Abstract
Various host systems have been employed to increase the yield of recombinant proteins. However, some recombinant proteins were successfully produced at high yields but with no functional activities. To achieve both high protein yield and high activities, molecular biological strategies have been continuously developed. This work describes the effect of signal peptide (SP) and co-expression of molecular chaperones on the production of active recombinant protein in Escherichia coli. Extracellular enzymes from Bacillus subtilis, including β-1,4-xylanase, β-1,4-glucanase, and β-mannanase constructed with and without their signal peptides and intracellular enzymes from Pseudomonas stutzeri ST201, including benzoylformate decarboxylase (BFDC), benzaldehyde dehydrogenase (BADH), and d-phenylglycine aminotransferase (d-PhgAT) were cloned and overexpressed in E. coli BL21(DE3). Co-expression of molecular chaperones with all enzymes studied was also investigated. Yields of β-1,4-xylanase (Xyn), β-1,4-glucanase (Cel), and β-mannanase (Man), when constructed without their N-terminal signal peptides, increased 1112.61-, 1.75-, and 1.12-fold, respectively, compared to those of spXyn, spCel, and spMan, when constructed with their signal peptides. For the natural intracellular enzymes, the chaperones, GroEL-GroES complex, increased yields of active BFDC, BADH, and d-PhgAT, up to 1.31-, 4.94- and 37.93-fold, respectively, and also increased yields of Man and Xyn up to 1.53- and 3.46-fold, respectively, while other chaperones including DnaK-DnaJ-GrpE and Trigger factor (Tf) showed variable effects with these enzymes. This study successfully cloned and overexpressed extracellular and intracellular enzymes in E. coli BL21(DE3). When the signal peptide regions of the secretory enzymes were removed, yields of active enzymes were higher than those with intact signal peptides. In addition, a higher yield of active enzymes was obtained, in general, when these enzymes were co-expressed with appropriate chaperones. Therefore, E. coli can produce cytoplasmic and secretory enzymes effectively if only the enzyme coding sequence without its signal peptide is used and appropriate chaperones are co-expressed to assist in correct folding.
Keywords: signal peptides, chaperone, heterologous proteins, productivity
1. Introduction
To produce recombinant proteins, various types of hosts, including eukaryotes such as plants, mammalian cells, insect cells, filamentous fungi, and yeast, or prokaryotes such as Escherichia coli and Bacillus sp. are employed [1,2,3]. To decide which production system is most suitable, the nature, origin, and final application of the target proteins must be considered. For example, if a downstream process modification such as glycosylation is not essential for bioactivity, bacterial expression systems are attractive for heterologous protein production because of their ability to grow rapidly and to reach high cell density using inexpensive substrates, as well as their genetics are well-characterized. Furthermore, a large number of cloning vectors and modified host strains are available [3]. In this study, we described the use of molecular biological strategies, including the alternation of secretory proteins to allow proper cytoplasmic folding and co-expression of molecular chaperones to assist the folding in order to improve the production of active recombinant proteins in the bacterial host, E. coli. It was reported that DnaK-DnaJ-GrpE or Trigger factor (Tf) assisted in increasing the solubility of some proteins at the early stages of the protein folding pathway [4], while GroEL-GroES was required at a later folding stage [5]. The chaperones were also reported to minimize protein aggregation by mediating the degradation of proteins that cannot be properly folded, as observed with some proteins when co-expressed with chaperones [6]. The recombinant proteins of interest in this study were enzymes involved in non-starch polysaccharide hydrolases; β-1,4-xylanase (Xyn; EC 3.2.1.8), β-1,4-glucanase (Cel; EC 3.2.1.4) and β-mannanase (Man; EC 3.2.1.78) and those involved in aromatic compound degradation pathway; benzoylformate decarboxylase (BFDC; EC 4.1.1.7) and benzaldehyde dehydrogenase (BADH; EC 1.2.1.28), as well as that involved in a reversible transamination reaction; d-phenylglycine aminotransferase (d-PhgAT; EC 2.6.1.72).
The non-starch polysaccharide hydrolases are those used in industrial processes such as bio-bleaching of pulp wood in the paper industry [7], reduction of gum content in the textile industry [8], preparation of animal feed as animal feed additives, preparation of certain compounds for the cosmetic and pharmaceutical industries [9], extraction of oils from leguminous seeds, reduction of viscosity in coffee manufacture [10], clarification of fruit juices, fabrication of sugar beet syrup and caramel and fruit flavors for numerous food industries [9,11,12]. The enzymes involved in the aromatic compound degradation pathway, BFDC, and BADH, were found to play roles in the catabolic pathways of mandelate [13,14] and d-phenylglycine [15]. They have been used for the synthesis of enantiomerically pure pharmaceutical and chemical compounds [14,16], which are important pharmaceutical agents for the production of, for example, anti-cancer drugs and analgesics such as aspirin and cocaine [17]. Lastly, d-PhgAT was reported to play a role in d-phenylglycine degradation pathway [15]. The enzyme has also been used for the synthesis of d-phenylglycine (d-Phg) or d-4-hydroxyphenylglycine (d-4-OHPhg) using L-glutamate as an amino-group donor, and itself is converted 2-oxoglutarate. d-Phg is the side chain of semisynthetic penicillins and cephalosporins such as ampicillin, cephalexin, cephaloglycine, and cefacro; whereas, d-4-OHPhg is the side-chain of amoxicillin, cephadroxil, and cefatrizine. Furthermore, d-PhgAT has been used to measure L-glutamate in food products [18], and recently, it has also been used to determine the vitamin B6 status by measuring pyridoxal-5′-phosphate in plasma samples [19].
Upon overexpression of heterologous proteins in E. coli, inclusion body formation and proteolytic degradation are commonly observed due to the differences in the cellular environment, folding machinery, and conformational quality control checkpoints of E. coli compared to those of the native hosts. To alleviate these problems, a number of approaches, including (1) reducing the rate of the target gene expression by using weaker promoters, (2) decreasing the concentration of inducer, (3) lowering the growth temperature so that transcription and translation rates are slowed down, and the strength of hydrophobic interactions that contribute to protein misfolding was reduced and (4) The use of plasmid with low copy number has been considered to attenuate the expression. However, these strategies cause a reduction in productivity. Newer approaches are the use of a hydrophilic affinity tag to decrease the inclusion body formation and the co-expression of molecular chaperones. In this study, co-expression of molecular chaperones was used to improve the yields of soluble proteins in the E. coli cytoplasm.
