Abstract
Herpes simplex virus type 1 immediate-early protein Vmw110 stimulates the onset of virus infection and is required for efficient reactivation from latency. In transfection assays, Vmw110 is a potent activator of gene expression, but its mode of action has yet to be determined. Previous work has shown that Vmw110 localizes to specific intranuclear structures known as ND10, PML bodies, or PODs and causes the disruption of these domains. The ability of Vmw110 to disrupt ND10 correlates with its biological activities in infected and transfected cells. It has also been found that Vmw110 binds strongly and specifically to a ubiquitin-specific protease known as HAUSP, itself a component of a subset of ND10. In this study we have investigated the role of HAUSP in Vmw110 activity; single amino acid residues of Vmw110 required for the interaction were identified, and the effects of mutation of these residues in infected and transfected cells were then assayed. The results indicate that the ability to bind to HAUSP contributes to the functional activities of Vmw110.
Herpes simplex virus type 1 (HSV-1) immediate-early (IE) protein Vmw110 (also known as ICP0) is an important regulator of viral gene expression which augments expression from transfected reporter genes and stimulates the onset of the viral lytic cycle (reviewed in reference 9). Failure to express functional Vmw110 decreases the probability that the virus will enter the lytic cycle after infection at low multiplicity of infection in cell culture (2, 8, 34). The genomes which fail to initiate lytic gene expression attain a quiescent state with similarities to latency, and provision of exogenous Vmw110 leads to their reactivation (18, 33). Consistent with the results obtained with cultured cells, Vmw110-deficient viruses reactivate poorly in mouse latency models (3, 24), and these data have led to the idea that Vmw110 has a role in determining whether the virus enters the lytic cycle or establishes a latent infection.
The mechanisms that underlie the functions of Vmw110 have not been clearly defined, although a recent study of Vmw110-activated gene expression ruled out posttranscriptional regulation (20). Mutagenesis experiments have identified several functional regions of Vmw110, including a characteristic zinc binding domain called a RING finger that lies in the N-terminal third of the protein, a nuclear localization signal, and sequences in the C-terminal 180 residues (reviewed in reference 9). Recent studies have explored the intermolecular interactions of Vmw110, and several candidate functional interactions have been identified. Two studies have suggested that Vmw110 might interact with Vmw175 (ICP4), the major transcriptional regulator expressed by HSV-1, although direct proof that such an interaction occurs in infected cells remains to be established (31, 36). Other studies have concentrated on the possible interactions between Vmw110 and cellular proteins and structures. It is now firmly established that at the earliest stages of infection Vmw110 migrates to specific nuclear structures termed ND10, PODs, or PML nuclear bodies, and in consequence the ND10 are disrupted (11, 26, 27). The ability of Vmw110 to interact normally with and disrupt ND10 appears to be functionally significant, since mutations that affect the biological activities of Vmw110 also affect its interactions with ND10 (11, 27). The initial observation that ND10 constitute a preferred site for the localization of parental viral genomes and the subsequent development of replication compartments (19, 28) has now been confirmed by other laboratories (25, 35).
At the molecular level, Vmw110 interactions with the translation factor elongation factor (EF) 1δ (21) and cyclin D3 have been observed in vitro and in the yeast two-hybrid system (22). The interaction that we have concentrated on in this laboratory is that between sequences in the C-terminal region of Vmw110 and a novel ubiquitin-specific protease named HAUSP (13, 29, 30). The binding of Vmw110 to HAUSP is both strong and specific in vitro and is readily observable by coimmune precipitation of the complex from infected cell extracts. An initial observation which suggested that the Vmw110-HAUSP interaction was functionally significant was the localization of the latter in a subset of ND10 in uninfected cells (13). Ubiquitin-specific protease (USP) enzymes are likely to have a role in the control of protein stability since they cleave ubiquitin adducts from substrate proteins, thereby protecting the substrate from proteasome-mediated degradation. In principle, Vmw110 could be inhibiting, activating or redirecting the natural enzymatic activity of HAUSP, and there are several examples of regulation of gene expression mediated by modulation of the stability of components of regulatory pathways (see reference 13 for further discussion). The association between Vmw110 and HAUSP has recently become particularly intriguing because of the finding that a major biochemical function of Vmw110 is indeed the control of the stability of a number of cellular proteins. For example, at early times during infection Vmw110 induces the degradation of certain isoforms of the major ND10 proteins PML (14) and sp100 (15), events which correlate with the disruption of ND10 (14). The induced degradation can be inhibited by inactivation of the ubiquitin-proteasome pathway (14), and under these conditions infection becomes stalled at the IE stage in a way that mirrors the requirement for Vmw110 (17). However, the ND10 proteins are not the sole targets for Vmw110-induced proteolysis since the inner centromere protein CENP-C (16) and the catalytic subunit of DNA-PK (23, 32) are also lost from the cell in a Vmw110-dependent manner.
The fact that Vmw110 binds to a ubiquitin-specific protease and induces the degradation of certain specific proteins is a compelling indication that the interaction with HAUSP is functionally significant. However, analysis of the importance of the interaction has hitherto been complicated by the complexity of the C-terminal region of Vmw110. For example, although the phenotypes of Vmw110 deletion mutants with lesions in the HAUSP binding region are consistent with a role for the interaction in virus infection (7, 30), the available deletions encroach closely on sequences required for migration of Vmw110 to ND10 and for self-multimerization (reference 30 and references therein). We wished to analyze the functional requirements for HAUSP binding by Vmw110 as rigorously as possible by conducting a fine-structure analysis of the Vmw110 sequences required. A number of mutants with single or double amino acid substitutions of Vmw110 which reduced the interaction to background levels in vitro were identified. The ability of Vmw110 to activate gene expression in transfection assays was significantly diminished by these mutations, and virus mutants carrying these substitutions grew poorly in tissue culture. We conclude that the ability of Vmw110 to bind to HAUSP makes a significant contribution to the ability of Vmw110 to activate gene expression and stimulate virus growth in cultured cells.
MATERIALS AND METHODS
Viruses and cells.
HSV-1 strain 17+ was the wild-type strain used in these studies. Virus dl1403, from which the majority of the Vmw110 coding region has been deleted, was derived from strain 17+ (34). The FXE, E52X, D12, and D14 viruses with defined deletions in Vmw110 have been described previously (8, 30). All viruses were propagated and titrated in baby hamster kidney (BHK) cells grown in Glasgow modified Eagle’s medium containing 100 U of penicillin/ml and 100 μg of streptomycin/ml and supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2.5% fetal calf serum, 2.5% newborn calf serum (NBCS), and antibiotics as described above. HEp-2 cells were grown in DMEM containing 10% fetal calf serum and antibiotics, and Cos7 cells were grown in DMEM containing 10% NBCS and antibiotics.
Construction of viruses with lesions in Vmw110.
The viruses with defined lesions in Vmw110 which were generated for this study included the truncation and deletion mutants E58X, A8X, and A78 and the substitution mutants M1, M2, and M4 (see below and Table 2 for details). Infectious dl1403 DNA was cotransfected with linearized DNA of plasmids carrying the relevant mutations (see below). The progeny plaques were screened by Southern blotting and plaque purified three times before preparation of stocks, all as described previously (8). The presence of the mutations was confirmed by restriction enzyme analysis and Southern blotting of viral DNA propagated from the final stocks (data not shown).
