Abstract
Serotype-cross-reactive dengue virus-specific cytotoxic T lymphocytes (CTL) induced during a primary dengue virus infection are thought to play a role in the immunopathogenesis of dengue hemorrhagic fever (DHF) during a secondary dengue virus infection. Although there is no animal model of DHF, we previously reported that murine dengue virus-specific CTL responses are qualitatively similar to human dengue virus-specific CTL responses. We used BALB/c mice to study the specificity of the CTL response to an immunodominant epitope on the dengue virus NS3 protein. We mapped the minimal H-2Kd-restricted CTL epitope to residues 298 to 306 of the dengue type 2 virus NS3 protein. In short-term T-cell lines and clones, the predominant CD8+ CTL to this epitope in mice immunized with dengue type 2 virus or vaccinia virus expressing the dengue type 4 virus NS3 protein were cross-reactive with dengue type 2 or type 4 virus, while broadly serotype-cross-reactive CTL were a minority population. In dengue type 3 virus-immunized mice, the predominant CTL response to this epitope was broadly serotype cross-reactive. All of the dengue virus-specific CTL clones studied also recognized the homologous NS3 sequences of one or more closely related flaviviruses, such as Kunjin virus. The critical contact residues for the CTL clones with different specificities were mapped with peptides having single amino acid substitutions. These data demonstrate that primary dengue virus infection induces a complex population of flavivirus-cross-reactive NS3-specific CTL clones in mice and suggest that CTL responses are influenced by the viral serotype. These findings suggest an additional mechanism by which the order of sequential flavivirus infections may influence disease manifestations.
The family Flaviviridae consists of over 65 arthropod-transmitted viruses that are known to infect humans. The dengue viruses, of which there are four serotypes, are the flaviviruses of greatest global health importance, causing an estimated 100 million infections each year (10, 28). Dengue hemorrhagic fever (DHF), the severe clinical form of dengue virus infection, has been shown to occur more frequently in individuals who have experienced a previous infection with a different dengue virus serotype (1, 37, 40), suggesting an immunopathologic basis for severe dengue illness.
Members of our laboratory have been studying memory dengue virus-specific cytotoxic T-lymphocyte (CTL) responses in order to understand the potential protective and immunopathologic effects of these CTLs during secondary dengue virus infections. CD4+ and CD8+ CTL responses induced by primary dengue virus infection have been characterized with bulk cultures and at the clonal level in a small number of adult human volunteers who were infected under experimental conditions with live tissue culture-passaged dengue viruses (8, 17, 18, 23, 27). Both serotype-specific and serotype-cross-reactive CTLs have been isolated from most subjects. Most serotype-cross-reactive CTLs have been directed at epitopes on nonstructural proteins, particularly the NS1-NS2A and NS3 proteins. Several different patterns of cross-reactivity have been observed; some CTL clones have recognized several but not all dengue virus serotypes, other clones have recognized all four dengue virus serotypes, and other clones have also recognized closely related flaviviruses, such as West Nile virus (WNV) (14). The frequencies of different CTL specificities have varied among the subjects studied; however, it has not been possible to relate these differences to host factors, such as HLA type, or differences in the viruses causing primary infections.
We previously reported that T-lymphocyte responses to primary dengue virus infection in mice are qualitatively similar to human T-lymphocyte responses to dengue virus in bulk cultures and at the clonal level (34–36). Both serotype-specific and serotype-cross-reactive dengue virus-specific CTL clones were isolated from BALB/c mice, and the serotype-cross-reactive clones recognized epitopes on the NS1-NS2A and NS3 proteins. One CD8+ CTL epitope, recognized in the context of H-2Kd, was mapped to amino acids 296 to 310 of the dengue type 2 virus NS3 protein (34). This epitope is completely conserved in dengue type 4 virus.
To define further the potential importance of these memory dengue virus-specific CD8+ CTLs in secondary dengue virus infection, we analyzed in more detail the specificity of the NS3-specific CTL clones. As reported here, we noted that all four serotypes of dengue viruses as well as closely related flaviviruses showed a high degree of amino acid homology at the epitope mentioned above. Specific single amino acid differences in this epitope had different effects on recognition by different CTL clones, accounting for both dengue virus subcomplex-specific and dengue virus complex-cross-reactive patterns of responses. We also explored the effects of the infecting virus on the patterns of CTL responses generated.
MATERIALS AND METHODS
Cells.
Target cell lines were the P815 murine mastocytoma line (H-2d), L929 (H-2k) cell lines transfected with Ld (T1.1.1) or Dd (T4.8.3) and provided by Carol Reiss of New York University (26), and an L929 cell line transfected with Kd(L-Kd-172) and obtained from Jack Bennick of the National Institutes of Health (5). The control L929 cell line (DAP) was a gift from Carol Reiss.
Viruses.
Mouse-adapted dengue type 2 virus (strain New Guinea C), type 3 virus (strain PR6), and type 4 virus (strain 814669) were kindly provided by Jack McCown of the Walter Reed Army Institute of Research. Tissue culture-adapted dengue type 2 virus (strain New Guinea C) was graciously donated by Walter Brandt, also of the Walter Reed Army Institute of Research, and was propagated in C6/36 cells. Recombinant vaccinia viruses expressing the dengue type 2 or type 3 virus NS3 protein were provided by Margo Brinton of Georgia State University (41), and a recombinant vaccinia virus expressing the dengue type 4 virus NS3 protein was provided by Ching-Juh Lai of the National Institutes of Health (6, 35).