2. Results and Discussion
2.1. Construction of Recombinant Plasmids
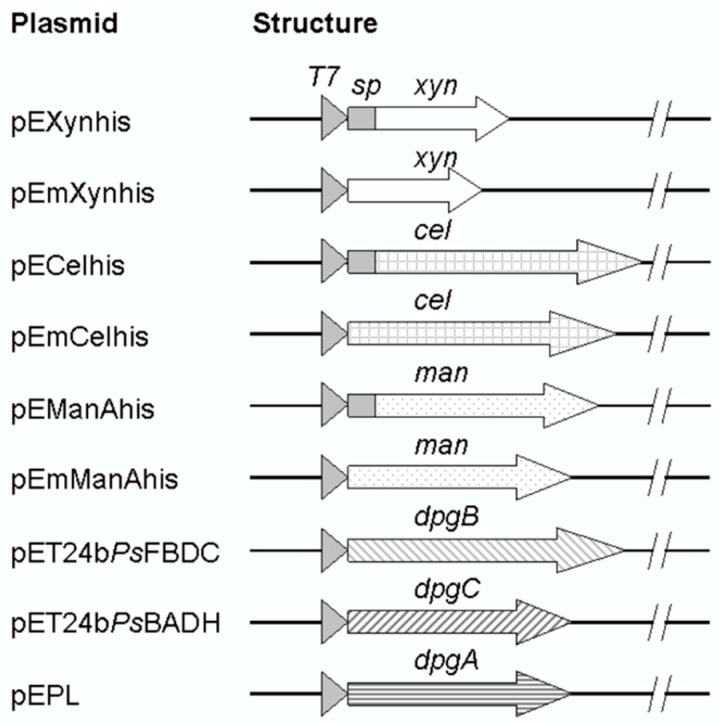
As shown in Figure 1, the full-length genes of B. subtilis R5 xylanase gene [GenBank:AB457186] (639 bp) and of B. subtilis I15 endo-1,4-glucanase gene [GenBank:FJ464332] (1497 bp) were synthesized and cloned into the E. coli expression vector pET24b(+) resulting in pEXynhis, which encoded for the xylanase (with signal peptide, spXyn) containing a C-terminal hexahistidine tag. The plasmid was used as a DNA template for amplification of the mature part of the xylanase corresponding region. The PCR product (555 bp) was then cloned into pET24(+), resulting in pEmXynhis, which encoded for the mature xylanase (Xyn) containing a C-terminal hexahistidine tag. Similarly, the 1497 bp synthetic DNA fragment containing the complete ORF of the glucanase gene was cloned into pET24b(+), resulting in pECelhis, which encoded for the full-length of the glucanase with signal peptide (spCel) with a C-terminal hexahistidine tag. The recombinant plasmid, pECelhis, was further used as a DNA template for amplification and cloning of the mature part of the glucanase corresponding region (1410 bp) by ligation into NdeI/XhoI site on pET24b(+) resulting in pEmCelhis which encoded for the mature glucanase (Cel) containing a C-terminal hexahistidine tag. Also, pEManAhis containing the full-length gene of a mannanase was used as a DNA template for amplification of the mature part of the mannanase corresponding region (1008 bp). The PCR fragment was cloned into pET24b(+), resulting in pEmManAhis, which encoded for the mature mannanase (Man) containing a C-terminal hexahistidine tag.
Figure 1.
Structures of recombinant plasmids. Secretory enzymes (xylanase, glucanase, and mannanase) corresponding genes were constructed to contain a full-length or only mature part as represented by arrows. DNA regions corresponding to signal peptides were depicted by a gray box located between the T7 promoter (triangles) and the mature part. β-1,4-xylanase gene; xyn, β-1,4-glucanase; cel, β-mannanase; man, benzoylformate decarboxylase; dpgB, benzaldehyde dehydrogenase; dpgC, and d-phenylglycine aminotransferase; dpgA.
All the recombinant plasmids (pEXynhis, pEmXynhis, pECelhis, pEmCelhis, pEManAhis, and pEmManAhis) and those which encoded for intracellular enzymes, including pET24bPsBFDC, pET19bPsBADH [15], and pEPL [20] were verified by restriction enzyme digestion and PCR amplification, and confirmed by nucleotide sequencing.
2.2. Expression of Enzymes from Bacillus and Pseudomonas in E. coli
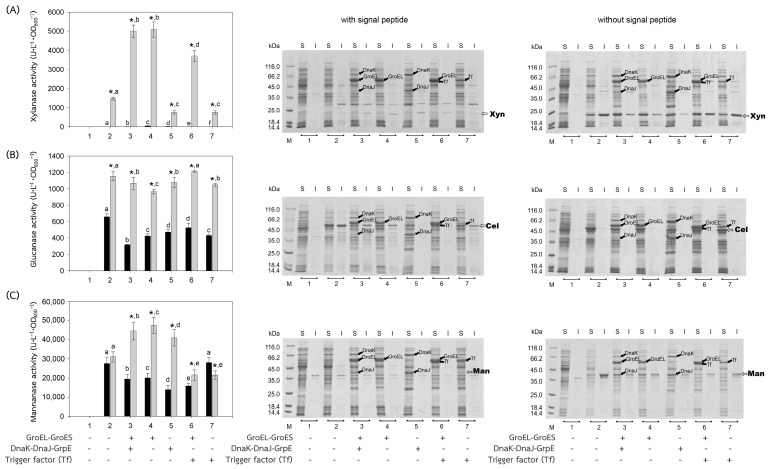
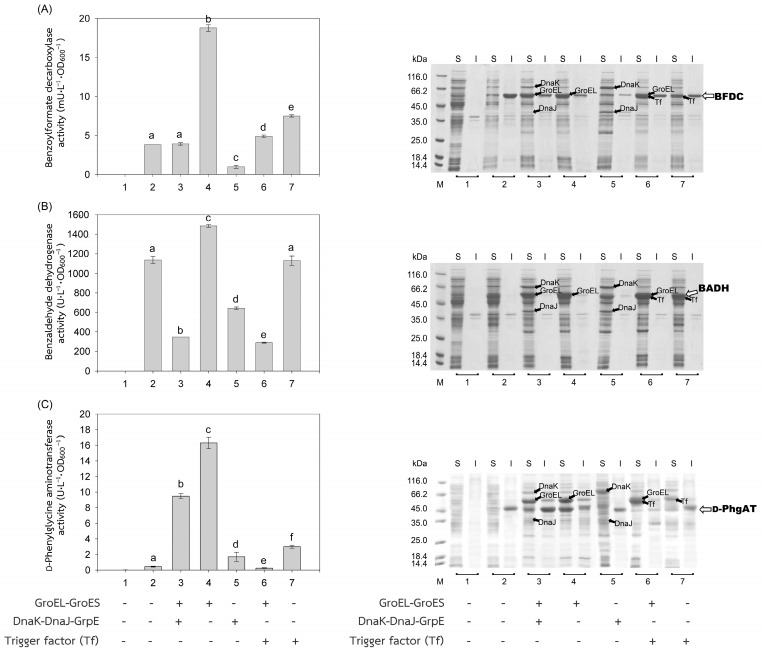
The recombinant plasmids, pEXynhis, pEmXynhis, pECelhis, pEmCelhis, pEManAhis and pEmManAhis, harboring genes of Bacillus secretory enzymes and pET24bPsBFDC, pET19bPsBADH, and pEPL harboring genes of Pseudomonas cytoplasmic enzymes were successfully cloned and overexpressed in E. coli BL21(DE3) (Table 1). Expression of the secretory enzyme genes whose signal peptide parts were removed was found to be more efficient than those containing the signal peptide parts; that was, yields of Xyn, Cel, and Man were 1468.64 U·L−1·OD600−1, 1154.95 U·L−1·OD600−1 and 30,898.45 U·L−1·OD600−1, which were 1112.61-, 1.75- and 1.12-fold higher than those of spXyn (1.32 U·L−1·OD600−1), spCel, (660.18 U·L−1·OD600−1) and spMan (27,620.03 U·L−1·OD600−1), respectively (Table 1 and Figure 2). The increase in yield of active enzymes might be directly due to the lack of signal peptide, which allows the protein to easily fold into its native form properly either by itself or with the assistance of the cytoplasmic chaperone(s) since the extra signal peptide may interfere both the folding of the protein and the function of the chaperone(s) [21,22]. In addition, the enzyme without signal peptide did not require further processing to be translocated to the cytoplasmic membrane, where its signal peptide was then cleaved to release the mature enzyme into the periplasmic space [23,24]. The results suggested that other extracellular enzymes besides xylanase, glucanase, and mannanase could be successfully folded into their native forms in the cytoplasm of E. coli if they were to produce no signal peptide. For intracellular enzymes, over-expression of benzoylformate decarboxylase (BFDC), benzaldehyde dehydrogenase (BADH), and d-phenylglycine aminotransferase (d-PhgAT), which have no signal peptide, were accomplished with the high level of active enzymes 3.80 mU·L−1·OD600−1, 1136.89 U·L−1·OD600−1 and 0.43 U·L−1·OD600−1, respectively (Table 1 and Figure 3).
Table 1.
The yield of enzymes expressed in E. coli BL21(DE3) and co-expressed with chaperones. Data were expressed as mean ± standard deviation from triplicate determinations. * represents a significant difference between the activities of the enzyme with and without signal peptide (SP) (p < 0.05). Different letters in the same row indicate significant differences (p < 0.05).
| Enzymes Activities (U·L−1·OD600−1) | |||||||
|---|---|---|---|---|---|---|---|
| Plasmid | Description | Chaperones Plasmid | |||||
| None | pG-KJE8 (GroEL-GroES, DnaK-DnaJ-GrpE) |
pGro7 (GroEL-GroES) |
pKJE7 (DnaK-DnaJ-GrpE) |
pG-Tf2 (GroEL-GroES, Tf) |
pTf16 (Tf) |
||
| pEXynhis | Xylanase with SP (spXyn) | 1.32 ± 0.04 a | 7.74 ± 0.15 b | 38.74 ± 1.17 c | 15.78 ± 0.39 d | 0.54 ± 0.01 e | 0.94 ± 0.02 f |
| pEmXynhis | Xylanase without SP (Xyn) | 1468.64 ± 90.66 *,a | 5002.20 ± 323.16 *,b | 5083.08 ± 409.19 *,b | 748.90 ± 87.68 *,c | 3705.46 ± 372.74 *,d | 753.91 ± 99.01 *,c |
| pECelhis | Glucanase with SP (spCel) | 660.18 ± 38.48 a | 318.51 ± 8.11 b | 423.05 ± 26.31 c | 472.55 ± 33.46 d | 525.53 ± 56.60 d | 428.72 ± 16.22 c |
| pEmCelhis | Glucanase without SP (Cel) | 1154.95 ± 58.66 *,a | 1068.83 ± 73.89 *,b | 964.79 ± 30.40 *,c | 1077.41 ± 63.79 *,b | 1211.92 ± 10.12 *,a | 1048.72 ± 18.19 *,b |
| pEManAhis | Mannanase with SP (spMan) | 27,620.03 ± 3029.29 a | 19,500.94 ± 2404.23 b | 20,113.83 ± 2402.98 c | 13,954.43 ± 1866.52 d | 15,864.20 ± 1422.00 e | 28,100.02 ± 2489.88 a |
| pEmManAhis | Mannanase without SP (Man) | 30,989.45 ± 2704.37 a | 44,512.17 ± 4575.60 *,b | 47,498.04 ± 4225.01 *,c | 40,933.50 ± 4275.97 *,d | 21,618.66 ± 2397.71 *,e | 21,575.18 ± 2139.69 *,e |
| pET24bPsBFDC | Benzoylformate decarboxylase (BFDC) # | 3.80 ± 0.01 a | 3.91 ± 0.15 a | 18.77 ± 0.43 b | 0.96 ± 0.15 c | 4.91 ± 0.13 d | 7.50 ± 0.15 e |
| pE19bPsBADH | Benzaldehyde dehydrogenase (BADH) | 1136.89 ± 36.76 a | 346.93 ± 1.06 b | 1486.04 ± 13.15 c | 641.93 ± 10.52 d | 288.11 ± 3.91 e | 1129.90 ± 47.30 a |
| pEPL | d-phenylglycine aminotransferase (d-PhgAT) | 0.43 ± 0.01 a | 9.49 ± 0.31 b | 16.31 ± 0.74 c | 1.70 ± 0.52 d | 0.25 ± 0.03 e | 3.00 ± 0.17 f |
# The activity of benzoylformate decarboxylase (BFDC) is shown in mU·L−1·OD600−1.
Figure 2.