TABLE 2.
Vmw110 mutant viruses used in this study
| Virus strain | Vmw110 residues, deletions, or substitution(s) | HAUSP bindinga | ND10 colocali-zationb | ND10 disruptionc | Multimerd |
|---|---|---|---|---|---|
| 17+ | 1–775 (wild type) | + | + | + | + |
| FXE | Δ106–149 | + | + | − | + |
| E52X | 1–593 | − | − | − | − |
| E58X | 1–632 | − | − | − | (−) |
| A8X | 1–646 | − | − | − | (−) |
| D12 | Δ594–632 | − | + | + | + |
| A78 | Δ592–647 | − | + | + | (+) |
| M1 | R623L/K624I | − | + | + | (+) |
| M2 | R623L | Reduced | + | + | (+) |
| M4 | K620I | − | + | + | (+) |
The ability to bind HAUSP in coimmunoprecipitation experiments from virus-infected cell extracts as shown in Fig. 4. The data for viruses FXE and D12 were taken from references 26 and 27.
The ability of the mutant protein to colocalize with PML in ND10 as shown in Fig. 6. The data for viruses not shown in Fig. 6 were taken from references 11, 24, and 27.
Disruption of ND10 in HEp-2 cells 5 h after the start of infection. See Fig. 6.
Plasmids. (i) GST fusion protein plasmids.
Glutathione S-transferase (GST) fusion proteins were expressed from plasmids derived from the vector pGEX2TN3; the C-terminal region of Vmw110 (residues 594 through 775) was expressed as a GST fusion protein from plasmid pGEXE52 (29). Truncation mutant derivatives of pGEXE52 were constructed as follows. Plasmids pGEXE4 and pGEXE9 contain the EcoRI-SalI coding region fragments of plasmids p110E4 and p110E9 (6) in place of the EcoRI-SalI fragment of pGEXE52, thus fusing residues 615 through 775 and 618 through 775 of Vmw110, respectively, to GST. In these cases a derivative of the pGEX2TN3 vector was used to maintain the reading frame. The integrity of the junction region was confirmed by DNA sequencing. Plasmids pGEXE52PmlI, pGEXE52RsaI, and pGEXE52AvaI are 3′ truncation derivatives of pGEXE52 which contain Vmw110 sequences from codon 594 in pGEXE52 to eponymous restriction sites located after codons 713, 680, and 646, respectively. They were constructed by cloning strategies incorporating multiple fragments of pGEXE52, and in each case the stop codons are located in vector sequences immediately downstream of the relevant restriction site. Plasmids pGEXE23X and pGEXE58X have stop codon oligonucleotides inserted into restriction sites located after codons 638 and 633, respectively. The stop codon linkers, which each contain an XbaI site, were initially inserted into the EcoRI sites in the insertion mutant plasmids p110E23 and p110E58 (6) to give plasmids p110E23X and p110E58X. Primers overlapping codon 594 and the SalI site at the 3′ end of the Vmw110 coding region were used in PCRs with the derivative plasmids. The 5′ primer was designed with additional sequences containing an EcoRI site so as to produce exactly the same sequence at the 5′ end of the fragment as that present at the EcoRI site linking GST to Vmw110 sequences in pGEXE52. In this way, cleavage of the PCR product with EcoRI and SalI produced a fragment which could be used to replace the equivalent fragment of pGEXE52, thereby creating plasmids pGEXE23X and pGEXE58X. The details for these plasmids are summarized in Table 1. The substitution-carrying mutant plasmids of the pGEXM series were transferred from the intermediate p110M series plasmids (see below) by the same PCR protocol as that used to create plasmids pGEXE23X and pGEXE58X. Expression of the expected fusion proteins was confirmed by Coomassie staining and Western blotting with monoclonal antibodies (MAbs) 10503 and 10810, which recognize epitopes in the regions downstream of position 633 and between residues 594 and 633, respectively (10).
TABLE 1.
Characteristics of the GST fusion proteins including C-terminal portions of Vmw110 used in this study
(ii) Plasmids expressing Vmw110 and mutant derivatives in eukaryotic cells.
Plasmid pCI110 was used to express Vmw110; this contains the NcoI-HpaI IE-1 fragment containing the Vmw110 coding region and IE-1 3′ processing signals from plasmid p111 (5) inserted downstream of the human cytomegalovirus (HCMV) promoter-enhancer in vector pCIneo (Promega). Restriction fragments containing the various mutations in the Vmw110 coding region were excised from plasmid p110E58X, plasmids of the p110M series (see below), and plasmids of the previously published p110 series (6), and the mutant fragments were then used to replace the corresponding wild-type fragment in pCI110.
Construction of amino acid substitution mutants.
The MluI-SalI fragment of p111, containing Vmw110 residues 519 through 768, was inserted into M13 mp19, and uridine-rich single-stranded template DNA was isolated from infected CJ236 Escherichia coli (ung dut mutant) in the presence of 100 μg of uridine/ml. Three mutagenesis primers which contained alterations in codons 621 and 622 were synthesized so as to produce a novel FspI site without changing the coding potential. This site was used to monitor transfer of mutagenized fragments. In addition, the three primers were synthesized so as to allow alterations (singly or in both) of the members of the codon pairs 619 and 620, 623 and 624, and 626 and 627 (for details, see Fig. 2). The primers were used to generate double-stranded forms of the template M13 DNA and transfected into the E. coli strain TG1, which detects and preferentially degrades the uracil-containing template strand. Progeny plaques were picked and screened for the presence of mutations by DNA sequencing. Replicative-form DNA from clones with desired mutations was prepared, and their AatII-BstEII fragments (containing Vmw110 codons 553 through 712) were excised and exchanged for the corresponding fragment in plasmid p111 to generate plasmids of the p110M series. Subsequently the mutant fragments were moved into the pCI110 series of Vmw110 expression plasmids (see above). Successful transfer was monitored by detection of the novel FspI site, by DNA sequence analysis of the mutagenized region and, for the mutants M1, M2, and M4, by extensive sequence analysis of the coding region of the C-terminal portion of Vmw110.
FIG. 2.
Mutagenesis of selected charged residues within the minimal HAUSP binding region of Vmw110. The upper three lines show the amino acid (aa) and coding sequences (seq.) of residues 616 through 629 of Vmw110. The next three lines show the relevant changes in the mutagenic oligonucleotides (oligo4, oligo5, and oligo6). Uppercase letters denote base changes which were present invariably, and lowercase letters indicate positions where equal proportions of normal and mutant nucleotide precursors were included during synthesis, so that individual single and double mutants could be isolated in the same mutagenesis experiment. The actual mutagenic oligonucleotides included greater lengths of flanking sequence than shown here, but these regions did not contain any substitutions. The line labelled Replace indicates the expected amino acid substitution if the mutagenic nucleotide change is present and also the presence of the FspI site (FspI) in residues 621 and 622. Below are shown the actual amino acid sequences of mutants that were isolated. The presence of the mutations was detected in the M13 isolates, then confirmed by DNA sequencing after transfer of the mutant fragment to plasmids of the p110 series (see Materials and Methods).
