Abstract
Transductional targeting of herpes simplex virus (HSV)-based gene therapy vectors offers the potential for improved tissue-specific delivery and can be achieved by modification of the viral entry machinery to incorporate ligands that bind the desired cell surface proteins. The interaction of nerve growth factor (NGF) with tropomyosin receptor kinase A (TrkA) is essential for survival of sensory neurons during development and is involved in chronic pain signaling. We targeted HSV infection to TrkA-bearing cells by replacing the signal peptide and HVEM binding domain of glycoprotein D (gD) with pre-pro-NGF. This TrkA-targeted virus (KNGF) infected cells via both nectin-1 and TrkA. However, infection through TrkA was inefficient, prompting a genetic search for KNGF mutants showing enhanced infection following repeat passage on TrkA-expressing cells. These studies revealed unique point mutations in envelope glycoprotein gH and in UL24, a factor absent from mature particles. Together these mutations rescued efficient infection of TrkA-expressing cells, including neurons, and facilitated the production of a completely retargeted KNGF derivative. These studies provide insight into HSV vector improvements that will allow production of replication-defective TrkA-targeted HSV for delivery to the peripheral nervous system and may be applied to other retargeted vector studies in the central nervous system.
Keywords: HSV, gene therapy, glycoprotein, vector targeting, TrkA, NGF
Graphical abstract
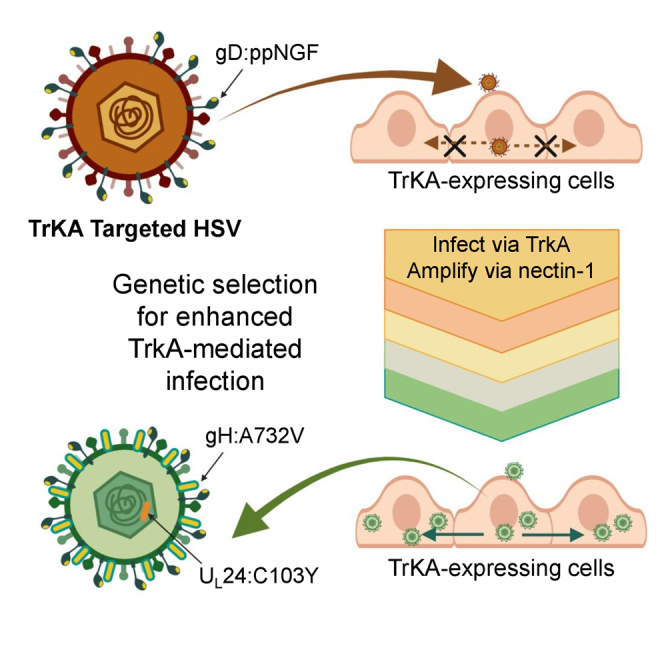
This article defines complementing mutations that enhance TrkA-retargeted virus infection and may be applicable to other retargeted vectors using different targeting ligands. These mutations in replication-defective vectors retargeted to TrkA neurons will provide gene delivery tools for studies of nerve cell function and treatment of chronic peripheral nerve pain.
Introduction
Transductional targeting of herpes simplex virus (HSV) vectors represents an important gene therapy strategy that can provide selective transgene expression in those cell populations and tissues where treatment is needed.1,2,3,4,5 The native HSV receptors are found on most cell types in the body and therefore do not allow transduction of specific subpopulations. Four HSV envelope glycoproteins, gB, gD, gH, and gL, are essential for viral entry and lateral cell-to-cell spread.6 Binding of gB and gC to the heparan sulfate (HS) components of proteoglycans on cell surfaces promotes interaction of gD with its cognate receptors consisting of the herpes virus entry mediator (HVEM or HveA), nectin-1 (HveC), and 3-O-sulfated HS. The receptor-gD interaction causes a conformational change in gD that activates the gH/gL heterodimer, which in turn activates the fusogen gB. gB mediates fusion between the virus envelope and the cell surface or endosomal membrane, ultimately releasing the nucleocapsid into the cytoplasm. Transductional targeting can be accomplished by genetic modification of gD to ablate the cognate entry receptor recognition elements (detargeting) in combination with incorporation of a new ligand that recognizes the target receptor (retargeting).
The majority of HSV retargeting studies have employed recombinant gD glycoproteins that are detargeted from nectin-1 and HVEM and contain specific ligands, such as single-chain antibodies, that enable binding to tumor-enriched receptors. These retargeted oncolytic HSV (oHSV) can mediate the specific destruction of tumor cells, while avoiding damage to neighboring normal cells that possess only the cognate HSV receptors on their surface.7,8,9,10,11,12,13,14,15,16,17,18,19 These studies have identified ligand-receptor interactions for targets such as EGFR, HER2, EpCAM, IL13R, uPAR, and CEA that were able to direct virus entry into specific tumor cells. However, the overall efficiency of infection was reduced compared with that observed with wild-type virus using the natural gD receptors. Genetic selection strategies have yielded virus isolates with improved retargeted infection and identified complementing mutations in HSV glycoproteins, including gB:D285N/A549T (gB:NT), that enhanced retargeted virus entry to levels observed for wild-type HSV.20
HSV can also be retargeted using growth factors as targeting ligands; growth factors are naturally occurring molecules and should be ideal for targeting infection to growth factor receptor-expressing cell populations such as tumors and neurons. For example, HSV amplicon vectors were targeted to neuronal subtypes via replacement of the signal peptide (SP) and HS binding domain of gC with pre-pro-(pp)GDNF or ppBDNF, enabling binding and entry into neurons bearing the cognate receptors GFRα1 or TrkB, respectively.21,22 However, these vectors were not detargeted from the natural gD receptors and entry was not restricted to the targeted receptors (GFRα1 or TrkB). More recently, ppGDNF was inserted in-frame at the N terminus of fully detargeted gD in the context of an oHSV backbone carrying the gB:NT entry-enhancing mutations.7 This retargeted virus was shown to specifically infect GFRα1-bearing breast cancer cells in culture and to effectively infect tumor tissue in animals, ultimately resulting in tumor regression.7 Here, we extend the use of growth factors for selective vector infection by employing nerve growth factor (NGF) as a targeting ligand for its receptor, tropomyosin receptor kinase A (TrkA). TrkA has been associated with inflammatory pain states,23,24,25 and most pain-sensing C-fibers in the PNS express TrkA on their surface.26,27,28 Thus, TrkA represents an interesting target for HSV vectors to both investigate the role of TrkA-expressing neurons in pain signaling and to deliver therapeutic genes to potentially block or alleviate inflammatory or neuropathic pain.
Results
HSV targeting to the NGF receptor
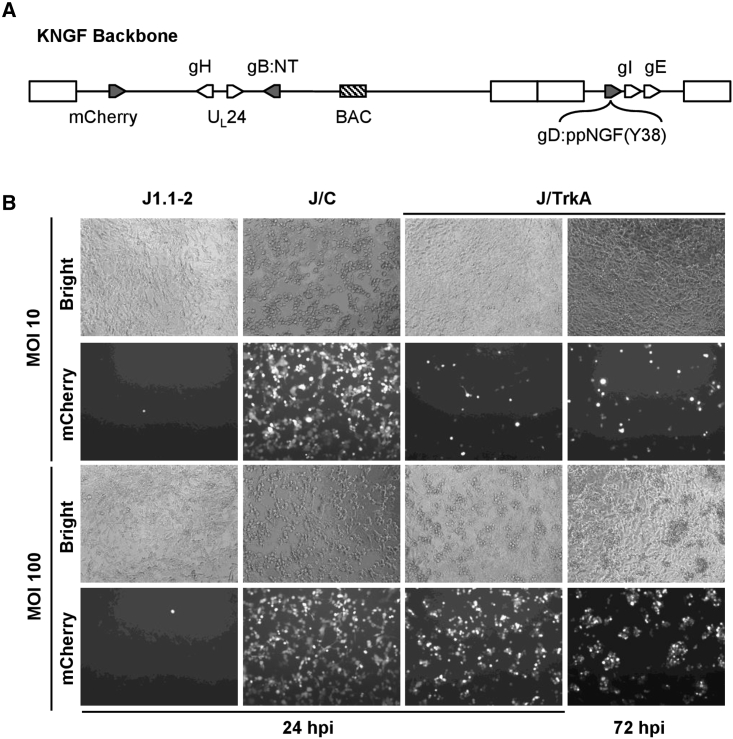
To generate a TrkA-targeted virus we used the previously described HSV-1 backbone, KNTc-ΔgD:GW,29 maintained as a bacterial artificial chromosome (BAC). This construct contains an mCherry reporter gene controlled by the ubiquitin C promoter, two mutations in the gB gene previously shown to enhance retargeted virus entry,20 and a Gateway (GW) cassette in place of the gD coding sequence. To target HSV infection to TrkA-expressing cells, we first modified the viral gD gene by replacing the region encoding the SP and HVEM-binding N-terminal domain with the mouse pre-pro-NGF (ppNGF) coding sequence to create gD:ppNGF(Y38). This design provided the ppNGF SP and processing sites and maintained gD residue Y38 to preserve the interaction with the viral entry receptor nectin-1. We introduced the recombinant gD:ppNGF(Y38) gene at the gD locus of KNTc-ΔgD:GW to create the KNGF genome (Figure 1A; Table 1), and produced virus by transfection and amplification on nectin-1-expressing U2OS cells.
