Summary
Mechanisms of infection and pathogenesis have predominantly been studied based on differential gene or protein expression. Less is known about posttranslational modifications, which are essential for protein functional diversity. We applied an innovative glycoproteomics method to study the systemic proteome-wide glycosylation in response to infection. The protein site-specific glycosylation was characterized in plasma derived from well-defined controls and patients. We found 3862 unique features, of which we identified 463 distinct intact glycopeptides, that could be mapped to more than 30 different proteins. Statistical analyses were used to derive a glycopeptide signature that enabled significant differentiation between patients with a bacterial or viral infection. Furthermore, supported by a machine learning algorithm, we demonstrated the ability to identify the causative pathogens based on the distinctive host blood plasma glycopeptide signatures. These results illustrate that glycoproteomics holds enormous potential as an innovative approach to improve the interpretation of relevant biological changes in response to infection.
Subject areas: Health sciences, Glycobiology, Immunology, Glycomics
Graphical abstract

Highlights
-
•
Proteome-wide site-specific glycopeptide analysis resulted in 3,682 unique features
-
•
96 glycopeptides were significantly different between bacterial and viral infection
-
•
Distinct host glycopeptide signatures found for different causative pathogen species
-
•
We demonstrated the diagnostic potential of the largely unexplored glycoproteome
Health sciences; Glycobiology; Immunology; Glycomics
Introduction
It has become increasingly clear that posttranslational modifications—such as glycosylation, methylation, acetylation, and phosphorylation—determine a large part of protein biology and are essential in regulation.1,2 Therefore, changes in protein modifications may provide a more specific representation of functional alterations rather than protein levels alone. Over 50% of all human proteins are glycosylated, and aberrant glycomics signatures have been associated with many genetic and acquired human diseases.3,4,5 Protein glycosylation modulates and controls multiple biological functions, including protein folding, trafficking, adhesion, stability, and regulation of cellular activities.6,7 Glycans also have major roles in control of both innate and adaptive immunity.8,9 Not only are cell surface glycans the first molecules that pathogens will encounter in the respiratory or gastrointestinal tract but host glycans are also involved in the recognition of foreign glycans from pathogens or serve as decoys to divert pathogens in many tissues and in the blood stream. Glycoproteomic analyses have shown distinct glycosylations during sepsis and even predicted patient outcome.10,11 We therefore hypothesize that aberrant glycosylation signatures during different types of infection will give us an unbiased insight into the host response and are potentially an important source for the discovery of biomarkers to discriminate the causative agents of infection.
The fields of translational glycomics and glycoproteomics are rapidly expanding and mass spectrometry has become the most prominent analytical method for glycoprotein characterization and clinical implementation.12 Most approaches are still based on the analysis of released glycans, where the N- or O-glycan moiety is released from the peptides or proteins by means of glycosidases.13 A large disadvantage of this approach is the loss of essential site-specific information to link the glycan to the peptide or protein. The most informative method to study protein glycosylations and their macroheterogeneity and microheterogeneity is by means of intact (native or top-down) glycoprotein analysis in combination with detailed characterization of individual glycosylation sites.14 However, given its complexity, this type of analysis is currently limited to less complex biological matrices and to specialized laboratories equipped with state-of-the-art mass spectrometry. We therefore propose intact glycopeptide analysis as the preferred approach to explore the proteome-wide site-specific glycosylation profile of complex biological samples. However, a comprehensive characterization of proteome glycosylation is challenging, as in an enzymatically digested proteome, only a small fraction of peptides are glycosylated. Furthermore, microheterogeneity and suppressed ionization efficiency complicate glycopeptide analysis.15 Recently, we developed an innovative method to profile intact N-glycopeptides in blood plasma samples.16 Through a combination of a glycopeptide enrichment prior to analysis and an in-source supercharging technique, both the yield and ionization efficiency of glycopeptides improved significantly, resulting in increased detection of glycopeptides in clinical samples.
Here, we applied this glycoproteomics method to blood plasma samples from 42 healthy individuals and 91 children with a diagnosed infection to identify significant site-specific glycosylation changes in response to five different bacterial and four different viral pathogens. We isolated 3,682 unique features present in at least 75% of the samples, of which over 400 features were significantly different between patients with a bacterial infection and patients with a viral infection after stringent statistical analyses, such as analysis of variance (ANOVA), principal component analysis (PCA), and partial least squares discriminant analysis (PLS-DA). Nearly a hundred unique significant glycopeptide features could be fully identified, which distinguish bacterial from viral infection. Supported by a machine learning algorithm, we demonstrated the ability to identify the causative pathogens on species level by analyzing distinctive host blood plasma glycopeptide signatures.
Results
Patient and healthy control demographics
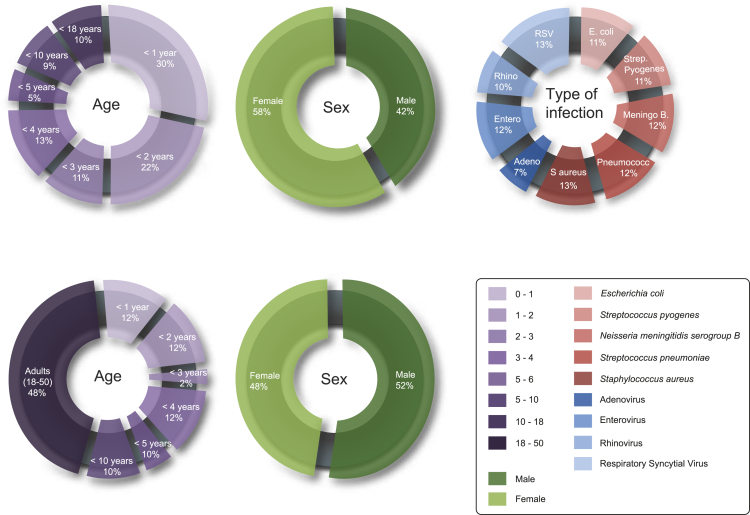
For this study, 42 controls and 91 patients were selected from five European medical centers, situated in the Netherlands, United Kingdom, and Spain. The pediatric patients had either a diagnosed bacterial (n = 53) or viral infection (n = 38), with a selection of the most frequent occurring infections, caused by: Escherichia coli, Streptococcus pyogenes, Neisseria meningitidis serogroup B, Streptococcus pneumoniae, Staphylococcus aureus, adenovirus, enterovirus, rhinovirus, or respiratory syncytial virus (RSV). The median age for the adult controls was 37 years, pediatric controls 3.0 years, and for the patients 2.1 years. Sex, age, and type of infection were equally distributed over all subgroups (Figure 1). Additional demographics for patients and controls are listed in Tables S1 and S2.
Figure 1.
Study group characteristics
Pie charts representing the distribution of age, sex, and type of infection in the patient and control groups, respectively. Additional clinical characteristics for the healthy individuals and patients are enlisted in Tables S1 and S2.
Data characteristics and exploratory data analysis demonstrate a high-quality dataset
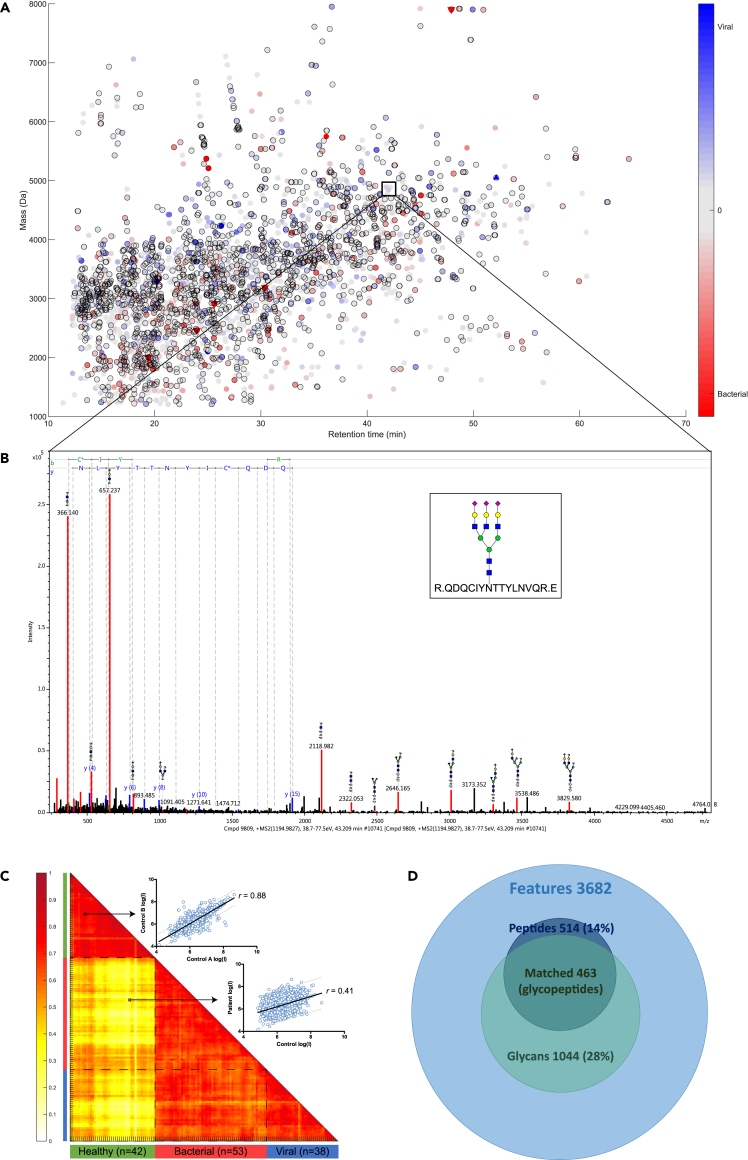
The plasma samples were analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). More than 30,000 compounds were detected per plasma sample as LC-MS features (MS1). Approximately 14,000 MS/MS spectra were recorded per sample, of which at least 2,000 MS/MS spectra were classified as an N-glycopeptide based on a predefined N-glycan oxonium ion signature and consecutive Y-ion saccharide mass distances. In total, we extracted 3,682 unique features from the whole dataset that were consistently detected in at least 75% of the samples from any group (bacterial, viral, control). This is depicted by the feature consensus map (Figure 2A), where each dot represents a unique (N-glyco)peptide feature (Figure 2B). These features displayed a normal distribution in retention time, precursor ion charge state, mass over charge (m/z), and feature mass (Figures S1A–S1D). Next, we investigated the similarity between the samples by calculating Pearson correlations within and between the control group (n = 42) and patient group (n = 91). We found a pooled Pearson’s correlation coefficient of r = 0.76 (within controls), r = 0.69 (within bacterial infection), r = 0.70 (within viral infection), r = 0.64 (between bacterial and viral infection), and r = 0.39 (between controls and patients). The correlation matrix for all individual samples and a representative single correlation plot for a control-control and a control-patient (bacterial) are presented in Figure 2C. This indicates that the within-group variation is smaller than the between-group variation and that the dataset can be explored for differential signatures.