Expression and purification of GST fusion proteins.
Bacteria harboring GST fusion protein expression plasmids were grown from freshly purified colonies in 100-ml cultures of yeast extract tryptone broth. When the optical density at 450 nm (OD450) reached about 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.1 mM to induce expression. After 2 h, cells were harvested by centrifugation and resuspended in 2 ml of phosphate-buffered saline (PBS). The bacteria were lysed with a soniprobe (Branson sonifier 450), and Triton X-100 was then added to a final concentration of 1%. After a 10-min incubation on ice, insoluble debris were removed by centrifugation (Sorvall SS34 rotor) at 9,500 rpm for 5 min. Aliquots (each, 300 μl) of supernatant were mixed with 50 to 100 μl of a 50% slurry of glutathione-agarose beads which had been preswollen for 1 h and washed in PBS. The beads were incubated in the extract at 4°C for 1 h with continuous mixing, harvested by brief centrifugation, washed three times in PBS, and stored on ice as a 50% slurry. In all GST pull-down experiments, the amounts of the fusion proteins bound to the beads were estimated by Coomassie staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel loaded with small samples of the bead preparations. Subsequent binding experiments used amounts of beads normalized for the amount of each fusion protein and maintenance of the total quantity of glutathione-beads by the addition of uncharged beads where necessary.
Analysis of cellular proteins bound to GST fusion proteins.
Freshly prepared glutathione-agarose beads with bound and normalized GST fusion proteins (30 μl of a 50% slurry) were mixed with 300 μl of labelled cell protein extract to which NaCl had been added to a final concentration of 0.5 M. Initially, all extracts were incubated for 1 h at 4°C with continuous mixing with beads linked to the GST protein expressed by vector plasmid pGEX2TN3 in order to reduce background. After removal of these beads by centrifugation, the precleared extracts were incubated with 30 μl of a 50% slurry of the appropriate GST fusion protein beads and a negative GST bead control, again for 1 h at 4°C with continuous mixing. The beads were harvested by brief centrifugation, then washed three times in a buffer containing 50 mM Tris-HCl (pH 8.0), 0.5 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40 (NP-40). Protease inhibitors phenylmethylsulfonyl fluoride, bestatin, and leupeptin were used in the wash buffer at concentrations of 1 mM, 40 μg/ml, and 0.5 μg/ml, respectively. Protein complexes were eluted from the beads with sequential washes (each, 20 μl) of 50 mM reduced glutathione in 0.25 M Tris-HCl (pH 7.0). Elution was at 25°C for 15 min. The supernatants containing fusion protein complexes were mixed with SDS-acrylamide gel loading buffer and boiled.
Virus infection and labelling of cellular proteins.
HeLa cells were seeded at 80% confluency 24 h before infection with viruses at a multiplicity of 5 PFU per cell. Extracts from virus-infected cells for immune precipitation experiments were prepared 16 h later as described below. Labelling of cellular proteins was conducted by removing the growth medium, washing the cells with PBS, and then adding PBS with [35S]methionine at a concentration of 100 μCi/ml (15 ml per 140-mm-diameter plate). The cells were harvested 2 h later for extract preparation.
Preparation of cell extracts.
Infected cells for immune precipitation experiments were washed in PBS and harvested in a buffer containing 50 mM Tris-HCl (pH 8.5), 200 mM NaCl, 0.1 mM zinc acetate, and 10 mM 2-mercaptoethanol (160 μl for the cells from an 80-mm-diameter plate). The cells were lysed with either 10 strokes of a small Dounce homogenizer or brief sonication in a sonibath, and the debris were pelleted by centrifugation (Sorvall SS35 rotor) at 10,000 rpm for 15 min. Uninfected cell extracts for use in binding experiments in vitro were prepared by resuspending the cells (either unlabelled or labelled with [35S]methionine, as appropriate and as described in the text) in 50 mM HEPES (pH 7.5)–50 mM NaCl–0.1% NP-40 (1 ml per 140-mm-diameter plate). The cells were sonicated in a sonibath, and the debris were pelleted by centrifugation with a Beckman bench top centrifuge at 3,000 rpm for 10 min.
Immune precipitations.
Crude extracts from 6-h-infection samples were made up to a volume of 0.6 ml, maintaining the concentration of NaCl at 200 mM and adding NP-40 to a final concentration of 0.2%. MAb 11060 (2 μl) and sheep anti-mouse immunoglobulin (Ig) serum (5 μl) were added and mixed on a rotary shaker at 4°C for 3 h. Then 60 μl of protein A-Sepharose (Sigma) equilibrated in the same buffer was added and the incubation was continued for a further hour. The Sepharose beads were pelleted and washed three times with 0.6 ml of buffer (50 mM Tris-HCl [pH 8.5], 0.2 M NaCl, 10 mM 2-mercaptoethanol, 0.1 mM zinc acetate, 0.2% NP-40). The beads were then taken up in SDS-acrylamide gel loading buffer and boiled.
SDS-gel electrophoresis and Western blotting.
Protein samples were loaded onto 7.5% or 10% 30:1 acrylamide–bis-acrylamide gels prepared for use in the BioRad MiniProtein II apparatus. Gels were either fixed and stained or used for Western blotting according to the methods recommended by the supplier. Nitrocellulose filters with immobilized blotted proteins were blocked with 5% dried milk in PBS containing 0.1% Tween 20, incubated with primary antibodies in PBS containing 0.1% Tween 20 with 2% dried milk for 2 to 4 h, and then washed thoroughly and incubated with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit Ig or sheep anti-mouse Ig; Sigma) prior to detection by the Amersham ECL method.
Immunofluorescence.
HEp2 cells were seeded at a density of 0.5 × 105 cells per ml into 24-well Linbro dishes containing glass coverslips. The cells were infected with wild-type and mutant viruses at the multiplicities indicated in the figure legends. After either 2 or 4 h, the cells were washed with PBS, fixed with formaldehyde (5% vol/vol of stock solution in PBS containing 2% sucrose) and permeabilized with 0.5% NP-40 in PBS with 10% sucrose. The primary antibodies were diluted in PBS containing 1% NBCS. Anti-Vmw110 monoclonal antibody 11060 was used at a dilution of 1/2,000, and anti-PML rabbit serum r8 was used at 1/1,000. Goat anti-mouse fluorescein isothiocyanate-labelled and goat anti-rabbit tetramethyl rhodamine isothiocyanate-labelled secondary antibodies (Sigma) were used at dilutions of 1/100. After staining, the coverslips were mounted and examined with a Zeiss LSM 510 confocal microscopy system with a ×63 NA 1.4 objective lens. Data collection was performed under conditions of no detectable channel overlap, and the images were processed by using Photoshop.
Antibodies.