Peptides.
Synthetic peptides were prepared by 9-fluorenylmethoxycarbonyl chemistry with a Symphony automated peptide synthesizer (Rainin Instruments, Woburn, Mass.) at the Peptide Core Facility at the University of Massachusetts Medical Center.
Mouse immunization and preparation of spleen cells.
Immunization of mice and preparation of clones from spleen cells were performed as previously described (34–36). Briefly, male BALB/c (H-2d) mice (Charles River Breeding Laboratories, Wilmington, Mass.) were immunized at 4 to 8 weeks of age with dengue virus (0.2 to 0.3 ml containing approximately 1 × 106 PFU) or with one to three doses of recombinant vaccinia virus expressing the dengue virus NS3 protein (0.1 ml containing approximately 2 × 107 PFU). Splenocytes were collected 4 to 8 weeks after immunization and incubated in RPMI 1640 medium with 5 × 105 M 2-mercaptoethanol, 10% fetal calf serum (Hyclone Laboratories, Logan, Utah), and 0.5 ml of dengue virus or 1 μM peptide. Approximately every 2 weeks, the cells were stimulated with either 2 × 106 gamma-irradiated dengue type 2 virus-infected P815 cells or 3 × 107 gamma-irradiated syngeneic nonimmune spleen cells plus 1 μM dengue virus NS3 peptide. Cells were fed twice weekly with medium containing 10% rat lectin-free T-cell growth factor as a source of interleukin 2.
Cytotoxicity assays.
Cytotoxicity assays were performed as previously described (34, 35). Briefly, P815 cells were radioactively labeled by incubation with Na251CrO4 for 1 h, washed extensively, and seeded at 0.5 × 104 cells per well in 96-well U-bottom plates. Serial dilutions of peptides were added directly to the target cells in the wells. The clones were then added at various effector/target cell ratios. The plates were incubated at 37°C for 4 h. The supernatant fluid was collected with a Supernatant Collecting System (Skatron, Inc., Sterling, Va.). A gamma counter detected the release of 51Cr. Minimum 51Cr release was measured by sampling supernatant fluid from labeled target cells incubated in medium alone. Maximum 51Cr release was determined from wells in which labeled target cells were incubated with Renex. Assays were performed in triplicate, and the mean of the samples was used to calculate percent specific lysis with the following formula: percent specific lysis = 100 × [(experimental 51Cr release − minimum 51Cr release)/(maximum 51Cr release − minimum 51Cr release)]. Minimum 51Cr release did not exceed 30% of maximum 51Cr release. Lysis of peptide-coated target cells was considered significant when it was ≥8% more than lysis of target cells in the absence of peptides and values were found significantly different by the Student t test.
RESULTS
Identification of the minimal epitope recognized by NS3-specific CTL clones.
We previously mapped a dominant epitope on the dengue type 4 virus NS3 protein recognized by H-2Kd-restricted CD8+ CTL clones derived from dengue type 2 virus-immunized BALB/c mice to a 15-amino-acid region between residues 296 and 310, ARGYISTRVEMGEAA (34). To define further the specific epitope recognized by these CTL clones, we tested for the recognition of target cells incubated with truncations of this 15-mer. Data obtained with clone 2D65 are shown in Table 1; similar data were obtained with clone 2D42 (data not shown). Initial data suggested that the minimal sequence was the 9-mer representing residues 298 (G) to 306 (M). Analysis of recognition of this 9-mer peptide and truncations thereof showed that the C-terminal methionine was required for recognition by these CTL clones. The 8-mer peptide representing residues 299 to 306 could be recognized by these clones, but recognition of this peptide was reproducibly less efficient than recognition of the 9-mer peptide representing residues 298 to 306.
TABLE 1.
Recognition of truncations of the dengue type 4 virus NS3 peptide (residues 296 to 310) by CD8+ CTL clone 2D65a
| Expt | Peptide residues | Sequence | % Specific lysis at peptide concn of:
|
|||
|---|---|---|---|---|---|---|
| 5 μM | 500 nM | 50 nM | 5 nM | |||
| 1 | 296–310 | ARGYISTRVEMGEAA | 56 | 23 | 3 | 2 |
| 298–310 | GYISTRVEMGEAA | 58 | 55 | 36 | 10 | |
| 300–310 | ISTRVEMGEAA | 1 | 2 | 2 | 2 | |
| 296–308 | ARGYISTRVEMGE | 58 | 34 | 6 | 5 | |
| 296–306 | ARGYISTRVEM | 59 | 32 | 5 | 3 | |
| 2 | 298–306 | GYISTRVEM | 25 | 27 | 27 | 18 |
| 298–305 | GYISTRVE | 6 | 3 | 11 | 5 | |
| 299–306 | YISTRVEM | 21 | 19 | 18 | 9 | |
| 300–306 | ISTRVEM | 9 | 2 | 4 | 9 | |
Recognition of synthetic peptide-pulsed target cells was tested at effector/target cell ratios of 4:1 in experiment 1 and 2:1 in experiment 2. Percentages of lysis of target cells in the absence of peptide were 2% in experiment 1 and 9% in experiment 2. Boldface values represent percent specific lysis significantly greater than that of the negative control.
CTL clones derived from dengue type 2 virus-immunized mice recognize the corresponding sequence of dengue type 4 but not dengue type 1 or dengue type 3 virus.
