Abstract
Among the nucleopolyhedroviruses (Baculoviridae), the occlusion-derived virus (ODV), which initiates infection in host insects, may contain only a single nucleocapsid per virion (the SNPVs) or one to many nucleocapsids per virion (the MNPVs), but the significance of this difference is unclear. To gain insight into the biological relevance of these different packaging strategies, we compared pathogenesis induced by ODV fractions enriched for multiple nucleocapsids (ODV-M) or single nucleocapsids (ODV-S) of Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) containing a β-galactosidase reporter gene. In time course experiments wherein newly molted fourth-instar Trichoplusia ni were challenged with doses of ODV-S or ODV-M that yielded the same final mortality (∼70%), we characterized viral foci as either being restricted to the midgut or involving tracheal cells (the secondary target tissue, indicative of systemic infection). We found that while the timing of primary infection by ODV-S and ODV-M was similar, ODV-S established significantly more primary midgut cell foci than ODV-M, but ODV-M infected tracheal cells at twice the rate of ODV-S. The more efficient establishment of tracheal infections by ODV-M decreased the probability that infections were lost by midgut cell sloughing, explaining why higher numbers of primary infections established by ODV-S within larvae were needed to achieve the same final mortality. These results showed that the multiple nucleocapsid packaging strategy of AcMNPV accelerates the onset of irreversible systemic infections and may indicate why MNPVs have wider individual host ranges than SNPVs.
Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the type species of the Nucleopolyhedrovirus genus of the family Baculoviridae. Baculoviruses are arthropod specific and generally produce fatal infections in their hosts. These pathogens are characterized by rod-shaped, enveloped nucleocapsids that contain supercoiled, circular, double-stranded DNA. The infection cycle of AcMNPV and many other baculoviruses includes two distinct phenotypes, an occluded form (occlusion-derived virus [ODV]) and a budded form (budded virus [BV]). ODV is embedded within a crystalline protein structure (the occlusion) which helps protect the infectious virions from environmental degradation. In the nucleopolyhedroviruses, ODV is composed of virions containing either a single nucleocapsid (SNPVs) or one to many nucleocapsids (MNPVs).
The ODV and BV phenotypes of AcMNPV play unique roles in the viral life cycle; ODV spreads infection among individual hosts, whereas BV transmits infection from cell to cell within a host. In larval lepidopterans, the hosts of AcMNPV, ODV is released after ingestion by the highly alkaline digestive juices of the midgut, which dissolve the occlusions (11). Infections are initiated when the ODV envelope fuses with the plasma membrane of a microvillus on a columnar epithelial cell that is exposed to the midgut lumen (12). The nucleocapsids subsequently enter the cell and move through the cytoplasm to the nucleus, which they enter via nuclear pores (11, 15, 25). During viral replication, BV buds from the basal plasma membrane as a single nucleocapsid that gains a loose-fitting envelope in the process (26, 27). The BV envelope contains spikes of gp64, a virally encoded protein that is essential for the infection to progress beyond the midgut in Trichoplusia ni (16). BV spreads infection among the host tissues until nearly the entire insect is infected, and in the final stages of pathogenesis, the host dies and liquefies in a dramatic “meltdown,” which releases millions of occlusions that can initiate infections when ingested by new hosts.
While a few NPVs are known to infect decapod crustaceans (3), the majority of described species have been isolated from insects, mostly Lepidoptera (28). The SNPVs also have been isolated from members of the Diptera, Hymenoptera, and Trichoptera, while the MNPVs have been isolated only from members of the Lepidoptera, the youngest of the insect orders (17). The restriction of the MNPVs to the Lepidoptera suggests that these viruses originated from the SNPVs and then radiated along with their hosts through evolutionary time (17). In this context, it is intriguing that the MNPVs have evolved the life history trait of packaging multiple nucleocapsids within a single envelope. Such a strategy might seem inefficient in vivo because fatal infections can be initiated within primary target cells of the host midgut epithelium by virions containing single nucleocapsids, as evidenced by the SNPVs. In addition to apparently being wasteful of infectious units, the multiple-nucleocapsid packaging strategy results in larger virions, which could pose physical constraints on movement through the peritrophic membrane of the midgut and/or the narrow microvilli of primary target cells (27). Why, then, would natural selection favor a redundant delivery system in which multiple nucleocapsids infect the same cell?
At least two hypotheses have been proposed to explain the multiple-nucleocapsid-per-virion packaging strategy of the MNPVs. First, it is well documented that viral occlusions rapidly lose viability when exposed to sunlight, presumably because of UV radiation damage to the viral DNA (8). By infecting primary target cells with multiple copies of the viral genome, it is possible that gene complementation compensates for damaged DNA and facilitates productive infections (17). The second hypothesis postulates that infection of primary target cells with multiple copies of the viral genome increases infection efficiency in vivo by accelerating the onset of secondary infection (1, 27). In this scenario, when multiple nucleocapsids from a single ODV particle infect the same cell, some of the nucleocapsids enter the nucleus and others remain in the cytoplasm. The viral genome(s) within the nucleus initiates gene expression and replication, whereas nucleocapsids that do not penetrate the nucleus are repackaged as BV and directly infect secondary target cells before viral replication is completed within the primary target.