Immune precipitations were conducted with MAb 11060, which recognizes and strongly interacts with an epitope between residues 20 and 105 of Vmw110. MAb 10503 recognizes an epitope in the C-terminal 140 residues of Vmw110 (10). Polyclonal rabbit serum 95 is directed against the RING finger domain of Vmw110 (10). Anti-PML rabbit serum r8 (1) was used to detect PML. Rabbit serum r201 was generated by using a branched peptide containing the 16 C-terminal residues of HAUSP (13).
Transfections and CAT assays.
The ability of Vmw110 and mutant derivatives to activate gene expression in transfected cells was determined by using plasmid pSS80 as a reporter (this plasmid carries the chloramphenicol acetyltransferase [CAT] gene linked to the ICP6 promoter region) essentially as described previously (12). In this series of experiments Cos7 cells were used, and Vmw110 and mutant derivatives were expressed from the pCI series of plasmids, as described above. Calcium phosphate-mediated transfection was used as described previously (12). The transfection experiments were repeated on at least four independent occasions in parallel with positive and negative controls.
RESULTS
Definition of the minimal HAUSP binding domain in Vmw110.
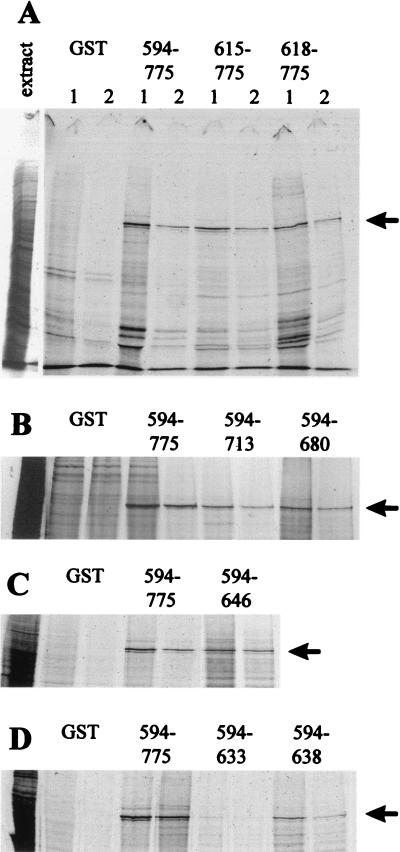
We have previously shown that a GST fusion protein including the C-terminal region of Vmw110 from residues 594 to 775 binds strongly to HAUSP in vitro and that shorter fragments (residues 633 to 775, 680 to 775, and 696 to 775) do not (29, 30). Therefore, the Vmw110 region between residue 594 and residue 632 contains residues that are essential for binding to HAUSP. To determine the nature of these residues and to define the minimal segment of Vmw110 required for HAUSP binding, plasmids were constructed that expressed GST fusion proteins with shorter segments of Vmw110, as summarized in Table 1. Extracts containing these proteins were prepared from induced bacteria, and GST pull-down assays were conducted with glutathione agarose beads charged with these proteins. The positive control was pGEXE52, expressing residues 593 to 775, and the negative control was GST alone. The results are summarized in Fig. 1. Shortening of the expressed segment from the N-terminal end (as in plasmids pGEXE4 and pGEXE9) showed that residues between 594 and 617 were dispensable for binding to HAUSP (Fig. 1A). Truncation of the Vmw110 sequences at the C-terminal end showed that sequences downstream of 681 were not essential for binding (Fig. 1B), and the shortest binding-positive segment in this assay contained residues 594 through 646 (pGEXE52AvaI; Fig. 1C). Further truncation of this segment to include only residues 594 to 638 (pGEXE23X) significantly weakened the binding, and no binding was observed with the segment containing 594 to 632 (pGEXE58X; Fig. 1D). Taken together, these data show that Vmw110 residues 594 to 646 (and probably 618 to 638) are sufficient for binding to HAUSP in vitro and that essential residues lie between residue 618 and residue 632. These data are consistent with results later obtained by coimmune precipitation of HAUSP and Vmw110 from virus-infected cell extracts, except that truncation at residue 646 significantly reduced HAUSP binding in the latter system (see below).
FIG. 1.
Binding of HAUSP to C-terminal fragments of Vmw110 in GST pull-down assays. Extracts of cellular proteins labelled with [35S]methionine were incubated with glutathione-agarose beads charged with GST or Vmw110 fusion protein derivatives as described in Materials and Methods. Bound proteins were eluted with reduced glutathione in two sequential steps (labelled 1 and 2 in panel A) and analyzed by SDS–7.5% polyacrylamide gel electrophoresis and autoradiography. Each set of experiments included a negative control with GST alone (GST) and the positive control fusion protein expressed by GEXE52 (594–775). The left-hand track in each panel is a sample of the extract, and the arrow points to the position of the HAUSP band. The identification of the HAUSP band at approximately 130 kDa has been described in detail previously (29, 30). (A) The results obtained with pGEXE4 (615–775) and pGEXE9 (618–775). (B, C, and D) Only the relevant parts of the gels obtained by using proteins expressed by the construct pGEXE52PmlI (594–713) and pGEXE52RsaI (594–680), the construct pGEXE52AvaI (594–646), and the constructs pGEXE23X (594–638) and pGEXE58X (594–633), respectively.
Disruption of HAUSP binding in vitro in mutants carrying single and double amino acid substitutions in Vmw110.
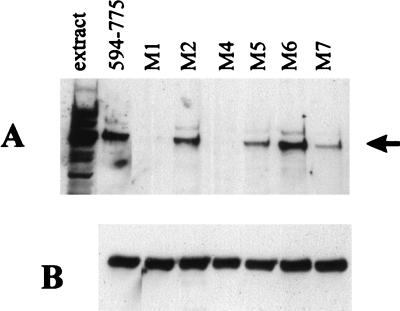
Of the 21 Vmw110 amino acids between residues 618 and 638, 7 are charged; in particular, there are three groups of positively charged doublets (Fig. 2). Comparison with the corresponding sequence from the HSV-2 homologue protein showed precise conservation of the charged and intervening residues (data not shown), so we considered that it was possible that these residues contributed to the HAUSP binding interface. A series of single and double amino acid substitutions in pGEXE52 were created by site-directed mutagenesis, as summarized in Fig. 2. The choice of replacement residues was determined so as to alter the charge but maintain the size of the residue as closely as possible. The mutant fusion proteins were used in GST pull-down assays, and in this experiment bound HAUSP was detected by Western blotting with rabbit serum r201. Probing of the gel with anti-Vmw110 MAb 10503 showed that the amounts of GST fusion proteins used were equivalent (Fig. 3B). The binding results (Fig. 3A) defined at least two residues, lysine 620 (mutant M4) and lysine 624 (mutant M1), that were crucial for HAUSP binding. Mutation of arginine 623 (mutant M2, also altered in mutant M1) indicated that this residue was not required for efficient binding. Histidine 627 was also not required for binding (mutant M6), while mutation of arginine residues 619 and 626 caused reproducible reductions in binding activity (mutants M5 and M7, respectively). These data do not give a complete definition of the Vmw110 residues that are required for HAUSP binding, but they do allow the construction of minimally mutated versions of Vmw110 which would be expected to be HAUSP binding deficient.
