Abstract
Poliovirus RNA genomes that contained deletions in the capsid-coding regions were synthesized in monkey kidney cells that constitutively expressed T7 RNA polymerase. These replication-competent subgenomic RNAs, or replicons (G. Kaplan and V. R. Racaniello, J. Virol. 62:1687–1696, 1988), were encapsidated in trans by superinfecting polioviruses. When superinfecting poliovirus resistant to the antiviral compound guanidine was used, the RNA replication of the replicon RNAs could be inhibited independently of the RNA replication of the guanidine-resistant helper virus. Inhibiting the replication of the replicon RNA also profoundly inhibited its trans-encapsidation, even though the capsid proteins present in the cells could efficiently encapsidate the helper virus. The observed coupling between RNA replication and RNA packaging could account for the specificity of poliovirus RNA packaging in infected cells and the observed effects of mutations in the coding regions of nonstructural proteins on virion morphogenesis. It is suggested that this coupling results from direct interactions between the RNA replication machinery and the capsid proteins. The coupling of RNA packaging to RNA replication and of RNA replication to translation (J. E. Novak and K. Kirkegaard, Genes Dev. 8:1726–1737, 1994) could serve as mechanisms for late proofreading of poliovirus RNA, allowing only those RNA genomes capable of translating a full complement of functional RNA replication proteins to be propagated.
The mechanism of packaging of picornavirus RNA has not been determined, and simple questions such as whether a packaging signal or sequence that confers specificity to the RNA packaging reaction have not been answered. Poliovirus positive-sense RNAs are specifically packaged in infected cells: cellular mRNAs, rRNAs, tRNAs, and negative-sense poliovirus RNAs are excluded from the capsids (34, 35). Sensitive trans-encapsidation assays have shown that replicating RNA genomes derived from poliovirus type 1 can be encapsidated by virion proteins encoded by any of the three poliovirus serotypes but not by capsid proteins from the related picornaviruses coxsackievirus A21 or B3 or enterovirus 70 (45). What determines the specificity of poliovirus RNA packaging? RNA genomes that contain deletions of most of the capsid-encoding region of the genome can be packaged in trans by coinfecting genomes (26, 39), arguing that a unique packaging signal, if it exists, does not lie between nucleotides (nt) 756 and 2956. Similarly, the ability of 5′ and 3′ noncoding sequences from other picornaviruses, which are not encapsidated by poliovirus capsids during coinfections, to replace the poliovirus 5′ and 3′ noncoding regions argues that nt 108 to 740 and 7385 to 7440 also do not contain specific packaging sites (1, 57).
One explanation (20, 38, 49) for the observed specificity of RNA packaging in poliovirus-infected cells is that, as first proposed by Baltimore (4), poliovirus RNA replication and packaging are directly coupled and only RNA genomes that are actively being replicated can be packaged. Several published observations are consistent with the hypothesis that only newly replicated poliovirus positive RNA strands are encapsidated. For example, when poliovirus-infected cells were pulse-labeled with [3H]uridine, radioactivity was detected in the mature virion peak within 5 min (5). However, newly synthesized 35S-labeled proteins required 30 min on average to move into the virion peak (5). By electron microscopy, RNA replication complexes and virion precursors colocalize to the membranous vesicles that proliferate during infection (40), and after pulse labeling with [3H]uridine, virions with the highest specific activity were found in direct association with the membranes on which RNA replication occurs (13).
Biochemical and genetic studies have suggested the possibility of direct interactions between proteins of picornavirus replication complexes and capsid proteins. For example, the virions of foot-and-mouth disease virus contain measurable amounts of 3D, the viral RNA-dependent RNA polymerase (32). Poliovirus protease 3CD shows higher specificity for capsid precursors than does 3C alone, arguing that the 3D sequences confer some binding energy to the capsid proteins (12, 63). Finally, RNAs that bear certain mutations in proteins 2C (28) and 3D (17) of the RNA replication complex can program the production of virions with altered cell entry properties.
The effects of compounds that inhibit poliovirus RNA replication and RNA packaging have also suggested a mechanistic link between these two processes. Low concentrations of guanidine hydrochloride specifically inhibit the poliovirus replicative cycle; both RNA synthesis and the formation of mature virus particles quickly cease following treatment of infected cells with 0.1 to 2.0 mM guanidine hydrochloride (21). Although originally it was thought that guanidine might have some effect on morphogenesis itself, it has since become clear that guanidine directly inhibits RNA replication (reviewed in reference 62). For example, mutations that confer resistance to guanidine map to the 2C region of the poliovirus genome (42) and mutations in the 2C coding region that cause defects in RNA replication (27) or suppress the cold sensitivity of wild-type virus (18) have been identified. Furthermore, the addition of guanidine to cell extracts that support poliovirus RNA replication was shown to inhibit the initiation of negative-strand synthesis (8) and to prevent the further packaging of positive-strand RNA (29). Therefore, either the inhibition of RNA packaging by guanidine derives from a requirement for nascent RNA in RNA packaging, or protein 2C is also directly involved in virion assembly, or both. Recently, hydantoin, a compound that inhibits the growth of both poliovirus and coxsackievirus A3, has been shown to inhibit poliovirus morphogenesis without affecting RNA replication (60). Mutations in the poliovirus genome that confer resistance to hydantoin map to the 2C region of the viral genome (60). Assuming that these mutations exert their effect through the change in the amino acid sequence of 2C, a direct role for 2C in virion assembly is suggested. Since 2C protein is firmly embedded in the intracellular membranes on which poliovirus RNA replication occurs (10, 11, 30, 40, 52), a physical interaction between poliovirus virion assembly and RNA synthesis is implied whether the role of the 2C protein in RNA packaging is direct or indirect.
Several review articles have described previously published data linking poliovirus RNA replication and RNA packaging (20, 49), attempts to identify RNA-packaging sites (2), and the use of trans-encapsidation to create poliovirus vectors (2). In the present study, we used RNase protection experiments to demonstrate the trans-encapsidation of poliovirus replicon RNAs in infected cells and to monitor this trans-encapsidation when such RNAs are associated with productive replication complexes and when they are not. Preaccumulated RNAs are not packaged in trans by capsids made from a coinfecting helper virus, even under conditions in which the helper virus is actively packaging its own RNAs and the preaccumulated RNA is stable. Therefore, only newly synthesized poliovirus RNAs are packaged. The requirement of new RNA synthesis for RNA packaging is likely to be due to a requirement for direct association with the RNA replication complex.
MATERIALS AND METHODS
Cell lines.
The KJT7 cell line, which constitutively expresses T7RNA polymerase, was constructed from CV-1 cells by the stable introduction of two plasmids: pSV2-neo (54), which confers resistance to the antibiotic G418, and pAR3126 (19), which contains the bacteriophage T7 gene 1 that does not contain a nuclear localization sequence. The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% calf serum, 1% penicillin-streptomycin, and 500 μg of G418 (Geneticin; GibcoBRL) per ml. HeLa cells were maintained as previously described (25).