While these two hypotheses are not mutually exclusive, several features of the baculovirus life history, as well as empirical evidence from studies of AcMNPV pathogenesis in vivo, are consistent with the second scenario. First, gp64 is unusual among viral structural proteins in that its synthesis is regulated by both an early and a late promoter (2, 14), allowing this essential glycoprotein to be produced and integrated into the plasma membrane of primary target cells hours before the de novo synthesis of progeny BV. Thus, AcMNPV has a built-in genetic mechanism for repackaging ODV. Second, in time course experiments aimed at elucidating the early events of AcMNPV pathogenesis in Spodoptera exigua, infection of secondary target cells, in this case midgut regenerative cells, was evident before late and very late gene expression had occurred in primary target cells, suggesting that regenerative cell infection had been mediated by repackaged parental nucleocapsids (9). Third, the loss of infected midgut cells is a key mechanism underlying developmental resistance in susceptible hosts such as T. ni and Heliothis virescens (7, 31), implying that sloughing primary target cells may be a common host response to baculovirus infection and one that might function as a powerful selection force for the rapid establishment of systemic infections.
To investigate the biological significance of the multiple nucleocapsid packaging of MNPVs in vivo, we isolated ODV fractions of AcMNPV carrying a β-galactosidase reporter gene (AcMNPV-hsp70/lacZ) enriched for virions containing either single (ODV-S) or multiple (ODV-M) nucleocapsids. We used these two inocula in time course experiments to compare the early events of viral pathogenesis, particularly the temporal onset of systemic infections, in fourth-instar T. ni. Our hypothesis was that if the nucleocapsids of ODV-M actually are repackaged as BV, insects challenged with this inoculum should exhibit increased rates of systemic infection than those challenged with ODV-S. The results demonstrated clearly that infections initiated by ODV-M moved more rapidly into secondary target cells of the tracheal epidermis of T. ni than did infections initiated by ODV-S. In addition, comparable mortality levels were achieved by ODV-M with far fewer primary midgut foci than those generated by ODV-S, implying that ODV-M was more efficient at establishing systemic infections.
MATERIALS AND METHODS
Virus preparation and assessment.
Occlusions of AcMNPV-hsp70/lacZ (5) used in this study were isolated from liquefied cadavers of T. ni and partially purified by centrifugation on sucrose gradients (22). ODV was subsequently liberated by exposure of occlusions to dilute alkaline saline (0.1 M Na2CO3, 0.1 M NaCl) and then banded by density equilibrium centrifugation on 25 to 59% sucrose gradients. The pale gray-blue bands of virus were collected individually with a 5-ml syringe and a 22-gauge needle. Samples from each fraction were negatively stained with 2% uranyl acetate and viewed by transmission electron microscopy (TEM) (JEOL model 100 CX). Fractions enriched with virions containing single nucleocapsids were identified and pooled (ODV-S), as were fractions enriched with virions containing multiple nucleocapsids (ODV-M). We examined over 500 negatively stained virions from both the ODV-S and ODV-M stock solutions by TEM to determine the distribution of nucleocapsids per virion. These stock solutions, aliquoted into Eppendorf tubes and stored at −80°C, were diluted to the appropriate concentration with Grace’s medium (JRH) plus 1% fetal bovine serum (Gemini Bio-products, Inc.) prior to use.
Viral protein and genome quantification.
Viral protein concentrations were estimated by the Bradford protein assay (Bio-Rad). Genomic copy numbers were estimated by Southern hybridization; known concentrations of pAcEcoRI-I DNA (4) and various dilutions of viral DNA were hybridized to a radiolabeled probe, and the signals were compared. The plasmid DNA was used to generate a standard curve because its concentration could be accurately determined by optical density, its size is known, and it contains a unique region of the AcMNPV genome. Equivalent copy numbers of viral genome and plasmid DNA should therefore register the same level of signal, and the plasmid copy number (and, by extrapolation, viral genome copy number) could be calculated for a given concentration of plasmid DNA determined by measuring the optical density at 260 nm. Experimentally, DNA was isolated from a known volume of our stock samples of ODV-S and ODV-M by proteinase K digestion followed by phenol-chloroform extraction. Both viral DNA and plasmid DNA were digested with BamHI, releasing a 927-bp fragment containing the AcMNPV open reading frame 1629 and polyhedrin genes. Dilutions of the digested plasmid and viral DNA were electrophoresed in a 1.2% agarose gel in Tris-acetate-EDTA (TAE) buffer and then transferred to a Nytran membrane. The BamHI fragment excised from pAcEcoRI-I also was used as the probe; for this purpose, it was purified from an agarose gel by using the QIAEX DNA gel extraction kit and then labeled with [α-32P]dATP by random priming. Hybridizing probe was quantified with a PhosphorImager (Molecular Dynamics) and ImageQuant software. Determinations of viral genome content for ODV-S and ODV-M were performed three times each, starting with extraction of viral DNA.