We previously reported that the sequence of the 15-mer recognized by the CTL clones was completely conserved between dengue type 2 and dengue type 4 viruses (34). We next examined the published sequences to determine the corresponding sequences of other dengue virus serotypes and related flaviviruses (Table 2) (2, 3, 7, 13, 22, 25, 30, 33, 39). This analysis showed that this region of the NS3 protein is highly conserved among flaviviruses. Compared to the dengue type 2 or type 4 virus sequence, there was only a single amino acid difference at peptide position 8 in dengue type 1 and dengue type 3 viruses, a single amino acid difference at position 9 in Kunjin virus, and two amino acid differences in Murray Valley encephalitis virus. WNV and Japanese encephalitis virus had three amino acid differences, and the less closely related yellow fever virus had seven amino acid differences.
TABLE 2.
Sequences of dengue virus and related flaviviruses at the CD8+ CTL epitope
| Virus | Sequence | Reference |
|---|---|---|
| Dengue type 4 | 298GYISTRVEM306 | 25 |
| Dengue type 1 | 299-------G-307 | 7 |
| Dengue type 2 | 298---------306 | 13 |
| Dengue type 3 | 299-------G-307 | 30 |
| Kunjin | 299--------L307 | 3 |
| Murray Valley encephalitis | 299---A----A307 | 22 |
| WNV | 299---A-K--L307 | 2 |
| Japanese encephalitis | 299---A-K--L307 | 39 |
| Yellow fever | 303-WAAH-ARA311 | 33 |
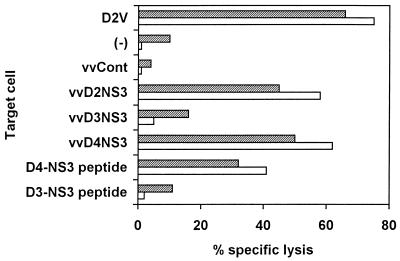
Since serotype cross-reactivity is most clinically relevant among the dengue viruses, we first tested for recognition of the dengue type 1 or type 3 virus sequence by the CTL clones (Fig. 1). Identical results were obtained with recombinant vaccinia viruses expressing the full-length NS3 proteins of dengue type 2, dengue type 3, and dengue type 4 viruses or synthetic peptides representing the dengue type 2 or type 4 virus or the dengue type 1 or type 3 virus sequences; none of six clones isolated from dengue type 2 virus-immunized mice recognized the dengue type 3 virus NS3 sequence.
FIG. 1.
Dengue virus serotype specificity of CD8+ CTL clones isolated from dengue type 2 virus-immunized mice. Clones 2D42 (hatched bars) and 2D65 (open bars) were isolated from dengue type 2 virus-immunized BALB/c mice and tested at an effector/target cell ratio of 4:1. P815 target cells were uninfected (−); infected with dengue type 2 virus (D2V), wild-type vaccinia virus (vvCont), or recombinant vaccinia viruses (vv) expressing the full-length dengue type 2, dengue type 3, or dengue type 4 virus NS3 proteins (vvD2NS3, vvD3NS3, and vvD4NS3, respectively); or pulsed with the dengue type 4 virus (residues 296 to 310) or dengue type 3 virus (residues 297 to 311) peptides (D4-NS3 peptide and D3-NS3 peptide, respectively) at 10 μM.
Dengue type 4 virus NS3-immunized mice do have serotype-cross-reactive CTL responses.
We next tested whether the dengue type 1 or type 3 virus NS3 sequences could be recognized by H-2Kd-restricted CTLs. Splenocytes from mice immunized with a recombinant vaccinia virus expressing the dengue type 4 virus NS3 protein were stimulated in bulk cultures with the dengue type 4 virus NS3 peptide (296 to 310) and tested for recognition of the dengue type 2 or type 4 virus and dengue type 1 or type 3 virus NS3 peptides (Table 3). At day 40 of culturing, the predominant response was specific for the dengue type 4 virus NS3 sequence, but there was significant, if less efficient, recognition of the dengue type 3 virus NS3 peptide. This result suggested that serotype-cross-reactive CTLs were present but at a lower level than CTLs specific for the dengue type 2 or type 4 virus peptide. The bulk cultures were then split and restimulated with either the dengue type 4 or the dengue type 3 virus NS3 peptide. The CTL lines were then tested for recognition of both peptides. The CTL line restimulated with the dengue type 4 virus NS3 peptide maintained recognition of the dengue type 4 virus NS3 peptide but showed little if any recognition of the dengue type 3 virus NS3 peptide at day 54 of culturing. In contrast, the CTL line restimulated with the dengue type 3 virus NS3 peptide showed a high level of cross-reactivity for both peptides. Both CTL lines recognized the target epitope in an H-2Kd-restricted manner (data not shown). These results suggest that serotype-cross-reactive CTLs were initially present at a low level but that they could be selectively expanded by stimulation with the dengue type 3 virus NS3 peptide.
TABLE 3.