Virus stocks.
Wild-type stocks of type 1 Mahoney poliovirus were amplified from individual plaque isolates obtained from transfection of HeLa cells with a plasmid, pPolio (17), containing the full-length poliovirus genome (46).
High-titer 3NC-202guaR virus stocks were amplified as described previously (24). This mutant virus strain contains several sequence alterations from wild-type virus. One mutation is an 8-bp insertion in the 3′ noncoding region (GGTTAACC following nt 7387); alone, this lesion is responsible for a temperature-sensitive defect in RNA synthesis (50). Additional mutations are responsible for the guanidine-resistant phenotype; two lesions have been identified within 2C (M187L and V250A) (24).
Plasmids.
Plasmids containing the T7 R2 and T7 R3 poliovirus constructs (22) were constructed by Janet Novak by deletion of sequences 1175 to 2956 and 1175 to 2471, respectively, in the T7pGempolio plasmid (51). The Z polymorphism was engineered into the T7R2 and T7R3 plasmids by replacing the DNA sequences from nt 675 to 702 with the following heterologous sequence: GGTCGACCAAATCTCTTCGAACTTTAGAAACACCGTGTCTGCCTTCGTAAAAGGTACCAGTCGACGGG CCAGCTGGTTTAGAGAAGCTTGAAATCTTTGTGGCACAGACGGAAGCATTTTCCATGGTCAGCTGCCC
The phenotype of virus that contains the Z substitution is indistinguishable from that of wild-type virus (53a).
The plasmid construct, pBSIISK+RZ, used for transcription of the probes for RNase protection contains the 68-bp Z sequence and sequences 702 to 742 from the wild-type poliovirus cDNA (47), cloned between a T3 promoter and a T7 promoter in the pBSIISK+ vector (Stratagene).
DNA transfection.
KJT7 cells were transfected with supercoiled plasmids as described previously (6). For the transfection mixture, 7 μg of DNA in 0.5 ml of OptiMEM was mixed with 70 μl (70 μg) of Lipofectin reagent (Gibco-BRL) in 1.5 ml of OptiMEM, allowed to sit at room temperature for 10 to 20 min, and placed on subconfluent cell monolayers (approximately 2 × 107 cells on a 100-mm plate) that had been washed twice with OptiMEM. An additional 2 ml of OptiMEM was then added, and the cells were placed in a CO2 incubator at 32.5 or 37.5°C. After 10 to 15 h, the lipofection mix was removed and 10 ml of DMEM containing 10% calf serum was added to each plate for the remainder of the incubation period.
Superinfections.
Transfected or mock-transfected cells were infected with virus stocks at multiplicities of infection of 20 to 100 PFU/cell, and cytoplasmic extracts were prepared as described previously (36). Virus stocks were made by resuspending cell pellets containing ∼2 × 107 cells in 0.5 ml of phosphate-buffered saline containing 0.01% CaCl2 and 0.01% MgCl2 (PBS+) and freeze-thawing the cells three times to lyse them. To make virus stocks that were free from full-length unpackaged RNAs, the virus stock was treated at room temperature with 17 μg of RNase A per ml for 20 min.
To infect HeLa cell monolayers (∼2 × 107 cells) with only encapsidated RNAs, plates of cells were exposed to 100 μl of the various treated virus stocks for 30 min at 37°C, medium containing DMEM and 10% calf serum was added, and incubation was continued either at 37°C for 5 h or at 32.5°C for 10 h as indicated.
Plaque assays.
Plaque assays on HeLa and KJT7 cell monolayers were performed as previously described (23). The virus was placed on the cells in PBS+ in a total volume of 200 μl. The mixture was incubated for 30 min at 32.5°C to allow the virus to adsorb to the cells, and then 5 ml of 1× DMEM–10% calf serum (Gibco-BRL)–1% agar was added to each plate. For virus that was grown in the presence of 0.5 mM guanidine-HCl, the drug was added to the agar overlays. After incubation for 3 days (32.5°C), the agar overlays were removed and the cells were stained with crystal violet.
Preparation of cytoplasmic and viral RNA.
To prepare RNA from frozen pellets of approximately 6 × 107 cells, the cells were thawed on ice and lysed as described previously (36). To prepare cytoplasmic RNA, 10 mM vanadate ribonucleoside complexes (New England Biolabs) was added to each cell aliquot. The lysates were clarified by centrifugation at 4°C for 8 min at 1,600 × g. The supernatants were transferred to fresh tubes, 5 μg of tRNA and sodium dodecyl sulfate (SDS) to 0.5% were added, and the lysates were extracted with buffer-saturated phenol. The aqueous phases were collected, EDTA was added to 1 mM, and the RNA was precipitated by the addition of 0.1 volume of 3 M sodium acetate and 3 volumes of ethanol, freezing on dry ice, and centrifugation.
To prepare encapsidated RNA, cell lysates were incubated with 12 U of DNase I (Promega) per ml, 30 μg of RNase A (Amersham/USB) per ml, and 12 U of RNase T1 (Gibco-BRL) per ml for 25 to 45 min at room temperature to digest the unprotected RNA and DNA and clarified by centrifugation at 4°C and 1,600 × g for 8 min. SDS to 0.1% was then added to the supernatant, and the virion particles were collected by centrifugation through sucrose step gradients. The gradients were composed of 1 ml of 45% sucrose in PBS+ that contained 0.1% SDS, 2.5 ml of 30% sucrose in PBS+, and 0.5 to 1.0 ml of virion preparation. Following centrifugation in an SW50.1 rotor (Beckman) for 4 h 10 min at 49,000 rpm at 4°C, the virion pellets were resuspended in 1 ml of RSB (10 mM TrisHCl, 10 mM NaCl, 1.5 mM MgCl2) with 10 U of RNase T1 per ml. After incubation at room temperature for 20 to 30 min, the virions were pelleted through a second sucrose step gradient as described above. After centrifugation, the virions were resuspended in 200 μl of TNE (10 mM TrisHCl, 10 mM NaCl, 1 mM EDTA) containing 10 mM vanadate ribonucleoside complexes, 10 μg of tRNA, 0.5% SDS, and 1% Brij 35. SDS was added to 1%, and the solution was extracted with phenol taking both the supernatant and the interface. The virions were then extracted twice with phenol-chloroform-isoamyl alcohol. Finally, the RNA was precipitated by the addition of 0.1 volume of 3 M sodium acetate and 3 volumes of ethanol and collected by centrifugation. To test whether the viral RNA preparations were free from contamination by unpackaged RNAs, one pellet of ∼6 × 107 mock-transfected, superinfected cells was deliberately contaminated with 31 fmol of in vitro-transcribed T7R2Z RNA before viral and cytoplasmic RNAs were prepared. RNase protection of the viral RNA from these cells shows no protected band corresponding to the R2Z sequence, indicating that the virions were successfully purified from the abundant contaminating transcript present in the cytoplasmic RNA (data not shown).
