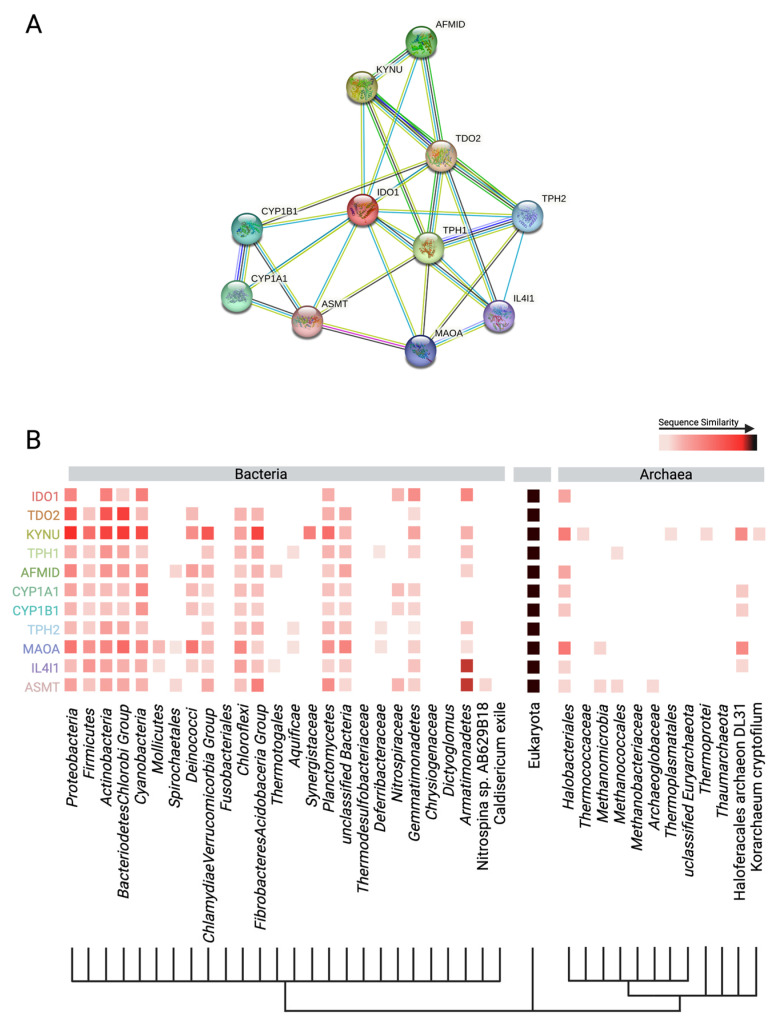
Figure 4.
STRING database output from the search for Escherichia coli K12 MG1655 indole amine 2,3-dioxygenase 1 (IDO1), one enzyme responsible for the gut conversion of tryptophan to kynurenine. Full protein names are tryptophan 2,3-dioxygenase (TDO2), kynureninase (KYNU), tryptophan hydroxylase (TPH1), kynurenine formamidase (AFMID), human cytochromes P450 1A1 (CYP1A1) P450 1B1 (CYP1B1), tryptophan hydroxylase 2 (TPH2), monoamine oxidase A (MAOA), IL4-induced gene 1 (IL4I1), and acetylserotonin O-methyltransferase (ASMT). (A) Protein interaction network with IDO1 as the central node. Light blue connections indicate known interaction from curated databases; pink lines indicate the interaction has been experimentally validated. Light green connections indicate the encoding genes are in the same gene neighborhood; red-orange lines indicate gene fusions, and dark blue indicate gene co-occurrence. Black lines indicate co-expression and light blue indicate protein homology. (B) Gene co-occurrence across Bacteria, Eukarya, and Archaea of IDO1- encoding genes and related protein-encoding genes. Color indicates highest sequence similarity within the taxa, with darker reds and black indicating higher similarity.