Recognition of dengue virus NS3 peptides by short-term CTL lines from dengue type 4 virus NS3-immunized micea
| Day | Dengue virus type used to stimulate cell lines | Target cellsb | % Specific lysis at peptide concn of:
|
|||
|---|---|---|---|---|---|---|
| 1 μM | 100 nM | 10 nM | — | |||
| 40 | 4c | D4-NS3 | 79 | 75 | 69 | 1 |
| D3-NS3 | 40 | 22 | 13 | |||
| 54 | 4c | D4-NS3 | 46 | 48 | 42 | 7 |
| D3-NS3 | 10 | 5 | 2 | |||
| 4→3d | D4-NS3 | 47 | 50 | 46 | 2 | |
| D3-NS3 | 41 | 36 | 25 | |||
BALB/c mice were immunized with a recombinant vaccinia virus expressing the dengue type 4 virus NS3 protein. Boldface values represent percent specific lysis significantly greater than that of the negative control (no peptide [—]).
P815 target cells were incubated with the corresponding peptides of dengue type 4 virus (D4-NS3) or dengue type 3 virus (D3-NS3).
Spleen cells from immunized mice were stimulated in vitro with the D4-NS3 peptide.
Spleen cells from immunized mice were initially stimulated in vitro with the D4-NS3 peptide for 40 days and then stimulated with the corresponding D3-NS3 peptide.
We then used this stimulation protocol to isolate dengue virus serotype-cross-reactive CD8+ CTL clones. Splenocytes from mice immunized with dengue type 4 virus were stimulated with the dengue type 3 virus NS3 peptide and cloned by limiting dilution. Five CTL clones were isolated; all recognized both the dengue type 4 and the dengue type 3 virus NS3 peptides in the context of H-2Kd (data not shown).
Dengue virus NS3-specific CTL from dengue type 3 virus-immunized mice are serotype cross-reactive.
We wanted to characterize further the serotype cross-reactivity of dengue virus NS3-specific CTL lines. We immunized mice with dengue type 3 virus to determine the pattern of CTL serotype cross-reactivity induced by the dengue type 3 virus NS3 epitope. Splenocytes from dengue type 3 virus-immunized mice were stimulated in vitro with the dengue type 3 virus NS3 peptide and tested for the recognition of the dengue type 1 or type 3 virus and dengue type 2 or type 4 virus NS3 peptides (Table 4). These CTLs lysed target cells pulsed with either peptide, indicating that the predominant CTL response in dengue type 3 virus-immunized mice is broadly dengue virus serotype cross-reactive. These CTLs also lysed target cells infected with recombinant vaccinia viruses expressing either the dengue type 2, dengue type 3, or dengue type 4 virus NS3 protein or target cells infected with dengue type 2 virus, indicating that these CTLs recognize the naturally processed epitope (Table 4). We isolated 14 CTL clones from this CTL line by limiting dilution; all the clones isolated recognized both the dengue type 3 and the dengue type 4 virus NS3 peptides in the context of H-2Kd (data not shown).
TABLE 4.
Serotype cross-reactivity of NS3-specific CTLs in dengue type 3 virus-immunized micea
| Day of culturing | Target cellsb | % Specific lysis |
|---|---|---|
| 27 | D4-NS3 (298–306) | 25 |
| D3-NS3 (299–307) | 27 | |
| WNV NS3 (299–307) | 4 | |
| 40 | D2V | 57 |
| Uninfected | −2 | |
| vvD2:NS3 | 31 | |
| vvD3:NS3 | 43 | |
| vvD4:NS3 | 25 | |
| vvControl | 1 |
Spleen cells from mice immunized with dengue type 3 virus were stimulated in vitro with the dengue type 3 virus NS3 (D3-NS3) peptide (residues 299 to 307) and tested for recognition of the indicated target cells. Boldface values represent percent specific lysis significantly greater than that of the negative control.
P815 target cells were incubated with the corresponding peptides of dengue type 4 virus, dengue type 3 virus, or WNV (day-27 effectors) or were infected with dengue type 2 virus (D2V), wild-type vaccinia virus (vvControl), or recombinant vaccinia viruses (vv) expressing the dengue type 2, dengue type 3, or dengue type 4 virus NS3 proteins (day-40 effectors) (e.g., vvD2:NS3 represents dengue type 2 virus NS3 protein expressed by a recombinant vaccinia virus).
Flavivirus specificity of dengue virus NS3-specific CTL clones.
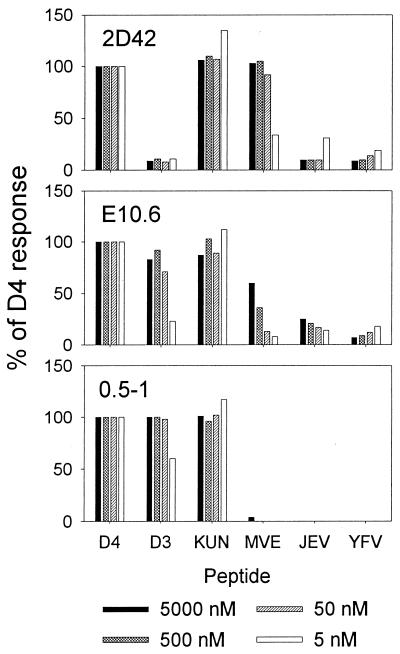
We next studied the specificity of the CTL clones for other closely related flaviviruses. Representative data obtained with CTL clones each corresponding to the different specificities are shown in Fig. 2. Clone 2D42, which was generated from dengue type 2 virus-immunized mice after stimulation with dengue type 2 virus-infected P815 cells, did not recognize the dengue type 1 or type 3 virus peptide but did recognize the homologous peptides of Kunjin virus and Murray Valley encephalitis virus. Clone E10.6, which was generated from a dengue type 4 virus-immunized mouse after stimulation with the dengue type 3 virus NS3 peptide, recognized the NS3 peptides from all dengue virus serotypes and Kunjin virus. This clone could recognize the Murray Valley encephalitis virus sequence but less efficiently than clone 2D42. Clone 0.5-1, which was generated from dengue type 3 virus-immunized mice after stimulation with the dengue type 3 virus NS3 peptide, was similarly dengue virus serotype cross-reactive and able to recognize the Kunjin virus peptide but did not show any recognition of the Murray Valley encephalitis virus peptide. None of the three clones recognized the homologous peptides of Japanese encephalitis virus, WNV, or yellow fever virus.