Maintenance of test larvae.
Eggs of T. ni were purchased from Ecogen Inc. (Langhorne, Pa.), and larval diet (Stoneville) was provided by the American Cyanamid Company (Princeton, N.J.). Larvae were reared in groups under constant light at 28 ± 3°C until the end of the third larval instar. Nonfeeding, quiescent third-instar larvae preparing to molt were culled and stored at various temperatures between 4 and 22°C for 4 to 12 h to synchronize developmental rates, making large numbers of test insects of the same age available for experiments (30).
Bioassay and time course experiments.
Virus, in 1.0-μl aliquots, was orally inoculated into the anterior midgut of newly molted (within 15 min of shedding the third-instar cuticle) fourth-instar larvae. The inoculum was delivered via a syringe fitted with a 32-gauge blunt-tip needle that was mounted onto a microapplicator (ISCO or Burkard). After inoculation, the larvae were maintained in a growth chamber at 28 ± 2°C under constant illumination in individual 25-ml plastic cups containing diet. For bioassays, cohorts of 25 or more larvae were inoculated and maintained until death or pupation. For time course experiments, ∼70% lethal doses (LD70) of ODV-S and ODV-M (determined from the bioassays) were inoculated into three discrete cohorts of larvae over a 3-week period. For each cohort, separate internal controls (bioassays) were used to confirm that mortality levels were consistent. In total, between 28 and 34 larvae were sacrificed at each of seven time points from 2 to 22 h postinoculation (p.i.). The intact midgut and associated tissues were removed from each larva and processed for lacZ elucidation as previously described (5, 30). The preparations were examined for lacZ expression by stereo (magnification, ×10 to ×50) and/or compound (magnification, ×100 to ×480) microscopy to quantify foci of infection and infected cell types (31). In these studies, several critical benchmarks of infection were established, including the timing and identity of the first host cells expressing lacZ, the onset of systemic infections in the host tracheal system, and the time point at which the proportion of lacZ-expressing insects was predictive of the mortality level obtained in bioassays. The analysis of AcMNPV pathogenesis presented here is based upon the distribution patterns of the reporter gene product among tissues from 199 and 219 insects challenged with the ODV-S and ODV-M inocula, respectively.
RESULTS
Characterization of ODV inocula.
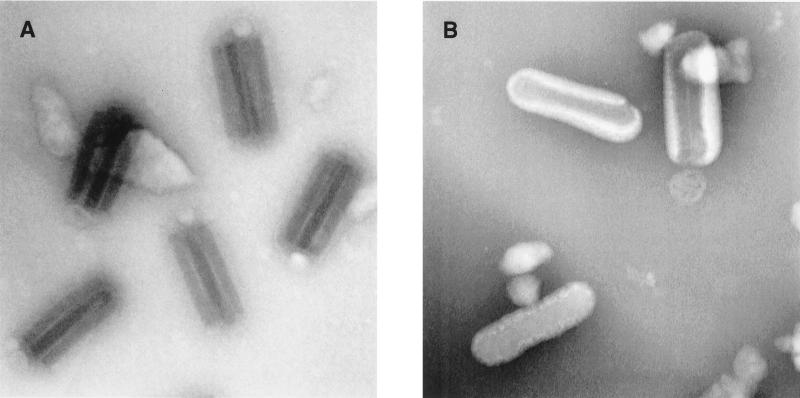
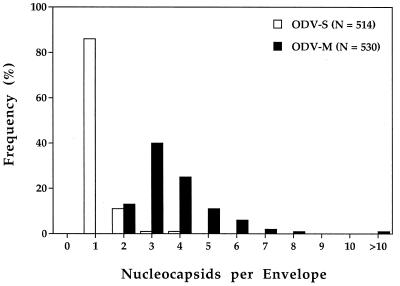
Examination of the ODV-S and ODV-M stocks by TEM confirmed that the ODV-S stock consisted primarily of virions with single nucleocapsids and that the ODV-M stock consisted exclusively of virions containing two or more nucleocapsids (Fig. 1). Evaluation of 530 virions from the ODV-M pool revealed that 78% contained two to four nucleocapsids per envelope and the balance contained five or more (Fig. 2). In contrast, 86% of the 514 virions evaluated from the ODV-S pool contained virions with only one nucleocapsid while the remaining 14% consisted of virions with two to four nucleocapsids (Fig. 1 and 2). While 14% contamination of the ODV-S stock with ODV-M was not ideal, we reasoned that it should not prohibit the detection of major differences in pathogenesis induced by the two virus populations.
FIG. 1.