Figure 2.
Overview of the data
(A) Charge deconvoluted consensus feature map, with a color gradient scale to visualize relative differences of all detected features between the bacterial and viral infection group. Differentials that are higher in the bacterial infection group are indicated by a red marker  , whereas differentials that were high in the viral infection group are indicated by a blue color
, whereas differentials that were high in the viral infection group are indicated by a blue color  . Differentials that were only detected in one of the two compared groups are indicated by a triangle▽. Significant univariate features determined by ANOVA are indicated by a black border ◯.
. Differentials that were only detected in one of the two compared groups are indicated by a triangle▽. Significant univariate features determined by ANOVA are indicated by a black border ◯.
(B) A representative MS/MS spectrum of a glycopeptide with typical oxonium B-ions masses, high mass glycan fragment Y-ions, and peptide fragment b- and y-ions.
(C) A Pearson correlation matrix of the healthy group (n = 42) and the patient group (bacterial infection n = 53 and viral infection n = 38) with pooled Pearson’s correlation coefficient of r = 0.76. An example of a linear regression plot (log intensity scale) is shown for the correlation of 3,682 features (blue dots) between two healthy individuals (r = 0.88) and a healthy individual versus an individual with a bacterial infection (r = 0.41).
(D) Venn diagram of the total amount of unique features (n = 3,682), of which 14% of peptide fractions were matched with the protein database and 28% of the N-glycan fractions were matched with the glycan database. Combined data resulted in 463 identified N-glycopeptides.
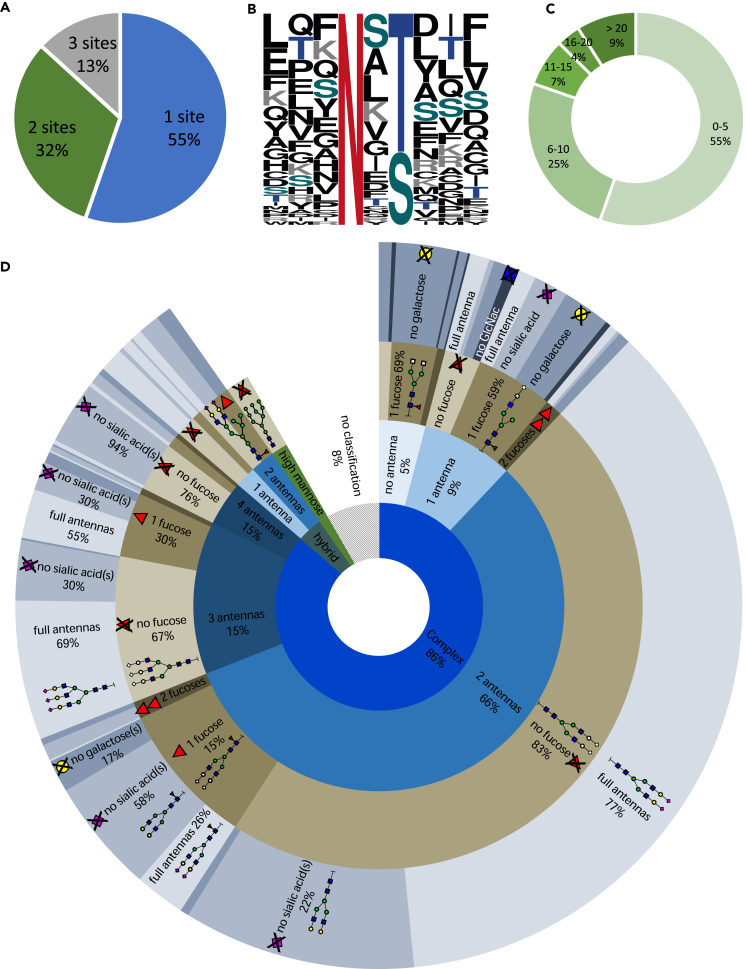
On average, 700 N-glycans and 200 peptides were identified per sample, through two independent database searches using the Consortium for Functional Glycomics and SwissProt databases, respectively. This enabled us to identify 514 peptide moieties (14%) and 1,044 N-glycan moieties (28%) from the 3,682 unique features in total. These identified moieties were mapped onto the consensus feature table based on retention time and mass by means of an in-house developed script at less than 1% false positive rate, resulting in 463 fully identified N-glycopeptides (Figure 2D). The majority of the identified N-glycopeptides originated from proteins with one identified glycosylation site (55%), although for multiple proteins two (32%) or even three (13%) N-glycosylation sites were elucidated (Figure 3A). Of these sites, 65% contained the N-X-T amino acid sequon and 35% the N-X-S sequon—where X can be any amino acid except proline (Figure 3B). Each of these N-glycosylation sites carried between 1 and over 20 different glycan forms (Figure 3C), comprising the main N-glycan classes ranging from high mannose and hybrid structures up to complex sialylated tetra-antennary glycans with a variable fucosylation status (Figure 3D), demonstrating an unbiased enrichment of the main N-glycoforms. In total, 60 unique peptides were identified, originating from 34 different proteins (Table S3). The identified proteins are primarily high abundant plasma glycoproteins, i.e. immunoglobulins (IgG, IgA, IgM), transferrin, fibrinogen, factor H, and complement C3.
Figure 3.
Overview of the dataset glycome
(A) The number of N-glycosylation sites that were identified per protein.
(B) The distribution (%) of each amino acid in the aligned sequence motif with either an N-X-T (65%) or N-X-S (35%) motif of all identified unique N-glycosylation sites (n = 55).
(C) The number of identified N-glycans per N-glycosylation site.
(D) The distribution of all 1,044 identified N-glycans over the defined classes: complex, hybrid or high mannose. The number of antennas is indicated for each class, subdivided into fucosylation grade and the percentage of missing sugars on the antenna(s).
PCA was employed on all detected features (n = 3,682) to explore the main sources of variance in the data; PC1 (29% explained variance) captures the variance of disease versus healthy, while PC2 (23% explained variance) captures the variance between bacterial and viral infection. No separation between samples could be observed for the first two principle components in score plots based on sex or age (accounting for 52% of total variance in the data) for both the control and patient group (Figures S1E and S1F). This indicates that age- or sex-related N-glycosylation differences are a subservient of the overall variation in the data. When comparing all features in a univariate approach, we found that 3.2% of all features in our dataset were significantly different between young and adult controls (Student’s t-test, 99.95%, p < 1.36e-05, after Bonferroni correction), of which the majority had an increment factor between 0.5 and 1.5 (Figure S1G). Of the identified glycopeptides (n = 463), 5.6% were significantly different (Student’s t-test, p < 1.08e-04, after Bonferroni correction) between pediatric and adult controls: with decreased fucosylation on IgG (peptide EEQ), IgM (peptide NNS), and α2-macroglobulin (peptide VSN), decreased high mannose on IgM (peptide NNS), and decreased fucosylated complex biantennary glycans at all these aforementioned peptides (Table S4). We did not find any significant sex differences between the pediatric and adult controls in either the whole dataset or the identified glycopeptides (Table S4). Based on these explorative analyses of the data, we concluded that the collected data were of high quality without any significant bias created by sample processing or by selective inclusion of the participants within this study, allowing for further in-depth analysis to study specific host responses.
