletion of nt 15 to 50 severely down-regulated replication but showed that replication still occurs, thereby defining a minimal replication promoter. Similarly, RNAs that retained fewer than the terminal 45 nt of Tr reduced RNA synthesis of the WT replicon progressively (Fig. 3), and a replicon that retained only the 5′-terminal 14 nt replicated, albeit extremely poorly. When these 5′ Tr deletions were examined in a template that exclusively replicated (DI-T), those which retained at least the terminal 22 nt were active templates for replication (almost 40% of wild-type levels), although maximal replication required the terminal 45 nt (52). Consistent with the location of these deletions in Tr, which is copied to become the negative-strand promoter (TrC), minus-strand RNA synthesis was reduced to a greater extent than plus-strand RNA synthesis (52).
Recently, the requirements for replication of DI-T were analyzed by engineering deletions into its termini (27). The authors concluded that the terminal 25 nt contained all of the signals necessary and sufficient for replication and that nt 25 to 46 of TrC act as a replication enhancer sequence (27). When nt 31 to 46 of this replication enhancer element were substituted into the Le of a replicon that contained the natural 3′ Le and 5′ Tr, a fourfold enhancement of replication was observed (27). However, as shown here, replicon Tr(LeC34–46) and the analogous infectious recombinant virus, rV1-Tr(LeC34–46), showed that nt 34 to 46 of Le function as well in replication as the proposed enhancer sequence of TrC. Thus, we conclude that the up-regulation of replication seen when nt 34 to 46 of TrC were placed in Le is, at least in part, a consequence of the down-regulation of transcription and an increase in the extent of terminal complementarity, rather than the presence of a specific replication enhancer sequence.
In summary, the data presented here show that the promoter for transcription comprises three separate elements: (i) the 3′ Le, (ii) the nontranscribed Le-N gene junction, and (iii) the N gene start signal. Additionally, distinct sequences within the Le region, nt 19 to 29 and 34 to 46, are shown to be required for transcription but not replication. These data also present further evidence that increasing the extent of complementarity between the natural genomic termini enhances replication. Finally, these data suggest that the functions of Le as a promoter for transcription and for replication are competing events, which helps explain the asymmetry of replication observed in infected cells.
ACKNOWLEDGMENTS
We thank Shawn Harmon for assistance in generating replicon Δ16–30, and we gratefully acknowledge the members of the G. W. Wertz and L. A. Ball laboratories for critical reviews of the manuscript.
This work was supported by PHS grant AI12464 from NIAID to G.W.W.
REFERENCES
- 1.Abraham G, Banerjee A K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnheiter H, Davis N L, Wetz G, Schubert M, Lazzarini R A. Role of the nucleocapsid protein in regulating vesicular stomatitis virus RNA synthesis. Cell. 1985;41:259–267. doi: 10.1016/0092-8674(85)90079-0. [DOI] [PubMed] [Google Scholar]
- 3.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr J N, Whelan S P, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr J N, Whelan S P, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown M, Dorson J W, Bollum F J. Terminal riboadenylate transferase: a poly(A) polymerase in purified vaccinia virus. J Virol. 1973;12:203–208. doi: 10.1128/jvi.12.2.203-208.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calain P, Roux L. The rule of six, a basic feature for efficient RNA replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong H K, Cheong C, Choi B S. Secondary structure of the panhandle RNA of influenza virus A studied by NMR spectroscopy. Nucleic Acids Res. 1996;24:4197–4201. doi: 10.1093/nar/24.21.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colonno R J, Banerjee A K. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell. 1978;15:93–101. doi: 10.1016/0092-8674(78)90085-5. [DOI] [PubMed] [Google Scholar]
- 11.Colonno R J, Banerjee A K. A unique RNA species involved in initiation of vesicular stomatitis virus RNA transcription in vitro. Cell. 1976;8:197–204. doi: 10.1016/0092-8674(76)90003-9. [DOI] [PubMed] [Google Scholar]
- 12.Conzelmann K-K, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowley J C, Dowling P C, Menonna J, Silverman J I, Schuback D, Cook S D, Blumberg B M. Sequence variability and function of measles virus 3′ and 5′ ends and intercistronic regions. Virology. 1988;164:498–506. doi: 10.1016/0042-6822(88)90564-8. [DOI] [PubMed] [Google Scholar]
- 14.De B P, Banerjee A K. Rescue of synthetic analogs of genome RNA of human parainfluenza virus type 3. Virology. 1993;196:344–348. doi: 10.1006/viro.1993.1486. [DOI] [PubMed] [Google Scholar]