Electron micrographs of AcMNPV-hsp70/lacZ ODV-M (A) and ODV-S (B) inocula used in this study.
FIG. 2.
Frequency distribution of the number of nucleocapsids per envelope of the AcMNPV-hsp70/lacZ ODV-S and ODV-M inocula used in this study. The frequencies were determined from counts of nucleocapsids within ODV particles from randomly chosen fields viewed by TEM.
After initial screening for activity of various dilutions of the ODV-S and ODV-M inocula in vivo, we selected one dilution of each that yielded approximately 70% mortality. We subsequently conducted four paired bioassay comparisons of these dilutions, resulting in mortalities of 72% ± 13% and 67% ± 12% (mean ± 1 standard deviation) for ODV-S and ODV-M, respectively; because the average mortalities were not significantly different (one-way analysis of variance of arcsine-transformed data; P = 0.57), we selected these dilutions for use in subsequent time course experiments.
The ODV-S and ODV-M inocula were compared with regard to protein content, genome copy number, and number of virions (based on the distribution of nucleocapsids per virion determined by TEM [above]) (Table 1). We found that the ODV-S inoculum contained 8-fold more protein than the ODV-M and 39% more genomic copies but that the ODV-S genomes were packaged in an estimated 2,070 virions compared to only 463 for ODV-M, a 4.5-fold difference. These findings suggested that the primary physical differences between the biologically equivalent dosages of the two inocula were manifested in protein content and in the number of infectious particles, not in the number of genomic copies.
TABLE 1.
Characterization of ODV-S and ODV-M inoculaa
| Inoculum | Mortality in T. ni (%) (n = 4) | Viral protein content (pg) (n = 3) | No. of genomic copies (n = 3) | No. of virions (n = 3) |
|---|---|---|---|---|
| ODV-S | 72 ± 13 | 64.7 ± 2.3 | 2,380 ± 682 | 2,070 ± 593 |
| ODV-M | 67 ± 12 | 8.0 ± 0.5 | 1,710 ± 300 | 463 ± 80 |
Results are given as mean ± 1 standard deviation.
Time course experiment.
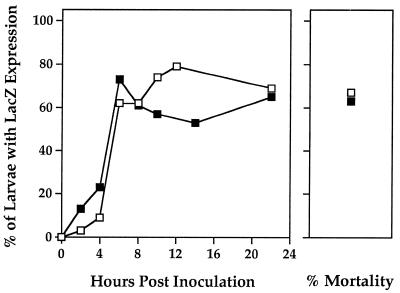
We found that the proportions of lacZ-positive T. ni larvae sampled at various time points following challenge with ODV-S and ODV-M were very similar (Fig. 3) and that primary infection of larval cohorts from both treatments occurred at comparable rates. lacZ expression was first detected at 2 h p.i. in 3 and 13% of the larvae in the ODV-S- and ODV-M-inoculated cohorts, respectively. In a separate test, no lacZ expression was detected in larvae sampled at 1 h after infection with either ODV preparation administered at dosages approximately 10 times an LD100 (data not shown). For both ODV populations, the proportions of infected larvae increased rapidly during the 4 h after first signaling (2 to 6 h p.i.) and, by 6 h p.i., were predictive of the average final mortalities in the companion bioassays (Fig. 3; ODV-S: 62% at 6 h p.i., LD67; ODV-M: 73% at 6 h p.i., LD63). Between 8 and 22 h p.i., the proportions of lacZ-positive larvae fluctuated but remained within ±12% of the final mortalities for ODV-S- and ODV-M-inoculated insects (Fig. 3).
FIG. 3.
Proportions of lacZ-positive T. ni larvae at various times after infection with AcMNPV-hsp70/lacZ ODV-S (open squares) or ODV-M (solid squares). Between 28 and 34 larvae were analyzed for each time point, with the exception of ODV-S at 8 h p.i., for which the sample size was 21. The mean percent mortalities from three bioassays for each inoculum conducted concurrently are shown.
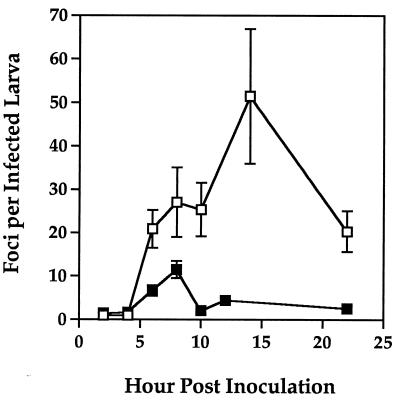
Our analysis of the number and cellular composition of viral foci and the temporal onset of systemic infections, however, revealed striking differences in the pathogenesis of ODV-S and ODV-M in T. ni. At all time points after 4 h p.i., there were more foci per infected larva in cohorts inoculated with ODV-S than in cohorts inoculated with ODV-M (Fig. 4). Among cohorts sampled at five time points between 6 and 22 h p.i., the ratio of foci between virus treatments varied from 2.4 (8 h p.i.) to 12.3 (10 h p.i.), showing that ODV-S established many more primary foci than did ODV-M, even though the final mortality rates were comparable. The maximum numbers of foci were observed at 8 and 16 h p.i. for ODV-M and ODV-S, respectively, and the subsequent decline in average numbers of foci within larvae from both virus treatments suggested that infected primary target cells were sloughed during the fourth instar between 8 and 22 h p.i. (Fig. 4), before the onset of quiescence preceding the molt to the fifth instar. Under our rearing conditions, larvae first initiated premolt quiescence 24 h into the fourth instar and molted to the fifth instar between 32 and 48 h.