Host response to an infection is reflected in glycopeptide levels
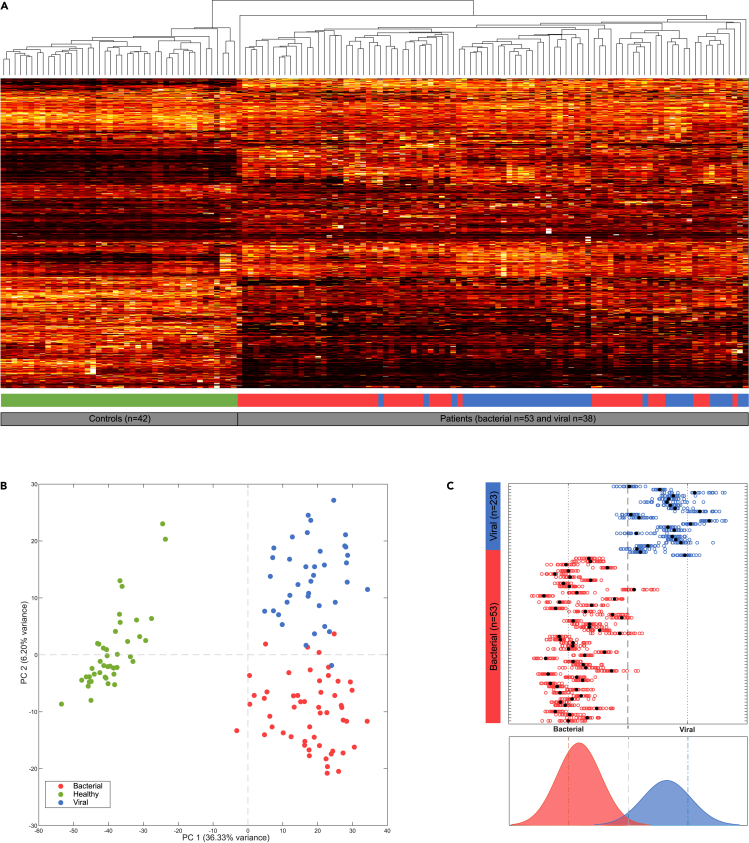
To explore how all features differ between patients and healthy individuals, an average linkage hierarchical clustering was performed. We observed that unsupervised data analysis of all 3,682 features led to two perfectly separated clusters for the patients and the healthy individuals. Within the patient group, a separation of patients with bacterial and viral infection is also visible, although less pronounced (Figure 4A). In order to obtain an overview of generic infection markers, we compared the glycopeptide intensities for healthy individuals to (both bacterial and viral) infected patients, using the conservative Welch t-test for unequal sample sizes (99.95%, p < 1.08e-4, after Bonferroni correction) (Table S5). This resulted in a list of 249 significant identified features. The top 10 significantly altered glycopeptides in the (both bacterial and viral) infected patients as compared to the control group originate from haptoglobin, α-1-acid glycoprotein 1 (A1AG1 or AGP), leucine-rich alpha-2-glycoproteinA, alpha-1-antichymotrypsin, fibrinogen (gamma), complement C3 (CO3), hemopexin (HEMO), and cullin-4B (Figure S2). Multiple glycans were altered, with an increase mainly observed in the patient group as compared to the control group, ranging from a factor 0.24 (CO3) to 15.0 (HEMO). Especially, the increase of unfucosylated complex biantennary glycans and (often fucosylated) hybrid glycans were a notable overall cumulative change.
Figure 4.
Data reduction and differential analysis indicate that the dataset contains multiple distinct signatures for infections
(A) Average linkage hierarchical clustering of 3,682 unique features for the individual samples, including the control (n = 42), bacterial (n = 53), and viral (n = 38) samples.
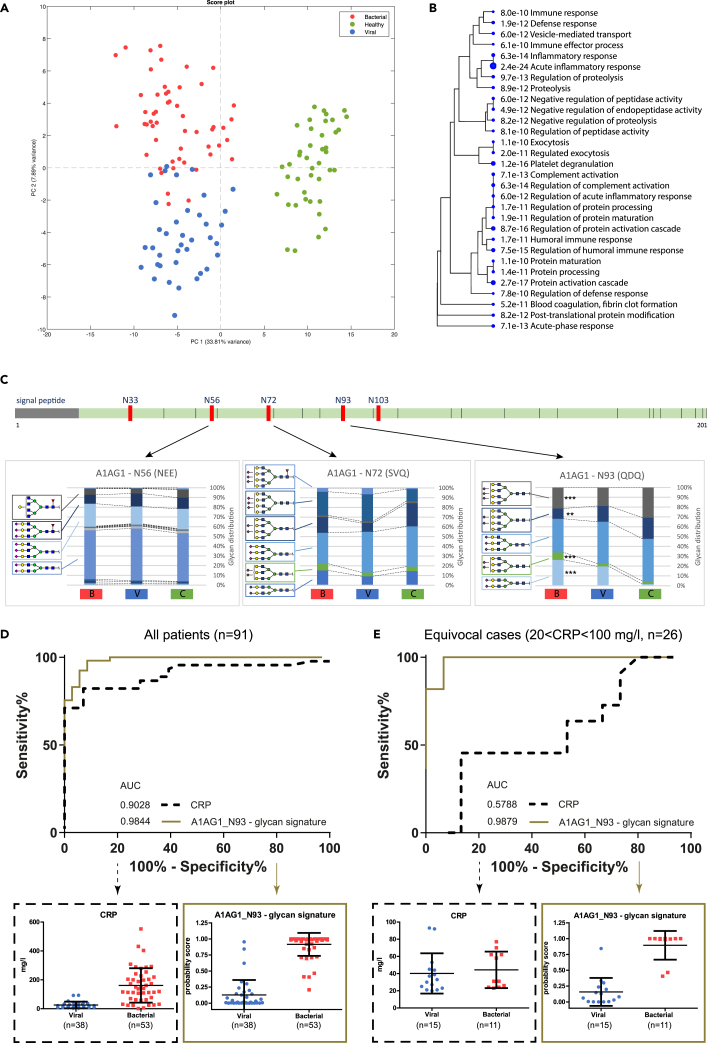
(B) Principal component analysis (PCA) score plot of all samples, based on 2,121 univariate significant features (selected using ANOVA, p < 4.5e-6 Bonferroni corrected), showing a complete separation between the control group and the whole patient group on PC1 (36% explained variance) and an almost full separation between the bacterial and viral group on PC2 (6.2% explained variance).
(C) PLS-DA classification result, based on a multivariate model of 447 features, for either a bacterial or viral infection (x axis), for each patient sample (y axis). Classification results for each of the 21 independent classification prediction models are indicated by the open circles ◯, where the final PLS-DA model is indicated by a closed circle  .
.
To further explore which features differed among the bacterial, viral, and healthy group, we performed an analysis of variance (ANOVA, 99.95%, p < 4.5e-06, after Bonferroni correction for multiple testing) and found 2,121 significantly different features among the three groups. A PCA score plot based on these differential features showed an improved separation between the bacterial and viral sample clusters (in PC2) (Figure 4B) as compared to the previous hierarchical clustering. In order to find the optimal (multivariate) combination of significant features to separate the bacterial infections from viral infections, a PLS-DA with a variable importance projection score of >1 and a false discovery rate < 1%, using 2000 permutations and 21 iterations with cross-validation, was performed on all features. Rhinovirus and adenovirus were excluded from the calculations as it is known from many studies that these viruses are often found as innocent bystanders, which could therefore interfere with the specificity of the classification model. This rendered a subset of 447 significant features that was used to successfully classify 97% of the samples either as a bacterial or viral infection (Figure 4C), with a precision of 0.98, sensitivity of 0.98, specificity of 0.98, and accuracy of 0.97. These excellent PLS-DA classification performance measures indicate that the dataset contains differential signatures that distinguish bacterial from a viral infection.
Identified N-glycopeptide signatures that discriminate bacterial from viral infection
Since not every feature in the dataset is a glycopeptide and as only 53% of the candidate features were fully annotated, we performed a separate ANOVA using only the features with fully characterized peptide and glycan moieties (n = 463). This yielded a reduced list of 96 highly significant N-glycopeptides (99.99% confidence; p < 7.2e-6, after Bonferroni correction), still showing a complete separation between the healthy individuals and patients (PC1, 33.8% explained variance) and a significant separation between bacterial and viral infection (PC2, 7.9% explained variance) in a PCA score plot (Figure 5A). We observed an increased overlap between the bacterial and viral groups, which suggests that some of the unidentified features that were left out were specific for either bacterial or viral infection. The list of identified glycopeptides that differed significantly between bacterial and viral infection comprises a combination of 43 unique glycans divided over 22 peptides from 18 different proteins (Table S3). The Gene Ontology terms show a clear enrichment for immune response proteins as compared to the reference proteome, with the highest significance for “acute inflammatory response” (p = 7.3e-15) (Figure 5B).
Figure 5.