RNase protection.
RNase protection assays were performed as previously described (35). To detect positive-strand R2Z, R3Z, wild-type poliovirus, and 3NC202 viral RNAs, the RNA probe was transcribed from BstUI-digested pBSIISK+RZ by bacteriophage T7 RNA polymerase (Promega). To detect negative strands, excess positive-strand RNAs were removed by a cycle of RNase protection in the absence of added probe (35) and then the probe was synthesized by T3 RNA polymerase from pBSIISK+RZ. Transcription reaction mixtures contained 570 Ci of [α-32P]UTP per mmol of UTP. The labeled probes were purified by electrophoresis through a 5% polyacrylamide gel and eluted. The samples were denatured at 85°C for 5 min and hybridized overnight at 60 to 61°C. RNase digestion was performed in an ice-water bath for 60 min in a high-salt RNase cocktail (500 mM NaCl, 10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 4.5 μg of RNase A per ml, 350 U of RNase T1 per ml). The samples were treated with proteinase K, extracted with phenol, precipitated with ethanol and 5 μg of tRNA, and loaded onto 8% polyacrylamide–8 M urea gels. The radioactivity in the bands was quantitated by phosphorimager analysis. The number of moles of RNA represented was determined from the protection signals of known amounts of RNA included in standard curves.
RESULTS
Packaging and superinfection assays.
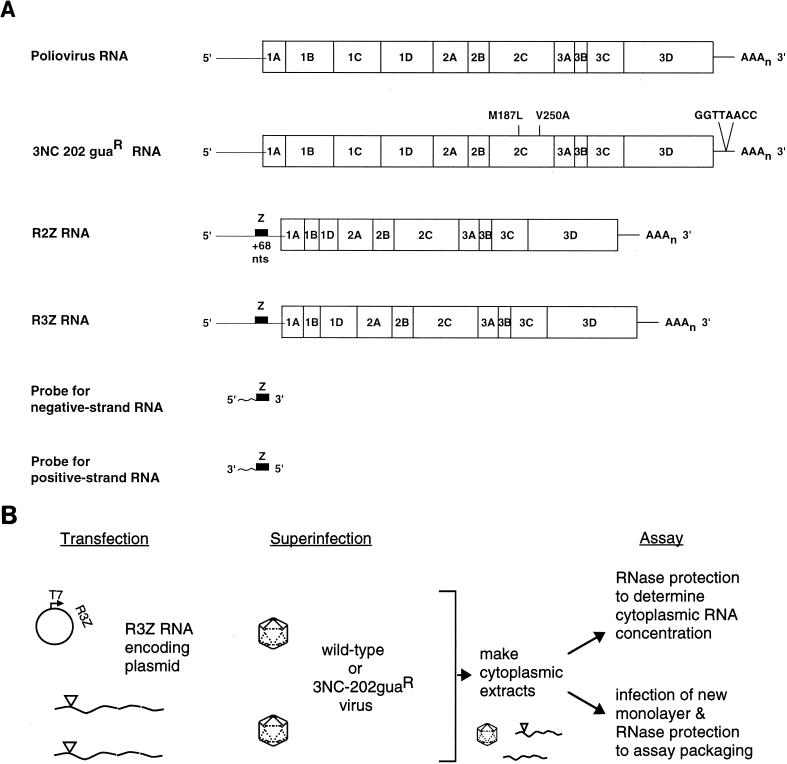
To measure encapsidation of one poliovirus RNA genome in trans by proteins encoded by another poliovirus genome, the RNA genomes diagrammed in Fig. 1A were used. R2Z and R3Z are RNA molecules which encode all of the viral proteins required for poliovirus RNA synthesis but contain, within their capsid coding regions, in-frame deletions of 1,791 and 1,296 nt, respectively (22). These replicon RNAs thus cannot be packaged by capsid proteins encoded by their own genomes and can be packaged only in trans by coinfecting virus. trans-encapsidation of replicon RNAs by capsid proteins expressed from recombinant vaccinia virus has been reported previously (3, 44). In the present study, however, the capsid proteins are provided by superinfecting poliovirus, more closely resembling the circumstance of naturally occurring defective interfering particle RNAs (15, 26, 39). To allow quantitative discrimination between replicon and helper viral RNAs, both R2Z and R3Z replicon RNAs bear a 68-nt insertion in their 5′ noncoding regions of the RNA (Z in Fig. 1A) that does not alter the phenotype of viruses or replicons that contain it (37).
FIG. 1.
Experimental design of trans-encapsidation assays. (A) RNA molecules used for infections, transfections, and RNase protection experiments are diagrammed. Wild-type poliovirus RNA, with the coding regions of the individual proteins within the polyprotein, is compared to the genome of 3NC-202guaR virus. Two point mutations in the coding region for 2C (24) and an 8-nt insertion in the 3′ noncoding region (50) of 3NC-202guaR are indicated. R2Z and R3Z RNAs contain deletions of nt 1215 to 2996 and 1215 to 2510, respectively (22). Both R2Z and R3Z also have 68 nt of heterologous sequence inserted into their 5′ noncoding region (53). RNA probes of 203 nt, used to detect positive- and negative-strand replicon and viral RNAs, are indicated. These probes contain 138 nt of sequence complementary to R2Z and R3Z and 70 nt of sequence complementary to sequences in both replicon and viral RNAs. (B) Scheme for detecting trans encapsidation of replicon RNAs. DNA plasmids that encode either R2Z or R3Z RNAs under the control of a T7 promoter were transfected into KJT7 cells, which constitutively express T7 RNA polymerase. After the R2Z or R3Z RNAs accumulated, cells were superinfected with wild-type or mutant poliovirus. Encapsidation of R2Z or R3Z RNA was detected either by using the cytoplasmic extracts to infect a new monolayer of HeLa cells or by quantifying the amount of replicon RNA in purified virions.