FIG. 4.
Number of foci per infected T. ni larva at various hours after inoculation with AcMNPV-hsp70/lacZ ODV-S (open squares) or ODV-M (solid squares). Between 28 and 34 larvae were analyzed for each time point, with the exception of ODV-S at 8 h p.i., for which the sample size was 21. Bars indicate 1 standard error.
At 2 h p.i., all viral foci (n = 5) consisted of single cells, one differentiating and four mature columnar cells in the midgut epithelium. In both ODV-S- and ODV-M-inoculated larvae, multicellular foci were first observed at 6 h p.i. but at significantly different frequencies; 28% of the ODV-M foci (41 of 146) were multicellular, compared to only 0.5% of ODV-S foci (2 of 376) (n = 552 foci; χ2 = 108.6, df = 1, P < 0.001). All multicellular foci from 6 h p.i. onward included lacZ-positive cells within the tracheal system, indicating the establishment of systemic infection. Without exception, the secondary targets for both viruses were tracheal epidermal cells on the distal side of the basal lamina supporting the midgut; these secondary targets were always immediately adjacent to lacZ-positive midgut cells within foci still containing primary target cells. We did not detect any multicellular foci that were restricted to the midgut epithelium in this study.
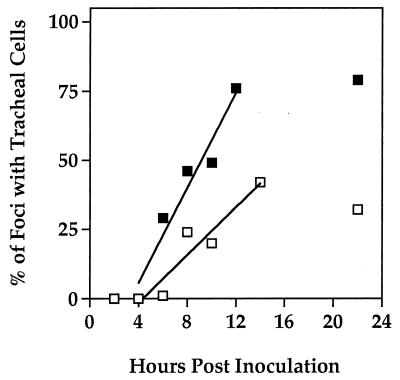
For each virus, the increase in the frequency of viral plaques with one or more infected tracheal cells reflected the temporal production of BV by infected midgut cells and the rate at which systemic infection was established (Fig. 5). These proportions increased rapidly and linearly between 6 and 14 h p.i.; at 22 h p.i., however, the percentage of foci containing tracheal cells was similar to the 12- and 14-h p.i. values for ODV-M and ODV-S, respectively. The slopes of the lines reflecting the rate of establishing systemic infection from 6 to 14 h p.i. were 8.60 for ODV-M and 4.31 for ODV-S, indicating that ODV-M BV established systemic infection twice as fast as ODV-S BV did (Fig. 5).
FIG. 5.
Percentages of foci containing one or more tracheal cells from T. ni inoculated with AcMNPV-hsp70/lacZ ODV-S (open squares) or ODV-M (solid squares) and sampled at various time points after inoculation. A total of 343 ODV-M and 2,910 ODV-S foci were used to calculate the frequencies of tracheal infections. The two lines were constructed by least-squares regression (ODV-S: y = 4.31x −18.81, r2 = 0.90; ODV-M, y = 8.60x − 28.81, r2 = 0.94) (see the text for discussion).
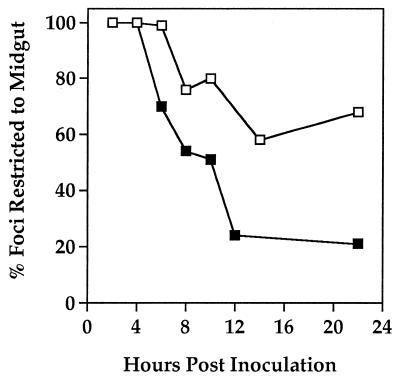
The proportions of foci restricted to the tracheal epidermis increased for both viruses throughout the fourth instar (data not shown). At 22 h p.i., for example, 25% of the ODV-M foci and 6% of the ODV-S foci consisted solely of tracheal cells, providing further evidence that underlying primary target cells were sloughed during the feeding stage of the fourth instar. On the other hand, within infected larvae from the cohorts sampled at 22 h p.i., 21% of the ODV-M foci and 68% of the ODV-S foci were still restricted to the midgut, suggesting that the inocula of both viruses continued to establish primary infection throughout the fourth instar. Because these cohorts were sampled within 2 h of onset of the premolt stage (during which time the larvae shed all infected midgut cells [30]), these values also were indicative of the frequencies with which primary infection failed to establish systemic infection. It can be estimated, therefore, that ODV-S primary infections were lost at the molt at threefold the frequency of ODV-M primary infections.