Activities and SDS-PAGE of Bacillus enzymes expressed in E. coli BL21(DE3). Bacillus enzymes, including (A) xylanase, (B) glucanase, and (C) mannanase, were successfully produced in E. coli BL21(DE3). Activities of the enzymes expressed with or without their signal peptides were represented by black or gray bars, respectively. (1) Non-induced E. coli BL21(DE3) harboring plasmid encoded the enzyme of interest; (2) induced E. coli BL21(DE3) harboring plasmid encoded the enzyme of interest, for 3–7, induced E. coli BL21(DE3) harboring both plasmids encoded the enzyme of interest and that encoded chaperones (3) GroEL-GroES and DnaK-DnaJ-GrpE, (4) GroEL-GroES, (5) DnaK-DnaJ-GrpE, (6) GroEL-GroES and Trigger factor (Tf), and (7) Trigger factor (Tf). In the case of SDS-PAGE, the enzyme was investigated both in soluble (S) and insoluble fractions (I). Lane M, molecular mass standard marker. Bars represent the standard deviations from triplicate determinations. * represents a significant difference between the activities of the enzyme with and without signal peptide (SP) (p < 0.05). Different letters in the same enzyme indicate significant differences (p < 0.05).
Figure 3.
Activities and SDS-PAGE of Pseudomonas enzymes expressed in E. coli BL21(DE3). Activities of (A) benzoylformate decarboxylase (BFDC), (B) benzaldehyde dehydrogenase (BADH), and (C) d-phenylglycine aminotransferase (d-PhgAT) were depicted by gray bars. Soluble (S) and insoluble proteins (I) were analyzed by SDS-PAGE. (1) Non-induced E. coli BL21(DE3) harboring plasmid encoded the enzyme of interest; (2) induced E. coli BL21(DE3) harboring plasmid encoded the enzyme of interest, for 3–7, induced E. coli BL21(DE3) harboring both plasmids encoded the enzyme of interest and that encoded chaperones (3) GroEL-GroES and DnaK-DnaJ-GrpE, (4) GroEL-GroES, (5) DnaK-DnaJ-GrpE, (6) GroEL-GroES and Trigger factor (Tf), and (7) Trigger factor (Tf). Lane M, molecular mass standard marker. Bars represent the standard deviations from triplicate determinations. Different letters in the same enzyme indicate significant differences (p < 0.05).
2.3. Effects of Chaperone(s) on Proteins Production in E. coli
In general, co-expression of the GroEL-GroES complex increased the yield of active forms of both intracellular enzymes; BFDC (4.94-fold), BADH (1.31-fold), and d-PhgAT (37.93-fold) (Table 1, Figure 3) and secretory enzymes which contained no signal peptides; xylanase (Xyn) (3.46-fold) and mannanase (Man) (1.53-fold) except that of glucanase where GroEL-GroES complex showed no positive effect (Figure 2). This is as expected since the GroEL-GroES complex function in assisting the correct folding of the protein. Overexpression of the GroEL-GroES complex would increase the helpers; thus, the target proteins will have a higher chance to fold into their native forms. Interestingly, when spMan with its intact signal peptide was co-expressed with the GroEL-GroES complex, the yield of active mannanase decreased from 27,620.03 U·L−1·OD600−1 to 20,113.83 U·L−1·OD600−1. This might be because the chaperones interfered with the normal mannanase maturation process by folding it in a way that decreased and/or delayed the process of translocation to the cell membrane and thereafter. The same negative effect of the GroEL-GroES complex was also seen with glucanase with its intact signal peptide but not that obvious with xylanase (spXyn) with its intact signal peptide since the small values made it less reliable. The exact reason for this is not known, but it was proposed that the nature of Cel, which was highly soluble, might be the reason for finding no inclusion body in the SDS-PAGE and/or the enzyme was relatively easy to renature. Renaturation was determined by heating all the enzyme solutions at 60, 70, and 80 °C for 10 min, then allowing them to renature by incubating them at room temperature for 100 min. The maximal activity of Cel was not diminished even after heating to 60 °C. However, Xyl retained more than 80% of its activity, Man, BFDC, and BADH retained more than 90%, while d-PhgAT lost the majority of its activity. The results are consistent with the prior discovery. After 2 h of incubation, the B. subtilis I15 cellulase maintained more than 90% of its maximal activity at 65 °C [25]. On the other hand, the xylanase from B. subtilis strain R5 reached 50% inactivation in 25 min at 60 °C and lost nearly all of its activity after 5 min incubation at 75 °C [26]. More than 80% of the mannanase activity from B. subtilis BCC41051 was retained after incubation at 60 °C, but it was totally lost following heating at 75 °C for 30 min [27]. The benzoylformate decarboxylase from P. putida is stable up to 60 °C for 2 h but rapidly inactivates at 80 °C [16]. The activity of P. putida MT53 benzaldehyde dehydrogenase decreased by 50% in 50 min of incubation at 60 °C [28]. d-phenylglycine aminotransferase activity of P. stutzeri ST-201 decreased significantly at 60 °C and was entirely inactivated at 70 °C for 10 min [29]. It was found that the activity of Cel recovered higher than the others at all temperatures indicating that Cel had the intrinsic ability to refold correctly better than the others. The other proteins (BFDC, BADH, d-PhgAT, Xyn, and Man), in contrast to Cel, were not only unable to fold into their native forms as easily, but several of them also failed to fold correctly in the absence of chaperones, as demonstrated by the development of inclusion bodies on SDS-PAGE. Hence, overexpression of GroEL-GroES was found to assist in correct protein folding in most cases and therefore increased the yield of active enzymes.