FIG. 2.
Flavivirus specificity of CD8+ CTL clones. Clones 2D42, E10.6, and 0.5-1 were tested in separate experiments. Virus sequences are designated as follows: D4, dengue type 4 (identical to dengue type 2); D3, dengue type 3 (identical to dengue type 1); KUN, Kunjin; MVE, Murray Valley encephalitis; JEV, Japanese encephalitis (identical to WNV); and YFV, yellow fever. Data are expressed as a percentage of the D4 response, calculated as 100 × [(percent specific lysis of target cells incubated with indicated peptide)/(percent specific lysis of target cells incubated with the dengue type 4 virus peptide)]. For clone 2D42, the effector/target cell ratio was 1:1, and percentages of specific lysis were as follows: no peptide, 3%; 5,000 nM, 36%; 500 nM, 35%; 50 nM, 36%; and 5 nM, 30%. For clone E10.6, the effector/target cell ratio was 2:1, and percentages of specific lysis were as follows: no peptide, 5%; 5,000 nM, 58%; 500 nM, 59%; 50 nM, 65%; and 5 nM, 55%. For clone 0.5-1, the effector/target cell ratio was 2:1, and percentages of specific lysis were as follows: no peptide, −3%; 5,000 nM, 29%; 500 nM, 30%; 50 nM, 28%; and 5 nM, 27%.
Identification of the critical contact residues for dengue virus-specific CTL clones.
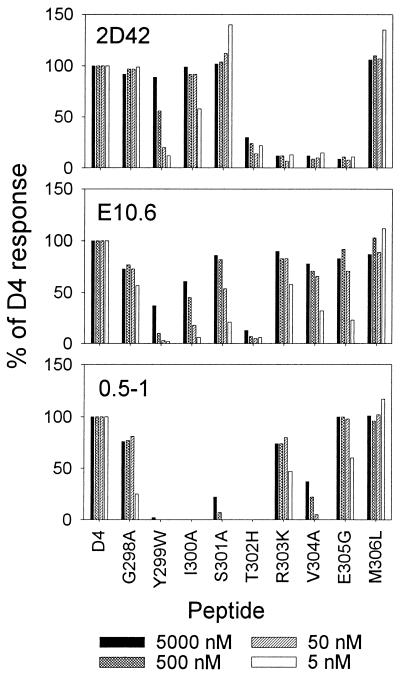
The different patterns of recognition of related flaviviruses suggested that the clones differed in the critical contact residues of the NS3 peptide. To characterize this idea further, we tested the three above-mentioned clones for the recognition of peptides with individual substitutions at each of the nine residues of the dengue type 4 virus NS3 sequence. To facilitate the comparison of the recognition of these peptides and the peptides of the related flaviviruses, we based the substitutions on the corresponding amino acids of other flaviviruses, except for position 1, where we substituted alanine for the conserved glycine. Recognition of these peptides by clones 2D42, E10.6, and 0.5-1 is shown in Fig. 3.
FIG. 3.
Effects of single amino acid substitutions on recognition by dengue virus-specific CTL clones. Clones 2D42, E10.6, and 0.5-1 were tested in separate experiments. Data are expressed as a percentage of the response to dengue type 4 virus, calculated as described in the legend to Fig. 2. The peptide designation specifies the amino acid and position number in the dengue type 4 virus NS3 sequence followed by the substituted amino acid. Peptides E305G and M306L are the dengue type 3 virus and Kunjin virus peptides, respectively.
The second and ninth residues are most important for binding to H-2Kd (32). Substitution of tryptophan for tyrosine at the second residue, as in yellow fever virus, abolished recognition by clones E10.6 and 0.5-1 and drastically inhibited recognition by clone 2D42. Substitution of leucine for methionine at the ninth position, as in Kunjin virus, did not impair recognition by any of the clones, in keeping with the known suitability of leucine at this position for binding to Kd (32). Substitution of histidine for threonine at the fifth residue, as in yellow fever virus, impaired recognition by all clones. Peptide recognition by clone 2D42 was also inhibited by substitutions at positions 6, 7, and 8, which included the single substitution in the dengue type 1 or type 3 virus peptide. In comparison, peptide recognition by clones E10.6 and 0.5-1 was inhibited most by substitutions at positions 3 and 4.
DISCUSSION
This study expands on our previous work showing that H-2Kd-restricted CD8+ CTL clones isolated from dengue type 2 virus-immunized BALB/c mice recognize a dengue type 2 and type 4 virus-cross-reactive epitope between amino acids 296 and 310 of the NS3 protein (34). We defined the minimal epitope as amino acids 298 to 306 of the dengue type 2 virus NS3 protein. We also generated additional CD8+ CTL clones that recognized the same epitope from mice immunized with dengue type 3 or dengue type 4 virus. We found that these CTL clones were either dengue virus subcomplex (dengue type 2 or type 4 virus) specific or broadly serotype cross-reactive. In addition, all of these CTL clones recognized the corresponding sequences of one or more other flaviviruses.