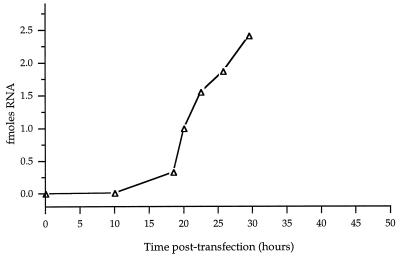
The strategy for quantifying the trans encapsidation of replicon RNAs is shown in Fig. 1B. R2Z and R3Z RNAs were engineered downstream of T7 promoters in DNA plasmids; therefore, they can be transcribed in any cells that express T7 RNA polymerase. The KJT7 cell line used in the present experiments was constructed by selecting stable transfectants of CV1 monkey kidney cells that constitutively expressed T7 polymerase (see Materials and Methods). Following transfection of KJT7 cells with supercoiled DNA plasmids that encoded the replicon RNAs, R2Z and R3Z transcripts were found to accumulate within the KJT7 cells. Despite having two extra G’s at their 5′ ends and undoubtedly heterogeneous 3′ ends, at least some of these RNAs were able to direct their own subsequent replication, as reported previously for transcripts of R2 and R3 synthesized in vitro and delivered to cells by transfection (22). Figure 2 shows the time course of accumulation of R3Z RNA following transfection of KJT7 cells with the T7R3Z plasmid. Under the conditions of these RNase protection experiments, no detectable RNA was seen following transfection with DNA plasmids encoding similar RNAs that contained out-of-frame deletions or when the incubations were performed in the presence of 0.5 mM guanidine, which specifically inhibits poliovirus RNA replication but not transcription by T7 RNA polymerase (data not shown). Therefore, the R3Z RNA accumulation shown in Fig. 2 was due predominantly to the ability of R3Z RNA to be amplified by the RNA replication proteins it encodes.
FIG. 2.
Time course of accumulation of R3Z RNA following transfection of KJT7 cells with T7R3Z DNA. RNase protection experiments show the amount of positive-strand R3Z RNA at various times posttransfection (see Materials and Methods). The amount of R3Z RNA in extracts prepared from 4 × 106 KJT7 cells was determined from standard curves of the RNase protection signal from known amounts of R3Z RNA transcripts.
R2Z and R3Z RNAs are encapsidated in trans.
To determine whether R2Z and R3Z RNAs could be encapsidated in trans by the capsid proteins provided by wild-type poliovirus, KJT7 cells transfected with plasmids encoding T7R2Z and T7R3Z were incubated at 32.5°C for 24 h, superinfected with wild-type poliovirus, and incubated for an additional 7 h at 32.5°C (Fig. 1B). Cytoplasmic extracts were prepared, and RNase protection was performed to display the total amounts of replicon and wild-type positive-strand RNAs in the cells (Fig. 3, lanes 6 to 10). A single RNA probe was used to detect both the replicon RNAs and the superinfecting viral RNAs. This RNA probe contained 76 nt of wild-type poliovirus sequence and the 68-nt Z insert present in the replicon RNAs (Fig. 1A). R2Z and R3Z replicon RNAs each protected 138 nt of the radioactive RNA probe, whereas RNAs from superinfecting viruses that did not contain the insertion protected only 76 nt of the labeled probe. The smaller size of the RNA protected by the full-length viral RNAs also allowed the use of hybridization conditions that were somewhat destabilizing to the 76-bp RNA duplex but not to the 138-bp duplex: this allowed the visualization of both the replicon RNAs, present in only the transfected cells, and the more abundant viral RNAs, present in all the cells in the population, in a single experiment. As shown in Fig. 3, R2Z and R3Z replicon RNAs accumulated in cells in both the presence and absence of superinfecting wild-type poliovirus RNA (lanes 6 to 10).
FIG. 3.
trans encapsidation of R2Z and R3Z RNAs. The RNase protection experiment shows the amounts of replicon and viral RNAs produced under various conditions. Total cytoplasmic RNA was extracted from 4 × 106 KJT7 cells that were transfected with plasmids that encode replicon RNAs and superinfected with wild-type poliovirus as indicated. The RNA was then subjected to RNase protection (lanes 6 to 10). The result of using the KJT7 cytoplasmic extracts to infect 4 × 106 HeLa cells for 5 h at 37°C is shown in lanes 11 to 20; only encapsidated RNAs should be capable of initiating infection in the HeLa cells. Lanes that display duplicate experiments are indicated by brackets. RNase protection conditions under which the 76-nt duplex that represents the wild-type RNA was relatively unstable were chosen; thus, the amounts of radioactivity in the “virus” and “replicon” bands cannot be compared directly. However, the use of standard curves of known amounts of replicon and wild-type RNAs (lanes 1 to 5 and data not shown) allowed the determination of the actual amounts of each RNA present in the cytoplasmic extracts.
To determine whether the R2Z and R3Z RNAs were packaged by the capsid proteins from the superinfecting wild-type poliovirus, the cytoplasmic extracts from the transfected KJT7 cells were used to infect monolayers of HeLa cells. The HeLa cells were incubated for 5 h at 37°C, and cytoplasmic extracts were prepared and probed for the presence of replicon and full-length poliovirus RNAs. Under these conditions, only RNAs which were successfully encapsidated in the KJT7 cells should be competent to initiate a replicative cycle in the HeLa cells. Figure 3 (lanes 11 to 20) shows a representation of the replicon and wild-type viral RNAs made in the infected HeLa cells. When R2Z- or R3Z-transfected KJT7 cells were superinfected with wild-type poliovirus, the replicon sequences could be transmitted to the HeLa cells to initiate an infection (lanes 13 and 14 and lanes 17 and 18). However, in the absence of superinfecting virus, similar amounts of R2Z and R3Z RNAs were not transmitted to the HeLa cells. Therefore, R2Z and R3Z could be encapsidated in trans by capsid proteins from the superinfecting wild-type virus.
To confirm the presence of R2Z and R3Z RNAs in purified virions from cells superinfected with wild-type virus, RNase protection of purified virion RNAs from the KJT7 cells was performed. Table 1 compares the amounts of R2Z and R3Z in the purified virions with their amounts in unfractionated cytoplasm. The use of standard curves in which known amounts of RNA were used for the RNase protection experiments allowed us to determine the amount of R2Z and R3Z RNA present in the transfected KJT7 cells and in the virions and the percentage of cytoplasmic RNA that was encapsidated. As shown in Table 1, 2 to 3% of the total cytoplasmic R2Z and R3Z RNA was packaged into virions when transfected KJT7 cells were superinfected with wild-type poliovirus. The percentage of wild-type poliovirus RNA that was simultaneously packaged was determined in the same RNase protection assays. For wild-type viral RNA, 25 to 31% of the cytoplasmic RNA was present in mature virions. The apparent reduction in packaging efficiency of the replicon RNA is likely to be due in part to the preaccumulation (Fig. 2) of much of the replicon RNA before superinfection at 24 h posttransfection. However, it is clear that R2Z and R3Z could be packaged into virions by the capsid proteins encoded by superinfecting poliovirus.
TABLE 1.