Separation between bacterial and viral class samples based on fully elucidated glycopeptide features
(A) PCA score plot of 96 identified highly significant features (ANOVA, 99.9% confidence, Bonferroni corrected p < 4.24E-7).
(B) The gene-ontology (GO) hierarchical clustering tree for biological processes summarizing the correlation among the top 30 significant pathways and the corresponding p-value. Pathways with many shared genes/proteins are clustered together. Larger dots indicate higher significant p-values.
(C) Schematic linear representation of α1-acid glycoprotein 1 (AGP or A1AG1) with its five N-glycosylation sites (N33, N56, N72, N93, N103), of which three sites were identified in the dataset. The corresponding cumulative bar graphs show for each N-glycosylation site the measured glycan distribution (%) (y axis), comparing the bacterial [B], viral [V], and healthy control group [C] (x axis), using ANOVA with Bonferroni correction, with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) ROC curve for all patients (n = 91) based on the clinical C-reactive protein (CRP) levels and the probability score of the model for the glycosylation signature at glycosylation site N93 (A1AG1_N93 – glycan signature). The boxplots below show all individuals for the viral and bacterial group, including mean and standard deviation, based on the CRP levels (left) and the probability score for the glycan signature at site N93 of A1G1 (right).
(E) ROC curve for patients with ambiguous CRP levels (>20 and <100 mg/L, n = 26) showing the clinical CRP and the probability score of the model for the glycosylation signature at glycosylation site N93. The boxplots below show all individuals for the viral and bacterial group, including mean and standard deviation, for the CRP concentration (left) and the probability score for the glycosylation signature at site N93 of A1AG1 (right).
Glycopeptide analysis enabled us to characterize multiple N-glycosylation sites on one protein (macroheterogeneity), as is demonstrated for A1AG1 or AGP, of which three out of five N-glycosylation sites were identified in this dataset, showing multiple site-specific glycan forms (microheterogeneity): N56 n = 20, N72 n = 7, and N93 with n = 5 different glycans (Figure 5C). Here, we primarily observed desialylation of three- and tetra-antennary complex structures during infection. The glycosylation signature at site N93 (“N93 – glycan signature”) had a higher AUC (0.98) as compared to the clinical C-reactive protein (CRP) (0.90) in the ROC curve (Figure 5D), in particular, for the cases with ambiguous CRP levels (n = 26 patients) as compared to the definite diagnosis by PCR or blood culture—i.e., patients displaying CRP levels above or below the standard cutoff level to distinguish viral from bacterial infection (>20 and <100 mg/L CRP, respectively). The AUC of the A1AG1_N93 glycopeptides model was significantly higher (AUC = 0.99) than CRP (AUC = 0.58) for these equivocal cases (Figure 5E). This demonstrates the potential of selecting candidates for glycopeptide signatures from this dataset for translational research.
Pathogen-specific host N-glycopeptide signatures
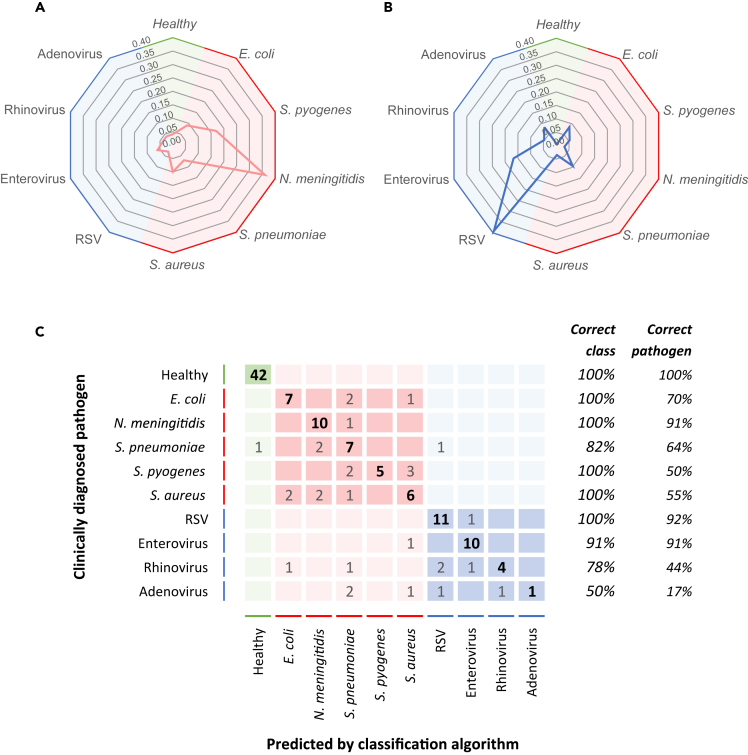
In general, we observed consistent glycopeptide levels for all types of pathogens within each of the two infection groups. However, for several features, a within-group difference was noticed, implying that those N-glycopeptide levels might exhibit a pathogen-specific response. In order to explore if we could use the features (n = 3,682) in this dataset to go beyond a bacterial/viral distinction and predict the causative agent of infection at species level, we employed a supervised machine learning classification method, Genetic Algorithm-Random Forest (GA-RF), to assign each sample in one of the classes associated with the causative pathogen: healthy (n = 42), E. coli (n = 10), N. meningitidis B (n = 11), S. pneumoniae (n = 11), S. pyogenes (n = 10), S. aureus (n = 11), RSV (n = 12), enterovirus (n = 11), rhinovirus (n = 9), or adenovirus (n = 6). We used the cumulative score of 2000 randomly selected training and validation sets for each patient to assess the quality of classification, as exemplified by two spider plots for a patient diagnosed with N. Meningitidis B or RSV, respectively (Figures 6A and 6B). A confusion matrix summarizes the main classification label for each sample in the clinically diagnosed pathogen class (Figure 6C). We found that all healthy subjects were assigned correctly. Whereas, for the bacterial infections, 96.4% was correctly classified in the broad group of bacterial infection and 65.8% of these samples were correctly classified at the pathogen species level, with the best score of 10/11 patients correctly classified for N. meningitidis B. For the viral etiology group, 79.7% was correctly classified as a viral infection, while 60.9% of these patients were correctly assigned to the causative pathogen species. It was, however, more difficult to correctly classify the rhinovirus (4/9) and adenovirus (1/6) pathogen class.
Figure 6.
Prediction of the type of pathogen
By means of a machine learning genetic algorithm random forest (GA-RF) analysis, each (control and patient) sample was classified using 2000 randomly selected training and validation sets from the total features dataset (n = 3,682).
(A) Representative spider plot of the classification scores for a patient who has been clinically diagnosed with N. meningitidis serogroup B (patient #102–1332).
(B) Representative spider plot of the classification scores for a patient who has been clinically diagnosed with respiratory syncytial virus (RSV) (patient #107–1113).
(C) The confusion matrix summarizes the main classification label (columns) for each sample in the clinically diagnosed pathogen class (rows). The numbers indicate the number of (control or patient) samples classified for each pathogen class label, with the correct pathogen classification in bold type.
Although this approach needs further optimization and out-of-sample validation, it highlights that the readily available information in this glycopeptide profiling dataset has the potential to correctly predict the causative pathogen species in patients with a bacterial and viral infection. Glycopeptide profiling thus presents a potential approach to reveal pathogen-specific host responses and adds alternative possibilities for innovative biomarker discovery and elucidation of the host response in different infectious disease classes.
Discussion
The role of posttranslational responses induced by infection, in particular the impact on glycosylation, is understudied and has so far primarily been focused on IgG only. Therefore, we applied an innovative and robust glycoproteomics method to measure the systemic proteome-wide site-specific glycosylation responses upon infection.
We were able to detect 3,682 unique features in at least 75% of the samples, including most highly abundant glycosylated proteins in blood17 with a normal distribution of the glycome18 and a robust reproducibility, which implied an adequate sample preparation, glycopeptide enrichment, chromatographic separation, and detection. From this dataset, we fully identify 463 unique intact glycopeptides. Using various statistical analyses, 96 glycopeptides were extracted that enabled significant differentiation between patients with a bacterial infection and patients with a viral infection. These glycopeptides comprised 43 different N-glycans divided over 22 N-glycosylation sites of 18 proteins that are involved in immune and inflammatory responses. The larger part is associated with an innate response, which is likely due to the fact that the samples are acquired during the acute phase of infection. Supported by a supervised machine learning classification algorithm, we additionally showed that this glycopeptide dataset contains potential signatures to even predict the correct causative pathogen species for 65% of the patients.
Although our dataset showed no significant overall bias for age or sex based on multivariate analysis, univariate analysis identified 3% of the features as significantly different between healthy children and healthy adult controls (with a ratio between 0.5 and 1.5). Age-related N-glycosylation differences thus seem limited to only a small subset of all plasma proteins, especially increased fucosylation of immunoglobulins in young children, which corresponds to previous reports.19,20 Differences between the sexes have been reported as well,19,21 but we did not observe any significant differences in our study. This might be due to the distinct detection methods used or may be a consequence of our smaller sample size of our healthy control group (22 males and 20 females). Also, sex differences are much smaller in children than in adults and are mainly present at the onset of puberty and again from pre-menopause to postmenopausal age.19,22 These age stages were both not included in the control group in this study—as our focus was on pediatric patients.