Figure 1.
Receptor-dependent infection with the KNGF retargeted virus
(A) The KNGF genome contains bacterial artificial chromosome (BAC) sequences between UL37 and UL38 for viral genome propagation and engineering in E. coli, a ubiquitin C promoter-mCherry expression cassette (mCherry) between UL3 and UL4 for visualization of infected cells, two viral entry-enhancing mutations in the gB gene (gB:NT), and the gD:ppNGF(Y38) retargeted gD gene. The positions of other viral genes mentioned in this study are also shown. (B) HSV receptor-deficient J1.1-2 cells, J/C cells (nectin-1), and J/TrkA cells (TrkA) were infected at the indicated multiplicities of infection (MOI) with KNGF virus. Virus infection was visualized with bright-field (bright) images and mCherry fluorescence at 24 or 72 hpi.
Table 1.
Engineered viruses
| Virus name | gDa | gH | gI | gE | UL24 |
|---|---|---|---|---|---|
| KNGF | Y38 | WT | WT | WT | WT |
| KNGFΔ38 | Δ38 | WT | WT | WT | WT |
| KNGF-Eʹ | Y38 | WT | WT | V154M | WT |
| KNGF-Iʹ | Y38 | WT | I286F | WT | WT |
| KNGF-EʹIʹ | Y38 | WT | I286F | V154M | WT |
| KNGF-Hʹ | Y38 | A732V | WT | WT | WT |
| KNGF-HʹEʹ | Y38 | A732V | WT | V154M | WT |
| KNGF-HʹIʹ | Y38 | A732V | I286F | WT | WT |
| KNGF-HʹIʹEʹ | Y38 | A732V | I286F | V154M | WT |
| KNGF-24ʹ | Y38 | WT | WT | WT | C103Y |
| KNGF-EʹIʹ24ʹ | Y38 | WT | I286F | V154M | C103Y |
| KNGF-Hʹ24ʹ | Y38 | A732V | WT | WT | C103Y |
| KNGF-HʹEʹIʹ24ʹ | Y38 | A732V | I286F | V154M | C103Y |
| J4HΔ38b | Δ38 | A732V | I286F | V154M | C103Y |
All viruses contain gB:NT (D285N/A549T).
Viruses contain either gD:ppNGF(Y38), which is able to enter cells via nectin-1, or gD:ppNGF(Δ38), which does not enter cells via nectin-1.
J4HΔ38 is a derivative of J4H.
To assess the ability of purified KNGF virus to enter cells by binding to TrkA, we derived a TrkA-expressing cell line from gD receptor-deficient J1.1-2 cells (J/TrkA; Figure S1). Unlike J1.1-2 cells, TrkA-transduced J1.1-2 cells formed aggregates once the cells reached confluence, indicative of anchorage-independent growth.30 J1.1-2 cells, a derivative expressing nectin-1 (J/C), or J/TrkA cells were infected with KNGF virus at a multiplicity of infection (MOI) of 10 or 100 PFU/cell (Figure 1B). Visualization of mCherry fluorescence at 24 h post infection (hpi) demonstrated no sign of KNGF infection in receptor-negative J1.1-2 control cells, whereas efficient infection of J/C cells was observed at both MOIs. The KNGF virus demonstrated MOI-dependent infection of J/TrkA cells at 24 hpi, although the overall infection was inefficient compared with that seen on J/C cells (Figure 1B). By 72 hpi, no evidence of subsequent virus spread was apparent in J/TrkA cells. As well, a nectin-1-detargeted BAC construct (KNGFΔ38), in which gD residue 38 was deleted (Table 1), did not produce virus upon transfection into TrkA-expressing cells (Table S1). Together these data suggested that the NGF ligand was able to mediate infection via TrkA, but this infection was inefficient.
Genetic selection of KNGF variants capable of enhanced TrkA-dependent infection
To select variants of KNGF with greater fitness for TrkA-mediated infection, we performed a genetic selection by repeat passage of the virus on J/TrkA cells. In brief, J/TrkA cells were infected with KNGF virus (MOI = 10 PFU/cell), viral supernatant was harvested at 3 days post infection (dpi) and amplified by nectin-1-mediated infection of U2OS cells to produce a new virus stock (KNGF-J). J/TrkA cells were infected with the KNGF-J selection pool at MOI 1 PFU/cell and observed over time for evidence of entry and cell-to-cell spread. Figure S2A shows that individual KNGF-J infected cells were apparent at 24 hpi, with no further spread observed over a 4-day period. Iterative passage (KNGF-J1 through -J4) revealed no improvement in infection on J/TrkA cells through the KNGF-J3 round of selection (data not shown). However, with the KNGF-J4 selection pool we observed individual infected cells at 24 hpi that spread to show robust infection within densely packed clusters of J/TrkA cells by 72 hpi (Figure S2A).
To quantify the observed difference in virus infection, J/TrkA cells were infected with the KNGF-J and KNGF-J4 selection pools and the total virus accumulated in the supernatant over a 7-day period was assessed by qPCR for viral genomes; the genome copy (gc)/PFU ratio for each virus stock is shown in Table S1. The KNGF-J virus pool failed to grow on J/TrkA cells, releasing no viral genomes into the supernatant relative to the initial input virus (Figure S2B). By comparison, supernatants from the KNGF-J4 infection contained 100-fold more viral genomes at 7 dpi than at 1 dpi (Figure S2B), demonstrating active virus replication and release. These results suggested that the KNGF-J4 virus stock had acquired one or more mutations that improved TrkA-dependent infection.
Identification of sequence variants in the KNGF-J4 virus pool
We expected the KNGF-J4 virus stock to represent a mixed population of viruses with potentially different genetic changes driving the ability to infect TrkA-expressing cells and enhance virus spread. We took advantage of the BAC region in the viral genome to rescue individual KNGF-J4 viral DNAs in E. coli. We converted clonal BAC isolates back into virus particles by transfection of U2OS cells and functionally characterized these viruses by infection of J/TrkA cells. Figure 2 shows the results of J/TrkA infection for nine representative isolates (J4A–J4I) at 2 and 5 dpi. Only J4H showed a phenotype similar to that observed with the KNGF-J4 stock, displaying significant spread in dense clusters of J/TrkA cells. Two other isolates, J4C and J4D, demonstrated increased spread, although to a lesser extent than J4H.
Figure 2.
Enhanced infectivity was observed for individual subclones isolated from the KNGF-J4 virus pool
J/TrkA cells were infected at MOI of 1 PFU/cell with either the KNGF-J4 selection pool or individual viral subclones isolated from the pool (J4A-J4I). Virus infection was visualized over time by expression of the mCherry fluorescent reporter, and representative images are shown for 2 and 5 dpi.
Full genome sequencing was performed for KNGF, and isolates J4H, J4C, and J4F (Table 2). These data revealed no mutations in isolate J4F, consistent with the observation that it infected J/TrkA cells with similar efficiency as the original KNGF virus. In the J4C isolate, we identified an alanine to valine substitution at position 732 of gH (gH:A732V). Localized sequencing revealed that the J4D isolate contained the same substitution mutation in gH. The J4H isolate contained mutations in the coding sequence of four genes resulting in (1) a valine to methionine substitution at residue 154 of gE (gE:V154M), (2) an isoleucine to phenylalanine substitution at residue 286 of gI (gI:I286F), (3) the same gH:A732V substitution as in J4C and J4D, caused by the same mutation, and (4) a cysteine to tyrosine substitution in UL24 at position 103 (UL24:C103Y). The gD:ppNGF(Y38) and gB:NT sequences remained unchanged in all isolates.
Table 2.