Quantitation of packaging of R2Z and R3Z RNAs by wild type capsid proteins following superinfection of R2Z- or R3Z-transfected cells with wild-type poliovirus
| Replicon | Virus | Measured amt (amol) of replicon RNA in 106 cellsa in:
|
% of replicon RNA encapsidated | % of viral RNA encapsidated | |
|---|---|---|---|---|---|
| Cytoplasmic extract | Virion | ||||
| R2Z | None | 160 | 0.5b | 0.3b | |
| R2Z | Wild type | 180 | 5.0 | 2.8 | 31 |
| R3Z | None | 99 | 0.2b | 0.2b | |
| R3Z | Wild type | 85 | 2.2 | 2.6 | 23 |
| None | Wild type | 25 | |||
The amounts (in attamoles [10−18 mol]) of protected R2Z, R3Z, and wild-type RNAs in cytoplasmic extracts or in purified virions was determined by comparing the RNase protection signal from the experimental samples, quantified by phosphoimager analysis, with that from known amounts of RNA in a standard curve.
Background level.
TABLE 2.
Quantitation of positive-strand R3Z RNA in cytoplasmic RNA from transfected and superinfected KJT7 cells and from HeLa cells after infection with cytoplasmic extracts from the KJT7 cells
| Virus | Guanidine | Amt (amol) of cytoplasmic RNA per 106 cells ina:
|
Ratio (HeLa/KJT7) | |
|---|---|---|---|---|
| KJT7 cells | HeLa cells | |||
| None | No | 7,200 | 40 | 0.006 |
| 3NC-202guaR | No | 7,900 ± 100 | 11,000 ± 2,000 | 1.4 |
| None | Yes | 2,100 | 65 ± 6 | 0.03 |
| 3NC-202guaR | Yes | 3,200 ± 150 | 250 ± 52 | 0.08 |
Errors given for measurements made in duplicate indicate one standard deviation from the mean. Amounts of radioactivity in protected RNAs were determined by phosphorimager analysis; the amount of RNA in attamoles (10−18 mol) was determined by comparison with the protected RNA signals from known amounts of R3Z replicon RNA.
Sensitivity of R3Z RNA replication to 0.5 mM guanidine is not efficiently rescued by superinfection with guanidine-resistant 3NC-202guaR virus.
To test the effect of ongoing replication of replicon RNAs on their ability to be packaged in trans, we needed a method to inhibit the replication of R3Z replicon RNA but not to inhibit the replication of superinfecting helper virus RNA. Mutations that confer resistance to guanidine have been characterized (41): most of them fall within the coding region for nonstructural protein 2C, and, curiously, many of them are cis dominant (24, 58, 59). That is, in coinfections between guanidine-sensitive wild-type viruses and certain guanidine-resistant mutant viruses, the wild-type virus is not rescued and the guanidine-resistant virus is not inhibited by the presence of guanidine. The guanidine-resistant allele used in the present study, present in the virus 3NC-202guaR, has been previously shown to be cis dominant in viral plaque assays (24). The 3NC-202guaR virus contains two mutations in the 2C coding region that together confer both guanidine resistance and temperature sensitivity (Fig. 1A) (24); an 8-nt insertion in the 3′ noncoding region also confers a temperature-sensitive phenotype (50).
To be useful in trans-encapsidation experiments, we needed a guaR allele that did not rescue the guanidine sensitivity of R3Z RNA replication in RNase protection assays or in the plaquing-efficiency experiments performed previously (24). To test whether R3Z RNA replication in the presence of guanidine was rescued by superinfecting 3NC-202guaR virus, KJT7 cells were transfected with T7R3Z DNA and incubated at 32.5°C for 24 h to allow R3Z RNA to accumulate in the absence of drug. Subsequently, 0.5 mM guanidine was added and incubation at 32.5°C was continued for an additional 13 h. RNase protection (35) was used to monitor the accumulation of R3Z-positive and -negative strands.
Similar to its effect on wild-type virus, the presence of 0.5 mM guanidine inhibits the accumulation of R3Z RNA (Fig. 4). Even in the presence of superinfecting 3NC-202guaR virus, guanidine significantly inhibited the accumulation of both positive-strand (Fig. 4A) and negative-strand (Fig. 4B) R3Z RNA, although a small increase in the amount of both R3Z RNA species was observed. Therefore, in the presence of 0.5 mM guanidine, conditions could be achieved in which a large amount of previously synthesized R3Z RNA was present but not efficiently replicating in the cytoplasm of cells in which the 3NC-202guaR virus was undergoing active RNA replication.
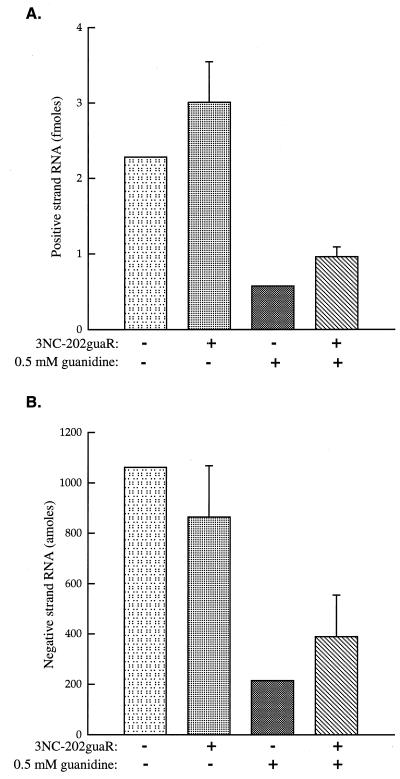
FIG. 4.
Test for complementation of the guanidine sensitivity of R3Z RNA synthesis by superinfecting 3NC-202guaR virus. KJT7 cells were transfected with R3Z-expressing DNA and incubated for 24 h before being superinfected or mock infected with 3NC-202guaR virus and treated with 0.5 mM guanidine. (A) RNase protection was used to quantify the amount of positive-strand R3Z RNA present in the cytoplasmic extracts. Comparison of the amount of labeled protected RNA to that protected by known amounts of RNA in a standard curve was used to determine the amount of positive-strand R3Z RNA in extracts from 1.6 × 107 cells. Error bars are shown for samples that were tested in duplicate in this particular experiment, one of several independent experiments. (B) The amounts of negative-strand R3Z RNA from 1.6 × 108 cells in the presence and absence of 0.5 mM guanidine and superinfecting 3NC-202guaR virus were analyzed by two-cycle RNase protection (35) and comparison to standard curves.
Testing the trans encapsidation of R3Z RNA in the presence and absence of guanidine.
To test whether preaccumulated R3Z RNAs could be packaged when they are not being actively replicated, KJT7 cells were transfected with T7R3Z DNA and incubated at 32.5°C for 24 h to accumulate R3Z RNA. They were then superinfected with 3NC-202guaR virus and incubated at 32.5°C for 13 h in the absence or presence of 0.5 mM guanidine. To determine the amounts of total R3Z and 3NC-202guaR RNA present in the KJT7 cells, RNase protection was performed on the total cytoplasmic extracts (Fig. 5A). Accumulated R3Z RNA was easily detectable in cytoplasmic extracts from guanidine-treated cells whether or not they had been superinfected with 3NC-202guaR virus (Fig. 5A).