Our analyses of viral foci revealed that the greater number of primary targets infected by ODV-S compensated for the lower rate of infection of the host tracheal system by BV (Fig. 6). This temporal lag in the progression of ODV-S-generated infection from primary to secondary targets was also reflected by the relative frequencies of larvae with tracheal infections at 6 h p.i., the time point when systemic infections were first observed in larvae from both virus treatments (Fig. 5); 73% of larvae infected by ODV-M supported one or more tracheal infections at 6 h p.i. compared to only 11% of larvae infected by ODV-S (n = 40 larvae; χ2 = 15.19, df = 1, P < 0.01). At subsequent time points, however, we found no significant differences between viral treatments in the proportions of larvae with tracheal infections.
FIG. 6.
Percentages of T. ni larvae with one or more lacZ-positive tracheal cells at various time points after inoculation with ODV-S (open squares) or ODV-M (solid squares). Between 28 and 34 larvae were analyzed for each time point, with the exception of ODV-S at 8 h p.i., for which the sample size was 21.
DISCUSSION
Results from our study provide compelling evidence that multiple nucleocapsid packaging of AcMNPV ODV had little effect on primary midgut infection but significantly facilitated secondary infection of tracheal epidermal cells in fourth-instar T. ni. These results are consistent with those of van Beek et al. (24), who found that an AcMNPV ODV fraction containing seven to nine nucleocapsids killed neonatal T. ni larvae significantly faster than an ODV fraction consisting of virions with single nucleocapsids. In our study, to realize mortality comparable to ODV-M in a cohort of T. ni, 4.5 times as many ODV-S particles had to be inoculated (Table 1). This difference in virion content between the ODV-S and ODV-M inocula was reflected by the increased number of primary foci per ODV-S-infected insect observed in time course experiments (Fig. 4). We saw no delay in the onset of lacZ expression in midgut cells infected by ODV-M relative to ODV-S; the larger physical size of ODV-M, therefore, apparently was not a significant impediment to viral passage from the midgut lumen to within the nuclei of primary target cells (27). The frequencies of foci with tracheal involvement at 6 h p.i. and later time points (Fig. 5) indicated that BV emitted from ODV-M-infected midgut cells established systemic infections twice as fast as did BV emitted from ODV-S-infected midgut cells. Moreover, at the last time point sampled before to the molt (22 h p.i.), threefold more ODV-S foci were restricted to the midgut. These data suggested that ODV-S foci probably incurred a much higher failure rate in establishing systemic infections because all infected midgut cells are sloughed before ecdysis to the fifth instar (7, 30). In addition, the decrease in the average number of foci per infected larva and the increase in the frequency of foci consisting exclusively of tracheal cells for both inocula throughout the time course demonstrated that sloughing of infected cells also occurred during the feeding stage of the fourth instar. It is highly unlikely that tracheal cells themselves were primary targets of infection, because these cells do not have direct access to the midgut lumen. Furthermore, ODV envelopes lack gp64, which is essential for infection to be transmitted beyond the midgut in this species (16). While midgut cell sloughing should reduce the efficiency of establishing systemic infections for both ODV-S and ODV-M, the reduction would be proportionately greater for ODV-S owing to its slower movement into the tracheal system.
In 1981, Granados and Lawler (12) published evidence suggesting that parental nucleocapsids of AcMNPV could bypass the columnar cell nucleus and bud directly through the peplomer-studded, basal plasma membrane and thereby establish systemic infection rapidly in T. ni. Subsequently, studies in which reporter gene recombinants of AcMNPV were used to track virus movement among host tissues revealed lags of less than 6 h between primary and secondary infection in larvae of T. ni (5), H. virescens (30, 31), Helicoverpa zea, Manduca sexta (31a), and S. exigua (9) orally infected with viral occlusions or ODV. While we do not know the time required for AcMNPV replication in T. ni midgut cells, the BV produced by T. ni-derived TN-368 cells inoculated with AcMNPV BV at a multiplicity of infection of 20 is not detected until after 10 h p.i. (29). In contrast, ODV-M-initiated infections in our study exhibited only a 4-h lag between the onset of lacZ expression in primary (2 h p.i.) and secondary (6 h p.i.) target cells. We also observed this to occur in 2 of 376 foci (0.5%) in ODV-S-inoculated insects, a finding that was inconsistent with our hypothesis. The most reasonable explanation for this observation, however, is the 14% contamination of the ODV-S inoculum with ODV-M (Fig. 2). Another possible explanation, although highly unlikely, is that the same midgut cell was infected by two or more ODV-S particles. We do not favor this explanation because of the small numbers of lacZ-positive cells in each infected larva (Fig. 4) and the fact that there are over 200,000 potential primary target cells within the midgut epithelium of a single fourth-instar T. ni (6).