The NS3 protein appears to be a dominant target for dengue virus-specific CD4+ and CD8+ T cells, and most dengue virus NS3-specific T cells are serotype cross-reactive (14, 27, 35). Multiple human CD4+ T-cell epitopes on the NS3 protein have been identified (15, 19, 21, 29, 41). Although few human CD8+ T-cell epitopes have been mapped on the NS3 protein, bulk-culture CD8+ CTL recognition of the NS3 protein was detected in six of eight dengue virus-immunized subjects in one study (27). CTLs from several mouse strains have been found to recognize the NS3 protein of dengue virus as well as other flaviviruses, including WNV, Murray Valley encephalitis virus, and Kunjin virus (11, 24, 31, 35). The abundance of T-cell epitopes on the flavivirus NS3 protein is not well explained. The relative conservation of the NS3 protein among flaviviruses suggests that the requirements for the enzymatic functions of this protein may restrict the ability of flaviviruses to survive mutations. The minimal peptide recognized by the H-2Kd-restricted CTLs that we studied, GYISTRVEM, contains the typical Kd binding motif, with tyrosine at position 2 and a hydrophobic amino acid at position 9 (32). This Kd binding motif is conserved in Kunjin virus, Murray Valley encephalitis virus, WNV, and Japanese encephalitis virus, suggesting that this epitope may be recognized by CTLs induced in mice by infection with these flaviviruses.
Memory dengue virus serotype cross-reactive CTLs may play a protective role in limiting viral replication, but they have also been proposed to contribute to the increased risk for DHF during secondary dengue virus infections (20). Levels of soluble CD8 in serum were found to be elevated in children with DHF compared to those with dengue fever, supporting an immunopathologic role for CD8+ CTLs (7a, 16). The NS3-specific CTL clones that we studied recognized one or more heterologous dengue virus serotypes and theoretically could be activated during a secondary dengue virus infection.
These results also demonstrate CTL cross-reactivity with other flaviviruses, specifically Kunjin and Murray Valley encephalitis viruses, which have the most homology with dengue viruses at the epitope in question. Similarly, Kurane et al. reported that some human dengue virus-specific CD4+ CTL clones could recognize WNV or yellow fever virus (14). Interestingly, not all flavivirus-cross-reactive CTL clones isolated from dengue virus-immunized humans and mice were able to recognize all four dengue virus serotypes (41). We used peptides incorporating single substitutions into the background of the dengue type 2 or type 4 virus NS3 peptide sequence to explain this unexpected pattern of specificity. Peptide recognition by clone 2D42, which recognized the dengue type 2 or type 4, Kunjin, and Murray Valley encephalitis virus sequences, was sensitive to the substitution at position 8 in the dengue type 1 or type 3 virus sequence, whereas substitutions at positions 4 and 9, as in Murray Valley encephalitis virus, had little effect. In contrast, clone 0.5-1, which was broadly dengue virus serotype cross-reactive but unable to recognize the Murray Valley encephalitis virus sequence, was unaffected by the substitution at position 8 but was unable to recognize the peptide incorporating the position 4 substitution.
CTL cross-reactivity with other flaviviruses suggests that protective and immunopathologic effects might occur during sequential infections with flaviviruses other than dengue virus. In a large field study of a Japanese encephalitis virus vaccine, Hoke et al. observed that the incidence of DHF was somewhat lower in immunized children than in control subjects over a 2-year period, although the difference was not statistically significant (12). Several studies have also found that the antibody response to candidate live dengue virus vaccines was enhanced in subjects previously immunized with a yellow fever virus vaccine (4, 38). Epidemiologic studies have not fully addressed this possibility, in part because the geographic distribution of dengue virus has historically overlapped that of only Japanese encephalitis virus. More recently, however, dengue virus infections have become more frequent in areas of the Western Hemisphere where yellow fever is endemic and in Australia where Kunjin and Murray Valley encephalitis viruses circulate (9). These epidemiologic changes increase the opportunities for sequential flavivirus infections.
We found that the patterns of dengue virus serotype cross-reactivity in short-term T-cell lines and clones differed in mice immunized with different dengue virus serotypes. In BALB/c mice immunized with dengue type 2 virus or a recombinant vaccinia virus expressing the dengue type 4 virus NS3 protein, the predominant CTL responses were dengue type 2 or type 4 virus cross-reactive, with a low level of CTL recognition of the dengue type 1 or type 3 virus sequence. In mice immunized with dengue type 3 virus, the CTL responses to this epitope were broadly serotype cross-reactive. Substantial variability in the levels of serotype-cross-reactive T-cell responses have been noted in studies of dengue virus-immunized humans, but an effect of the viral serotype on these responses has not been apparent. In studies of murine CTL responses to WNV and Kunjin virus, Hill et al. also noted nonreciprocal CTL flavivirus cross-reactivity (11). Although a detailed analysis of the levels of CTLs with different specificities, such as by use of intracellular gamma interferon staining in response to peptide stimulation, would be necessary to provide a definitive demonstration of this effect, our data suggest that there is nonreciprocal CTL cross-reactivity among the dengue virus serotypes as well.