Mutations identified in selected KNGF subclones
| KNGF subclonesa | gH | gI | gE | UL24 |
|---|---|---|---|---|
| J4C | A732V | WT | WT | WT |
| J4D | A732V | WT | WT | WT |
| J4F | WT | WT | WT | WT |
| J4H | A732V | I286F | V154M | C103Y |
All sequenced subclones contained gB:NT (D285N/A549T) and gD:ppNGF(Y38).
UL24 is the only viral protein other than a glycoprotein that is known to acquire syncytial mutations. UL24 null mutants and substitution mutants (G121A and E99A/K101A) demonstrated a syncytial phenotype on Vero cells that was only observed at 39°C.31 To determine if our J4H mutant possessed a similar phenotype, we assessed plaque morphology of J4H and KNGF on Vero cells at both 37°C and 39°C. The results (Figure S3) demonstrated that neither KNGF nor J4H produce syncytial plaques at either temperature.
Contribution of the substitution mutations to the enhanced spread phenotype
The J4C, J4D, and J4H isolates all conferred an infection advantage to the KNGF backbone, with J4H showing the greatest enhancement of infection. All three mutants contained the gH:A732V substitution, suggesting that this substitution is responsible for the altered phenotype of the first two and contributes to that of the third. To determine the relative contributions of the gH, gI, gE, and UL24 substitutions to the phenotype of J4H, we introduced each mutation into the parental KNGF backbone, individually and in combination (Table 1). These KNGF-derived mutant viruses were compared with KNGF and J4H by infection of J/TrkA cells (MOI = 1 PFU/cell) followed by fluorescent microscopy for mCherry expression; representative images are shown at 2 and 4 dpi (Figure 3A). Infection with the control KNGF virus resulted in few mCherry-positive cells at both 2 and 4 dpi, demonstrating an overall lack of infection. Infection with KNGF-IʹEʹ resulted in few mCherry-positive cells at 2 dpi and demonstrated a slight increase in mCherry-positive cells by 4 dpi. The individual KNGF-I′ and KNGF-E′ mutants displayed a similar phenotype to KNGF-IʹEʹ (Figure S4). Infection with KNGF-H′ or KNGF-24′ resulted in a significant increase in mCherry-positive cells at 2 dpi relative to KNGF, with a further increase between 2 and 4 dpi; the addition of gE:V154M and gI:I286F did not further enhance infection (KNGF-H′ compared with KNGF-HʹIʹE′ and KNGF-24′ compared with KNGF-EʹIʹ24′). While KNGF-Hʹ and KNGF-24′ individually demonstrated enhanced virus infection, they did not completely reproduce the level of infection observed for J4H. The combination of gH:A732V and UL24:C103Y (KNGF-H′24′) was sufficient to reproduce the level of mCherry expression observed for J4H; the addition of gE:V154M and gI:I286F (KNGF-HʹIʹE′24′) did not further improve virus infection or cell-to-cell virus spread.
Figure 3.
The gH A732V and UL24 C103Y mutations are essential for enhanced infectivity on J/TrkA cells
(A) J/TrkA cells were infected with KNGF, J4H, and the indicated mutant viruses at MOI of 1 PFU/cell and virus infection was evaluated by mCherry fluorescent imaging at 2 and 4 dpi. (B) Virus yield for KNGF, J4H, and the indicated mutant viruses was evaluated on J/TrkA cells (MOI = 0.75 PFU/cell) at 2 to 6 dpi. At each time point, the total gc were quantified in the supernatant and data are presented as the mean ± SD (n = 3). Two-way ANOVA was used to compare the difference between groups. A statistically significant difference (∗∗∗∗p < 0.0001) was observed between: KNGF and -24′, -H′, -H′24′, -HʹEʹI′24′, and J4H; KNGF-24′ and -H′, -H′24′, -HʹEʹI′24′, and J4H. No statistically significance difference in virus yield was observed between: KNGF-H′24 vs. KNGF-HʹEʹI′24ʹ; KNGF-H′24 vs. J4H; KNGF-HʹEʹI′24′ vs. J4H.
To quantify the differences observed among the viral recombinants, J/TrkA cells were infected at low MOI and the total virus accumulated in the supernatant was measured over a 6-day period (Figure 3B). Statistically significant differences (∗∗∗p < 0.0001) were seen between the parental KNGF virus and the single-mutant viruses KNGF-H′ and KNGF-24′, confirming that the enhanced infection phenotype observed via mCherry expression correlates with increased overall virus production. KNGF-H′24′ and KNGF-HʹEʹI′24′ both demonstrated an additional enhancement in virus production, increasing virus production relative to the KNGF-H′ and KNGF-24′ single-mutant viruses. However, KNGF-HʹEʹI′24′ and KNGF-H′24′ displayed similar levels of virus production, confirming that the addition of the gI and gE mutations did not contribute significantly to the overall phenotype. Of note, KNGF-H′24′ and J4H did not statistically differ in virus production. Together these data suggested that the combination of gH:A732V and UL24:C103Y was necessary and sufficient to recapitulate the observed increase in virus infectivity via the targeted NGF receptor and that the addition of gE:V154M and gI:I286F did not significantly contribute to this phenotype.
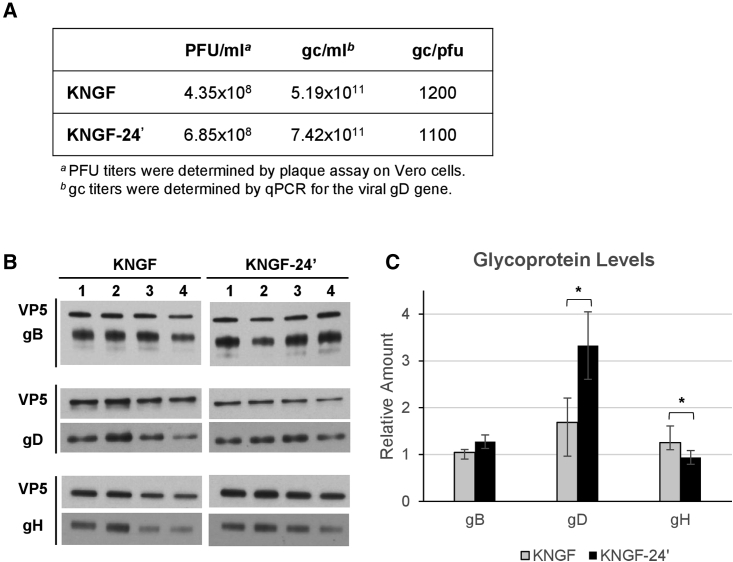
UL24 contribution to incorporation of gD in the viral particle
UL24 is not present in the viral particle but has proposed roles in nuclear egress and in glycoprotein distribution.32,33 We performed western blot analysis to determine if glycoprotein incorporation is altered in KNGF-24′ virus particles compared with the parental KNGF virus. We first produced high-titer virus stocks for KNGF and KNGF-24′ that had comparable gc/PFU ratios (Figure 4A). Then the relative amounts of gB, gD, and gH present in purified virus particles were quantified by immunoblotting using VP5 capsid protein for normalization (Figure 4B). These data demonstrated that UL24:C103Y statistically increased the amount of gD in the viral envelope, producing virus particles with approximately twice as much gD glycoprotein when normalized to VP5 (Figure 4C). The amount of gH in the viral envelope was significantly reduced by approximately 25% in KNGF-24′ relative to KNGF, and the relative amount of gB was not statistically different between the two viruses (Figure 4C). These observed differences in glycoprotein envelope incorporation may ultimately contribute to the enhanced infectivity observed on J/TrkA cells in the presence of UL24:C103Y.
Figure 4.
Western blot analysis of glycoproteins gB, gD, and gH in KGNF and KGNF-24ʹ viral particles
(A) KGNF and KGNF-24ʹ virus stocks used for western blot analysis were characterized for PFU titer, gc titer, and gc/PFU ratio. (B) Equivalent gc of each virus were loaded per lane (1–4); 5 × 107 gc/lane for gH and gB quantification or 7 × 107 gc/lane for gD quantification. Each glycoprotein blot is shown below its corresponding VP5 control. (C) The relative amount of each glycoprotein was determined using ImageJ software, normalized to VP5, and presented relative to KNGF lane 1 (mean ± SD). Statistical differences between groups were performed with the Welch’s t test, ∗p < 0.05.