Several lines of empirical evidence suggest that sloughing of infected midgut cells is the first line of defense against AcMNPV infection and has therefore been a major force shaping AcMNPV infection strategies. The ability to slough cells with primary infections is the principal mechanism underlying developmental resistance within the fourth instar of highly susceptible hosts such as T. ni and H. virescens (7, 31). Additionally, stilbene optical brighteners (e.g., M2R) that greatly enhance fatal infection by many baculoviruses (both SNPVs and MNPVs) (13, 18–21, 23, 32, 33) improve AcMNPV infectivity specifically by preventing the host from sloughing infected midgut cells (31). Moreover, because M2R also acts as a synergist for pathogens within the Poxviridae and Reoviridae (13, 18, 19), midgut cell sloughing may be a widespread response by lepidopteran larvae to insect viruses in general. Finally, data from this and previous studies show that highly susceptible hosts, such as T. ni and H. virescens, succumb to AcMNPV when BV infects even a single tracheal epidermal cell (see, e.g., references 7 and 30). For AcMNPV, the payoff for circumventing midgut cell sloughing is extremely high, and it is not surprising that this pathogen has evolved complementary traits that facilitate the rapid establishment of irreversible, fatal secondary infections. The packaging of redundant nucleocapsids within a single virion by the MNPVs also may explain why the MNPVs, in general, have broader individual host ranges than the SNPVs (27). However, this strategy would work only if the major surface glycoprotein of the BV phenotype were driven by an early promoter. This character, in fact, has been described for several MNPVs (10).
ACKNOWLEDGMENTS
Financial support for this project was provided by USDA grant NRICG 95-37302-1835 and by Federal HATCH funds.
We thank T. Jong, M. Vu, and S. Wong for technical assistance and The American Cyanamid Company for providing the larval diet used in this study.
REFERENCES
- 1.Blissard G W. Baculovirus-Insect Cell Interactions. Cytotechnology. 1996;20:79–93. doi: 10.1007/BF00350390. [DOI] [PubMed] [Google Scholar]
- 2.Blissard G W, Rohrmann G F. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugatamulticapsid nuclear polyhedrosis virus. Virology. 1989;170:537–555. doi: 10.1016/0042-6822(89)90445-5. [DOI] [PubMed] [Google Scholar]
- 3.Blissard G W, Rohrmann G F. Baculovirus diversity and molecular biology. Annu Rev Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 4.Cochran M A, Carstens E B, Eaton B T, Faulkner P. Molecular cloning and physical mapping of restriction endonuclease fragments of Autographa californicanuclear polyhedrosis virus DNA. J Virol. 1982;41:940–946. doi: 10.1128/jvi.41.3.940-946.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelhard E K, Kam-Morgan L N W, Washburn J O, Volkman L E. The insect tracheal system: a conduit for the systemic spread of Autographa californicaM nuclear polyhedrosis virus. Proc Natl Acad Sci USA. 1994;91:3224–3227. doi: 10.1073/pnas.91.8.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelhard E K, Keddie B A, Volkman L E. Isolation of third, fourth, and fifth instar larval midgut epithelia of the moth, Trichoplusia ni. Tissue Cell. 1991;23:917–928. doi: 10.1016/0040-8166(91)90041-q. [DOI] [PubMed] [Google Scholar]
- 7.Engelhard E K, Volkman L E. Developmental resistance within fourth instar Trichoplusia ni orally inoculated with Autographa californicaM nuclear polyhedrosis virus. Virology. 1995;209:384–389. doi: 10.1006/viro.1995.1270. [DOI] [PubMed] [Google Scholar]
- 8.Evans H F. Ecology and epizootiology of baculoviruses. In: Granados R R, Federici B A, editors. The biology of baculoviruses. II. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 89–132. [Google Scholar]
- 9.Flipsen J T M, Martens J W M, Van Oers M M, Vlak J M, Van Lent J W M. Passage of Autographa californica nuclear polyhedrosis virus through the midgut epithelium of Spodoptera exigualarvae. Virology. 1995;208:328–335. doi: 10.1006/viro.1995.1156. [DOI] [PubMed] [Google Scholar]
- 10.Funk C J, Braunagel S C, Rohrmann G F. Baculovirus structure. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 7–32. [Google Scholar]
- 11.Granados R R. Early events in the infection of Heliothis zeamidgut cells by a baculovirus. Virology. 1978;90:170–174. doi: 10.1016/0042-6822(78)90347-1. [DOI] [PubMed] [Google Scholar]
- 12.Granados R R, Lawler K A. In vivo pathway of Autographa californicabaculovirus invasion and infection. Virology. 1981;108:297–308. doi: 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- 13.Hamm J J, Shapiro M. Polyhedrovirus enhanced by a fluorescent brightener. J Econ Entomol. 1992;85:2149–2152. [Google Scholar]