FIG. 5.
Test of encapsidation of R3Z RNAs in the presence and absence of guanidine treatment and superinfection by 3NC-202guaR virus. (A) Accumulation of R3Z and 3NC-202guaR positive-strand RNAs in the cytoplasm of transfected and superinfected cells in the presence and absence of 0.5 mM guanidine. Lane P contains 0.35 fmol of full-length probe RNA, and lanes 1 to 14 contain labeled probe RNA protected by 5 μg of tRNA (lane 1), known amounts of R3Z RNA to generate a standard curve (lanes 2 to 6), and the cytoplasmic extracts indicated (lanes 7 to 14). Brackets indicate duplicate experiments. (B) Accumulation of R3Z and 3NC-202guaR positive-strand RNAs in HeLa cells infected with extracts from the transfected and superinfected KJT7 cells shown in panel A. HeLa cells were infected with cytoplasmic extracts and incubated for 10 h at 32.5°C, and cytoplasmic extracts were prepared. RNase protection identifies bands protected by R3Z and 3NC-202guaR RNAs. Experiments were performed in duplicate or quadruplicate as indicated by the brackets.
To determine whether R3Z positive-strand RNAs were packaged, cytoplasmic extracts from the transfected KJT7 cells were used to infect HeLa cell monolayers. After incubation for 10 h at 32.5°C to allow one infectious cycle, the HeLa cells were harvested and RNase protection experiments were performed (Fig. 5B). Clearly, a much higher percentage of R3Z RNA present in the KJT7 cells was packaged after superinfection with 3NC-202guaR virus in the absence of 0.5 mM guanidine than in its presence. The ratios of the amounts of cytoplasmic R2Z RNA in KJT7 cells to the amounts in HeLa cells after infection with the R3Z supernatants are given in Table 2. Thus, although the controls showed that substantial amounts of R3Z RNA were present in the cytoplasm of KJT7 cells with or without guanidine and that R3Z RNA could be packaged by the capsid proteins provided in trans by 3NC-202guaR virus, R3Z RNAs were not packaged when they were not replicating.
DISCUSSION
Encapsidation of poliovirus RNAs in trans has been frequently documented as the mechanism by which defective interfering particles are propagated (15, 26, 33), as a technique to test the specificity of packaging (39, 45), and as a tool to propagate poliovirus-derived RNAs that contain exogenous sequences (39, 44). Poliovirus genomes can be trans-encapsidated by capsids of the same serotype provided by helper viruses, by other serotypes of poliovirus, and by capsid proteins expressed by other viruses such as vaccinia virus (reviewed in references 2 and 20). Here, we used a trans-encapsidation system to test the effect of active RNA replication on the ability of subgenomic poliovirus RNAs to be packaged.
trans encapsidation of poliovirus replicon R2Z and R3Z genomes, which contain deletions of nt 1175 to 2470 and 1175 to 2956, respectively, in the capsid-coding region (22), was readily observed (Fig. 3; Table 1). When RNA replication of R3Z replicon RNA in KJT7 cells was inhibited by the addition of guanidine, preaccumulated R3Z RNA remained in the cytoplasm for several hours (Fig. 5A). When these cells were superinfected with 3NC-202guaR virus, whose RNA replication was not inhibited by guanidine, little rescue of either positive- or negative-strand R3Z RNA synthesis was seen (Fig. 4), even though the replication and packaging of the 3NC-202guaR virus continued unabated (Fig. 5B). Therefore, the guanidine-inhibited replication complex associated with the R3Z RNA is not a trans-dominant inhibitor of 3NC-202guaR RNA packaging.
The two populations of poliovirus-derived RNAs were apparently recognized quite differently by the viral capsid proteins encoded by the 3NC-202guaR virus. The 3NC-202guaR viral RNAs, associated with functional replication complexes, were efficiently packaged, but the R3Z replicon RNAs, either free in the cytoplasm or associated with guanidine-inhibited RNA replication complexes, were not (Fig. 5; Table 2). Why would the ability of these RNAs to be packaged be so different? The coupling between RNA replication and RNA packaging could arise by several possible mechanisms.
If some transient physical property of newly synthesized RNA, such as a short-lived RNA structure or linkage to VPg, were required for packaging, only newly synthesized RNAs in the immediate vicinity of the replication complex would be packaged. However, no evidence exists for such a transient structure or linkage, and this explanation would not account for the effect of mutations in 2C (28, 60) and 3D (17) on virion function.
The possible direct physical interaction between the RNA replication complex and the assembling virion particles is shown schematically in Fig. 6. If the RNA replication complex needed to be actively replicating RNA to allow a productive association with virion precursors, a good explanation for the coupling between RNA synthesis and RNA packaging would be provided. The direct involvement of viral proteins 2C and 3D in this interaction would rationalize the effect of mutations in these coding regions on the capsid structure (17, 28, 60). If specific interactions between the proteins of the RNA replication complex and capsid proteins were required for successful RNA packaging, it is possible that they would even be sufficient for the specificity of RNA packaging (20, 38, 49). That is, even though interactions between the capsid proteins and specific RNA sequences may yet be identified, they would not be required for the specificity of RNA encapsidation if the coupling of RNA replication and RNA packaging is accomplished by specific protein-protein interactions (Fig. 5).
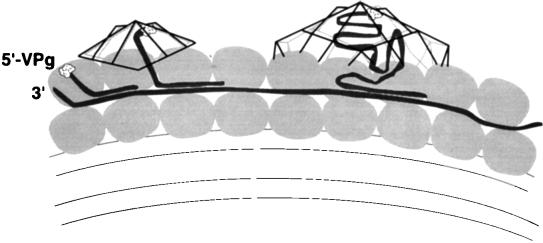
FIG. 6.
Model for coupling between virion assembly and RNA synthesis. Viral RNA strands are hypothesized to be coated with proteins of the viral RNA replication complex on the surface of the double cytoplasmic membranes (52) on which the RNA replication complexes assemble. Interactions between the 14S pentamers, subviral particles that accumulate in poliovirus-infected cells, and the proteins of the replication complex are proposed to position these subviral particles to encapsidate single-stranded RNA as it emerges from the replication complex. The presence of the RNA facilitates the addition of additional pentamers to the growing virion, possibly by increasing the rate of assembly about the threefold symmetry axis (9). Specificity for poliovirus RNA is dictated primarily by the proteins in the RNA replication complex, not by a particular sequence in the viral RNA. This image was also suggested by the electron microscopic images of Pfister et al. (40).