Receptor-dependent infection of fully retargeted J4HΔ38
As mentioned above, a nectin-1-detargeted derivative of KNGF (KNGFΔ38; Table 1) did not yield virus upon transfection into TrkA-expressing cells (Table S1). To determine whether the improved infectivity of the J4H isolate on TrkA-expressing cells would allow the production of a nectin-1-detargeted derivative, we deleted gD residue 38 from the J4H genome, creating BAC construct J4HΔ38 (Table 1), and found that we could produce the corresponding J4HΔ38 virus in J/TrkA cells (Table S1). This result demonstrated that the J4H backbone supported efficient TrkA-mediated virus growth. To confirm the receptor specificity of J4HΔ38, we infected J1.1-2, J/C, and J/TrkA cells with 10 gc/cell of J4H and J4HΔ38 and recorded mCherry expression at 4 dpi. These results showed that, while J4H infected both J/TrkA and J/C cells efficiently, the J4HΔ38 virus was only able to infect J/TrkA cells and mCherry expression was not observed in J/C cells (Figure 5). Consistent with our previous work,7,29 these data demonstrated that nectin-1-mediated virus infection was effectively eliminated in the J4HΔ38 virus.
Figure 5.
Receptor-dependent infection with the KNGFΔ38 retargeted virus
J1.1-2 cells, J/C cells, and J/TrkA cells were infected at 10 gc/cell with J4H or J4HΔ38 virus and the infection was visualized at 4 dpi by mCherry fluorescent imaging.
Neuronal subtype transduction using TrkA-retargeted J4HΔ38
The neuronal subtype specificity of J4HΔ38 was examined using dorsal root ganglion (DRG) neurons isolated from embryonic day 15 (E15) rat embryos cultured in the presence of NGF and BDNF to maintain the survival of TrkA- and TrkB-expressing cells. Neurons were either mock-infected or infected with J4HΔ38 at 1,500 gc/cell, and 4 hpi immune-reactive (IR) staining for the HSV ICP4 immediate-early (IE) gene product was compared with IR staining for cell surface markers TrkA and TrkB. Negative control, mock-infected neurons exhibited no IR staining with the antibody recognizing ICP4, while clear staining was observed with the antibody recognizing TrkA (Figure S4). In J4HΔ38-infected cultures, significant overlap was observed between ICP4-positive and TrkA-positive cells (Figure 6A). In contrast, minimal overlap was observed between ICP4-positive and TrkB-positive cells (Figure 6B). Quantification of cells that stained positive for both ICP4 and TrkA or ICP4 and TrkB relative to total ICP4-positive cells is shown in Figure 6C. These data illustrated that 78.1% ± 8.0% of ICP4-positive cells were also positive for TrkA, while only 9.6% ± 8.5% of ICP4-positive cells demonstrated TrkB expression (Figure 6C). These data suggested that J4HΔ38 readily transduced TrkA-positive neurons compared with those expressing TrkB (∗∗∗∗p < 0.0.001).
Figure 6.
Infection of DRG neuronal subtypes by J4HΔ38
(A and B) Representative images for entry comparison of J4HΔ38 (1,500 gc/cell) on primary rat E15 DRG neurons at 4 hpi using antibody to HSV ICP4 for virus detection and antibodies to (A) TrkA (n = 8; scale bar 20 µm)or (B) TrkB (n = 6; scale bar 50 µm) for neuronal cell subtype detection; overlaid images (merge) were used to determine percent overlap in (C). (C) Cell counts were determined for three images per stained panel and data are presented as the percent overlap in fluorescent signals for J4HΔ38 entry into the DRG neuronal subtypes (ICP4+ and neuronal marker+/total ICP4+) shown as the mean ± SD (∗∗∗∗p < 0.0001 by two-tailed unpaired t test).
Discussion
Targeting HSV-mediated gene delivery to the desired cell type will maximize the impact of therapeutic gene expression and avoid potentially deleterious off-target effects. However, targeting of HSV vectors is complicated by the complex mechanism of virus attachment and entry, which involves multiple envelope glycoproteins and a cascade of reactions leading to envelope fusion with the cell membrane.6 Here, we explored the use of NGF as a ligand to target HSV infection to TrkA-bearing cells. By replacing the SP and HVEM binding domain of gD with ppNGF we generated a recombinant gD protein, gD:ppNGF(Y38), that demonstrated TrkA-specific entry but not lateral cell-to-cell spread. Previous genetic selection studies identified the gB:NT variant that enhanced the rate of HSV entry and allowed efficient infection of an EGFR-retargeted virus.8 However, for the TrkA-retargeted virus, gB:NT was already present in the viral backbone and was not sufficient to support efficient gD:ppNGF(Y38)-initiated infection.
We exploited the inefficient TrkA-retargeted virus to select for secondary mutations that would enhance retargeted virus entry and spread. Repeated rounds of retargeted virus infection of J/TrkA cells resulted in selection of a mutant virus pool that demonstrated enhanced infection via TrkA. Independent analysis of genetic variants present within the mixed population identified two substitution mutations, gH:A732V and UL24:C103Y, that in combination increased virus infectivity via the targeted receptor. HSV gH forms a heterodimer with gL (gH/gL) and is essential for viral entry and spread.34,35 Following receptor binding, receptor-bound gD interacts with the gH/gL heterodimer to trigger activation of the gB viral fusogen. We previously selected a unique gH variant (gH:KV; N753K and A778V) that facilitated virus spread between cells that lacked canonical entry receptors.36 It was therefore not unexpected that a mutation in the same domain of gH, such as gH:A732V, would be identified in a viral screen aimed at enhancing virus spread. Of interest, while UL24 is not present within mature virions,37,38 and is therefore not directly involved in viral entry, the UL24:C103Y variant contributed to enhanced TrkA-targeted infection.
UL24 is a multifunctional late viral protein that is conserved throughout the Herpesviridae family.38 UL24 localizes to both the nucleus and cytoplasm of infected cells,38 and UL24-deficient virus has been associated with defects in the nuclear egress of viral capsids, and with an overall reduction in infectious particles produced during infection.33 UL24 has been described as a virulence factor in which deletion mutants displayed reduced pathogenesis in animal models,39 and reduced productive infection in neurons of the trigeminal ganglia (TG).40,41 Of interest, it has been shown that UL24-deficient virus has a cell-type-dependent defect in the subcellular distribution of the essential viral glycoproteins gB and gD.32 In contrast to the UL24-deficient virus, the unique UL24:C103Y substitution mutation identified here provides a gain of function, enhancing virus infection efficiency. Our western blot data suggest that, in the presence of the UL24:C103Y mutation, incorporation of recombinant gD into the viral envelope is significantly enhanced, pointing to a potential mechanism of action. The contribution of UL24 to glycoprotein localization, nuclear egress, and production of infectious virus particles could suggest other roles in different steps along this complex pathway. Further investigation will be required to better understand this unique UL24 gain-of-function mutation and how it impacts these other processes.
Ultimately, the combined action of gH:A732V and UL24:C103Y created an enhanced backbone in which the fully TrkA-retargeted virus could be produced and evaluated. To assess TrkA specificity outside of engineered J/TrkA cells, we isolated primary DRG neurons from E15 rat embryos. This cell culture system provided a model for assessing virus retargeting of neuronal C-fibers that express TrkA or TrkB.28 The level of infection in neurons that express TrkA, compared with those expressing TrkB, reaffirms the ability of the ppNGFgD(Δ38) recombinant gD to obviate entry via nectin-1 and mediate subtype specific binding and entry via TrkA and not TrkB. Prior studies have shown little to no overlap between TrkA and TrkB expression patterns in rat DRG neurons.28 These staining patterns were comparable with those observed using HSV replication-defective amplicon vectors targeted to TrkB or the GDNF receptor GFRα1.21,22 Of note, TrkA was not only detected on small C-fibers, but also on some larger diameter Aδ-fiber cell bodies present within the images (Figure 6A). TrkA can be expressed at lower levels on these neuronal subtypes that may be sufficient to allow entry of the retargeted virus.42,43,44
The PNS neurons of the DRG and TG are ideal targets for HSV-mediated gene therapy since HSV naturally establishes life-long latency in these ganglia.45,46,47 PNS neurons are a diverse population of subtypes that can be classified by their morphology, the expression of various neuronal markers, and their function as nociceptors, mechanoreceptors, or proprioceptors.48,49,50,51,52,53 The NGF receptor TrkA has been associated with inflammatory pain states,23,24,25 and most pain-sensing C-fibers in the PNS express TrkA on their surface.26,27,28 Thus, TrkA represents an interesting neuronal subtype target for the delivery of potentially therapeutic genes to block or alleviate inflammatory or neuropathic pain.