The other chaperones, DnaK-DnaJ-GrpE, increased the production of active Man (1.32-fold) (Figure 2C), which lacks a signal peptide, and d-PhgAT (3.95-fold) (Figure 3C) but decreased the production of Xyn (Figure 2A), BFDC (Figure 3A) and BADH (Figure 3B), whereas no appreciable effects on the production of active Cel (Figure 2B). On the other hand, the Trigger factor (Tf) increased the production of BFDC (1.97-fold) (Figure 3A) and d-PhgAT (6.98-fold) (Figure 3C) but decreased the production of Xyn (Figure 2A) and Man (Figure 2C), and had no effect on the production of active Cel (Figure 2B). Moreover, co-expression of the chaperones GroEL-GroES and DnaK-DnaJ-GrpE (pG-KJE8) or GroEL-GroES and Trigger factor (Tf) (pG-Tf2) did not result in higher yields than those of GroEL-GroES alone, as shown in Table 1. GroEL-GroES decreased in activity when combined with other chaperones compared to when they were produced alone, possibly as a result of an imbalanced number of folding modulators that could also lead to unfavorable proteolytic activities. Previous studies revealed negative consequences for protein productivity and quality when certain chaperones were co-expressed. DnaK was involved in the degradation of aggregation-prone but functional polypeptides by targeting them to proteases such as Lon and ClpP. Due to the proteolysis stimulation caused by DnaK, the yield of recombinant protein was reduced. Additionally, DnaK is a negative regulator of the heat shock response; an increase in DnaK concentration above physiological levels might cause other heat shock proteins to be down-regulated. [30]. On the other hand, the amount of active N-acyl-d-aspartate amidohydrolase (d-AAase) was reduced by the co-expression of GroEL-GroES and Tf [31]. Therefore, the results implied that there were differences in the substrate recognition ability among chaperones as reported data [32,33,34,35,36,37]. Hence, the appropriate selection of chaperones would enhance the yield of active enzymes.
3. Materials and Methods
3.1. Bacterial Strains, Plasmids, and Culture Conditions
E. coli DH5α and E. coli BL21(DE3), used for plasmid propagation and over-expression, respectively, were purchased from Novagen (Madison, WI, USA). pET24b(+) was obtained from Novagen (Madison, WI, USA) and was used as a cloning and expression vector. pET24bPsBFDC and pET19bPsBADH containing benzoylformate decarboxylase gene (dpgB) and benzaldehyde dehydrogenase gene (dpgC) of Pseudomonas stutzeri ST201, respectively, were obtained from our previous study [15], and also pEPL containing d-phenylglycine aminotransferase gene (dpgA) was obtained from our previous study [20]. Chaperone plasmids pG-KJE8, pGro7, pKJE7, pG-Tf2, and pTf16 were purchased from Takara Bio Inc. (Shiga, Japan). GFXTM PCR DNA and Gel Band Purification Kit were products of GE Healthcare Inc. (Buckinghamshire, UK). Different strains of E. coli were cultivated at 37 °C in Luria Bertani (LB) medium (Difco, Tucker, GA, USA).
3.2. Synthesis and Cloning of Gene Encoding Full-Length (with Signal Peptide) and Mature Part of Xylanase
An open reading frame (ORF) of a xylanase gene was synthesized by GenScript USA Inc. (Piscataway, NJ, USA) based on the B. subtilis R5 xylanase gene [GenBank:AB457186]. The synthesized fragment was designed to contain point mutations from thymine to cytosine at nucleotide position 87 and from adenine to guanine at position 396 in order to eliminate NheI and NdeI recognition sites in the ORF, respectively. In addition, the synthesized fragment was designed to contain oligonucleotide linkers with NdeI and XhoI recognition sites immediately upstream and downstream of the xylanase gene, respectively. The DNA fragment was ligated into the NdeI/XhoI linearized pET24b(+), resulting in pEXynhis, which encoded for xylanase (with a signal peptide, spXyn) with a C-terminal hexahistidine tag. Following propagation in E. coli DH5α, the recombinant plasmid was then transformed into E. coli BL21(DE3) using the standard procedure [38] to allow gene expression.
To construct a xylanase gene containing only the mature protein coding part, primers Xyn-ms-F1 (5′-GTACGCCATATGGCCAGCACAGACTACTGGC-3′, containing a NdeI site as underlined) and Xyn-ms-R1 (5′-GTACGCCTCGAGCCACACTGTTACGTTAGAAC-3′, containing an XhoI site as underlined) were used to amplify the mature xylanase corresponding region from pEXynhis. The PCR was initially denaturized for 2 min at 95 °C, then underwent 30 cycles of denaturation for 30 s at 95 °C, annealing for 30 s at 50 °C, extension for 1 min at 68 °C, and final extension for 10 min at 68 °C. The PCR fragment was digested with NdeI and XhoI and then ligated into the corresponding site of the vector pET24(+). The resulting plasmid, pEmXynhis, was expressed for the production of the mature part of xylanase (Xyn) with a C-terminal hexahistidine tag. Each recombinant plasmid was transformed and expressed in E. coli BL21(DE3).
3.3. Synthesis and Cloning of Gene Encoding Full-Length (with Signal Peptide) and Mature Part of Glucanase
A coding region of the endo-1,4-β-glucanase gene was also synthesized by GenScript USA Inc. (USA) based on the B. subtilis I15 endo-1,4-β-glucanase gene [GenBank:FJ464332]. Silent mutation was carried out to change nucleotide residue at position 786 of the ORF from cytosine to thymine, resulting in no MluI site in the sequence, facilitating future DNA cloning. The synthesized DNA fragment containing the glucanase gene was designed to have NdeI and XhoI sites upstream and downstream of the ORF, respectively, cloned into the corresponding sites of pET24b(+). The resulting recombinant plasmid, pECelhis, encoded for the glucanase enzyme (with a signal peptide, spCel) with a C-terminal hexahistidine tag.
The mature part of the glucanase corresponding sequence was amplified from pECelhis using oligonucleotides Cel-ms-F1 (5′-GTACGCCATATGGCAGGGACAAAAACGCCAGTAG-3′, containing a NdeI site as underlined) and Cel-ms-R1 (5′-GTACGCCTCGAGATTTGGTTCTGTTCCCCAAATC-3′, containing a XhoI site as underlined) as primers. The PCR cycling parameters were as follows: initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 68 °C for 2 min and 50 s before the process is finished with extension at 68 °C for 10 min. After digestion with NdeI and XhoI, the PCR product was then ligated into the corresponding site of the vector pET24b(+), resulting in pEmCelhis, which encoded for the mature part of the glucanase (Cel) enzyme with a C-terminal hexahidine tag. Each recombinant plasmid was transformed and expressed in E. coli BL21(DE3).