Overall, we observed desialylation and decreased fucosylation of plasma proteins upon infection. This agrees with reduced sialylation observed in the serum N-glycome of bacteremic patients.10 Considering that we have measured samples that were taken during the acute phase of infection, i.e. within 24 h upon hospital admission, the contribution of de novo glycan synthesis to the differences in glycosylation that we measured might be limited. We therefore speculate that a biological explanation for the observed desialylation is that the glycans have been altered in situ, thus at the site of infection. Desialylation is a well-known biological “danger signal,” as many pathogens trim terminal sialic acids from host glycans.23 Since it is not very likely that the high degree of desialylation we measured is solely caused by the activity of pathogen-derived sialidases, considering their low abundance, it is conceivable that an evolutionary determined host mechanism is the major cause for these changes. Vertebrate sialidases are however unstable in extracellular fluids, possibly to prevent unintended danger signals.24 Yet, the mammalian sialidase neuraminidase 1 (NEUR1 protein, NEU1 gene) is able to translocate to the cell surface and desialylate membrane-bound Toll-like receptors (TLRs) and integrins in reaction to inflammation.25 In a recent study on the transcriptome of patients with a bacterial or viral infectious disease,26 more than 500 glycogenes were found significantly upregulated during acute infection, including NEU1. It could thus well be that other nearby acute proteins are also desialylated in the process and in this way augments the danger signal into the fluid phase or blood stream. For example, factor H, a specific sialic acid-binding protein, is then no longer able to bind to the desialylated glycans on cell surface proteins, with the consequence that the complement system is activated.6 Additional studies are required to further elucidate glycosylation mechanisms and pathways in infectious diseases. Extending glycopeptide analysis to other infectious diseases, like COVID-19, ZIKA, HIV, HBV, or HBC infections, especially longitudinal studies, could provide further understanding of the mechanisms involved in infection. In particular, in vitro studies could aid in in this respect.
Various types of host biomarkers have been targeted to distinguish viral from bacterial infections: RNA markers,26,27,28,29 host protein markers,30,31,32 host plasma lipids,33 and the (released) host N-glycome.34 To the best of our knowledge, we are the first to find pathogen-specific host markers by analysis of intact glycopeptides in full plasma. We observed site-specific differences in N-glycosylation, of which several are highly significant signatures to distinguish bacterial from viral infection. This essential information cannot be obtained from released glycans or protein levels only. The example of AGP or A1AG1, an acute phase protein, showing that glycosylation is site specific (microheterogeneous), adds an extra dimension to the analysis allowing to further elucidate the function of specific glycans and improve the biological interpretation.
Moreover, using a supervised machine learning classification algorithm (GA-RF), we showed that these glycopeptide data contain potential signatures specific for different types of pathogens. It was however more difficult to classify rhinovirus and adenovirus infections. This may be the result of the smaller sample size of these pathogen subgroups. Another explanation might be related to the fact that rhinovirus and adenovirus are often found to cause sub-clinical infections,35,36 which can evolve into co-infections, evoking a mixed host response that interferes with the specificity of the classification. In the PLS-DA model, we also observed less pronounced responses for adenovirus- and rhinovirus-infected patients. They were therefore excluded from the PLS-DA training model to only train on distinct patterns. It is thus of importance to test for co-infections when selecting patients for these types of studies.
In conclusion, this study shows that N-glycans, more specifically N-glycopeptides, are significantly affected upon infection. Bacterial infections cause significantly different patterns of glycosylation as compared to viral infections. Moreover, we have shown that causative pathogen species can be identified based on unique blood plasma glycopeptide signatures. This demonstrates that glycoproteomic methodologies hold enormous potential to improve the interpretation of biological changes in response to infection and that the glycoproteome is a relatively unexplored source for biomarkers, which could support the development of innovative diagnostics for infectious diseases.
Limitations of the study
Ideally, the control and patient group are age-matched. It is, however, very difficult to obtain ethical approval and informed consent from parents to draw blood samples from healthy individuals at the average age of 2 years old. To increase statistical power, and after careful consideration, we therefore decided to add the adult controls (n = 20) to the child control group (n = 22), because an age difference was not affecting variation in the first two principle components of our multivariate model and age-dependent glycopeptides were not the leading differential features for infection in our dataset. Age-matched groups would be of particular importance for subsequent biomarker validation, which is beyond the scope of this work.
The identification of only 14% of the peptide sequences and 28% of the glycans seems rather low. However, our method analyzed full plasma which is a highly complex matrix, whereas our method did not involve any fractionation, depletion of high abundant proteins, or specific protein enrichment pre-treatments. Moreover, the duration of the chromatographic separation was only 60 min. Thus, in order to study the glycoproteome in more detail, multiple enrichments strategies could be employed and the chromatographic separation step could be elongated. An alternative stationary or mobile phase combination or a different enzymatic sample preparation would have allowed for inclusion of very small or highly hydrophilic glycopeptides that might have been missed as they are not retained on the C18 reversed-phase column. Furthermore, with the method used in this study, we cannot resolve linkage and positional isomers, which would require a preceding linkage-specific sialic acid derivatization,37 capillary electrophoresis hyphenation,38 or ion mobility separation.39
Although these preliminary results of the used classification models demonstrate the high potential of many features of this dataset, the limitation of this study is that the training set was also partially used for the validation set due to the small size of the cohort. For additional research, we therefore recommend a larger sample cohort to enable selection of an independent training set and validation set.
We were not able to identify all features. The dataset is made publicly available to allow re-analysis when more advanced computational tools become available.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Patient plasma samples | Radboud UMC, the Netherlands | N/A |
| Healthy plasma samples | Radboud UMC, the Netherlands | N/A |
| Deposited data | ||
| Raw datafiles | This paper | http://www.proteomexchange.org/dataset/PXD028437 |
| Software and algorithms | ||
| DataAnalysis v4.2 | Bruker Daltonik GmbH, Germany | https://www.bruker.com |
| BioPharma Compass v3.0 | Bruker Daltonik GmbH, Germany | https://www.bruker.com/en/products-and-solutions/mass-spectrometry/ms-software/biopharma-compass.html |
| KNIME v3.2.1 | KNIME, USA | https://www.knime.com |
| KNIME OpenMS nodes v2.7.0 | KNIME, USA | https://www.knime.com/community/bioinf/openms |
| OpenMS workflow | Wessels et al.16 | http://www.proteomexchange.org/dataset/PXD034214 |
| MATLAB 2014b | MatWorks, USA | https://www.mathworks.com/products/matlab.html |
| PERL | Perl, USA | https://www.perl.org/get.html |
| PLS-DA and GA-RF codes | Wessels et al.16 | http://www.proteomexchange.org/dataset/PXD034214 |
| ShinyGO v0.61 (Gene Ontology enrichment analysis) | Ge et al.40 | http://bioinformatics.sdstate.edu/go/ |
| Other | ||
| Patient metadata | This paper | Table S2, PXD028437 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marien de Jonge (Marien.deJonge@radboudumc.nl)).
Materials availability
This study did not generate new unique reagents. Patient material used in this study may be made available only when the purpose is described in the informed consent and upon reasonable request to the lead contact, Marien de Jonge (Marien.deJonge@radboudumc.nl).
Experimental model and study participant details
For this study a group of 91 children (0-18 years) diagnosed with either a definite bacterial infection (n=53), or a definite viral infection (n=38) were selected. These patients were hospitalized in a medical centre situated in the Netherlands, United Kingdom, or Spain. Most patients have European ethnicity. Bacterial or viral infection were determined by a positive blood culture or PCR, respectively. Of all subjects, plasma samples taken within 24 hours post-admission to the hospital were used. All samples were part of the EUCLIDS41,42 and IRIS28 studies, which were approved by the Medical Ethical Committees of the academic hospitals involved in these studies. Parents or guardians and children above 12 years old provided written informed consent. The selection was based on an equal distribution of the type of causative pathogen, sex, and age. Additional clinical data from the patients enrolled in this study are summarized in Table S2.
Furthermore, plasma samples from 22 pediatric healthy subjects (0-5 years old) from a previous study43 were included. These blood sample were collected for medical reasons not related to inflammatory or infectious diseases. The selection criteria for these pediatric controls and approval by Medical Ethical Committee have previously been described.43 Additionally, 20 adult healthy volunteers (23-55 years) donated their blood for this study after informed consent and were collected according to the guidelines of the Human-related Research Committee Arnhem-Nijmegen. Exclusion criteria were: fever (>38.5°C), symptoms of infection (bacterial or viral), chronic illness and immune suppressive medication. The healthy controls have European ethnicity. Additional clinical data from all healthy subjects enrolled in this study are summarized in Table S2.
Method details
Plasma sample collection and preparation
Patient plasma samples were collected and frozen as described previously (EUCLIDS41,42 and IRIS28). The selected samples were shipped on dry ice and stored at -80°C upon arrival. Plasma samples from the pediatric healthy subjects43 and healthy adult volunteers were placed on ice immediately after collection and were processed within one hour (10 min, 2500g, 4°C). Aliquots were stored at -80°C.