Overall, these experiments have defined complementing mutations that can enhance TrkA-retargeted virus infection and may be applicable to other retargeted vectors using different targeting ligands. Introduction of these mutations into replication-defective vectors retargeted to TrkA neurons will provide gene delivery tools for studies of nerve cell function in brain and treatment of chronic peripheral nerve pain. Furthermore, we are currently exploiting these mutations in oHSV retargeted to other tumor-associated surface markers such as EGFR in anticipation that these vector modifications will greatly improve virus infectivity and spread, ultimately enhancing oncolytic vector killing of tumor cells.
Materials and methods
Cell culture
U2OS (ATCC HTB-96; Manassas, VA), Vero (ATCC CCL-81), J1.1-2,54 and J/C cells,55 have been previously described. All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Corning, Durham, NC) supplemented with penicillin/streptomycin (Corning-Mediatech, Manassas, VA) and 5%–10% (vol/vol) fetal bovine serum (FBS) (Sigma, St Louis, MO). J/Trk-A cells were generated by infection of J1.1-2 cells with a retrovirus expressing TrkA, produced by co-transfection of 293T cells (ATCC CRL-3216) with plasmids pCX4-bsr-TrkA, pCL-gag-pol, and pHCMV-VSVG. Transduced J1.1-2 cultures were selected for resistance to blasticidin (10 mg/mL), single clones were isolated and screened for TrkA expression by immunofluorescent staining with rabbit anti-Pan Trk antibody (EP1058Y; Abcam, Waltham, MA, ab76291).
Plasmids
The rat TrkA cDNA was provided by Susan Meakin (University of Western Ontario, Ontario, Canada), pCL-GagPol and pHCMV-VSV-G retroviral packaging plasmids were provided by Nobutaka Kiyokawa, Hajime Okita, and Akihiro Umezawa (NRICHD, Tokyo, Japan), and murine NGF was provided by William Rutter.56,57
The pCX4-TrkA-bsr plasmid was derived by first subcloning an EcoRI-SalI fragment containing the rat TrkA cDNA into pENTR1A at EcoRI and XhoI sites to obtain pENTR_TrkA, followed by LR Clonase II-mediated recombination between pENTR_TrkA and pCX4-GW-bsr.7,58
To create pENTR-gD:ppNGF(Δ38) and pENTR-gD:ppNGF(Y38), the N-terminal coding sequence of gD, up to and including amino acid 24, was replaced with murine ppNGF by PCR amplification of ppNGF with primers ppNGF-F and ppNGF-R (Table S2) and insertion between DraI and NotI sites of either pENTR-gD:Δ224/Δ38 or pENTR-gD:Δ224/38Y.7 All constructs were confirmed by DNA sequencing.
Viruses
KNGF (Figure 1) and KNGFΔ38 (Table 1) were derived from KNTc-ΔgD:GW,29 by LR Clonase II-mediated recombination with pENTR-gD:ppNGF(Δ38) and pENTR-gD:ppNGF(Y38). To create J4HΔ38 we first generated J4H-ΔgD:GW by replacing the gD coding sequence of J4H with a GW cassette, as described previously.29 Then gD:ppNGF(Δ38) was introduced by LR Clonase II (Thermo Fisher, Pittsburgh, PA)-mediated recombination between J4H-ΔgD:GW and pENTR-gD:ppNGF(Δ38).
The genes coding for gI (US7) and gE (US8) are adjacent, US8 overlaps with US8A, and US8A overlaps with US9. Because of this genomic organization, we first replaced the region spanning from the ATG of US7 to the STOP codon of US9 with a GW cassette in KNGF, generating the intermediate backbone KNGF-ΔUS7-9:GW. This change facilitated introduction of point mutations in gE and/or gI. The US7- US9 regions from KNGF and J4H were amplified with US7-F and US9-R primers, containing EcoRV and XbaI sites, respectively (Table S2), and subcloned into pENTR1A at XmnI/XbaI sites to produce pENTR-IE and pENTR-IʹEʹ. We exchanged mutant and wild-type gI between these two plasmids to generate pENTR-IʹE and pENTR-IEʹ, using AlfII and AgeI restriction sites. These pENTR plasmids were used for LR Clonase II-mediated recombination to introduce the corresponding mutations into KNGF-ΔUS7-9:GW, yielding KNGF-I′, KNGF-E′, and KNGF-IʹEʹ (Table 1).
The gH A732V mutation was introduced by Scarless Red recombination, as described.59 The necessary pBAD-IsceI and pEPkan-S2 plasmids were kindly provided by Nikolaus Osterrieder (Free University of Berlin, Berlin, Germany). The gH A732V mutation was amplified from the J4H BAC isolate with primers gH-F and gH-R, containing XbaI and NheI sites, respectively (Table S2). The gH A732V amplicon was subcloned into pcDNA3.1 at corresponding NheI and XbaI sites to create pcDNA-732VgH. The I-SceI-containing kanamycin-resistance gene (KanR) from pEPkan-S2 was amplified by PCR with gH-Kan-F and gH-Kan-R primers containing BstX1 sites (Table S2) and subcloned into the BstXI restriction site in pcDNA-gHA732V to produce pcDNA-732VgH-Kan. The NheI/XbaI fragment from pcDNA-732VgH-Kan was inserted into KNGF and KNGF-ΔUS7-9:GW using Scarless Red recombination to generate KNGF-Hʹ (Table S1) and KNGF-H′-ΔUS7-9:GW. The pENTR plasmids, pENTR-IʹEʹ, pENTR-IʹE, and pENTR-IEʹ, were used for LR Clonase II-mediated recombination to introduce the corresponding mutations into KNGF-H′-ΔUS7-9:GW, yielding KNGF-HʹIʹ, KNGF-HʹEʹ, and KNGF-HʹIʹEʹ (Table 1).
For UL24 modification we used S. pyogenes Cas9 nuclease (NEB, Ipswich, MA) in combination with sgRNAs generated using the sgRNA Synthesis Kit (NEB). sgRNA were designed using the online tool CHOPCHOP (http://chopchop.cbu.uib.no/).60 KNGF, KNGF-H′, KNGF-IʹEʹ, and KNGF-HʹIʹE′ BACs were purified using the Plasmid Midi Kit (QIAGEN, Germantown, MD) with protocol QP01 for isolation of BAC DNA, and digested with sgRNA-UL24a and sgRNA-UL24b (Table S2) to delete the UL24 region between nucleotides 272 and 322 of the UL24 coding sequence (corresponding to positions 47,949 –47,999 of GenBank: JQ673480.1). A KanR fragment containing the intended UL24 point mutation (UL24′) was amplified from pEPkan-S2 by PCR with primers UL24′-Kan-F and UL24′-Kan-R (Table S2). We introduced the UL24′-KanR fragment into the sgRNA-digested BAC by Gibson reaction (NEBuilder HiFi DNA Assembly Master Mix, NEB). To remove the KanR selectable marker, the BAC-UL24′-Kan DNA was digested with sgRNA-KAN(1) and sgRNA-KAN(2) (Table S2) that recognize the 5′ and 3′ boundaries of the KanR cassette, and repaired by Gibson reaction as described above.
All constructs were confirmed by PCR analysis, field inversion gel electrophoresis, and targeted DNA sequencing.
Virus growth and purification
BAC-containing viruses were converted to infectious virus by transfection into U2OS cells (nectin-1 competent viruses) or J/TrkA cells (J4HΔ38 virus). Biological titers of virus stocks were established on Vero cells (PFU/mL) and physical titers were determined by quantitative PCR (qPCR) for the viral gD gene as described (gc/mL) (Table S1).61
Virus genetic selection
J/TrkA cells (1 × 107) were plated in a 10 cm dish and the next day confluent monolayers were infected with KNGF virus at 10 PFU/cell. Viral supernatant was harvested at 3 dpi and amplified by nectin-1-mediated infection of U2OS cells to produce a virus stock (KNGF-J, -J1, -J2, -J3, -J4). Serial 10-fold dilutions of selection supernatant were used to infect J/TrkA cells plated at 1 × 105 cells/well in a 48-multiwell plate (Corning) and observed over time for evidence of cell-to-cell spread. The same J/TrkA-based infection and U2OS amplification was repeated with each stock produced until evidence of enhanced infection efficiency on J/TrkA cells was observed.
BAC rescue
U2OS cells were infected with KNGF-J4 at MOI 5 PFU/cell in the presence of 200 μg/mL phosphonoacetic acid (Sigma). At 3 hpi, cells were rinsed with 1× PBS and DNA was extracted by Proteinase K (Promega, Madison, WI) digestion for 1 h at 50°C, followed by extraction with phenol/chloroform/isoamyl alcohol, 25:24:1 (v/v) (Sigma), and precipitation with ethanol. DNA pellets were resuspended in 50 μL of 1× Tris-EDTA solution at pH 8 (Thermo Fisher Scientific), electroporated into ElectroMAX DH10B electrocompetent cells (Thermo Fisher Scientific), and plated on Luria-Bertani agar plates supplemented with 15 μg/mL chloramphenicol.