FIG. 3.
Binding of HAUSP to GST-Vmw110 fusion proteins with substitution mutations with the minimal HAUSP binding region. GST pull-down experiments were conducted as described in the legend to Fig. 1 and in Materials and Methods by using unlabelled extracts of cellular proteins. Proteins remaining bound to the beads were separated by SDS–7.5% polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes by Western blotting. Bound HAUSP was detected by probing with rabbit serum r201 (which detects a number of other bands in addition to the major band of HAUSP). (A) The left-hand track contains a sample of the extract, and the adjacent lane shows the result obtained by using the fusion protein expressed by pGEXE52. The results obtained with the mutants of the M series, whose details are given in Fig. 2, are shown with the position of HAUSP indicated by the arrow. (B) The relevant portion of the same blot reprobed with MAb 10503 to compare the quantities of the fusion protein used.
Construction of viruses expressing proteins with deletions and substitutions in the C-terminal region of Vmw110.
To explore the functional significance of Vmw110-HAUSP interaction, a series of plasmids was constructed, which contained a selection of the deletion and substitution mutations in the C-terminal region of Vmw110 as used in the GST fusion protein experiments described above. A number of these plasmids were cotransfected with infectious DNA of the Vmw110 deletion mutant dl1403 in order to construct viruses which expressed mutant Vmw110 proteins with defined lesions in the HAUSP binding region. The desired mutant viruses were identified by Southern blotting, and stocks were prepared after three rounds of plaque purification. The viruses expressed Vmw110 proteins of the expected sizes (see Fig. 4 and 5) and had the predicted restriction enzyme fragment patterns (data not shown). A summary of the properties of these viruses is given in Table 2.
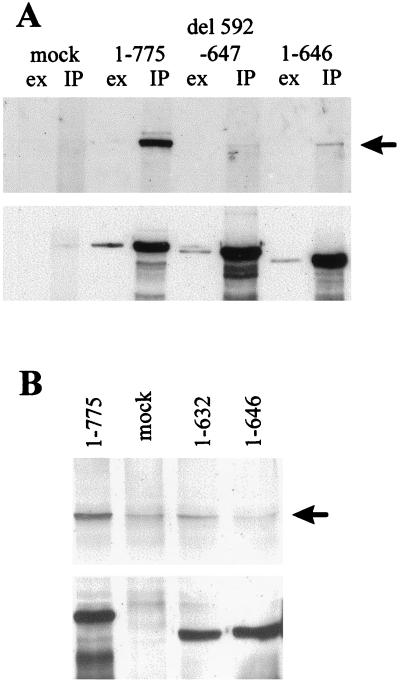
FIG. 4.
Coimmune precipitation of HAUSP with Vmw110 from extracts of cells infected with wild-type and Vmw110 mutant viruses. HeLa cells were mock infected (mock) or infected with wild-type virus (1–775) and with the deletion-carrying mutant viruses A78 (del 592–647) and A8X (1–646) (A) and with the deletion mutants A8X and E58X (1–632) (B). Extracts were prepared and used for immune precipitation of Vmw110 as described in Materials and Methods. Panel A shows precipitated proteins (IP) analyzed alongside samples from the corresponding extracts (ex) by Western blotting. In panel B, only the precipitated proteins are shown. The upper part of each panel shows the relevant portion of the filter probed with anti-HAUSP serum r201, while the lower part shows the same filter probed with anti-Vmw110 serum r95. The arrows point to HAUSP.
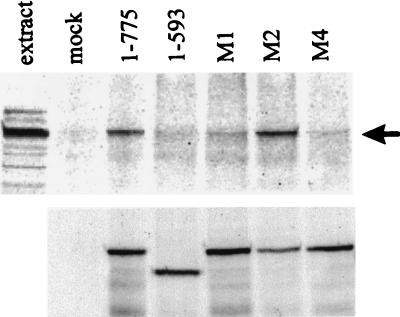
FIG. 5.
Coimmune precipitation of HAUSP with Vmw110 from extracts of cells infected with wild-type and Vmw110 substitution mutant viruses. An experiment similar to that illustrated in Fig. 5 was conducted with wild-type virus positive control (1–775), the mutant E52X negative control (1–593), and the substitution-carrying mutant viruses M1, M2, and M4. The upper part of the figure shows proteins detected by Western blotting by using anti-HAUSP serum r201 in a sample of cell extract and in anti-Vmw110 immune precipitates from mock-infected (mock) and virus-infected cells as indicated. The arrow indicates the position of HAUSP. The lower part shows the presence of Vmw110 proteins in the immune precipitates after reprobing of the filter with anti-Vmw110 serum r95.
Extracts from cells infected with these mutant viruses were prepared and used in immune precipitation reactions to determine whether mutations caused defects in HAUSP binding similar to those observed in the GST pull-down experiments described above. The results were largely consistent both with previously published observations (29, 30) and with the in vitro binding data (see Fig. 1 and 3). The deletion mutant A78 (Δ592–647) reduced HAUSP binding to background levels (Fig. 4A). A quantitative difference between the in vitro binding and the immune precipitation results was that the viral truncation mutant A8X exhibited substantially reduced binding (Fig. 4A and B) while its equivalent GST fusion protein (expressed by pGEXE52AvaI) appeared to bind almost as efficiently as the complete C-terminal region expressed by pGEXE52 (Fig. 1C). It is possible that the large amounts of fusion protein used in the in vitro assays mask reductions in binding affinity that are revealed in the more sensitive immune precipitation assay. The truncation mutant E58X had minimal binding activity (Fig. 4B). Perhaps most importantly, the substitution mutations M1 and M4, which decreased binding in vitro to background levels, resulted in substantial reductions in HAUSP binding in infected cell extracts (Fig. 5). In contrast, the M2 mutation had little effect in both assays. These results confirm the importance of this region of Vmw110 for HAUSP binding. The plasmids and viruses constructed in these experiments allow a thorough analysis of the contribution of HAUSP binding to Vmw110 function.
The ability to bind to HAUSP contributes to the activation of gene expression by Vmw110.