- 14.Jarvis D L, Garcia A., Jr Biosynthesis and processing of the Autographa californicanuclear polyhedrosis virus gp64 protein. Virology. 1994;205:300–313. doi: 10.1006/viro.1994.1646. [DOI] [PubMed] [Google Scholar]
- 15.Knudson D L, Harrap K A. Replication of a nuclear polyhedrosis virus in a continuous cell culture of Spodoptera frugiperda: microscopy study of the sequence of events of the virus infection. J Virol. 1976;17:254–268. doi: 10.1128/jvi.17.1.254-268.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monsma S A, Oomens A G P, Blissard G W. The gp64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohrmann G F. Evolution of occluded baculoviruses. In: Granados R R, Federici B A, editors. The biology of baculoviruses. I. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 203–215. [Google Scholar]
- 18.Shapiro M. Use of optical brighteners as radiation protectants for gypsy moth (Lepidoptera: Lymantriidae) nuclear polyhedrosis virus. J Econ Entomol. 1992;85:1682–1686. [Google Scholar]
- 19.Shapiro M, Dougherty E M. Enhancement in activity of homologous and heterologous viruses against the gypsy moth (Lepidoptera: Lymantriidae) by an optical brightener. J Econ Entomol. 1994;87:361–365. [Google Scholar]
- 20.Shapiro M, Robertson J L. Enhancement of gypsy moth (Lepidoptera: Lymantriidae) baculovirus activity by optical brighteners. J Econ Entomol. 1992;85:1120–1124. [Google Scholar]
- 21.Shapiro M, Vaughn J L. Enhancement in activity of homologous and heterologous baculoviruses infectious to cotton bollworm (Lepidoptera: Noctuidae) by an optical brightener. J Econ Entomol. 1995;88:265–269. doi: 10.1603/0022-0493-93.3.572. [DOI] [PubMed] [Google Scholar]
- 22.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agricultural Experiment Station bulletin 1555. College Station: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 23.Vail P V, Hoffmann D F, Tebbets J S. Effects of a fluorescent brightener on the activity of Anagrapha falcifera(Lepidoptera: Noctuidae) nuclear polyhedrosis virus to four noctuid pests. Biol Control. 1996;7:121–125. [Google Scholar]
- 24.van Beek N A M, Wood H A, Hughes P R. The numbers of nucleocapsids of enveloped Autographa californica nuclear polyhedrosis virus particles affects the survival time of neonate Trichoplusia nilarvae. J Invertebr Pathol. 1988;52:185–186. [Google Scholar]
- 25.Vaughn J L, Dougherty E M. The replication of baculoviruses. In: Maramorosch K, Sherman K E, editors. Viral insecticides for biological control. New York, N.Y: Academic Press, Inc.; 1985. pp. 569–633. [Google Scholar]
- 26.Volkman L E. The gp64 envelope protein of budded Autographa californicanuclear polyhedrosis virus. Curr Top Microbiol Immunol. 1986;131:103–118. doi: 10.1007/978-3-642-71589-1_6. [DOI] [PubMed] [Google Scholar]
- 27.Volkman L E. Nucleopolyhedrovirus interactions with their insect hosts. Adv Virus Res. 1997;48:313–348. doi: 10.1016/s0065-3527(08)60291-2. [DOI] [PubMed] [Google Scholar]
- 28.Volkman L E, Blissard G W, Friesen P, Keddie B A, Possee R, Theilmann D. Family Baculoviridae. In: Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Taxonomy. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. pp. 104–113. [Google Scholar]
- 29.Volkman L E, Summers M S, Hsieh C H. Occluded and nonoccluded nuclear polyhedrosis virus grown in Trichoplusia ni: comparative neutralization, comparative infectivity, and in vitro growth studies. J Virol. 1976;19:820–832. doi: 10.1128/jvi.19.3.820-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washburn J O, Kirkpatrick B A, Volkman L E. Comparative pathogenesis of Autographa californica M nuclear polyhedrosis virus in larvae of Trichoplusia ni and Heliothis virescens. Virology. 1995;209:561–568. doi: 10.1006/viro.1995.1288. [DOI] [PubMed] [Google Scholar]
- 31.Washburn J O, Kirkpatrick B A, Haas-Stapleton E, Volkman L E. M2R enhances Autographa californica M nucleopolyhedrovirus infection of Trichoplusia ni and Heliothis virescensby preventing sloughing of infected midgut epithelial cells. Biol Control. 1998;11:58–69. [Google Scholar]
- 31a.Washburn, J. O., and L. E. Volkman. Unpublished data.
- 32.Zou Y, Young S Y. Enhancement of nuclear polyhedrosis virus activity in larval pests of lepidoptera by a stilbene fluorescent brightener. J Entomol Sci. 1994;29:130–133. [Google Scholar]
- 33.Zou Y, Young S Y. Use of a fluorescent brightener to improve Pseudoplusia includens(Lepidoptera: Noctuidae) nuclear polyhedrosis virus activity in the laboratory and field. J Econ Entomol. 1996;89:92–96. [Google Scholar]