Whole-genome sequencing
We performed whole-genome sequencing on KNGF and the KNGF mutant subclones to identify mutations occurring after genetic selection (Table 2). In brief, purified viral DNA was harvested using the DNeasy blood and tissue kit (QIAGEN). Sequencing was performed by the University of Pittsburgh’s Health Sciences Sequencing core as previously described,62 to ensure a 40× genome coverage. Illumina sequencing reads (150 × 150 paired-end sequencing reads) were mapped to the parental in-silico-derived KNGF sequence using CLC Genomics Workbench (QIAGEN). All sequencing data are available from the authors upon request.
Virus growth curves
Cells were plated at a density of 1 × 105 cell/well in 48-well plates (Corning), and infected at the indicated MOI when cells reached confluence and began to aggregate; MOI was determined by the cell count at the time of plating. Supernatants were collected every 24 h for up to 7 dpi, as indicated in the figure legends. DNA was collected using the DNeasy blood and tissue kit (QIAGEN) and total genome copies per sample were determined by real-time qPCR as described previously.61 Data are presented as the average total gc (n = 3) ± SD.
gc quantification
Viral gc titers were determined using qPCR for the viral gD gene as described previously.7,29,61 The portion of the gD gene amplified in this assay corresponded to an unchanged region roughly 100 codons downstream of codon 38.
Western blot analyses
Whole-cell lysates were collected in 1× RIPA buffer (Millipore-Sigma, Burlington, MA) plus protease inhibitor cocktail (Millipore-Sigma, Roche) and samples were diluted in 1× Laemmli sample buffer (Bio-Rad, Hercules, CA). Viruses were diluted in 1× Laemmli sample buffer (Bio-Rad) to the indicated gc per lane (Figures 4B and S3). Lysates were heated for 5 min at 100°C, proteins were subjected to electrophoresis on precast 4%–15% SDS-PAGE gels (Bio-Rad) and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA). Membranes were blocked for 1 h in 5% nonfat dry milk in PBS +0.05% Tween (PBS-T) (Sigma) and incubated sequentially with primary antibody and horseradish peroxidase-conjugated secondary antibody (anti-mouse IgG; Abcam, Cambridge, UK) diluted to 1:50,000 in 5% nonfat milk/PBS-T. Primary antibodies: VP5 (3B6; Virusys Corporation, Randallstown, MD) 1:1,000 in PBS-T; gB (10B7 Virusys Corporation) 1:5,000 in 5% nonfat dry milk/PBS-T; gD (DL6, Santa Cruz Biotechnology, Dallas, TX) 1:500 in 5% nonfat dry milk/PBS-T; gH (H6, Virusys Corporation) 1:1,000 in PBS-T; VP16 (1–21, Santa Cruz Biotechnology) 1:2,000 in PBS-T. To ensure accurate quantification, four independent dilutions were made for each virus and filters were cut horizontally to detect both the intended glycoproteins and the VP5 loading control from the same lane. Band intensities were calculated using ImageJ software.63
DRG isolation and culture
DRG were isolated from E15 rat embryos per an Institutional Animal Care and Use Committee-approved protocol (1110488) and dissociated with 3 mg of collagenase A (Boehringer-Mannheim, Indianapolis, IN) per mL in Leibowitz-15 medium (L-15) (Thermo Fisher Scientific) containing 10% FBS (Sigma-Millipore, St. Louis, MO) and 20,000 U of penicillin and streptomycin (Thermo Fisher Scientific) for 30 min at 37°C with constant shaking. The cells were triturated further to disrupt any visible clumps of cells remaining after the enzymatic treatment to provide for better dissociation. After being washed four times in fresh L-15-10% FBS, the cells are plated on poly-D-lysine-coated (Sigma) coverslips at roughly 5 × 104 cells/well in 24-well plates (Corning) in 500 μL of defined Neurobasal medium containing B27, GlutaMAX I, AlbuMAX II, and penicillin/streptomycin, supplemented with 100 ng/mL of 7.0S NGF (Sigma), 10 ng/mL BDNF (Sigma), and 10 ng/mL NT-3 (Sigma) to stimulate the survival of TrkA-, TrkB-, and TrkC-expressing DRG neurons, respectively,64,65,66 and 10 μM uridine (Sigma) and 10 μM fluoro-deoxyuridine (Sigma) were added to inhibit the growth of nonneuronal cells. Cells were observed daily by light microscopy and medium was replenished every 3 days.
DRG infection and immunochemistry
At 15 days following DRG plating, when unwanted dividing cells had been reduced or eliminated, the cells were infected with J4HΔ38 (1,500 gc/cell) for 1 h, washed with fresh medium, and incubated for an additional 4 h in complete medium. Cells were then fixed in 4% buffered formalin (Thermo Fisher Scientific) for 10 min, washed three times in PBS, and blocked in PBS +5% normal goat serum (Sigma) and 0.2% Tween 20 (Thermo Fisher Scientific) for 1 h at room temperature (RT). Cells were incubated with antibodies recognizing either HSV ICP4 (1:100, Santa Cruz Biotechnology, sc-56986), TrkA (1:250, Thermo Fisher Scientific, 06–574), NF200 (1:250, Abcam, ab8135), TRPV1 (1:250, Santa Cruz Biotechnology, sc-286759), or TrkB (1:250, Santa Cruz Biotechnology, sc-12-G) for 24 h at 4°C. Cells were washed three times in PBS then incubated with a 1:1,000 dilution of the appropriate secondary antibody, Alexa Fluor 594-labeled goat anti-mouse antibody (Thermo Fisher Scientific, A11005), Alexa Fluor 488-labeled donkey anti-goat antibody (Thermo Fisher Scientific, A11055), or Alexa Fluor 488-labeled goat anti-rabbit antibody (Thermo Fisher Scientific, A11008) for 1h at RT. Cells were washed three times in PBS, once in DI water, and mounted onto glass slides with Aqua Poly/Mount (Polysciences, Warrington, PA). All images were acquired on a Zeiss Axiovert 200 microscope using an Axiocam MRC5 high-resolution camera and Axiovision software. Scale bars were added to images using ImageJ software version 1.53a (National Institutes of Health).
Statistical analyses
GraphPad Prism software was used for all statistical analyses. Averages for each experiment are shown ± SD. As noted in the relevant figure legends, Welch’s t test and one-way or two-way ANOVA were used to determine the statistical significance of differences observed between groups, and significant differences are indicated in the figures (∗p < 0.05, ∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant).
Acknowledgments
This work was supported by grants R01-CA222804 (to J.C.G.) and R01-NS064988 (to J.C.G.) from the NIH.
Author contributions
Conceptualization, M.M., B.L.H., J.B.C., and J.C.G.; methodology, M.M., B.L.H., J.B.C., and J.C.G.; investigation, M.M. and M.Z.; writing – original draft, M.M. and B.L.H.; writing – review & editing, B.L.H., W.F.G., and J.C.G.; funding acquisition, J.C.G.; supervision, W.F.G., J.B.C., and J.C.G.
Declaration of interests
J.B.C. and J.C.G. are inventors of intellectual property licensed to Oncorus, Inc. (Cambridge, MA). J.C.G. is a consultant and Chair of the Scientific Advisory Board of Oncorus, Inc.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2023.06.012.
Supplemental information
Data availability
Data and materials described in this article will be available upon reasonable request to the corresponding author.