Epidemiologic studies have suggested that the order of acquisition of dengue virus infections is important; specifically, having a sequence of infections in which dengue type 2 virus is the agent of secondary dengue virus infection increases the odds of DHF or dengue shock syndrome (37, 40). Differences in virulence between the different dengue virus serotypes have been proposed to explain this finding. Our results raise another possible explanation, that CTLs induced by other serotypes may recognize dengue type 2 virus to a greater extent than the reverse.
The full story on the importance of T-cell immunity in DHF pathogenesis is not yet known, but clarifying the specificity and cross-reactivity of T-cell clones could give insights into how cell-mediated immunity might work in nature. Although dengue virus-infected mice do not manifest DHF, our results suggest that the immune response to sequential dengue virus infection in mice may provide information useful for directing further human studies.
ACKNOWLEDGMENTS
We thank Jurand Janus, Kim West, Anita Leporati, and Lichen Dai for technical assistance.
This work was supported by grants K11 AI00971 and T32 AI07272 from the NIAID.
REFERENCES
- 1.Burke D S, Nisalak A, Johnson D E, Scott R M. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 2.Castle E, Leidner U, Nowak T, Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986;149:10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- 3.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 4.Eckels K H, Kliks S C, Dubois D R, Wahl L M, Bancroft W H. The association of enhancing antibodies with seroconversion in humans receiving a dengue-2 live-virus vaccine. J Immunol. 1985;135:4201–4203. [PubMed] [Google Scholar]
- 5.Eisenlohr L C, Yewdell J W, Bennink J R. A transient transfection system for identifying biosynthesized proteins processed and presented to class I MHC restricted T lymphocytes. J Immunol Methods. 1992;154:131–138. doi: 10.1016/0022-1759(92)90220-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falgout B, Pethel M, Zhang Y M, Lai C J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu J, Tan B H, Yap E H, Chan Y C, Tan Y H. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90) Virology. 1992;188:953–958. doi: 10.1016/0042-6822(92)90560-c. [DOI] [PubMed] [Google Scholar]
- 7a.Green, S. Personal communication.
- 8.Green S, Kurane I, Edelman R, Tacket C O, Eckels K H, Vaughn D W, Hoke C H, Jr, Ennis F A. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J Virol. 1993;67:5962–5967. doi: 10.1128/jvi.67.10.5962-5967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubler D J, Clark G G. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis. 1995;1:55. doi: 10.3201/eid0102.952004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halstead S B. The XXth century dengue pandemic: need for surveillance and research. World Health Stat Q. 1992;45:292–298. [PubMed] [Google Scholar]
- 11.Hill A B, Mullbacher A, Parrish C, Coia G, Westaway E G, Blanden R V. Broad cross-reactivity with marked fine specificity in the cytotoxic T cell response to flaviviruses. J Gen Virol. 1992;73:1115–1123. doi: 10.1099/0022-1317-73-5-1115. [DOI] [PubMed] [Google Scholar]
- 12.Hoke C H, Nisalak A, Sangawhipa N, Jatanasen S, Laorakapongse T, Innis B L, Kotchasenee S, Gingrich J B, Latendresse J, Fukai K, Burke D S. Protection against Japanese encephalitis by inactivated vaccines. N Engl J Med. 1988;319:608–614. doi: 10.1056/NEJM198809083191004. [DOI] [PubMed] [Google Scholar]
- 13.Irie K, Mohan P M, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain) Gene. 1989;75:197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 14.Kurane I, Brinton M A, Samson A L, Ennis F A. Dengue virus-specific, human CD4+ CD8− cytotoxic T-cell clones: multiple patterns of virus cross-reactivity recognized by NS3-specific T-cell clones. J Virol. 1991;65:1823–1828. doi: 10.1128/jvi.65.4.1823-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurane I, Dai L C, Livingston P G, Reed E, Ennis F A. Definition of an HLA-DPw2-restricted epitope on NS3, recognized by a dengue virus serotype-cross-reactive human CD4+ CD8− cytotoxic T-cell clone. J Virol. 1993;67:6285–6288. doi: 10.1128/jvi.67.10.6285-6288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurane I, Innis B L, Nimmannitya S, Nisalak A, Meager A, Janus J, Ennis F A. Activation of T lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J Clin Investig. 1991;88:1473–1480. doi: 10.1172/JCI115457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurane I, Innis B L, Nisalak A, Hoke C, Nimmannitya S, Meager A, Ennis F A. Human T cell responses to dengue virus antigens. Proliferative responses and interferon gamma production. J Clin Investig. 1989;83:506–513. doi: 10.1172/JCI113911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurane I, Meager A, Ennis F A. Dengue virus-specific human T cell clones. Serotype crossreactive proliferation, interferon gamma production, and cytotoxic activity. J Exp Med. 1989;170:763–775. doi: 10.1084/jem.170.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurane I, Okamoto Y, Dai L C, Zeng L L, Brinton M A, Ennis F A. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8− cytotoxic T lymphocyte clones. J Gen Virol. 1995;76:2243–2249. doi: 10.1099/0022-1317-76-9-2243. [DOI] [PubMed] [Google Scholar]