- 15.Desselberger U, Racaniello V R, Zazra J J, Palese P. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 16.Dimock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emerson S U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 18.Emerson S U, Wagner R R. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fodor E, Pritlove D C, Brownlee G G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu M-T, Parvin J D, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keene J D, Thornton B J, Emerson S U. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc Natl Acad Sci USA. 1981;78:6191–6195. doi: 10.1073/pnas.78.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiley M P, Wagner R R. Ribonucleic acid species of intracellular nucleocapsids and released virions of vesicular stomatitis virus. J Virol. 1972;10:244–255. doi: 10.1128/jvi.10.2.244-255.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim H J, Fodor E, Brownlee G G, Seong B L. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J Gen Virol. 1997;78:353–357. doi: 10.1099/0022-1317-78-2-353. [DOI] [PubMed] [Google Scholar]
- 26.Leppert M, Rittenhouse L, Perrault J, Summers D F, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Pattnaik A K. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 28.Luo G, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellon M G, Emerson S U. Rebinding of transcriptase components (L and NS proteins) to the nucleocapsid template of vesicular stomatitis virus. J Virol. 1978;27:560–567. doi: 10.1128/jvi.27.3.560-567.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy S K, Ito Y, Parks G D. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol. 1998;72:10–19. doi: 10.1128/jvi.72.1.10-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park K H, Huang T, Correia F F, Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci USA. 1991;88:5537–5541. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattnaik A K, Ball L A, LeGrone A, Wertz G W. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology. 1995;206:760–764. doi: 10.1016/S0042-6822(95)80005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 34.Patton J T, Davis N L, Wertz G W. N protein alone satisfies the requirement for protein synthesis during RNA replication of vesicular stomatitis virus. J Virol. 1984;49:303–309. doi: 10.1128/jvi.49.2.303-309.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritlove D C, Fodor E, Seong B L, Brownlee G G. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J Gen Virol. 1995;76:2205–2213. doi: 10.1099/0022-1317-76-9-2205. [DOI] [PubMed] [Google Scholar]
- 36.Raju R, Kolakofsky D. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J Virol. 1989;63:122–128. doi: 10.1128/jvi.63.1.122-128.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schincariol A L, Howatson A F. Replication of vesicular stomatitis virus. II. Separation and characterization of virus-specific RNA species. Virology. 1972;49:766–783. doi: 10.1016/0042-6822(72)90533-8. [DOI] [PubMed] [Google Scholar]
- 39.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonsen C C, Batt-Humphries S, Summers D F. RNA synthesis of vesicular stomatitis virus-infected cells: in vivo regulation of replication. J Virol. 1979;31:124–132. doi: 10.1128/jvi.31.1.124-132.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smallwood S, Moyer S A. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology. 1993;192:254–263. doi: 10.1006/viro.1993.1028. [DOI] [PubMed] [Google Scholar]
- 42.Soria M, Little S P, Huang A S. Characterization of vesicular stomatitis virus nucleocapsids. I. Complementary 40 S RNA molecules in nucleocapsids. Virology. 1974;61:270–280. doi: 10.1016/0042-6822(74)90261-x. [DOI] [PubMed] [Google Scholar]
- 43.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tapparel C, Roux L. The efficiency of Sendai virus genome replication: the importance of the RNA primary sequence independent of terminal complementarity. Virology. 1996;225:163–171. doi: 10.1006/viro.1996.0584. [DOI] [PubMed] [Google Scholar]
- 46.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarreal L P, Breindl M, Holland J J. Determination of molar ratios of vesicular stomatitis virus induced RNA species in BHK21 cells. Biochemistry. 1976;15:1663–1667. doi: 10.1021/bi00653a012. [DOI] [PubMed] [Google Scholar]
- 48.Wertz G W. Isolation of possible replicative intermediate structures from vesicular stomatitis virus-infected cells. Virology. 1978;85:271–285. doi: 10.1016/0042-6822(78)90431-2. [DOI] [PubMed] [Google Scholar]
- 49.Wertz G W, Whelan S, LeGrone A, Ball L A. Extent of terminal complementarity modulates the balance between transcription and replication of vesicular stomatitis virus RNA. Proc Natl Acad Sci USA. 1994;91:8587–8591. doi: 10.1073/pnas.91.18.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan S P, Ball L A, Barr J N, Wertz G T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whelan, S. P., J. N. Barr, and G. W. Wertz. Unpublished observations.
- 52.Whelan S P, Wertz G W. The 5′ terminal trailer region of vesicular stomatitis virus contains a position-dependent cis-acting signal for assembly of RNA into infectious particles. J Virol. 1999;73:307–315. doi: 10.1128/jvi.73.1.307-315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Q, Hardy R W, Wertz G W. Functional cDNA clones of the human respiratory syncytial (RS) virus, N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J Virol. 1995;69:2412–2419. doi: 10.1128/jvi.69.4.2412-2419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]