References
- 1.Mata M., Zhang M., Hu X., Fink D.J. HveC (nectin-1) is expressed at high levels in sensory neurons, but not in motor neurons, of the rat peripheral nervous system. J. Neurovirol. 2001;7:476–480. doi: 10.1080/135502801753170336. [DOI] [PubMed] [Google Scholar]
- 2.Simpson S.A., Manchak M.D., Hager E.J., Krummenacher C., Whitbeck J.C., Levin M.J., Freed C.R., Wilcox C.L., Cohen G.H., Eisenberg R.J., Pizer L.I. Nectin-1/HveC Mediates herpes simplex virus type 1 entry into primary human sensory neurons and fibroblasts. J. Neurovirol. 2005;11:208–218. doi: 10.1080/13550280590924214. [DOI] [PubMed] [Google Scholar]
- 3.Haarr L., Shukla D., Rødahl E., Dal Canto M.C., Spear P.G. Transcription from the gene encoding the herpesvirus entry receptor nectin-1 (HveC) in nervous tissue of adult mouse. Virology. 2001;287:301–309. doi: 10.1006/viro.2001.1041. [DOI] [PubMed] [Google Scholar]
- 4.Richart S.M., Simpson S.A., Krummenacher C., Whitbeck J.C., Pizer L.I., Cohen G.H., Eisenberg R.J., Wilcox C.L. Entry of herpes simplex virus type 1 into primary sensory neurons in vitro is mediated by Nectin-1/HveC. J. Virol. 2003;77:3307–3311. doi: 10.1128/JVI.77.5.3307-3311.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoj S., Jogger C.R., Myscofski D., Yoon M., Spear P.G. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc. Natl. Acad. Sci. USA. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilterbrand A.T., Heldwein E.E. Go go gadget glycoprotein!: HSV-1 draws on its sizeable glycoprotein tool kit to customize its diverse entry routes. PLoS Pathog. 2019;15:e1007660. doi: 10.1371/journal.ppat.1007660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall B.L., Leronni D., Miyagawa Y., Goins W.F., Glorioso J.C., Cohen J.B. Generation of an oncolytic herpes simplex viral vector completely retargeted to the GDNF receptor gfrα1 for specific infection of breast cancer cells. Int. J. Mol. Sci. 2020;21:8815. doi: 10.3390/ijms21228815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uchida H., Marzulli M., Nakano K., Goins W.F., Chan J., Hong C.-S., Mazzacurati L., Yoo J.Y., Haseley A., Nakashima H., et al. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 2013;21:561–569. doi: 10.1038/mt.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G., Roizman B. Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13alpha2 receptor. Proc. Natl. Acad. Sci. USA. 2006;103:5508–5513. doi: 10.1073/pnas.0601258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G., Roizman B. Characterization of a recombinant herpes simplex virus 1 designed to enter cells via the IL13Ralpha2 receptor of malignant glioma cells. J. Virol. 2005;79:5272–5277. doi: 10.1128/JVI.79.9.5272-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamiyama H., Zhou G., Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13:621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 12.Menotti L., Cerretani A., Hengel H., Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J. Virol. 2008;82:10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menotti L., Avitabile E., Gatta V., Malatesta P., Petrovic B., Campadelli-Fiume G. HSV as A Platform for the Generation of Retargeted, Armed, and Reporter-Expressing Oncolytic Viruses. Viruses. 2018;10:352. doi: 10.3390/v10070352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okubo Y., Uchida H., Wakata A., Suzuki T., Shibata T., Ikeda H., Yamaguchi M., Cohen J.B., Glorioso J.C., Tagaya M., et al. Syncytial Mutations Do Not Impair the Specificity of Entry and Spread of a Glycoprotein D Receptor-Retargeted Herpes Simplex Virus. J. Virol. 2016;90:11096–11105. doi: 10.1128/JVI.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanni P., Gatta V., Menotti L., De Giovanni C., Ianzano M., Palladini A., Grosso V., Dall’ora M., Croci S., Nicoletti G., et al. Preclinical therapy of disseminated HER-2+ ovarian and breast carcinomas with a HER-2-retargeted oncolytic herpesvirus. PLoS Pathog. 2013;9:e1003155. doi: 10.1371/journal.ppat.1003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leoni V., Vannini A., Gatta V., Rambaldi J., Sanapo M., Barboni C., Zaghini A., Nanni P., Lollini P.-L., Casiraghi C., Campadelli-Fiume G. A fully-virulent retargeted oncolytic HSV armed with IL-12 elicits local immunity and vaccine therapy towards distant tumors. PLoS Pathog. 2018;14:e1007209. doi: 10.1371/journal.ppat.1007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata T., Uchida H., Shiroyama T., Okubo Y., Suzuki T., Ikeda H., Yamaguchi M., Miyagawa Y., Fukuhara T., Cohen J.B., et al. Development of an oncolytic HSV vector fully retargeted specifically to cellular EpCAM for virus entry and cell-to-cell spread. Gene Ther. 2016;23:479–488. doi: 10.1038/gt.2016.17. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda H., Uchida H., Okubo Y., Shibata T., Sasaki Y., Suzuki T., Hamada-Uematsu M., Hamasaki R., Okuda K., Yamaguchi M., et al. Antibody Screening System Using a Herpes Simplex Virus (HSV)-Based Probe To Identify a Novel Target for Receptor-Retargeted Oncolytic HSVs. J. Virol. 2021;95:e01766-20. doi: 10.1128/JVI.01766-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovic B., Gianni T., Gatta V., Campadelli-Fiume G. Insertion of a ligand to HER2 in gB retargets HSV tropism and obviates the need for activation of the other entry glycoproteins. PLoS Pathog. 2017;13:e1006352. doi: 10.1371/journal.ppat.1006352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida H., Chan J., Goins W.F., Grandi P., Kumagai I., Cohen J.B., Glorioso J.C. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J. Virol. 2010;84:12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H., Zhang G.r., Wang X., Kong L., Geller A.I. Enhanced nigrostriatal neuron-specific, long-term expression by using neural-specific promoters in combination with targeted gene transfer by modified helper virus-free HSV-1 vector particles. BMC Neurosci. 2008;9:37. doi: 10.1186/1471-2202-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X., Kong L., Zhang G.r., Sun M., Geller A.I. Targeted gene transfer to nigrostriatal neurons in the rat brain by helper virus-free HSV-1 vector particles that contain either a chimeric HSV-1 glycoprotein C-GDNF or a gC-BDNF protein. Brain Res. Mol. Brain Res. 2005;139:88–102. doi: 10.1016/j.molbrainres.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantyh P.W., Koltzenburg M., Mendell L.M., Tive L., Shelton D.L. Antagonism of nerve growth factor-TrkA signaling and the relief of pain. Anesthesiology. 2011;115:189–204. doi: 10.1097/ALN.0b013e31821b1ac5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMahon S.B., Bennett D.L., Priestley J.V., Shelton D.L. The biological effects of endogenous nerve growth factor on adult sensory neurons revealed by a trkA-IgG fusion molecule. Nat. Med. 1995;1:774–780. doi: 10.1038/nm0895-774. [DOI] [PubMed] [Google Scholar]
- 25.Ugolini G., Marinelli S., Covaceuszach S., Cattaneo A., Pavone F. The function neutralizing anti-TrkA antibody MNAC13 reduces inflammatory and neuropathic pain. Proc. Natl. Acad. Sci. USA. 2007;104:2985–2990. doi: 10.1073/pnas.0611253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krüttgen A., Heymach J.V., Kahle P.J., Shooter E.M. The role of the nerve growth factor carboxyl terminus in receptor binding and conformational stability. J. Biol. Chem. 1997;272:29222–29228. doi: 10.1074/jbc.272.46.29222. [DOI] [PubMed] [Google Scholar]
- 27.Ibáñez C.F., Ilag L.L., Murray-Rust J., Persson H. An extended surface of binding to Trk tyrosine kinase receptors in NGF and BDNF allows the engineering of a multifunctional pan-neurotrophin. EMBO J. 1993;12:2281–2293. doi: 10.1002/j.1460-2075.1993.tb05882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi K., Fukuoka T., Obata K., Yamanaka H., Dai Y., Tokunaga A., Noguchi K. Distinct expression of TRPM8, TRPA1, and TRPV1 mRNAs in rat primary afferent neurons with adelta/c-fibers and colocalization with trk receptors. J. Comp. Neurol. 2005;493:596–606. doi: 10.1002/cne.20794. [DOI] [PubMed] [Google Scholar]