Ten microliter of plasma was denaturated in 10 μl urea (8 M, pH 8.0) and reduced with 15 μl dithiothreitol for 30 min at RT. Reduced cysteines were alkylated through incubation with 15 μl 2-chloroacetamide (CAA) in the dark for 20 min at RT. Next, proteins were subjected to LysC digestion (1 μg LysC/50 μg protein) by incubating the sample at RT for 3 hours. Then, samples were diluted with 3 volumes of 50 mM ammonium bicarbonate and trypsin was added (1 μg trypsin /50 μg protein) for overnight digestion at 37°C. The glycopeptides were enriched according to Wessels et al.16 Briefly, the glycopeptides were captured using 100 μl Sepharose CL-4B beads slurry (Sigma) per sample well in a filter plate (AcroPrep Advance, VWR). The beads were washed three times with 20% ethanol and 83% acetonitrile, respectively. The sample was then incubated on the beads for 20 min at RT on a shaking plate (Eppendorf). The filter plate was then centrifuged and the beads were first washed three times with 83% acetonitrile and then three times with 83% acetonitrile in 0.1% TFA. Next, the glycopeptides were eluted from the beads by incubation with milliQ water for 5 min at RT, followed by centrifugation. The eluates were stored at -80°C until analysis.
Liquid chromatography mass spectrometric analysis
Samples were analyzed in randomized order by means of liquid chromatography with online tandem mass spectrometry (maXis Plus, Bruker Daltonics) interfaced via a vacuum-assisted axial desolvation nanoflow electrospray ionization source (captive sprayer, Bruker Daltonics) for data-dependent acquisition. Five microliters of sample were loaded onto the trapping column (Acclaim PepMap RSLC, 100μm×2cm, nanoViper, 5μm 100Å C18 particles, Thermo Scientific) using 0,1% formic acid at a flow rate of 7000 nl/min for 3 minutes at room temperature. Next, peptides were separated on a C18 reversed phase 15cm length × 75μm internal diameter C18RP analytical column (eluteFIFTEEN, C18 ReproSil AQ, 1.9μm particles, 120Å pore size, Bruker Daltonics) at 45°C using a linear gradient of 3–45% acetonitrile (ACN) 0.1% formic acid in 60 min at a flow rate of 500 nl/min. Electrospray ionization conditions were 3 L/min 180°C N2 drying gas, 1500 V capillary voltage and 0.2 Bar N2 for gas phase supercharging (nanobooster) using acetonitrile as dopant. Data dependent acquisition (AutoMSn) was performed using a 3s duty cycle at 2 Hz acquisition rate for full MS spectra and a variable number of MS/MS experiments at precursor intensity scaled acquisition rate (4 Hz MS/MS spectral rate at 10.000 counts, 9 Hz MS/MS spectral rate at 75.000 counts). Precursor ions above 700 m/z with charge state z=2+ or higher (preferred charge state range of z=2+ to z=6+) were selected for MS/MS analysis with active exclusion enabled (excluded after one spectrum, released after 0.5 min, reconsidered precursor if current intensity/previous intensity ≥4, smart exclusion disabled). Collision induced dissociation energy stepping was applied to acquire glycan- and peptide-moiety fragmentation data within a merged MS/MS spectrum recorded at 100% collision energy for glycan moiety fragmentation and 200% collision energy for peptide moiety fragmentation in a 3:7 stoichiometry. Collision cell tuning was respectively stepped for 50% of each collision energy MS/MS step time interval from 1200 to 1800 Vpp collision cell RF and 64 to 110 μsec transfer time to ensure optimal transmission over the full 300-3700 m/z mass range.
Quantification and statistical analysis
Raw data were processed using DataAnalysis 4.2: post-acquisition mass calibration (lock-mass and sodium-acetate clusters) and MS/MS data were exported to XML file format. Mass calibrated raw data files were converted to mzML using compass Xport (Bruker Daltonics) and subsequently processed by Knime OpenMS nodes v2.7.0 (Knime v3.2.1) to generate a consensus feature map that contains intensities and associated metadata for aligned features from all analysis files, as described previously by Wessels et al.16 The MS/MS xml-files were processed in BioPharma Compass 3.0 to classify glycopeptide fragmentation spectra by HexNAc-Hex-Hex (N-glycan trimannosyl chitobiose core) mass distance pattern matching and to calculate peptide- and glycan-moiety masses, respectively. Classified glycopeptide fragmentation MS/MS spectra were searched against the Consortium for Functional Glycomics (2019-03-01) glycan structure database and glycan-spectrum matches with GlycoQuest (part of BioPharma Compass v3.0); score>10, >10% spectral intensity coverage and >10% theoretical fragment ion coverage (B-,Y- and internal-fragment ions) were accepted for subsequent processing. Classified and non-classified fragmentation spectra were searched against the human SwissProt protein sequence database (2016-06, fasta file) by means of the MASCOT search engine (v2.5.1), using calculated peptide-moiety precursor masses for classified glycopeptide spectra using the following settings: trypsin digestion, a precursor mass tolerance of 20.0 ppm, MS/MS tolerance of 0.05 Da, allowing for 1 missed cleavage and a fixed carbamidomethyl modification (cys), and variable deamidated (NQ), oxidation (M), acetyl (N-term), HExNac (N) and pyro-carbamidomethyl (N-term) modifications, with percolator enabled to achieve an FDR of ≤1.0%.
In-house developed Matlab (R2014b) and Perl (v5.36.1) scripts were used to map the identification data to the consensus features within the consensus feature map and to perform subsequent data pre-processing and downstream analyses (heatmaps, hierarchical clustering, ANalysis Of VAriance (ANOVA), Principle Component Analysis (PCA), Partial Least Squares - Discriminant Analysis (PLS-DA), and Genetic Algorithm – Random Forest (GA-RF). PLS-DA was performed using a repeated double cross validation procedure (VIP>1, FDR<1%, 2000 permutations, 21 iterations with number of latent variables optimized in a Leave-One-Out inner cross validation loop and classification models build using a 3-fold outer cross validation loop resulting in predicted class labels), as described previously by Wessels et al.16 The t-tests, ANOVA, and ROC curves were performed and created using IBM SPSS Statistics v25.0.0.1 and GraphPad Prism v5.03. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Post-hoc multiple testing correction was performed by multiplying the P-value with a Bonferroni correction factor, depending on the number of features and groups tested, specified for each corrected P-value in the results section and supplemental information. Statistical approaches were based on recommendations on chemometric and statistical aspects in biomarker discovery by Suppers et al.44 Gene Ontology (GO) enrichment analysis was performed using ShinyGO v0.61.40
Acknowledgments
This research, part of the PERFORM project, has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 668303. The samples collected previously were funded by: the European Seventh Framework Programme for Research and Technological Development (FP7) under EUCLIDS Grant Agreement No. 279185.
We would like to thank all the patients and the healthy controls for donating their blood for the EUCLIDS, IRIS, and Westra studies. We would like to thank the Radboud Consortium for Glycoscience for their advice for data interpretation. We also thank the PERFORM consortium (see supplemental material for all participants) for their collaboration and fruitful discussions.
Author contributions
Conceptualization, E.W., H.W., and M.J.; Methodology, E.W., M.D., H.W., and M.J.; Investigation, E.W.; Software, A.S. and H.W.; Formal Analysis, E.W. and H.W.; Visualization, E.W. and H.W.; Resources, M.F., L.H., N.K., R.P., M.K., V.W., J.H., and F.M.; Writing – Original Draft, E.W. and M.J.; Writing – Review & Editing, E.W., J.G., A.S., M.F., L.H., N.K., R.P., M.D., M.K., V.W., J.H., F.M., M.L., R.G., A.G., D.L., H.W., and M.J.; Funding Acquisition, M.J., R.G., and M.L.; Supervision, H.W., D.L., and M.J.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 4, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107257.
Contributor Information
Esther Willems, Email: esther.willems1@radboudumc.nl.