- 20.Kurane I, Rothman A L, Livingston P G, Green S, Gagnon S J, Janus J, Innis B L, Nimmannitya S, Nisalak A, Ennis F A. Immunopathologic mechanisms of dengue hemorrhagic fever and dengue shock syndrome. Arch Virol. 1994;9:59–64. doi: 10.1007/978-3-7091-9326-6_7. [DOI] [PubMed] [Google Scholar]
- 21.Kurane I, Zeng L, Brinton M A, Ennis F A. Definition of an epitope on NS3 recognized by human CD4+ cytotoxic T lymphocyte clones cross-reactive for dengue virus types 2, 3, and 4. Virology. 1998;240:169–174. doi: 10.1006/viro.1997.8925. [DOI] [PubMed] [Google Scholar]
- 22.Lee E, Fernon C, Simpson R, Weir R C, Rice C M, Dalgarno L. Sequence of the 3′ half of the Murray Valley encephalitis virus genome and mapping of the nonstructural proteins NS1, NS3, and NS5. Virus Genes. 1990;4:197–213. doi: 10.1007/BF00265630. [DOI] [PubMed] [Google Scholar]
- 23.Livingston P G, Kurane I, Dai L C, Okamoto Y, Lai C J, Men R, Karaki S, Takiguchi M, Ennis F A. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J Immunol. 1995;154:1287–1295. [PubMed] [Google Scholar]
- 24.Lobigs M, Arthur C E, Mullbacher A, Blanden R V. The flavivirus nonstructural protein NS3 is a dominant source of cytotoxic T cell peptide determinants. Virology. 1997;202:195–201. doi: 10.1006/viro.1994.1335. [DOI] [PubMed] [Google Scholar]
- 25.Mackow E, Makino Y, Zhao B T, Zhang Y M, Markoff L, Buckler-White A, Guiler M, Chanock R, Lai C J. The nucleotide sequence of dengue type 4 virus: analysis of genes coding for nonstructural proteins. Virology. 1987;159:217–228. doi: 10.1016/0042-6822(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 26.Margulies D H, Evans G A, Ozato K, Camerini-Otero R D, Tanaka K, Appella E, Seidman J G. Expression of H-2Dd and H-2Ld mouse major histocompatibility antigen genes in L cells after DNA-mediated gene transfer. J Immunol. 1983;130:463–470. [PubMed] [Google Scholar]
- 27.Mathew A, Kurane I, Rothman A L, Zeng L L, Brinton M A, Ennis F A. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J Clin Investig. 1996;98:1684–1694. doi: 10.1172/JCI118964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monath T P. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci USA. 1994;91:2395–2400. doi: 10.1073/pnas.91.7.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto Y, Kurane I, Leporati A M, Ennis F A. Definition of the region on NS3 which contains multiple epitopes recognized by dengue virus serotype-cross-reactive and flavivirus-cross-reactive, HLA-DPw2-restricted CD4+ T cell clones. J Gen Virol. 1998;79:697–704. doi: 10.1099/0022-1317-79-4-697. [DOI] [PubMed] [Google Scholar]
- 30.Osatomi K, Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990;176:643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- 31.Parrish C R, Coia G, Hill A, Mullbacher A, Westaway E G, Blanden R V. Preliminary analysis of murine cytotoxic T cell responses to the proteins of the flavivirus Kunjin using vaccinia virus expression. J Gen Virol. 1991;72:1645–1653. doi: 10.1099/0022-1317-72-7-1645. [DOI] [PubMed] [Google Scholar]
- 32.Rammensee H G, Friede T, Stevanovic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 33.Rice C M, Lenches E M, Eddy S R, Shin S J, Sheets R L, Strauss J H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 34.Rothman A L, Kurane I, Ennis F A. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus. J Virol. 1996;70:6540–6546. doi: 10.1128/jvi.70.10.6540-6546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothman A L, Kurane I, Lai C J, Bray M, Falgout B, Men R, Ennis F A. Dengue virus protein recognition by virus-specific murine CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:801–806. doi: 10.1128/jvi.67.2.801-806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothman A L, Kurane I, Zhang Y M, Lai C J, Ennis F A. Dengue virus-specific murine T-lymphocyte proliferation: serotype specificity and response to recombinant viral proteins. J Virol. 1989;63:2486–2491. doi: 10.1128/jvi.63.6.2486-2491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sangkawibha N, Rojanasuphot S, Ahandrik S, Viriyapongse S, Jatanasen S, Salitul V, Phanthumachinda B, Halstead S B. Risk factors for dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am J Epidemiol. 1984;120:653–669. doi: 10.1093/oxfordjournals.aje.a113932. [DOI] [PubMed] [Google Scholar]
- 38.Scott R M, Eckels K H, Bancroft W H, Summers P L, McCown J M, Anderson J H, Russell P K. Dengue 2 vaccine: dose response in volunteers in relation to yellow fever immune status. J Infect Dis. 1983;148:1055–1060. doi: 10.1093/infdis/148.6.1055. [DOI] [PubMed] [Google Scholar]
- 39.Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H, Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 40.Thein S, Aung M M, Shwe T N, Aye M, Zaw A, Aye K, Aye K M, Aaskov J. Risk factors in dengue shock syndrome. Am J Trop Med Hyg. 1997;56:566–572. doi: 10.4269/ajtmh.1997.56.566. [DOI] [PubMed] [Google Scholar]
- 41.Zeng L, Kurane I, Okamoto Y, Ennis F A, Brinton M A. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J Virol. 1996;70:3108–3117. doi: 10.1128/jvi.70.5.3108-3117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]