Direct coupling between genome replication and packaging has been documented for a few other viruses that are not closely related to poliovirus. When synthesis of the single-stranded DNA genome of minute virus of mice (a parvovirus) is inhibited, assembly of the virus ceases immediately (48). Furthermore, no free single-stranded DNA can be found in infected cells (56). It has been proposed that the viral DNA is either stripped from its template by the process of encapsidation or driven off the template by new DNA synthesis and directly incorporated into empty capsids (reviewed in reference 16). For hepatitis B virus, a ribonucleoprotein complex that includes the viral reverse transcriptase is required for specific encapsidation of the pregenomic RNA (7, 14, 43, 61). Thus, the polymerase protein is a key participant in both reverse transcription and genome packaging, and although its role in packaging is not thought to involve its enzymatic activity (7), this role may help ensure that it is included in the newly formed capsids for the subsequent reverse transcription. Although poliovirus has not been reported to encapsidate its polymerase, detectable amounts of the RNA-dependent RNA polymerase of foot-and-mouth disease virus have been reported to copurify with virions (32).
Direct transfer of RNA between cytoplasmic macromolecular complexes has been documented in the case of the “channeling” of cellular tRNAs. Specifically, attempts to detect or create a free cytoplasmic pool of aminoacylated tRNAs have not been successful. Instead, there appears to be direct transfer of charged tRNAs from aminoacyl synthetases to the A site of the ribosome (31) and from the P site back to the synthetases (55). As we have suggested for the transfer of nascent poliovirus positive-strand RNA from the replication complex to the capsid precursors, the protein-protein interactions may help provide some of the specificity to the recognition of charged amino-acylated tRNAs by the ribosome.
For an RNA genome with a mutation rate estimated to be as high as 6 × 10−4 mutations/nucleotide synthesized (reviewed in reference 62), coupling of RNA encapsidation to RNA synthesis and of RNA synthesis to translation (36) could be of evolutionary significance. Specifically, a requirement for successful translation of the poliovirus genome before it is used as a template for RNA synthesis should prevent the replication of RNA genomes that contain frameshift and nonsense mutations (36). Then the coupling between RNA synthesis and RNA packaging imposes a requirement that RNAs to be packaged must emerge from active replication complexes, ensuring that the templates for those RNAs were competent to form such complexes. Thus, the coupling between translation and RNA replication and the coupling between RNA replication and RNA packaging can impose a form of late proofreading to the error-prone RNA genome of poliovirus.
ACKNOWLEDGMENTS
We thank Eric Simoes for providing viruses and plasmids containing insertions in the 5′ noncoding region, Janet Novak for assistance with RNase protection experiments, Dana Dodd for critical reading of the manuscript, and John Lyle for help with phosphorimager quantitation.
This work was supported by NIH grant AI25166 to K.K., NIH grant AI25105 to P.S., and a fellowship from the Colorado Institute for Research in Biotechnology to C.I.N.
REFERENCES
- 1.Alexander L, Lu H H, Wimmer E. Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA. 1994;91:1406–1410. doi: 10.1073/pnas.91.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansardi D C, Porter D C, Anderson M J, Morrow C D. Poliovirus assembly and encapsidation of genomic RNA. Adv Virus Res. 1996;46:1–68. [PubMed] [Google Scholar]
- 3.Ansardi D C, Porter D C, Morrow C D. Complementation of a poliovirus defective genome by a recombinant vaccinia virus which provides P1 capsid precursor in trans. J Virol. 1993;67:3684–3690. doi: 10.1128/jvi.67.6.3684-3690.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltimore D. The replication of picornaviruses. In: Levy H B, editor. The biochemistry of viruses. New York, N.Y: Marcel Dekker, Inc.; 1969. pp. 101–176. [Google Scholar]
- 5.Baltimore D, Girard M, Darnell J E. Aspects of the synthesis of poliovirus RNA and the formation of virus particles. Virology. 1966;29:179–189. doi: 10.1016/0042-6822(66)90024-9. [DOI] [PubMed] [Google Scholar]
- 6.Bandyopadhyay P K, Wang C, Lipton H L. Cap-dependent translation by the 5′ untranslated region of Theiler’s murine encephalomyelitis virus. J Virol. 1992;66:6249–6256. doi: 10.1128/jvi.66.11.6249-6256.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartenschlager R, Junker-Niepmann M, Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton D J, Flanegan J B. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basavappa R, Syed R, Flore O, Icenogle J P, Filman D J, Hogel J M. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bienz K, Egger D, Pfister T, Troxler M. Structural and functional characterization of the poliovirus replication complex. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bienz K, Egger D, Troxler M, Pasamontes L. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J Virol. 1990;64:1156–1163. doi: 10.1128/jvi.64.3.1156-1163.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns C C, Lawson M, Semler B L, Ehrenfeld E. Effects of mutations in poliovirus 3Dpol on RNA polymerase activity and on polyprotein cleavage. J Virol. 1989;63:4866–4874. doi: 10.1128/jvi.63.11.4866-4874.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caliguiri L A, Compans R W. The formation of poliovirus particles in association with the RNA replication complexes. J Gen Virol. 1973;21:99–108. doi: 10.1099/0022-1317-21-1-99. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Robinson W S, Marion P L. Naturally occurring point mutation in the C terminus of the polymerase gene prevents duck hepatitis B virus RNA packaging. J Virol. 1992;66:1282–1287. doi: 10.1128/jvi.66.2.1282-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole C N, Baltimore D. Defective interfering (DI) particles of poliovirus. II. Nature of the defect. J Mol Biol. 1973;76:325–361. doi: 10.1016/0022-2836(73)90508-1. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore S F, Tattersall P. Parvovirus DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 799–813. [Google Scholar]
- 17.Diamond S E, Kirkegaard K. Clustered chared-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dove A W, Racaniello V R. Cold-adapted poliovirus mutants bypass a postentry replication block. J Virol. 1997;71:4728–4735. doi: 10.1128/jvi.71.6.4728-4735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn J J, Krippl B, Bernstein K E, Westphal H, Studier F W. Targeting bacteriophage T7 RNA polymerase to the mammalian cell nucleus. Gene. 1988;68:259–266. doi: 10.1016/0378-1119(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 20.Hellen C U T, Wimmer E. Maturation of poliovirus capsid proteins. In: Rotbart H A, editor. Human enterovirus infections. Washington, D.C: ASM Press; 1995. pp. 155–174. [Google Scholar]