PERFORM consortium:
Amina Abdulla, Christoph Aebi, Koen van Aerde, Rachel Agbeko, Philipp Agyeman, Umberto D’alessandro, Ladan Ali, Wynand Alkema, Karen Allen, Fernando Álvez González, Suzanne Anderson, Imran Ansari, Tasnim Araf, Tanja Avramoska, Bryan Baas, Natalija Bahovec, Cristina Balo Farto, Anda Balode, A.M. Barendregt, Ruth Barral-Arca, María Barreiro Castro, Arta Bārzdiņa, David Bath, Sebastian Bauchinger, Lucas Baumard, Hinrich Baumgart, Frances Baxter, Ashley Bell, Kathryn Bell, Xabier Bello, Evangelos Bellos, Martin Benesch, Mirian Ben García, Joshua Bennet, Christoph Berger, J.M. van den Berg, Sara Bernhard-Stirnemann, Sagida Bibi, Christoph Bidlingmaier, Alexander Binder, Vera Binder, Kalifa Bojang, Dorine M. Borensztajn, Ulrich von Both, Karen Brengel-Pesce, Bryan van den Broek, Judith Buschbeck, Leo Calvo-Bado, Sandra Carnota, Enitan D. Carrol, Michael J. Carter, Miriam Cebey-López, Samba Ceesay, Astrid Ceolotto, Adora Chan, Elizabeth Cocklin, Kalvin Collings, Stephen Crulley, Aubrey Cunnington, María José Curras-Tuala, Katharina Danhauser, Saffiatou Darboe, Sarah Darnell, Tisham De, Dārta Deksne, Kirsty Devine, Juan Emmanuel Dewez, Julia Dudley, Carlos Durán Suárez, Ernst Eber, Irini Eleftheriou, Marieke Emonts, Daniel Fabian, Tobias Feuchtinger, Katy Fidler, Colin Fink, Michiel van der Flier, A.M. van Furth, Rachel Galassini, Siegfried Gallistl, Luisa García Vicente, Dace Gardovska, J. Geissler, G.P.J.M. Gerrits, Eric Giannoni, Ilona van der Giessen, Jolein Gloerich, Alberto Gómez-Carballa, Jose Gómez Rial, Alain J. van Gool, Gunther Gores, Dagne Grāvele, Matthias Griese, Ilze Grope, Ronald de Groot, Meeru Gurung, L. de Haan, Nikolaus Haas, Dominic Habgood-Coote, Nienke N. Hagedoorn, Harald Haidl, Shea Hamilton, Almuthe Hauer, J. Heidema, Ulrich Heininger, Stefanie Henriet, Jethro Herberg, Clive Hoggart, Susanne Hösele, Sara Hourmat, Christa Hude, Martijn Huijnen, Heather Jackson, Rebecca Jennings, Joanne Johnston, Marien I. de Jonge, Ilse Jongerius, Rikke Jorgensen, Myrsini Kaforou, Christian Kahlert, Rama Kandasamy, Matthias Kappler, Julia Keil, Markus Keldorfer, Dominic F. Kell, Eunjung Kim, Sharon King, Lieke Kloosterhuis, Daniela S. Kohlfürst, Benno Kohlmaier, Laura Kolberg, Mojca Kolnik, Larissa Krenn, Taco Kuijpers, M. van der Kuip, Pilar Leboráns Iglesias, Simon Leigh, Manuel Leitner, M. van Leur, Emma Lim, Naomi Lin, Ching-Chuan Liu, Sabine Löffler, Eberhard Lurz, Ian Maconochie, Christine Mackerness, François Mallet, Federico Martinón-Torres, Antonis Marmarinos, Alex Martin, Mike Martin, José María Martinón Sánchez, Nazareth Martinón-Torres, Federico Martinón-Torres, Paul McAlinden, Anne McDonnell, Sam McDonald, C.J. Miedema, Anija Meiere, Stephanie Menikou, G. van Mierlo, Alec Miners, Ravi Mistry, Henriëtte A. Moll, Marine Mommert, Belén Mosquera Pérez, David R. Murdoch, Sobia Mustafa, Giancarlo Natalucci, C. Neeleman, Karen Newall, Samuel Nichols, Tobias Niedrist, Anita Niederer-Loher, Ruud Nijman, Ieva Nokalna, Urzula Nora Urbāne, Gudrun Nordberg, C.C. Obihara, Daniel O'Connor, Wilma Oosthoek, Veronika Osterman, Alexandre Pachot, D. Pajkrt, Jacobo Pardo-Seco, Stéphane Paulus, Jana Pavāre, Ivonne Pena Paz, Salina Persand, Andreas Pfleger, Klaus Pfurtscheller, Ria Philipsen, Ailsa Pickering, Benjamin Pierce, Heidemarie Pilch, Lidia Piñeiro Rodríguez, Sara Pischedda, Tina Plankar Srovin, Marko Pokorn, Andrew J. Pollard, Lena Pölz, Klara M. Posfay-Barbe, Petra Prunk, Zanda Pučuka, Glorija Rajic, Aqeela Rashid, Lorenzo Redondo-Collazo, Christa Relly, Irene Rivero Calle, Sara Rey Vázquez, Mathew Rhodes, Vivien Richmond, Thomas Riedel, Anna RocaIsatou Sarr, Siegfried Rödl, Carmen Rodríguez-Tenreiro, Sam Romaine, Emily Rowlands, Miguel Sadiki Ora, Manfred G. Sagmeister, Momodou Saidykhan, Antonio Salas, Luregn J. Schlapbach, D. Schonenberg, Fatou Secka, Katrīna Selecka, Sonia Serén Fernández, Cristina Serén Trasorras, Priyen Shah, Ching-Fen Shen, Shrijana Shrestha, Aleksandra Sidorova, Andrea Skrabl-Baumgartner, Giselle D’Souza, Matthias Sperl, Evelien Sprenkeler, Nina A. Schweintzger, Laura Stampfer, Molly Stevens, Martin Stocker, Volker Strenger, Dace Svile, Kelly Syggelou, Maria Tambouratzi, Chantal Tan, Emma Tavliavini, Evelyn Thomson, Stephen Thorson, Holger Till, G.A. Tramper-Stranders, Andreas Trobisch, Maria Tsolia, Effua Usuf, Lucille Valentine, Clementien L. Vermont, Marisol Vilas Iglesias, Katarina Vincek, Marie Voice, Gabriella de Vries, Diane Wallia, Shih-Min Wang, Esther Willems, Clare Wilson, Amanda Wood, Phil Woodsford, Victoria Wright, Marietta Xagorari, Shunmay Yeung, Joany Zachariasse, Dace Zavadska, Syed M.A. Zaman, Judith Zandstra, Werner Zenz, Christoph Zurl, and Manuela Zwerenz
Supplemental information
Data and code availability
All data and codes used in this paper are publicly available.
-
•
The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028437.
-
•
The de-identified metadata and processed data matrix (feature map) have also been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028437.
-
•
The original codes used to process the data have been published previously16 and have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD034214.
References
- 1.Minguez P., Parca L., Diella F., Mende D.R., Kumar R., Helmer-Citterich M., Gavin A.C., Van Noort V., Bork P. Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. 2012;8:599. doi: 10.1038/msb.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltrao P., Bork P., Krogan N.J., van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol. Syst. Biol. 2013;9:714. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebrilla C.B., An H.J. The prospects of glycan biomarkers for the diagnosis of diseases. Mol. Biosyst. 2009;5:17–20. doi: 10.1039/b811781k. [DOI] [PubMed] [Google Scholar]
- 4.Van Scherpenzeel M., Willems E., Lefeber D.J. Clinical diagnostics and therapy monitoring in the congenital disorders of glycosylation. Glycoconj. J. 2016;33:345–358. doi: 10.1007/s10719-015-9639-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reily C., Stewart T.J., Renfrow M.B., Novak J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019;15:346–366. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varki A. Biological roles of glycans. Glycobiology. 2017;27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varki A., Cummings R.D., Esko J.D., Freeze H.H., Hart G.W., Marth J.D. third Edition. Cold Spring Harbor Laboratory Press; 2017. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 8.Rabinovich G.A., Van Kooyk Y., Cobb B.A. Vol. 1253. Annals of the New York Academy of Sciences; 2012. Glycobiology of Immune Responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnaar R.L. Glycobiology simplified: diverse roles of glycan recognition in inflammation. J. Leukoc. Biol. 2016;99:825–838. doi: 10.1189/jlb.3RI0116-021R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeCoux A., Tian Y., DeLeon-Pennell K.Y., Nguyen N.T., de Castro Brás L.E., Flynn E.R., Cannon P.L., Griswold M.E., Jin Y.-F., Puskarich M.A., et al. Plasma glycoproteomics reveals sepsis outcomes linked to distinct proteins in common pathways. Crit. Care Med. 2015;43:2049–2058. doi: 10.1097/CCM.0000000000001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joenvaara S., Saraswat M., Kuusela P., Saraswat S., Agarwal R., Kaartinen J., Järvinen A., Renkonen R. Quantitative N-glycoproteomics reveals altered glycosylation levels of various plasma proteins in bloodstream infected patients. PLoS One. 2018;13:e0195006. doi: 10.1371/journal.pone.0195006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan N., Wuhrer M., Ruhaak L.R. Mass spectrometry in clinical glycomics: The path from biomarker identification to clinical implementation. Clin. Mass Spectrom. 2020;18:1–12. doi: 10.1016/j.clinms.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang L., Jia L., Zhi Y., Li P., Zhao T., Zhu B., Lan R., Hu Y., Zhang H., Sun S. Mapping human N-linked glycoproteins and glycosylation sites using mass spectrometry. Trends Anal. Chem. 2019;114:143–150. doi: 10.1016/j.trac.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Franc V., Heck A.J.R. Glycoproteomics: a balance between high-throughput and in-depth analysis. Trends Biotechnol. 2017;35:598–609. doi: 10.1016/j.tibtech.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Yu A., Zhao J., Peng W., Banazadeh A., Williamson S.D., Goli M., Huang Y., Mechref Y. Advances in mass spectrometry-based glycoproteomics. Electrophoresis. 2018;39:3104–3122. doi: 10.1002/elps.201800272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wessels H.J., Kulkarni P., van Dael M., Suppers A., Willems E., Zijlstra F., Kragt E., Gloerich J., Schmit P.-O., Pengelley S. Plasma glycoproteomics delivers high-specificity disease biomarkers by detecting site-specific glycosylation abnormalities. bioRxiv. 2022 doi: 10.1101/2022.05.31.494121. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson N.L., Anderson N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y., Furukawa J.i., Naito S., Higashino K., Numata Y., Shinohara Y. Quantitative analysis of total serum glycome in human and mouse. Proteomics. 2016;16:2747–2758. doi: 10.1002/pmic.201500550. [DOI] [PubMed] [Google Scholar]