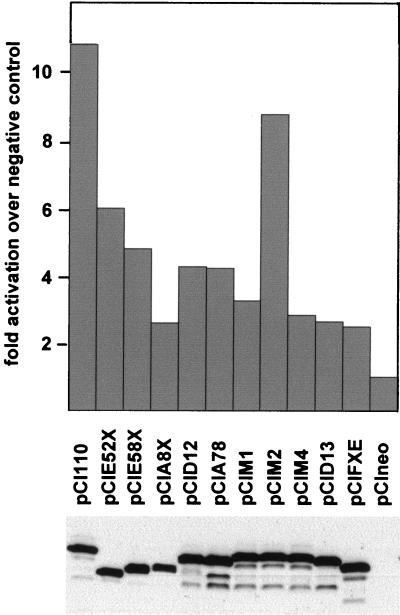
Once simple mutations which disrupt HAUSP binding by Vmw110 in vitro were defined, it was possible to determine the effects of these mutations on the ability of the protein to activate gene expression in transfected cells. The results were compared with those obtained by using other more extensive alterations of the C-terminal region of Vmw110. Cotransfection of Cos7 cells with a reporter plasmid (pSS80, which expresses CAT from the ICP6 promoter) and pCI110 (which expresses wild-type Vmw110 from the HCMV promoter in the vector pCIneo) resulted in a 9.6-fold increase in CAT activity (Fig. 6). This was reduced to only 2.3-fold by deletion of the RING finger region in plasmid pCIFXE; this region has been shown previously to be of prime importance for Vmw110 activity in this type of assay (7). On the basis of the HAUSP binding data presented above and previously published mapping of the self-multimerization domain of Vmw110 (4, 30), the proteins expressed by the various C-terminal region Vmw110 deletion mutants can be classified as follows: HAUSP binding competent but multimerization deficient (those expressed by pCIA8X and pCID13); HAUSP binding deficient but multimerization competent (pCID12 and pCIA78); and neither HAUSP binding competent nor multimerization competent (pCIE52X and pCIE58X). All of these deletions reduced activation of gene expression by Vmw110 (Fig. 6), suggesting that the abilities of Vmw110 to bind to HAUSP and to multimerize contribute to its activity. However, the reductions caused by these deletions were not as great as that resulting from loss of the RING finger. In contrast, the substitution-carrying mutants pCIM1 and pCIM4 had substantially reduced activities, while mutant pCIM2 as expected activated gene expression as efficiently as wild-type pCI110. Western blotting of extracts of cells transfected with this series of plasmids indicated that the mutant proteins were expressed to similar levels (Fig. 6, bottom).
FIG. 6.
Activation of gene expression by Vmw110 and derivatives with mutations in the C-terminal region. Cos7 cells were cotransfected with reporter plasmid pSS80 (ICP6 promoter linked to CAT) and Vmw110 expression plasmids. The negative control is the vector pCIneo, and all CAT activities are given as fold activation over this basal level. The data are averages for at least four independent transfection assays. The nature of the mutations carried by E52X, E58X, A8X, D12, A78, M1, M2, M4, and FXE is shown in Table 2. The deletion carried by D13 (deletion of residues 633 through 680) affects the multimerization and ND10 binding of Vmw110 but not its ability to bind to HAUSP (29). The lower part of the figure shows a Western blot of total proteins of cells, transfected in parallel with the same plasmids, probed with anti-Vmw110 MAb 11060.
A puzzling feature of these results from transfection assays is the relatively modest effect of complete deletion of the C-terminal region of Vmw110 (pCIE52X) compared to the M1 and M4 point mutations and other smaller deletions. There is no clear explanation of this result, but the following considerations of transfection assays and the effects of Vmw110 may be relevant. First, the viral E52X deletion mutation causes a defect in PML isoform degradation in the early stages of infection, but this is a kinetic defect in that at later times at least some E52X-induced degradation occurs (14). If this process is directly related to Vmw110 activity, the reduced rate of E52X activity might be sufficient in the much longer timescale of a transfection assay to induce significant activation of gene expression. On the other hand, the E52X mutant protein is actually as efficient as wild-type virus in inducing the loss of CENP-C, whereas the M1 mutant appears to be less efficient than the wild type (16). Although the significance of the effect of Vmw110 on CENP-C in terms of gene expression remains unknown, this finding raises the possibility that the effect of Vmw110 on any particular cellular target may be varied by deletion or point mutation in unpredictable ways. Whatever the explanation of the unexpectedly high activity of the E52X mutation in this transfection assay system, the results obtained with the M-series point mutant plasmids are consistent with the hypothesis that the ability to bind to HAUSP contributes to the activation of gene expression induced by Vmw110 in transfected cells.
The relationship between HAUSP binding and ND10 disruption by Vmw110.
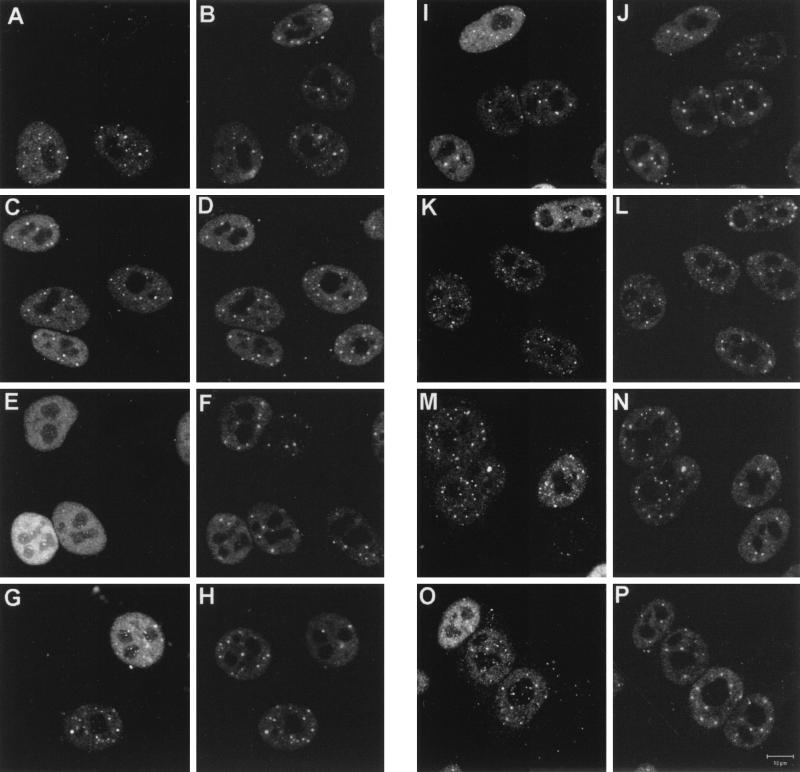
Previous data have shown that many deletion mutations in the C-terminal region of Vmw110 result in failure to localize to and disrupt ND10 (11, 30), but mutations that specifically affect HAUSP binding have not been studied in this respect in any detail. Therefore, HEp-2 cells were infected with a panel of deletion- and substitution-carrying mutant viruses, and the localization of Vmw110 and PML (to detect ND10) was examined by immunofluorescence at 2 and 4 h postinfection. After 2 h of infection (Fig. 7), the localization of wild-type and FXE mutant proteins was as previously reported (27); discrete punctate accumulations were seen within a diffusely distributed background (Fig. 7A and C). At this stage of infection, neither the wild type nor the FXE mutant caused any obvious systematic change in the distribution of PML in ND10 (compare the infected and uninfected cells in Fig. 7B and D), and many of the localized accumulations of Vmw110 corresponded to the sites of ND10 (compare horizontal pairs of panels). Mutants D12, A78, M1, M2, and M4 all gave results similar to that for the wild-type protein at 2 h. The mutants which differed were A8X (Fig. 7E) and E58X (not shown), which gave a diffuse nuclear distribution similar to those of previously described viruses E52X, D13, and D14 (27, 30). Since the A78 and D12 deletion mutations and the M1 and M4 substitution mutations remove sequences required for HAUSP binding, it can be concluded that the ability of Vmw110 to bind to HAUSP is not essential for its localization to ND10.