- 21.Jacobsen M F, Baltimore D. Morphogenesis of poliovirus. I. Association of viral RNA with coat protein. J Mol Biol. 1968;33:369–378. doi: 10.1016/0022-2836(68)90195-2. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan G, Racaniello V R. Construction and characterization of poliovirus subgenomic replicons. J Virol. 1988;62:1687–1696. doi: 10.1128/jvi.62.5.1687-1696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirkegaard K. Mutations in VP1 of poliovirus specifically affect both encapsidation and release of viral RNA. J Virol. 1990;64:195–206. doi: 10.1128/jvi.64.1.195-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkegaard K, Baltimore D. The mechanism of RNA recombination in poliovirus. Cell. 1986;47:433–443. doi: 10.1016/0092-8674(86)90600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkegaard K, Nelsen B. Conditional poliovirus mutants made by random deletion mutagenesis of infectious cDNA. J Virol. 1990;64:185–194. doi: 10.1128/jvi.64.1.185-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuge S, Saito I, Nomoto A. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J Mol Biol. 1986;192:473–487. doi: 10.1016/0022-2836(86)90270-6. [DOI] [PubMed] [Google Scholar]
- 27.Li J-P, Baltimore D. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J Virol. 1988;62:4016–4021. doi: 10.1128/jvi.62.11.4016-4021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J-P, Baltimore D. An intragenic revertant of a poliovirus 2C mutant has an uncoating defect. J Virol. 1990;64:1102–1107. doi: 10.1128/jvi.64.3.1102-1107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molla A, Paul A V, Wimmer E. Cell-free, de novo synthesis of poliovirus. Science. 1991;254:1647–1651. doi: 10.1126/science.1661029. [DOI] [PubMed] [Google Scholar]
- 30.Moscufo N, Simons J, Chow M. Myristoylation is important at multiple stages in poliovirus assembly. J Virol. 1991;65:2372–2380. doi: 10.1128/jvi.65.5.2372-2380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negrutskii B S, Deutscher M P. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc Natl Acad Sci USA. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman J F E, Piatti P G, Gorman B M, Burrage T G, Ryan M D, Flint M, Brown F. Foot-and-mouth disease virus particles contain replicase protein 3D. Proc Natl Acad Sci USA. 1994;91:733–737. doi: 10.1073/pnas.91.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomoto A, Jacobson A, Lee Y F, Dunn J, Wimmer E. Defective interfering particles of poliovirus: mapping of deltion and evdience that the deletions in the genome of DI (1), (2) and (3) are located in the same region. J Mol Biol. 1975;128:179–196. doi: 10.1016/0022-2836(79)90125-6. [DOI] [PubMed] [Google Scholar]
- 34.Nomoto A, Kitamura N, Golini F, Wimmer E. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci USA. 1977;74:5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 37.Nugent C I. Analysis of poliovirus assembly and genome encapsidation. Ph.D. thesis. Boulder: University of Colorado; 1995. [Google Scholar]
- 38.Nugent C I, Kirkegaard K. RNA binding properties of poliovirus subviral particles. J Virol. 1995;69:13–22. doi: 10.1128/jvi.69.1.13-22.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percy N, Barclay W S, Sullivan M, Almond J W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfister T, Pasamontes L, Troxler M, Egger D, Bienz K. Immunocytochemical localization of capsid-related particles in subcellular fractions of poliovirus-infected cells. Virology. 1992;188:676–684. doi: 10.1016/0042-6822(92)90522-q. [DOI] [PubMed] [Google Scholar]
- 41.Pincus S E, Rohl H, Wimmer E. Guanidine-dependent mutants of poliovirus: identification of three classes with different growth requirements. Virology. 1987;157:83–88. doi: 10.1016/0042-6822(87)90316-3. [DOI] [PubMed] [Google Scholar]
- 42.Pincus S E, Wimmer E. Production of guanidine-resistant and -dependent poliovirus mutants from cloned dDNA: mutations in polypeptide 2C are directly responsible for altered guanidine sensitivity. J Virol. 1986;60:793–796. doi: 10.1128/jvi.60.2.793-796.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Porter D C, Ansardi D C, Morrow C D. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J Virol. 1995;69:1548–1555. doi: 10.1128/jvi.69.3.1548-1555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter D C, Ansardi D C, Wang J, McPherson S, Moldoveanu Z, Morrow C D. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology. 1998;243:1–11. doi: 10.1006/viro.1998.9046. [DOI] [PubMed] [Google Scholar]
- 46.Racaniello V R, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science. 1981;214:916–919. doi: 10.1126/science.6272391. [DOI] [PubMed] [Google Scholar]
- 47.Racaniello V R, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards R, Linser P, Armentrout R W. Kinetics of assembly of a parvovirus, minute virus of mice, in synchronized rat brain cells. J Virol. 1977;22:778–793. doi: 10.1128/jvi.22.3.778-793.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rueckert R R. Picornaviridae and their replication. In: Fields B, editor. Virology. New York, N.Y: Raven Press; 1996. pp. 507–548. [Google Scholar]
- 50.Sarnow P, Bernstein H D, Baltimore D. A poliovirus temperature-sensitive RNA synthesis mutant located in a noncoding region of the genome. Proc Natl Acad Sci USA. 1986;83:571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel A, Giddings T H, Jr, Ladinsky M S, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simoes E A, Sarnow P. An RNA hairpin at the extreme 5′ end of the poliovirus RNA genome modulates viral translation in human cells. J Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Simoes, E. A., and P. Sarnow. Unpublished data.
- 54.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 55.Stapulionis R, Deutscher M P. A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci USA. 1995;92:7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tattersall P, Ward D C. Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature. 1976;263:106–109. doi: 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 57.Todd S, Towner J S, Brown D M, Semler B L. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolskaya E, Romanova L, Kolesnikova M, Agol V I. Intertypic recombination in poliovirus: genetic and biochemical studies. Virology. 1983;124:121–132. doi: 10.1016/0042-6822(83)90295-7. [DOI] [PubMed] [Google Scholar]
- 59.Tolskaya E A, Romanova L I, Kolesnikova M S, Gmyl A P, Gorbalenya A E, Agol V I. Genetic studies on the poliovirus 2C protein, an NTPase. A plausible mechanism of guanidine effect on the 2C function and evidence for the importance of 2C oligomerization. J Mol Biol. 1994;236:1310–1323. doi: 10.1016/0022-2836(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 60.Vance L M, Moscufo N, Chow M, Heinz B A. Poliovirus 2C region functions during encapsidation of viral RNA. J Virol. 1997;71:8759–8765. doi: 10.1128/jvi.71.11.8759-8765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Y, Tavis J E, Ganem D. Relationship between viral DNA synthesis and virion envelopment in hepatitis B viruses. J Virol. 1996;70:6455–6458. doi: 10.1128/jvi.70.9.6455-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wimmer E, Hellen C U T, Cao X. Poliovirus genetics. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- 63.Ypma-Wong M F, Dewalt P G, Johnson V H, Lamb J G, Semler B L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]