- 19.Pučić M., Mužinić A., Novokmet M., Škledar M., Pivac N., Lauc G., Gornik O. Changes in plasma and IgG N-glycome during childhood and adolescence. Glycobiology. 2012;22:975–982. doi: 10.1093/glycob/cws062. [DOI] [PubMed] [Google Scholar]
- 20.De Haan N., Reiding K.R., Driessen G., van der Burg M., Wuhrer M. Changes in healthy human IgG Fc-glycosylation after birth and during early childhood. J. Proteome Res. 2016;15:1853–1861. doi: 10.1021/acs.jproteome.6b00038. [DOI] [PubMed] [Google Scholar]
- 21.Ding N., Nie H., Sun X., Sun W., Qu Y., Liu X., Yao Y., Liang X., Chen C.C., Li Y. Human serum N-glycan profiles are age and sex dependent. Age Ageing. 2011;40:568–575. doi: 10.1093/ageing/afr084. [DOI] [PubMed] [Google Scholar]
- 22.Knežević A., Gornik O., Polašek O., Pučić M., Redžić I., Novokmet M., Rudd P.M., Wright A.F., Campbell H., Rudan I., Lauc G. Effects of aging, body mass index, plasma lipid profiles, and smoking on human plasma N-glycans. Glycobiology. 2010;20:959–969. doi: 10.1093/glycob/cwq051. [DOI] [PubMed] [Google Scholar]
- 23.Wei M., Wang P.G. Desialylation in physiological and pathological processes: New target for diagnostic and therapeutic development. Prog. Mol. Biol. Transl. Sci. 2019;162:25–57. doi: 10.1016/bs.pmbts.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Varki A., Gagneux P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amith S.R., Jayanth P., Franchuk S., Finlay T., Seyrantepe V., Beyaert R., Pshezhetsky A.V., Szewczuk M.R. Neu1 desialylation of sialyl α-2, 3-linked β-galactosyl residues of TOLL-like receptor 4 is essential for receptor activation and cellular signaling. Cell. Signal. 2010;22:314–324. doi: 10.1016/j.cellsig.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 26.Kaforou M., Habgood-Coote D. RNA-sequencing of Whole Blood Samples from Children with a Range of Acute Febrile Illnesses. 2022. https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-11671
- 27.Mahajan P., Kuppermann N., Suarez N., Mejias A., Casper C., Dean J.M., Ramilo O., Febrile Infant Working Group for the Pediatric Emergency Care Applied Research Network PECARN RNA transcriptional biosignature analysis for identifying febrile infants with serious bacterial infections in the emergency department: a feasibility study. Pediatr. Emerg. Care. 2015;31:1–5. doi: 10.1097/PEC.0000000000000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herberg J.A., Kaforou M., Wright V.J., Shailes H., Eleftherohorinou H., Hoggart C.J., Cebey-López M., Carter M.J., Janes V.A., Gormley S., et al. Diagnostic test accuracy of a 2-transcript host RNA signature for discriminating bacterial vs viral infection in febrile children. JAMA. 2016;316:835–845. doi: 10.1001/jama.2016.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gliddon H.D., Herberg J.A., Levin M., Kaforou M. Genome-wide host RNA signatures of infectious diseases: discovery and clinical translation. Immunology. 2018;153:171–178. doi: 10.1111/imm.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oved K., Cohen A., Boico O., Navon R., Friedman T., Etshtein L., Kriger O., Bamberger E., Fonar Y., Yacobov R., et al. A novel host-proteome signature for distinguishing between acute bacterial and viral infections. PLoS One. 2015;10:e0120012. doi: 10.1371/journal.pone.0120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Houten C.B., de Groot J.A.H., Klein A., Srugo I., Chistyakov I., de Waal W., Meijssen C.B., Avis W., Wolfs T.F.W., Shachor-Meyouhas Y., et al. A host-protein based assay to differentiate between bacterial and viral infections in preschool children (OPPORTUNITY): a double-blind, multicentre, validation study. Lancet Infect. Dis. 2017;17:431–440. doi: 10.1016/S1473-3099(16)30519-9. [DOI] [PubMed] [Google Scholar]
- 32.Tan C.D., van den Broek B., Womersley R.S., Kaforou M., Hagedoorn N.N., van der Flier M., Jackson H., Moll H.A., Snijder R., de Jonge M.I., et al. A Novel Combination of Host Protein Biomarkers to Distinguish Bacterial From Viral Infections in Febrile Children in Emergency Care. Pediatr. Infect. Dis. J. 2023;42:e235–e242. doi: 10.1097/INF.0000000000003952. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Nijman R., Camuzeaux S., Sands C., Jackson H., Kaforou M., Emonts M., Herberg J.A., Maconochie I., Carrol E.D., et al. Plasma lipid profiles discriminate bacterial from viral infection in febrile children. Sci. Rep. 2019;9:17714–17813. doi: 10.1038/s41598-019-53721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee S., Kawahara R., Tjondro H.C., Shaw D.R., Nenke M.A., Torpy D.J., Thaysen-Andersen M. Serum N-Glycomics Stratifies Bacteremic Patients Infected with Different Pathogens. J. Clin. Med. 2021;10:516. doi: 10.3390/jcm10030516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassoun A., Huff M.D., Weisman D., Chahal K., Asis E., Stalons D., Grigorenko E., Green J., Malone L.L., Clemmons S., Lu S. Seasonal variation of respiratory pathogen colonization in asymptomatic health care professionals: A single-center, cross-sectional, 2-season observational study. Am. J. Infect. Control. 2015;43:865–870. doi: 10.1016/j.ajic.2015.04.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Man W.H., van Houten M.A., Mérelle M.E., Vlieger A.M., Chu M.L.J.N., Jansen N.J.G., Sanders E.A.M., Bogaert D. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir. Med. 2019;7:417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Haan N., Yang S., Cipollo J., Wuhrer M. Glycomics studies using sialic acid derivatization and mass spectrometry. Nat. Rev. Chem. 2020;4:229–242. doi: 10.1038/s41570-020-0174-3. [DOI] [PubMed] [Google Scholar]
- 38.Kammeijer G.S.M., Jansen B.C., Kohler I., Heemskerk A.A.M., Mayboroda O.A., Hensbergen P.J., Schappler J., Wuhrer M. Sialic acid linkage differentiation of glycopeptides using capillary electrophoresis–electrospray ionization–mass spectrometry. Sci. Rep. 2017;7:3733–3810. doi: 10.1038/s41598-017-03838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M., Lee K.K. Site-specific mapping of sialic acid linkage isomers by ion mobility spectrometry. Anal. Chem. 2016;88:5212–5217. doi: 10.1021/acs.analchem.6b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeddha N.P., Schlapbach L.J., Driessen G.J., Herberg J.A., Rivero-Calle I., Cebey-López M., Klobassa D.S., Philipsen R., de Groot R., Inwald D.P., et al. Mortality and morbidity in community-acquired sepsis in European pediatric intensive care units: a prospective cohort study from the European Childhood Life-threatening Infectious Disease Study (EUCLIDS) Crit. Care. 2018;22:143–213. doi: 10.1186/s13054-018-2052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinón-Torres F., Salas A., Rivero-Calle I., Cebey-López M., Pardo-Seco J., Herberg J.A., Boeddha N.P., Klobassa D.S., Secka F., Paulus S., et al. Life-threatening infections in children in Europe (the EUCLIDS Project): a prospective cohort study. Lancet. Child Adolesc. Health. 2018;2:404–414. doi: 10.1016/S2352-4642(18)30113-5. [DOI] [PubMed] [Google Scholar]
- 43.Westra D., Volokhina E.B., van der Molen R.G., van der Velden T.J.A.M., Jeronimus-Klaasen A., Goertz J., Gracchi V., Dorresteijn E.M., Bouts A.H.M., Keijzer-Veen M.G., et al. Serological and genetic complement alterations in infection-induced and complement-mediated hemolytic uremic syndrome. Pediatr. Nephrol. 2017;32:297–309. doi: 10.1007/s00467-016-3496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suppers A., van Gool A.J., Wessels H.J.C.T. Integrated chemometrics and statistics to drive successful proteomics biomarker discovery. Proteomes. 2018;6:20. doi: 10.3390/proteomes6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and codes used in this paper are publicly available.
-
•
The raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028437.
-
•
The de-identified metadata and processed data matrix (feature map) have also been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD028437.
-
•
The original codes used to process the data have been published previously16 and have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD034214.