- 29.Tuzmen C., Cairns T.M., Atanasiu D., Lou H., Saw W.T., Hall B.L., Cohen J.B., Cohen G.H., Glorioso J.C. Point Mutations in Retargeted gD Eliminate the Sensitivity of EGFR/EGFRvIII-Targeted HSV to Key Neutralizing Antibodies. Mol. Ther. Methods Clin. Dev. 2020;16:145–154. doi: 10.1016/j.omtm.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyker-Snowman K., Hughes R.M., Yankaskas C.L., Cravero K., Karthikeyan S., Button B., Waters I., Rosen D.M., Dennison L., Hunter N., et al. TrkA overexpression in non-tumorigenic human breast cell lines confers oncogenic and metastatic properties. Breast Cancer Res. Treat. 2020;179:631–642. doi: 10.1007/s10549-019-05506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertrand L., Leiva-Torres G.A., Hyjazie H., Pearson A. Conserved residues in the UL24 protein of herpes simplex virus 1 are important for dispersal of the nucleolar protein nucleolin. J. Virol. 2010;84:109–118. doi: 10.1128/JVI.01428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben Abdeljelil N., Rochette P.-A., Pearson A. The UL24 protein of herpes simplex virus 1 affects the sub-cellular distribution of viral glycoproteins involved in fusion. Virology. 2013;444:263–273. doi: 10.1016/j.virol.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Lymberopoulos M.H., Bourget A., Ben Abdeljelil N., Pearson A. Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress. Virology. 2011;412:341–348. doi: 10.1016/j.virol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Connolly S.A., Jackson J.O., Jardetzky T.S., Longnecker R. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 2011;9:369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atanasiu D., Saw W.T., Cohen G.H., Eisenberg R.J. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 2010;84:12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchida H., Chan J., Shrivastava I., Reinhart B., Grandi P., Glorioso J.C., Cohen J.B. Novel mutations in gB and gH circumvent the requirement for known gD Receptors in herpes simplex virus 1 entry and cell-to-cell spread. J. Virol. 2013;87:1430–1442. doi: 10.1128/JVI.02804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loret S., Guay G., Lippé R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson A., Coen D.M. Identification, localization, and regulation of expression of the UL24 protein of herpes simplex virus type 1. J. Virol. 2002;76:10821–10828. doi: 10.1128/JVI.76.21.10821-10828.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blakeney S., Kowalski J., Tummolo D., DeStefano J., Cooper D., Guo M., Gangolli S., Long D., Zamb T., Natuk R.J., Visalli R.J. Herpes simplex virus type 2 UL24 gene is a virulence determinant in murine and guinea pig disease models. J. Virol. 2005;79:10498–10506. doi: 10.1128/JVI.79.16.10498-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobson J.G., Chen S.H., Cook W.J., Kramer M.F., Coen D.M. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology. 1998;242:161–169. doi: 10.1006/viro.1997.9012. [DOI] [PubMed] [Google Scholar]
- 41.Rochette P.-A., Bourget A., Sanabria-Solano C., Lahmidi S., Lavallée G.O., Pearson A. Mutation of UL24 impedes the dissemination of acute herpes simplex virus 1 infection from the cornea to neurons of trigeminal ganglia. J. Gen. Virol. 2015;96:2794–2805. doi: 10.1099/vir.0.000189. [DOI] [PubMed] [Google Scholar]
- 42.Averill S., McMahon S.B., Clary D.O., Reichardt L.F., Priestley J.V. Immunocytochemical localization of trkA receptors in chemically identified subgroups of adult rat sensory neurons. Eur. J. Neurosci. 1995;7:1484–1494. doi: 10.1111/j.1460-9568.1995.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fünfschilling U., Ng Y.-G., Zang K., Miyazaki J.-I., Reichardt L.F., Rice F.L. TrkC kinase expression in distinct subsets of cutaneous trigeminal innervation and nonneuronal cells. J. Comp. Neurol. 2004;480:392–414. doi: 10.1002/cne.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Priestley J.V., Michael G.J., Averill S., Liu M., Willmott N. Regulation of nociceptive neurons by nerve growth factor and glial cell line derived neurotrophic factor. Can. J. Physiol. Pharmacol. 2002;80:495–505. doi: 10.1139/y02-034. [DOI] [PubMed] [Google Scholar]
- 45.Cabrera J.R., Charron A.J., Leib D.A. Neuronal Subtype Determines Herpes Simplex Virus 1 Latency-Associated-Transcript Promoter Activity during Latency. J. Virol. 2018;92:e00430-18. doi: 10.1128/JVI.00430-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flowerdew S.E., Wick D., Himmelein S., Horn A.K.E., Sinicina I., Strupp M., Brandt T., Theil D., Hüfner K. Characterization of neuronal populations in the human trigeminal ganglion and their association with latent herpes simplex virus-1 infection. PLoS One. 2013;8:e83603. doi: 10.1371/journal.pone.0083603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thellman N.M., Triezenberg S.J. Herpes simplex virus establishment, maintenance, and reactivation: in vitro modeling of latency. Pathogens. 2017;6:28. doi: 10.3390/pathogens6030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moqrich A., Earley T.J., Watson J., Andahazy M., Backus C., Martin-Zanca D., Wright D.E., Reichardt L.F., Patapoutian A. Expressing TrkC from the TrkA locus causes a subset of dorsal root ganglia neurons to switch fate. Nat. Neurosci. 2004;7:812–818. doi: 10.1038/nn1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan Y., Wessel J.R. Two types of motor inhibition after action errors in humans. J. Neurosci. 2022;42:7267–7275. doi: 10.1523/JNEUROSCI.1191-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lechner S.G., Frenzel H., Wang R., Lewin G.R. Developmental waves of mechanosensitivity acquisition in sensory neuron subtypes during embryonic development. EMBO J. 2009;28:1479–1491. doi: 10.1038/emboj.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dykes I.M., Lanier J., Eng S.R., Turner E.E. Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation. Neural Dev. 2010;5:3. doi: 10.1186/1749-8104-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marmigère F., Montelius A., Wegner M., Groner Y., Reichardt L.F., Ernfors P. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat. Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zylka M.J., Rice F.L., Anderson D.J. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Nakano K., Kobayashi M., Nakamura K.i., Nakanishi T., Asano R., Kumagai I., Tahara H., Kuwano M., Cohen J.B., Glorioso J.C. Mechanism of HSV infection through soluble adapter-mediated virus bridging to the EGF receptor. Virology. 2011;413:12–18. doi: 10.1016/j.virol.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frampton A.R., Stolz D.B., Uchida H., Goins W.F., Cohen J.B., Glorioso J.C. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J. Virol. 2007;81:10879–10889. doi: 10.1128/JVI.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott J., Selby M., Urdea M., Quiroga M., Bell G.I., Rutter W.J. Isolation and nucleotide sequence of a cDNA encoding the precursor of mouse nerve growth factor. Nature. 1983;302:538–540. doi: 10.1038/302538a0. [DOI] [PubMed] [Google Scholar]
- 57.Selby M.J., Edwards R., Sharp F., Rutter W.J. Mouse nerve growth factor gene: structure and expression. Mol. Cell Biol. 1987;7:3057–3064. doi: 10.1128/mcb.7.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akagi T., Sasai K., Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. USA. 2003;100:13567–13572. doi: 10.1073/pnas.1834876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miyagawa Y., Marino P., Verlengia G., Uchida H., Goins W.F., Yokota S., Geller D.A., Yoshida O., Mester J., Cohen J.B., Glorioso J.C. Herpes simplex viral-vector design for efficient transduction of nonneuronal cells without cytotoxicity. Proc. Natl. Acad. Sci. USA. 2015;112:E1632–E1641. doi: 10.1073/pnas.1423556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazzacurati L., Marzulli M., Reinhart B., Miyagawa Y., Uchida H., Goins W.F., Li A., Kaur B., Caligiuri M., Cripe T., et al. Use of miRNA response sequences to block off-target replication and increase the safety of an unattenuated, glioblastoma-targeted oncolytic HSV. Mol. Ther. 2015;23:99–107. doi: 10.1038/mt.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson J.W., Hall B.L., Marzulli M., Shah V.K., Bailey L., Chiocca E.A., Goins W.F., Kohanbash G., Cohen J.B., Glorioso J.C. Treatment of glioblastoma with current oHSV variants reveals differences in efficacy and immune cell recruitment. Mol. Ther. Oncolytics. 2021;22:444–453. doi: 10.1016/j.omto.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goins W.F., Lee K.A., Cavalcoli J.D., O’Malley M.E., DeKosky S.T., Fink D.J., Glorioso J.C. Herpes simplex virus type 1 vector-mediated expression of nerve growth factor protects dorsal root ganglion neurons from peroxide toxicity. J. Virol. 1999;73:519–532. doi: 10.1128/jvi.73.1.519-532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malcangio M., Garrett N.E., Cruwys S., Tomlinson D.R. Nerve growth factor- and neurotrophin-3-induced changes in nociceptive threshold and the release of substance P from the rat isolated spinal cord. J. Neurosci. 1997;17:8459–8467. doi: 10.1523/JNEUROSCI.17-21-08459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de León A., Gibon J., Barker P.A. NGF-Dependent and BDNF-Dependent DRG Sensory Neurons Deploy Distinct Degenerative Signaling Mechanisms. eNeuro. 2021;8 doi: 10.1523/ENEURO.0277-20.2020. ENEURO.0277-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials described in this article will be available upon reasonable request to the corresponding author.