3.4. Cloning of Gene Encoding Full-Length (with Signal Peptide) and Mature Part of Mannanase
Plasmid pEManAhis containing the full-length of a β-mannanase gene from Bacillus subtilis was previously constructed [27] and was used as template DNA for amplification of a region encoding for the mature part of the enzyme. The amplification was performed using primers Man-ms-F1 (5′-GTACGCCATATGCATACTGTGTCGCCTGTGAATCC-3′, containing a NdeI site as underlined) and Man-CHR (5′-GTACGCCTCGAGTTCAACGATTGGCGTTAAAGAATC-3′, containing an XhoI site as underlined) with the PCR parameter as follow: initial denaturation of the PCR took place at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, extension at 68 °C for 2 min, and final extension at 68 °C for 10 min. The PCR product was digested with NdeI and XhoI and then ligated into the corresponding site of the vector pET-24b(+). The resulting plasmid pEmManAhis was expressed for the mature part of the mannanase (Man) with a C-terminal hexahistidine tag. Each recombinant plasmid was transformed and expressed in E. coli BL21(DE3).
3.5. Transformation of pET24bPsBFDC, pET19bPsBADH and pEPL
Plasmids pET24bPsBFDC and pET19bPsBADH containing Pseudomonas stutzeri ST-201 benzoylformate decarboxylase (dpgB) gene, benzaldehyde dehydrogenase (dpgC) gene, respectively [15], and pEPL containing d-phenylglycine aminotransferase (dpgA) gene were previously constructed [20]. Each of these plasmids was transformed and expressed in E. coli BL21(DE3).
3.6. Co-Transformation with the Chaperone Plasmid
Plasmids pG-KJE8 (encoded for GroEL-GroES and DnaK-DnaJ-GrpE), pGro7 (encoded for GroEL-GroES), pKJE7 (encoded for DnaK-DnaJ-GrpE), pG-Tf2 (encoded for GroEL-GroES and Trigger factor), and pTf16 (encoded for Trigger factor) [4,39] each was transformed into E. coli BL21(DE3) harboring pEXynhis, pEmXynhis, pECelhis, pEmCelhis, pEManAhis, pEmManAhis, pET24bPsBFDC, pET19bPsBADH, and pEPL. Each of the transformants was grown on LB agar plates supplemented with 34 μg·ml−1 of chloramphenicol (for selection of the chaperone plasmid) and 30 μg·ml−1 of kanamycin (for selection of the expression plasmids pEXynhis, pEmXynhis, pECelhis, pEmCelhis, pEManAhis, pEmManAhis, and pET24bPsBFDC) or 100 μg·ml−1 of ampicillin (for selection of the expression plasmid pET19bPsBADH and pEPL). To verify the presence of both recombinant plasmids in the selected E. coli transformants, the plasmids were extracted and digested with appropriate restriction enzymes.
3.7. Gene Expression
To induce co-expression, a single colony of each transformant was inoculated in 3 mL of LB broth supplemented with appropriate antibiotic(s) and grown overnight at 37 °C with 200 rpm shaking. The overnight pre-cultured was transferred into 50 mL of fresh LB medium in a 250-mL flask with a starting OD600 of 0.03. The culture was incubated at 30 °C with 200 rpm shaking until 0.4 OD600 was reached. Chaperone(s) expression was induced by adding L-arabinose and/or tetracycline at the final concentration of 0.5 mg·mL−1 and/or 5 ng·mL−1, respectively, according to the instruction manual provided by Takara Bio Inc. (Shiga, Japan). After 1-h of chaperone(s) induction, synthesis of the target proteins (xylanase, glucanase, mannanase, benzoylformate decarboxylase, benzaldehyde dehydrogenase, and d-phenylglycine aminotransferase) was induced by adding IPTG at a final concentration of 0.4 mM, and the cultures were further incubated (at 30 °C, 200 rpm) for additional 4 h. The cells were collected by 8000 g of centrifugation and washed with 20 mM Tris-HCl buffer (pH 8.0). The cell pellet was resuspended in the same buffer and disrupted by sonication on ice (Vibra-CellTM, Sonics & Materials, Newtown, CT, USA). Cell debris and insoluble aggregates were sedimented at 16,000 g for 30 min at 4 °C. Cell-free extracts containing soluble proteins were used to determine the enzyme activities. Pellets containing insoluble proteins were washed twice with the same buffer. The amount of target proteins, either in soluble or insoluble fractions, was investigated by SDS-PAGE [40].
3.8. Protein Determination and SDS-PAGE Analysis
The Bradford method [41] was used to determine the protein content using bovine serum albumin (BSA) as the reference. Protein samples were loaded onto a polyacrylamide gel (12% separating gel and 4% stacking gel). Coomassie Blue staining was applied to the gel following 1.5 h of electrophoresis at 150 V [40].
3.9. Enzyme Assays
Xylanase activity; after incubating 0.1 mL of the diluted enzyme sample with 0.9 mL of 0.5% (w/v) xylan solution in 50 mM sodium phosphate buffer, pH 6.0 at 42 °C for 20 min, the amount of reducing sugars liberated from oat spelt xylan was measured using the dinitrosalicylic acid (DNS) method [26,42]. Absorbance at 540 nm was measured using a spectrophotometer (Unicam Helios Alpha, Thermo, Cambridge, UK). One unit of xylanase activity is defined as the amount of the enzyme required to liberate 1 μmole of reducing sugar equivalent to xylose per min under the assay condition.
Glucanase and mannanase activities were determined using 5% (w/v) carboxymethyl-cellulose (CMC) (Sigma, Livonia, MI, USA) [25] and 0.5% (w/v) locust bean gum (LBG) (Sigma, USA) [27] as substrates, respectively, and the DNS method was used to measure the released reducing sugars [42]. One unit of glucanase or mannanase activity was defined as the amount of the enzyme that released 1 μmole of glucose equivalent or mannose equivalent per min under the assay condition, respectively.
Benzoylformate decarboxylase activity was determined using the direct decarboxylase assay by incubating 20 μL of the enzyme sample with 980 μL of the substrate solution containing 100 mM sodium phosphate buffer, pH 6.0, 8.5 mM benzoylformate, 1 mM MgSO4, and 40 μM thiamine diphosphate (ThDP). For three minutes at 25 °C, the change in absorbance at 340 nm as benzaldehyde was formed from benzoylformate was continually observed [15,16,43]. One unit of benzoylformate decarboxylase activity is defined as the amount of enzyme that catalyzes the decarboxylation of 1 μmole of benzoylformate per min under the assay condition. The reaction rate was determined with the specific extinction coefficient (ε) of 32 μM−1·cm−1 for benzoylformate at 340 nm [16].