FIG. 7.
Distribution of wild-type and mutant Vmw110 proteins in HEp-2 cells 2 h after the start of infection. Cells on coverslips were infected with the HSV-1 strain 17+ (A and B) and with mutant derivatives FXE (C and D), A8X (E and F), D12 (G and H), A78 (I and J), M1 (K and L), M2 (M and N), and M4 (O and P). Each pair of panels shows the same field of cells stained for Vmw110 (MAb 11060) (left) and PML (r8, to indicate ND10) (right). The bar in P corresponds to 10 μm.
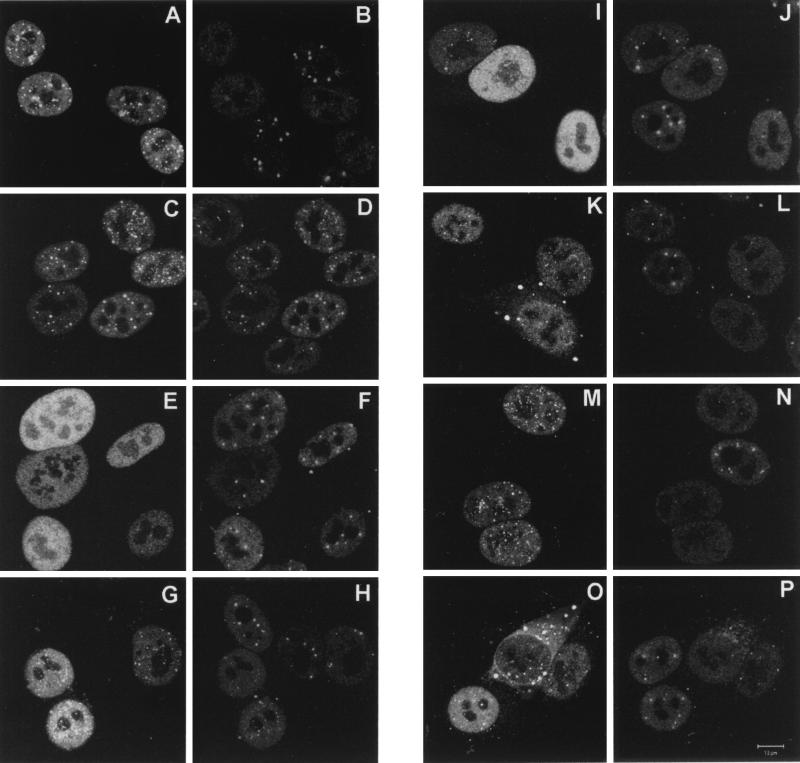
The differences between the mutants were more marked after 4 h of infection (Fig. 8). By that time, wild-type Vmw110 had caused the complete disruption of ND10 (Fig. 8A and B), while the RING finger mutant FXE remained colocalized with PML (C and D). The Vmw110 deletion mutant proteins expressed by viruses A8X (E) and E58X (data not shown) again gave diffuse nuclear staining patterns, and ND10 was extensively retained in the infected cells (F). By 4 h after the start of infection, virus A78 gave an intermediate phenotype, with some punctate staining in some cells (I) and some infected cells retaining some ND10 (J). Virus D12 (G) gave a greater degree of punctate staining than A78, but again some cells retained a few ND10 (H). The HAUSP-binding-proficient M2 point mutant behaved exactly like the wild type, inducing the complete loss of PML in ND10 at 4 h (M and N), but the HAUSP-binding-negative mutants M1 and M4 showed two interesting differences in this assay. First, although many infected cells had lost all ND10 by this time, it was noticeable that some infected cells retained at least some ND10 (L and P). It should be noted that the results of this type of experiment vary with time, from cell to cell, and with cell type (see below), so it is difficult to compare the relative kinetics of ND10 disruption with precision. However, it is clear that the HAUSP-binding-negative mutants retain the ability to disrupt ND10. The second difference from the wild type exhibited by the M1 and M4 proteins was their accumulation in large cytoplasmic foci in many cells (K and O). This effect became more marked as infection progressed (data not shown).
FIG. 8.
Disruption of ND10 by wild-type and mutant Vmw110 proteins. Coverslips were processed for immunofluorescence 4 h after the start of infection. All other details are exactly as described for Fig. 7.
The ability of Vmw110 to bind to HAUSP contributes to the efficiency of virus replication in tissue culture.
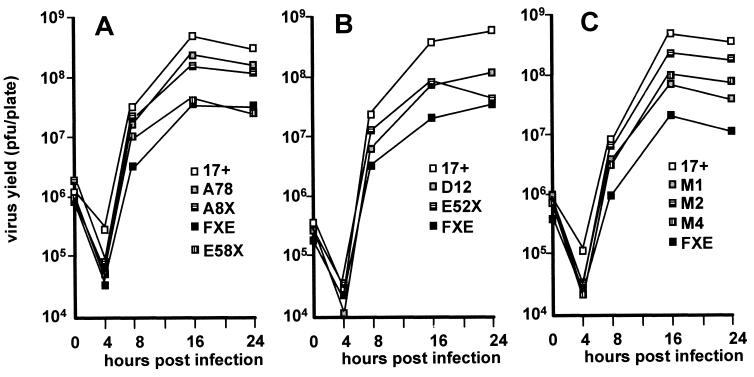
In the experiments described above we have shown that the C-terminal region of Vmw110 is complex and responsible (at least in part) for a number of phenomena, including localization to ND10, disruption of ND10, binding to HAUSP, and (as deduced by comparison with previous data) self-multimerization. The conclusions that emerge from this study of multiple deletion and substitution mutations in this region are that localization to ND10 requires sequences that have not been separated from the mapped self-multimerization domain and that efficient disruption of ND10 requires the ability of Vmw110 to localize to ND10 but not to bind to HAUSP. However, all the mutations are to some extent deleterious for the ability of Vmw110 to activate gene expression in transfected cells. To investigate the effects of these mutations on virus replication, single-step growth curve experiments were conducted with BHK cells and all the available virus mutants and the results were compared with those for strain 17+ (wild type) and the RING-finger-deletion-carrying mutant FXE. The results obtained with the mutant viruses (Fig. 9) were similar to those obtained in previous studies; in particular, the moderate growth defect caused by the A78 mutation parallels that seen previously with the D12 deletion mutation (30), while the E58X truncation mutation had an effect similar to that of the more extensive E52X deletion. However, the most important conclusion is that the M series of substitution mutations caused growth defects which reflected their reduction of HAUSP binding by Vmw110. That a single amino acid substitution at Vmw110 residue 620 can reduce HAUSP binding to background levels and cause a significant reduction in virus growth provides compelling evidence that the ability of Vmw110 to bind to HAUSP contributes to its biological properties.
FIG. 9.
Growth curves of the HSV-1 strain 17+ and derivatives with lesions in Vmw110. BHK cells were infected with the viruses at 1 PFU per cell, replicate plates were harvested 4, 8, 16, and 24 h later, and progeny virus was titrated on BHK cells. Panels A and C show the results obtained with the viruses constructed for this study, with wild-type virus, and with the Vmw110 RING finger deletion mutant virus FXE. For comparison and completeness, panel B shows the results obtained with the complete C-terminal region deletion mutant, E52X, and the HAUSP-binding-region-deletion mutant, D12 (taken from reference 30). The genotypes of the viruses are given in Table 2. The data are averages of two independent experiments.
DISCUSSION
This paper presents a thorough analysis of the role of HAUSP binding in Vmw110 activity in transfected and infected cells. We have identified single amino acid residues within a small region of Vmw110 which are required for HAUSP binding, and we have shown that removal of this region or alteration of specific residues within it reduces Vmw110 activity. The results obtained with the substitution mutants M1 and M4 are particularly striking. However, it is clear that other factors are also involved in Vmw110 activity.
An understanding of the role of Vmw110 in virus infection is made difficult by both the complexity of the phenotypes of Vmw110 mutants and the presence of multiple domains within the protein that contribute to its functions. The defect caused by lack of functional Vmw110 is cell type, cell cycle, and multiplicity dependent (2, 34). Therefore, by definition, the effect of Vmw110 varies among individual cells in a single experiment, because each cell varies in its stage in the cell cycle and the dose of virus that it receives. Since the defect can be overcome at high multiplicity, infection with a sufficient amount of virus to enable viral gene expression to be readily detectable (by Western or Northern blotting, for example) circumvents the requirement for Vmw110, at least to some extent. Furthermore, once the lytic cycle has been established the role of Vmw110 appears to be dispensable since equal numbers of progeny virus particles are produced (8). Indeed, growth curves of the type shown in Fig. 9 in reality reflect the subsequent probability of initiation of plaque formation by the progeny virus rather than the total amount of virus particles produced. For these reasons, simple measurement of viral gene expression in productive infection is a problematic method of gauging Vmw110 activity.
Dissecting this complex phenotype is further complicated by the nature of Vmw110 itself. It contains sequences required for self-multimerization, for transport to the nucleus, for localization at ND10, for disruption of ND10, and for binding to HAUSP. Of these, the RING finger domain appears to be the most important, since RING finger mutants do not disrupt ND10, do not alter the stability of a number of cellular proteins, are very poor activators of gene expression in transfection assays, and form plaques as inefficiently as a null mutant. This paper shows that the HAUSP binding region of Vmw110 also affects activation of gene expression in transfection assays and plaque forming efficiency (as indicated by a single-step growth curve), but in HEp-2 cells the HAUSP binding mutants colocalize with PML and disrupt ND10 almost as well as the wild type. At first sight, this finding presents an anomaly since in all other respects ND10 disruption correlates with Vmw110 activity, and it is an attractive hypothesis that the mechanisms which result in the disruption of ND10 underlie the biological activity of Vmw110. A possible explanation of this paradox lies in the cell type specificity of the Vmw110 mutant phenotype. We have previously shown that, compared to the results in HEp-2 cells (Fig. 8), the HAUSP-binding-defective mutant D12 disrupted ND10 inefficiently in BHK cells (30). Recent studies on the correlation between ND10 disruption and the Vmw110-induced loss of the PML isoforms have confirmed that in BHK cells mutant D12 is indeed defective in ND10 disruption, and this correlates with inefficient degradation of the PML isoforms (14) and its reduced growth in BHK cells (Fig. 9). Furthermore, the D12 mutant disrupts ND10 more efficiently in HFL cells, and this correlates with improved growth in this cell type (14).
The picture emerging from this paper and other recent work is that a major biochemical function of Vmw110 is the control of the stability of a number of specific cellular proteins. A likely scenario is that one or more of the target proteins is involved in a pathway that promotes the establishment of a quiescent state of the incoming viral genome, and therefore its Vmw110-induced loss would increase the probability of the onset of the lytic cycle. The conclusion of this paper that the ability to bind to HAUSP contributes to the biological functions of Vmw110 is, in principle, consistent with the idea that protein stability pathways play a key role in control of HSV-1 infection. The next goals must be to catalogue the cellular proteins which are affected by Vmw110 and then to determine which of them lie at the heart of Vmw110 activity.
ACKNOWLEDGMENTS
We are grateful for the helpful criticism of the manuscript by Duncan McGeoch and for the supply of antibodies by Paul Freemont (ICRF, London, United Kingdom) and Roel van Driel (E. C. Slater Institute, Amsterdam, The Netherlands).
This work was supported by the Medical Research Council.
REFERENCES
- 1.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. PIC1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 2.Cai W, Schaffer P A. A cellular function can enhance gene expression and plating efficiency of a mutant defective in the gene for ICP0, a transactivating protein of herpes simplex virus type 1. J Virol. 1991;65:4078–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai W, Astor T D, Liptak L M, Cho C, Coen D, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciufo D M, Mullen M-A, Hayward G S. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J Virol. 1994;68:3267–3282. doi: 10.1128/jvi.68.5.3267-3282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett R D. Transactivation of transcription by herpes virus products: requirements for two HSV-1 immediate-early gene polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett R D. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1986;6:2069–2076. doi: 10.1002/j.1460-2075.1987.tb02472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett R D. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87–96. doi: 10.1016/0022-2836(88)90521-9. [DOI] [PubMed] [Google Scholar]
- 8.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 9.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 50–76. [Google Scholar]
- 10.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D, Maul G G. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett R D, Orr A, Elliott M. The equine herpesvirus 1 gene 63 RING finger protein partially complements Vmw110, its herpes simplex virus type 1 counterpart. J Gen Virol. 1995;76:2369–2374. doi: 10.1099/0022-1317-76-9-2369. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110 and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., T. Sternsdorf, and H. Will. Unpublished data.
- 16.Everett, R. D., P. Lomonte, and W. C. Earnshaw. Herpes simplex virus immediate-early protein Vmw110 associates with centromeres, induces the proteasome-dependent destruction of CENP-C and causes abnormal cell division. Unpublished data.
- 17.Everett, R. D., A. Orr, and C. M. Preston. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 18.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishov A M, Maul G G. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan R, Schaffer P A. Activation of gene expression by herpes simplex virus type 1 ICP0 occurs at the level of mRNA synthesis. J Virol. 1997;71:6850–6862. doi: 10.1128/jvi.71.9.6850-6862.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate-early gene 1 product ICP0. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 27.Maul G G, Everett R D. The nuclear location of PML, a cellular member of the C3HC4 zinc binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 28.Maul G G, Ishov A, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type 1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 29.Meredith M R, Orr A, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135 kD cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 30.Meredith M R, Orr A, Elliott M, Everett R D. Separation of the sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135 kD cellular protein. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 31.Mullen M-A, Gerstberger S, Ciufo D M, Mosca J D, Hayward G S. Evaluation of the colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J Virol. 1995;69:476–491. doi: 10.1128/jvi.69.1.476-491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptice Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 35.Uprichard S L, Knipe D M. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 36.Yao F, Schaffer P A. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J Virol. 1995;69:6249–6258. doi: 10.1128/jvi.69.10.6249-6258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]