Benzaldehyde dehydrogenase activity was determined by incubating 20 μL of the enzyme sample with 980 μL of substrate solution containing 100 mM KCl, 100 mM [(2-hydroxy-1,1-bis[hydroxymethyl]ethyl)amino]-1-propanesulfonic acid (TAPS) buffer, pH 8.5, 1 mM benzaldehyde, 1 mM DTT, and 1 mM NAD+. For three minutes at 25 °C, the spectrophotometric monitoring of the increase in absorbance at 340 nm as the reduction of NAD+ to produce NADH [13,15]. According to the definition of benzaldehyde dehydrogenase activity, one unit is the amount of the enzyme that is necessary to produce 1 μmol of NADH per minute under the assay conditions. For NADH at 340 nm, the reaction rate was calculated using a molar extinction coefficient (ε) of 6220 M−1·cm−1 [13].
d-phenylglycine aminotransferase activity was determined by measuring the amount of 4-hydroxy benzoylformic acid (4-OHBZF) formed upon transamination of d-4-hydroxyphenylglycine (d-4-OHPhg) using 2-oxoglutarate as an amino acceptor [18,44]. A 20 μL of d-PhgAT solution was incubated with 980 μL of the reaction mixture containing 50 mM 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid buffer, pH 9.5, 10 mM d-4-OHPhg, 10 mM α-ketoglutarate, 5 μM PLP, and 5 μM EDTA. The rate of 4-OHBZF formation was monitored by observing the rise in UV absorption at 340 nm for 3 min at 25 °C. One unit of d-PhgAT activity was defined as the amount of enzyme that catalyzes the formation of 1 μmole of 4-OHBZF per min under the assay condition. The molar absorption coefficient of 4-OHBZF at 340 nm (ε340nm) is 2.4 × 104 M−1·cm−1 [44].
3.10. Statistical Analysis
The mean values of enzyme activities were compared using the statistical t-test (SPSS statistics 26.0). The p-value of 0.05 was used to determine statistical significance.
4. Conclusions
A new approach for producing secretory enzymes in E. coli was developed. The enzymes tested included xylanase, glucanase, and mannanase, whose yields could be increased when the genes were constructed without their signal peptides corresponding region and expressed in E. coli BL21(DE3). Furthermore, when co-expressed with chaperones, especially GroEL-GroES complex, xylanase, and mannanase coding sequences lacking the N-terminal signal peptide regions yielded even higher active enzymes than that with no chaperone co-expression. The benefit of chaperone co-expression was not only with the structural genes of secretory enzymes but also for all the tested intracellular enzymes, BFDE, BADH, and d-PhgAT, whose yields were accomplished in particular, with the GroEL-GroES complex.
Acknowledgments
We wish to acknowledge Walailak University for facility support.
Author Contributions
Conceptualization, J.J. and P.S.; methodology, J.J., S.S. and P.S.; software, J.J. and P.S.; validation, J.J. and P.S.; investigation, J.J., S.S. and P.S.; resources, J.J. and P.S.; data curation, J.J., S.S. and P.S.; writing—original draft preparation, J.J., S.S. and P.S. writing—review and editing, J.J., S.S. and P.S.; funding acquisition, J.J. and P.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
Funding Statement
This research was financially supported by a WU grant (Grant No. WU-IRG-63-036) from Walailak University, Thailand.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Larrick J.W., Thomas D.W. Producing proteins in transgenic plants and animals. Curr. Opin. Biotechnol. 2001;12:411–418. doi: 10.1016/S0958-1669(00)00236-6. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt F.R. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 2004;65:363–372. doi: 10.1007/s00253-004-1656-9. [DOI] [PubMed] [Google Scholar]
- 3.Terpe K. Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 4.Nishihara K., Kanemori M., Yanagi H., Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 2000;66:884–889. doi: 10.1128/AEM.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewalt K.L., Hendrick J.P., Houry W.A., Hartl F.U. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell. 1997;90:491–500. doi: 10.1016/S0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 6.Huang H.C., Sherman M.Y., Kandror O., Goldberg A.L. The molecular chaperone DnaJ is required for the degradation of a soluble abnormal protein in Escherichia coli. J. Biol. Chem. 2001;276:3920–3928. doi: 10.1074/jbc.M002937200. [DOI] [PubMed] [Google Scholar]
- 7.Paice M.G., Gurnagul N., Page D.H., Jurasek L. Mechanism of hemicellulose-directed prebleaching of kraft pulps. Enzym. Microb. Technol. 1992;14:272–276. doi: 10.1016/0141-0229(92)90150-M. [DOI] [Google Scholar]
- 8.Bruhlmann F., Leupin M., Erismann K.H., Fiechter A. Enzymatic degumming of ramie bast fibers. J. Biotechnol. 2000;76:43–50. doi: 10.1016/S0168-1656(99)00175-3. [DOI] [PubMed] [Google Scholar]
- 9.Polizeli M.L., Rizzatti A.C., Monti R., Terenzi H.F., Jorge J.A., Amorim D.S. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005;67:577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- 10.Heck J.X., de Barros Soares L.H., Ayub M.A.Z. Optimization of xylanase and mannanase production by Bacillus circulans strain BL53 on solid-state cultivation. Enzym. Microb. Technol. 2005;37:417–423. doi: 10.1016/j.enzmictec.2005.02.015. [DOI] [Google Scholar]
- 11.Minic Z., Jouanin L. Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol. Biochem. 2006;44:435–449. doi: 10.1016/j.plaphy.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Moreira L.R., Filho E.X. An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 2008;79:165–178. doi: 10.1007/s00253-008-1423-4. [DOI] [PubMed] [Google Scholar]
- 13.McLeish M.J., Kneen M.M., Gopalakrishna K.N., Koo C.W., Babbitt P.C., Gerlt J.A., Kenyon G.L. Identification and characterization of a mandelamide hydrolase and an NAD(P)+-dependent benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633. J. Bacteriol. 2003;185:2451–2456. doi: 10.1128/JB.185.8.2451-2456.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprenger G.A., Pohl M. Synthetic potential of thiamin diphosphate-dependent enzymes. J. Mol. Catal. B Enzym. 1999;6:145–159. doi: 10.1016/S1381-1177(98)00107-6. [DOI] [Google Scholar]
- 15.Saehuan C., Rojanarata T., Wiyakrutta S., McLeish M.J., Meevootisom V. Isolation and characterization of a benzoylformate decarboxylase and a NAD+/NADP+-dependent benzaldehyde dehydrogenase involved in d-phenylglycine metabolism in Pseudomonas stutzeri ST-201. Biochim. Biophys. Acta. 2007;1770:1585–1592. doi: 10.1016/j.bbagen.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Iding H., Dunnwald T., Greiner L., Liese A., Muller M., Siegert P., Grotzinger J., Demir A.S., Pohl M. Benzoylformate decarboxylase from Pseudomonas putida as stable catalyst for the synthesis of chiral 2-hydroxy ketones. Chemistry. 2000;6:1483–1495. doi: 10.1002/(SICI)1521-3765(20000417)6:8<1483::AID-CHEM1483>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 17.Wildermuth M.C. Variations on a theme: Synthesis and modification of plant benzoic acids. Curr. Opin. Plant Biol. 2006;9:288–296. doi: 10.1016/j.pbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Khampha W., Meevootisom V., Wiyakrutta S. Spectrophotometric enzymatic cycling method using L-glutamate dehydrogenase and d-phenylglycine aminotransferase for determination of L-glutamate in foods. Anal. Chim. Acta. 2004;520:133–139. doi: 10.1016/j.aca.2004.05.044. [DOI] [Google Scholar]
- 19.Jomrit J., Isarangkul D., Summpunn P., Wiyakrutta S. A kinetic spectrophotometric method for the determination of pyridoxal-5’-phosphate based on coenzyme activation of apo-d-phenylglycine aminotransferase. Enzyme Microb. Technol. 2018;117:64–71. doi: 10.1016/j.enzmictec.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Kongsaeree P., Samanchart C., Laowanapiban P., Wiyakrutta S., Meevootisom V. Crystallization and preliminary X-ray crystallographic analysis of d-phenylglycine aminotransferase from Pseudomonas stutzeri ST201. Acta Crystallogr. D Biol. Crystallogr. 2003;59:953–954. doi: 10.1107/S0907444903006498. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhuri T.K., Verma V.K., Maheshwari A. GroEL assisted folding of large polypeptide substrates in Escherichia coli: Present scenario and assignments for the future. Prog. Biophys. Mol. Biol. 2009;99:42–50. doi: 10.1016/j.pbiomolbio.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Tjalsma H., Bolhuis A., Jongbloed J.D., Bron S., van Dijl J.M. Signal peptide-dependent protein transport in Bacillus subtilis: A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 2000;64:515–547. doi: 10.1128/MMBR.64.3.515-547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freudl R. Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Factories. 2018;17:52. doi: 10.1186/s12934-018-0901-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsirigotaki A., De Geyter J., Šoštarić N., Economou A., Karamanou S. Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 25.Yang D., Weng H., Wang M., Xu W., Li Y., Yang H. Cloning and expression of a novel thermostable cellulase from newly isolated Bacillus subtilis strain I15. Mol. Biol. Rep. 2010;37:1923–1929. doi: 10.1007/s11033-009-9635-y. [DOI] [PubMed] [Google Scholar]
- 26.Jalal A., Rashid N., Rasool N., Akhtar M. Gene cloning and characterization of a xylanase from a newly isolated Bacillus subtilis strain R5. J. Biosci. Bioeng. 2009;107:360–365. doi: 10.1016/j.jbiosc.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Summpunn P., Chaijan S., Isarangkul D., Wiyakrutta S., Meevootisom V. Characterization, gene cloning, and heterologous expression of [beta]-mannanase from a thermophilic Bacillus subtilis. J. Microbiol. 2011;49:86–93. doi: 10.1007/s12275-011-0357-1. [DOI] [PubMed] [Google Scholar]
- 28.Chalmers R.M., Scott A.J., Fewson C.A. Purification of the benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase encoded by the TOL plasmid pWW53 of Pseudomonas putida MT53 and their preliminary comparison with benzyl alcohol dehydrogenase and benzaldehyde dehydrogenases I and II from Acinetobacter calcoaceticus. Microbiology. 1990;136:637–643. [Google Scholar]
- 29.Wiyakrutta S., Meevootisom V. A stereo-inverting d-phenylglycine aminotransferase from Pseudomonas stutzeri ST-201: Purification, characterization and application for d-phenylglycine synthesis. J. Biotechnol. 1997;55:193–203. doi: 10.1016/S0168-1656(97)00075-8. [DOI] [PubMed] [Google Scholar]
- 30.Martínez-Alonso M., García-Fruitós E., Ferrer-Miralles N., Rinas U., Villaverde A. Side effects of chaperone gene co-expression in recombinant protein production. Microb. Cell Factories. 2010;9:64. doi: 10.1186/1475-2859-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimune K., Ninomiya Y., Wakayama M., Moriguchi M. Molecular chaperones facilitate the soluble expression of N-acyl-D-amino acid amidohydrolases in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2004;31:421–426. doi: 10.1007/s10295-004-0163-4. [DOI] [PubMed] [Google Scholar]
- 32.Baneyx F., Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 33.Deuerling E., Patzelt H., Vorderwulbecke S., Rauch T., Kramer G., Schaffitzel E., Mogk A., Schulze-Specking A., Langen H., Bukau B. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 34.Hartl F.U., Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 35.Teter S.A., Houry W.A., Ang D., Tradler T., Rockabrand D., Fischer G., Blum P., Georgopoulos C., Hartl F.U. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/S0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 36.Eicholt L.A., Aubel M., Berk K., Bornberg-Bauer E., Lange A. Heterologous expression of naturally evolved putative de novo proteins with chaperones. Protein Sci. 2022;31:e4371. doi: 10.1002/pro.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haacke A., Fendrich G., Ramage P., Geiser M. Chaperone over-expression in Escherichia coli: Apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr. Purif. 2009;64:185–193. doi: 10.1016/j.pep.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 38.Pope B., Kent H.M. High efficiency 5 min transformation of Escherichia coli. Nucleic Acids Res. 1996;24:536–537. doi: 10.1093/nar/24.3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. Chaperone coexpression plasmids: Differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 1998;64:1694–1699. doi: 10.1128/AEM.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.Ghose T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987;59:257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- 43.Hegeman G.D. Benzoylformate decarboxylase (Pseudomonas putida) Meth. Enzymol. 1970;17:674–678. [Google Scholar]
- 44.Summpunn P., Jomrit J., Panbangred W. Improvement of extracellular bacterial protein production in Pichia pastoris by co-expression of endoplasmic reticulum residing GroEL–GroES. J. Biosci. Bioeng. 2018;125:268–274. doi: 10.1016/j.jbiosc.2017.09.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.